Abstract
The retinoblastoma protein (pRB) is best known for regulating cell proliferation through E2F transcription factors. In this report, we investigate the properties of a targeted mutation that disrupts pRB interactions with the transactivation domain of E2Fs. Mice that carry this mutation endogenously (Rb1ΔG) are defective for pRB-dependent repression of E2F target genes. Except for an accelerated entry into S phase in response to serum stimulation, cell cycle regulation in Rb1ΔG/ΔG mouse embryonic fibroblasts (MEFs) strongly resembles that of the wild type. In a serum deprivation-induced cell cycle exit, Rb1ΔG/ΔG MEFs display a magnitude of E2F target gene derepression similar to that of Rb1−/− cells, even though Rb1ΔG/ΔG cells exit the cell cycle normally. Interestingly, cell cycle arrest in Rb1ΔG/ΔG MEFs is responsive to p16 expression and gamma irradiation, indicating that alternate mechanisms can be activated in G1 to arrest proliferation. Some Rb1ΔG/ΔG mice die neonatally with a muscle degeneration phenotype, while the others live a normal life span with no evidence of spontaneous tumor formation. Most tissues appear histologically normal while being accompanied by derepression of pRB-regulated E2F targets. This suggests that non-E2F-, pRB-dependent pathways may have a more relevant role in proliferative control than previously identified.
INTRODUCTION
The retinoblastoma tumor suppressor protein (pRB) has a central role in the regulation of the G1-to-S-phase transition. Inactivation of its control over cell cycle progression is one of the most common events in cancer (1). The RB protein is thought to regulate entry into S phase through its ability to repress E2F-dependent transcription (2). In the G1 phase of the cell cycle, a direct interaction between the large pocket domain of pRB (RBLP) and the transactivation domain of E2Fs blocks transcription and recruits chromatin regulators that maintain the cell in G1 (3). Activation of cyclin-dependent kinases (CDKs) results in the phosphorylation of pRB and the release of E2F transcription factors (4). Free E2Fs then activate a transcriptional program that drives the cell into S phase (3). This model of pRB regulation of E2F dominates our understanding of G1-to-S-phase control. Much of our knowledge of this model was derived from studies using viral oncoproteins encoded by small DNA tumor viruses (5, 6). Of particular note, the human papillomavirus E7 protein has been shown to compete for pRB-E2F interactions to deregulate proliferation (7, 8). However, E7 must also target pRB for degradation in order to induce proliferation (8). Thus, the experimental system that gave rise to the pRB-E2F regulatory axis in cell cycle control also suggests that pRB may engage other growth-suppressing activities beyond E2F regulation. By comparison with the pRB-E2F pathway, we know very little about pRB's non-E2F-dependent growth control mechanisms and their relative contribution to cell cycle regulation and tumor suppressor activities.
The minimal growth-suppressive region of pRB has been mapped to the A, B, and C regions of its open reading frame, a domain called the “large pocket” that includes amino acids 379 to 928 (3). This is also the minimal domain needed for stable interaction with E2Fs and to repress their transcription (9–12). E2Fs are a family of transcription factors, and each of E2F1, E2F2, E2F3, and E2F4 is capable of binding to pRB at endogenous levels through its transactivation domain; this is termed the “general” interaction (13, 14). E2F1 is unique among E2Fs in that it has roles outside transcriptional activation of cell cycle genes, including the regulation of apoptotic targets (15, 16) and DNA replication (17–19). E2F1 is also capable of making a protein interaction with pRB qualitatively different from that of the other E2Fs (13, 20), and this interaction is mediated by separate protein-protein contacts (13, 20–22). This E2F1 “specific” interaction has been suggested to allow it to regulate apoptotic target genes independently of E2F transcriptional control during the cell cycle (13, 15, 20, 23). One reason that the specific interaction with pRB is distinct from the general interaction is because E2F1 bound to pRB through this site is unable to efficiently bind the consensus E2F promoter element (13) but contributes to regulation of apoptotic target genes such as TA-p73 (15, 23). Furthermore, the regulation of this interaction is distinct, as the specific pRB-E2F1 interaction is resistant to disruption by CDK phosphorylation (21, 24). Thus, recent structural and functional insights into pRB-E2F interactions indicate that pRB's relationship with E2F transcription factors may be more complex than simply silencing their activity during cell cycle arrest. This background highlights the difficulty in understanding how individual biochemical aspects of pRB function contribute to its complete role as a cell cycle regulator and tumor suppressor.
In order to investigate pRB-dependent functions in cell cycle control that are independent of canonical E2F transcriptional control, we generated a gene-targeted mouse allele whose encoded protein is selectively deficient for the general interaction. We call this allele Rb1ΔG because it disrupts the interaction between the transactivation domain of E2Fs and pRB. Our analysis indicates that this mutant protein is defective for pRB-E2F interactions at cell cycle promoters and is unable to regulate E2F transcriptional activity in reporter assays. Primary fibroblast cultures and tissues from Rb1ΔG/ΔG mice exhibit derepression of direct pRB-E2F transcriptional targets and yet maintain the ability to control proliferation in response to serum deprivation, p16 expression, and gamma irradiation. Furthermore, Rb1ΔG/ΔG mice are relatively normal in development and remain cancer free throughout their lives. This study suggests that pRB functions that are independent of E2F transcriptional control can contribute to its tumor suppressor activity in a meaningful way.
MATERIALS AND METHODS
Protein interaction analysis and Western blotting.
To generate extracts for glutathione S-transferase (GST) pulldowns and gel shifts, cells were washed twice with phosphate-buffered saline (PBS) and collected in 1 ml of gel shift extract (GSE) buffer (20 mM Tris, pH 7.5, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 0.1 mM Na3VO4, 0.5 mM NaF, and 1 mM dithiothreitol [DTT]) per 15-cm dish of cells. Cells were frozen at −80°C and thawed rapidly to lyse them; cellular debris was removed by centrifugation at 14,000 rpm. For GST pulldowns, extracts were diluted approximately 2-fold in wash buffer without NaCl (20 mM Tris, pH 7.5, 1.5 mM MgCl2, 0.2 mM EDTA, 25 mM DTT, and 0.1% NP-40) to approximately physiological salt concentrations. Beads and fusion proteins were added and incubated with rocking for 1 h at 4°C. The protein G-Sepharose beads and associated proteins were washed twice with immunoprecipitation (IP) wash buffer and then resuspended in 1× SDS-PAGE sample buffer and boiled at 95°C for 5 min to elute the bound proteins. The eluted material was resolved by SDS-PAGE and transferred to a nitrocellulose membrane by standard techniques.
Nuclear extracts were prepared from mouse embryonic fibroblasts (MEFs) as previously described (25). A sheep anti-pRB antibody was generated using the C terminus of murine pRB, and antibodies were purified using a peptide corresponding to amino acids 867 to 881 of murine pRB coupled to agarose using the Sulfolink immobilization kit (Pierce). Five micrograms of antibody, which had previously been covalently coupled to protein G Dynabeads using bis(sulfosuccinimidyl suberate) (BS3; Pierce), was used to immunoprecipitate pRB from 1 mg of nuclear extract. Precipitated proteins were detected in Western blot assays using the following antibodies: pRB was detected using G3-245 (BD Pharmingen), E2F1 with C-20 (Santa Cruz), E2F2 with TFE-25 (Santa Cruz), E2F3 with PG37 (Upstate) or C-18 (Santa Cruz), and E2F4 with C-20 (Santa Cruz). Other antibodies used for Western blotting included PCNA F2 (Santa Cruz), p107 C-18 (Santa Cruz), Mcm3 4012S (Cell Signaling), cyclin E M20 (Santa Cruz), and cyclin A H432 (Santa Cruz). Rabbit anti-SP1 H225 (Santa Cruz) was used as a loading control.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed using DNA probes described in the work of Seifried et al. (26). These probes were labeled with 50 μCi of [α-32P]dCTP with Klenow fragment for 15 min at room temperature. The labeled probes were purified on a G25 spin column. Extracts were prepared from confluent MEFs in GSE buffer as described above. Five micrograms of nuclear extract was diluted into EMSA buffer (20 mM Tris, pH 7.5, 4% Ficoll 400-DL [Sigma], 2.5 mM MgCl2, 40 mM KCl, 0.1 mM EGTA, 2 mM spermine, 0.5 mM DTT, 0.25 μg salmon sperm DNA, 10 μg bovine serum albumin) in a 20-μl total volume. Samples with cold competitors were first incubated with 40 ng of wild-type or mutant unlabeled oligonucleotides for 10 min on ice. Four hundred picograms of labeled probe was then added to each reaction mixture and incubated on ice for 10 min. For antibody supershifts, antibodies were added and the samples were incubated on ice for a further 25 min. For supershifts, 1 μg of anti-pRB 21C9 (a kind gift from Sibylle Mittnacht, London, United Kingdom) and anti-CDK2 (Upstate) was used. Samples were loaded onto a 4% polyacrylamide gel (containing 0.25× Tris-borate-EDTA and 2.5% glycerol) and electrophoresed at 4°C for 4 h at 180 V. Gels were dried, and protein-DNA complexes were detected by autoradiography. Gel shifts for determining differences of affinity were carried out essentially as described above, except that C33A nuclear extracts containing overexpressed hemagglutinin (HA)-tagged E2F1, E2F2, E2F3, and E2F4 with HA-DP1 proteins were used. Anti-HA antibodies (12CA5 hybridoma supernatant) were used to shift HA-E2F/DP1 complexes, and the indicated amounts of GST or GST-RBLP proteins were added.
Gene targeting and phenotypic analysis of animals.
Embryonic stem (ES) cell culture, transfection, and selection were performed using standard methods. Correctly targeted ES cells were identified by Southern blotting. Genomic DNA was digested with KpnI, and the indicated probes outside the 5′ and 3′ arms of homology were used to detect homologous recombinants. MscI digestion was also performed to cut DNA within the neomycin resistance gene and outside the 5′ arm of homology. A probe specific to the neomycin resistance gene was used to probe this Southern blot to ensure that the targeted clones contained only a single site of integration of the targeting vector. Correctly targeted ES clones were grown and injected into blastocysts to generate chimeric mice. Male chimeras were mated with B6.FVB EIIa-cre transgenic mice to remove the PGK-Neo selectable marker that was flanked by LoxP sites. Progeny were then intercrossed to generate mice that had excised the selectable marker and did not contain EIIa-cre. Rb1ΔG mice were genotyped by amplification of a genomic sequence that surrounds the remaining LoxP site. Using L-F (5′ CTGCAATCTGCGCATTTTTA 3′) and L-R (5′ CGATGCTGCAGGCCTATAAT 3′) primers, a 250- or a 330-bp fragment that corresponds to the wild-type or mutant allele, respectively, is produced.
E2f1−/− mice (B6; 129S4-E2f1tm1Meg/J) (27) were obtained from the Jackson Laboratory and genotyped as recommended by the distributor. All animals were housed and handled as approved by the Canadian Council on Animal Care. Mice were monitored throughout their lives, and animals were euthanized after the development of signs of tumor burden or at defined ages as indicated in the figure legends. Survival data were subjected to Kaplan-Meier analysis, and significant differences were compared using a log rank test.
Euthanized animals were subjected to a necropsy where tissues of interest and tumors were fixed in formalin. Tumors and tissues were fixed in formalin for at least 72 h, washed in phosphate-buffered saline (PBS), and then transferred to 70% ethanol. The tissues were embedded in paraffin, and 5-μm sections were cut from superficial and deep sections of the tissue blocks. Sections were subsequently stained with hematoxylin and eosin (H&E), and images were captured on a Zeiss Axioskop 40 microscope and Spot Flex camera using EyeImage software (Empix Imaging, Mississauga, Ontario, Canada) or similar system.
Luciferase reporter assays.
Luciferase reporter assays were performed as described previously (28). Saos-2 cells were plated at 5 × 105 per well of a 6-well dish. Transfection mixtures contained 100 ng of a luciferase reporter and 200 ng of cytomegalovirus (CMV)–β-galactosidase (β-Gal). CMV-RB expression plasmids were included up to a total of 100 ng, and where indicated, 15 ng of CMV-HA-E2F and 15 ng of CMV-HA-DP1 were included. Total CMV plasmid DNA was normalized with the addition of CMV-CD20. Cells were lysed 36 h after transfection in reporter lysis buffer (Promega). Luciferase activity was determined using the luciferase assay system (Promega) and normalized to β-Gal activity. Each data point is the average of three independent transfections, and the error bars indicate 1 standard deviation from the mean.
Mouse embryonic fibroblasts (MEFs), retroviral infections, and myogenic differentiation.
Wild-type, Rb1ΔG/ΔG, and Rb1−/− fibroblasts were derived from E13.5 embryos, and experiments were carried out using passage 3 to 5 MEFs. Asynchronous cell populations were cultured according to standard methods. Cell culture was carried out in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, penicillin (50 U/ml), and streptomycin (50 μg/ml). Growth curves were generated by plating MEFs at low density followed by trypsinization and counting for 5 consecutive days. Cells deprived of serum were cultured for 60 h in medium as described above except with 0.1% FBS. For serum restimulation assays, 10% FBS was added following 60 h of serum deprivation. Gamma irradiation was performed by exposing cells to a cobalt-60 radiation source until a dose of 15 Gy was received.
Retroviral infections of MEFs were undertaken with pBabe-p16 and pBabe-MyoD constructs at passage 3. Infections were performed as described in the work of Pear et al. (29). BOSC or Phoenix-Eco packaging cells were plated at a density of 10 million cells per 15-cm plate on the day before the transfections. On the following day, the cells were transfected with 60 μg of pBabe plasmid or pBabe containing p16 using calcium phosphate, and on the next morning, the medium was replaced. The medium was removed 48 h later, filtered through a 0.45-μm filter, and supplemented with 4 μg/ml of Polybrene. The filtered viral supernatant was placed directly on MEFs that had been plated the previous day at 8 × 105 cells in a 10-cm dish. Fresh medium was added to the transfected packaging cells for another 12 h. After 12 h, the medium from the MEFs was removed and a second round of infection was performed by once again adding the filtered viral supernatant with Polybrene to the MEFs. The viral supernatant was incubated on the MEFs for a further 8 to 12 h and then replaced with medium containing 5 μg/ml of puromycin for 4 days. The infected MEFs were then replated in puromycin-containing medium for subsequent analysis. Myogenic differentiation was carried out using MEFs infected with a pBABE-MyoD-expressing retrovirus and by following the cell culture methods of Novitch et al. (30). Once differentiated, cells were restimulated with 15% serum and labeled with bromodeoxyuridine (BrdU) for 24 h. Cells were fixed and stained for BrdU, myosin heavy chain (MHC), and DNA with 4′,6-diamidino-2-phenylindole (DAPI) and analyzed by fluorescence microscopy. BrdU incorporation in MHC-positive and -negative cells was quantitated as previously reported (31).
Chromatin immunoprecipitation (ChIP) experiments.
Asynchronously growing Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− MEFs were washed twice with PBS followed by incubation with 2 mM ethylene glycol bissuccinimidylsuccinate (EGS) diluted in PBS for 1 h with shaking at room temperature (RT). Formaldehyde was then added to a final concentration of 1% and incubated at RT for 15 min without shaking. Glycine was then added to a final concentration of 0.125 M to quench the reaction. Cells were washed twice and then collected in PBS. Cell pellets were resuspended in buffer 1 (10 mM HEPES, pH 6.5, 10 mM EDTA, 0.5, EGTA, 0.25% Triton X-100), incubated on ice for 5 min, and pelleted at 600 × g at 4°C. Pellets were then resuspended twice in buffer 2 (10 mM HEPES, pH 6.5, 1 mM EDTA, 0.5, EGTA, 200 mM NaCl), incubated on ice for 5 min, and pelleted at 600 × g at 4°C between each wash. Cells were then suspended in SDS-lysis buffer (1% SDS, 1 mM EDTA, 50 mM Tris-HCl, pH 8.0, and protease inhibitors) and incubated for 15 min on ice followed by 35 min of sonication in a Bioruptor (Diagenode). Seventy-five micrograms of chromatin was diluted 10-fold in dilution buffer (0.01% SDS, 0.4% Triton X-100, 1 mM EDTA, 20 mM Tris, pH 8, 200 mM NaCl, and protease inhibitors) and precleared for 1.5 h with IgG prebound to Dynabeads. IPs were performed by rotation at 4°C for 16 h using 5 μg each of the following antibodies mixed together and prebound to Dynabeads: M-153 and C-15 (both from Santa Cruz), Rb 4.1 (Developmental Studies Hybridoma Bank), an affinity-purified sheep antibody raised against amino acids 867 to 881 of mouse pRB, and an affinity-purified rabbit antibody raised against amino acids 847 to 859 of mouse pRB. Beads were then serially washed for 5 min with rotation at 4°C for the following washes: twice in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 150 mM NaCl), once in high-salt buffer (0.1% SDS 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 500 mM NaCl), and twice in TE (10 mM Tris-HCl, pH 8, 1 mM EDTA). Protein was eluted with 2 serial incubations with 150 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C for 10 min each. Cross-links were reversed by adding NaCl to a final concentration of 200 mM and incubating the mixture at 65°C for 4 h. RNase was added and incubated at 37°C for 30 min followed by addition of 50 μg/ml protease K and 10 mM EDTA and incubation at 45°C for 1 h. DNA was then isolated using a PCR purification kit (Invitrogen).
Real-time PCR analysis of ChIP.
DNA isolated from ChIPs was analyzed using iQ Sybr green Super Mix (Bio-Rad). Primer sets used for analysis were as indicated below, and their locations were chosen based on human pRB ChIP sequencing data previously published by Chicas et al. (32) visualized on the UCSC genome browser. Primers included Rbl1 TSS Fwd (5′ CTT CGG GGT TTT CTT TTC CCT C 3′), Rbl1 TSS Rev (5′ TAG AGT CCG AGG TCC ATC TTC TTA T 3′), Rbl1 Neg Fwd (5′ AGT CGT TTC AGG AAT AGA GAT GGT C 3′), Rbl1 Neg Rev (5′ TAC CTG GTG CAT CTG AAT GCT ATT A 3′), Mcm3 TSS Fwd (5′ ATC CAG GAA GTC CAA GTA GTC TCT C 3′), Mcm3 TSS Rev (5′ TTG AAG TGG TTA GCC AAT CAT AAC G 3′), PCNA TSS Fwd (5′ CAG AGT AAG CTG TAC CAA GGA GAC 3′), PCNA TSS Rev (5′ CGT TCC TCT TAG AGT AGC TCT CAT C 3′), PCNA Neg Fwd (5′ CAT CAG TGA ATA CGT CTC TGT TCC A 3′), PCNA Neg Rev (5′ CTG CTT CTC AGT TGT TTT AGG AAG G 3′), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Fwd (5′ GAG CCA GGG ACT CTC CTT TT 3′), and GAPDH Rev (5′ CTG CAC CTG CTA CAG TGC TC 3′). Percent inputs were calculated as follows: 2−(antibody CT − input CT) × % input used, where CT is threshold cycle. Values were then normalized to percent input of GAPDH.
Cell cycle analysis.
Cell cycle analysis of MEFs was performed by pulse-labeling cells with bromodeoxyuridine (BrdU; Amersham Biosciences) for 1.5 h before harvesting cells. The cells were fixed in ethanol and immunostained with anti-BrdU antibodies (BD Biosciences), along with propidium iodide (PI) as reported in the work of Cecchini et al. (25). Cell populations were analyzed by flow cytometry on a Beckman-Coulter Epics XL-MCL instrument, and the relative abundance of each phase was determined. Gates were used to quantitate the proportion of cells with 8N DNA content, and the average forward scatter was determined on G1-phase cells where indicated.
mRNA quantitation.
Total RNA was extracted from cells or minced tissues using TRIzol reagent according to the manufacturer's instructions (Invitrogen). RNA from tissues was isolated using an RNeasy fibrous tissue kit (Invitrogen). Expression levels of the E2F target genes, Pcna, Ccne1 (cyclin E1), Ccna2 (cyclin A2), Tyms (thymidylate synthase), Mcm3, and Rbl1 (p107), were determined using the Quantigene Plex 2.0 reagent system from Affymetrix (Santa Clara, CA) and a BioPlex200 multiplex analysis system. Expression levels were normalized to the expression of actin.
Microarray analysis.
Total RNA was extracted from MEFs following serum starvation using TRIzol reagent. RNA was quality controlled using an Agilent 2100 Bioanalyzer before labeling and hybridization onto an Affymetrix mouse 1.0 ST gene array in the London Regional Genomics Centre. Affymetrix.cel files were normalized with robust multiarray averaging (33–35) using BRB-Array Tools (http://linus.nci.nih.gov/BRB-ArrayTools.html). Log ratios of Rb1ΔG/ΔG RNA compared with wild type were determined by subtracting the average wild-type log signal value from each replicate of Rb1ΔG/ΔG. The three log ratios were subjected to hierarchical clustering using Euclidian distance and average linkage using MeV (http://www.tm4.org/mev.html). In the figures, positive log ratios are colored red, and negative ratios are colored green.
Isolation of intestinal villi.
Isolation of villi was carried out essentially as described previously (36). Mice were sacrificed by cervical dislocation, and the small intestine was immediately removed, measured, and cut into thirds. The middle section, corresponding roughly to the jejunum, was opened using blunt-end scissors washed 3 times in PBS and placed in 25 ml of PBS supplemented with 1 mM DTT and 5 mM EDTA for 30 min. The intestine was then transferred to a 15-ml tube with 10 ml of Release buffer (PBS, 1 mM DTT) and shaken gently to release intact villi from the intestine. Five sequential fractions were obtained with increasingly more vigorous shaking. To release the intestinal crypts, the intestine was placed in 25 ml of PBS supplemented with 1 mM DTT and 9 mM EDTA and incubated for 20 min at room temperature. The intestine was then placed once again in a 15-ml tube with 10 ml of release buffer and mixed to release the crypts from the intestines. This was repeated with 5 tubes to sequentially release the crypts from the intestine. During the fractionation process, the release of cells was monitored under a dissecting microscope to quantify the proportion of villi and crypts isolated. Fractions were further enriched for villi by allowing samples to settle and decanting the supernatant, which contains largely crypts. In contrast, crypts were enriched by filtering samples through 100-μm nylon cell strainers (Fisher Scientific). Only samples with significant enrichment were utilized for subsequent analysis.
BrdU staining of intestinal tissue sections.
To analyze proliferation, mice were injected with 200 μl of 16-μg/ml BrdU (Sigma) 2 h before sacrifice. Intestines were then isolated, fixed in formalin, embedded in paraffin, and sectioned according to standard protocols. Paraffin was removed, and sections were rehydrated using a series of xylene extractions and ethanol washes. The sections were brought to a boil in sodium citrate buffer and then maintained at 95°C for 10 min. The cooled sections were rinsed in water three times for 5 min each time and then rinsed in PBS for 5 min. The sections were blocked in phosphate-buffered saline (PBS) supplemented with 2.5% horse serum and 0.3% Triton X-100 for 1 h. The sections were incubated with anti-BrdU antibodies (BD Biosciences) in blocking buffer overnight at 4°C and then rinsed in PBS three times for 5 min each time. The slides were incubated with horse anti-mouse immunoglobulin G-fluorescein isothiocyanate (Vector) for 1 h and then rinsed in PBS. The slides were mounted with Vectashield plus 4′,6-diamidino-2-phenylindole (DAPI) (Vector). Fluorescent images were captured on a Zeiss Axioskop 40 microscope and Spot Flex camera and colored using EyeImage software (Empix Imaging, Mississauga, Ontario, Canada) or a similar system.
BrdU staining of embryos.
To analyze proliferation in the embryos, pregnant mice at embryonic day 18.5 (E18.5) were injected with 10 μg of BrdU/g of body weight, 2 h before sacrifice. Embryos were collected, fixed in formalin, embedded in paraffin, and sectioned according to standard protocols. Paraffin was removed, and sections were rehydrated using a series of xylene extractions and ethanol washes. The sections were incubated in 2 N HCl at 37°C for 60 min followed by neutralization in 0.1 M Na2BO4 two times for 5 min each. Sections were rinsed in PBS twice for 5 min. Endogenous peroxidases were blocked by incubating the sections in 3% H2O2 for 20 min followed by washing in PBS twice for 5 min. The sections were blocked in phosphate-buffered saline (PBS) supplemented with 5% horse serum for 1 h at room temperature. The sections were incubated with anti-BrdU antibodies (BD Biosciences) in blocking buffer for 1 h at room temperature and then rinsed in PBS three times for 5 min each. The slides were incubated with biotinylated anti-mouse immunoglobulin (Vector Laboratories) for 1 h and then rinsed in PBS three times for 5 min each. The slides were incubated with streptavidin-horseradish peroxidase (HRP) (Vector Laboratories) for 30 min and then rinsed in PBS before incubation with ImmPACT diaminobenzidine (DAB) substrate (Vector Laboratories). The slides were counterstained with hematoxylin, dehydrated in ethanol, and mounted with Vectamount (Vector Laboratories) for analysis. Microscopic examination and photography of slides were performed as described above.
RESULTS
Characterization of an Rb1 mutant that is defective for E2F transactivation domain binding.
To better understand pRB-dependent functions outside E2F transcriptional control, we sought to develop a gene-targeted mutant mouse line that is deficient for this function. The interaction between pRB and E2F transcription factors is complex (Fig. 1A), as pRB possesses a well-studied mechanism in which it interacts with the C-terminal transactivation domain of E2Fs, but it also forms an alternate interaction with E2F1 that has little effect on canonical E2F transcriptional regulation (13, 21, 22). We set about designing an Rb1 allele that disrupts only pRB's contact site with the transactivation domain of E2F1, E2F2, E2F3, and E2F4.
FIG 1.
Characterization of an RB mutant that is defective for binding the E2F transactivation domain. (A) The domain structures of pRB and E2F proteins are depicted. Mapped interaction sites for the different classes of E2F interaction are shown. DBD, DNA binding domain; DMZ, dimerization (with DP) domain; MB, marked box domain. The E2F transactivation domain is shown in red, and the minimal RB interaction site within it is indicated. (B) Sequence alignment of the pRB binding region of E2F1, E2F2, E2F3, and E2F4 created using ClustalW. Three conserved aspartate residues in E2Fs make contacts with distinct basic amino acids on pRB. Codon positions use human numbering. (C) Locations of basic amino acids that were mutagenized to disrupt E2F binding are shown in red in a crystal structure of the small pocket of pRB (PDB entry 1O9K) (63). RB is shown in blue, and an E2F1-derived peptide is shown in green. (D) Electrophoretic mobility shift assays utilizing extracts expressing HA-E2F2 and HA-DP1 were combined with a radiolabeled probe containing a canonical E2F binding site. Migration positions for free E2F bound to the probe and E2Fs bound to GST-RBLP proteins are shown on the right of the gel. 100× WT and 100× MT denote samples with excess cold oligonucleotide. The α-HA lane denotes the addition of the 12CA5 antibody recognizing the HA epitope on the exogenous E2F/DP proteins. The GST lane contains 1 μg of GST as a negative control, and the remaining 12 lanes consist of a 10-fold dilution series of recombinant GST-WT-RBLP and ΔG-RBLP (R467E and K548E) from 1 μg to 10 pg. (E) To compare the affinities of WT and ΔG for each E2F, the proportion of RB bound relative to residual free E2F was determined for each quantity of GST-RBLP used. A graph of percent RB bound versus GST-RBLP quantity is shown for E2F2. (F) The table shows the half-maximal binding quantity for WT-RBLP and ΔG-RBLP in EMSAs for each of E2F1, E2F2, E2F3, and E2F4. (G) Increasing amounts of CMV-RB expression vector were cotransfected along with a constant level of a p107(promoter)-luciferase reporter construct in Saos-2 cells. Extracts were prepared to assess relative luciferase activity and determine transcriptional repression by the overexpressed ΔG-pRB mutant. Error bars indicate 1 standard deviation from the mean (n = 3). An asterisk indicates a statistically significant difference from E2F/DP1 transfection alone (t test, P < 0.05).
We focused on conserved acidic side chains used by E2F1, E2F2, E2F3, and E2F4 to interact with basic residues in the groove formed by the A and B domains of the pRB pocket (Fig. 1B and C). We generated a charge reversal mutant in which R467 and K548 on pRB were changed to glutamate to antagonize E2F binding (21). This mutant was titled ΔG because it disrupts the general interaction that pRB makes with the E2F transactivation domain (13, 21). To determine the severity of this mutation, we utilized electrophoretic mobility shift assays to quantitate the defect in pRB-E2F interactions. HA-tagged E2F1, E2F2, E2F3, and E2F4 were produced by transfection and mixed with a 32P-labeled double-stranded oligonucleotide containing an E2F binding site. E2F binding was then tested by titrating increasing amounts of GST-RBLP proteins, a domain that contains both E2F binding sites. Figure 1D demonstrates analysis for HA-E2F2 and reveals the inability of the GST-ΔG-RBLP protein to form a complex with HA-E2F2 on DNA. The percentage of HA-E2F2 bound to wild-type or mutant GST-RBLP was determined for each lane of the gel and was plotted in Fig. 1E. From these data, we determined half-maximal binding quantities for GST-RBLP and the ΔG mutant for each of E2F1, E2F2, E2F3, and E2F4 (Fig. 1F). This indicates that the ΔG substitutions have a strong effect on E2F binding, as the interactions with E2F2, E2F3, and E2F4 are nearly undetectable. As stated above, E2F1 has the ability to form an alternate complex with pRB that is reported to have reduced binding to the E2F sequence element in this probe (13). For this reason, E2F1's apparent affinity for GST-RBLP is lower and is relatively unaffected by the ΔG mutations (Fig. 1F).
In order to assess the effects of ΔG-pRB on E2F transcriptional control, we transfected cells with CMV-RB expression vectors and a p107-luciferase reporter (Fig. 1G). This experiment demonstrates that ΔG-pRB is unable to repress transcription of this reporter when relying on endogenous E2Fs. In addition, we have also tested the ability of ΔG-pRB to block transcriptional activation of an E2F-responsive reporter when individual E2Fs are overexpressed. As shown in Fig. 2A, wild-type pRB is capable of repressing transcription by each of E2F1, E2F2, or E2F3. Conversely, ΔG-pRB has a similar defect in controlling each E2F, even though it is still capable of binding E2F1 in interaction assays. These results are consistent with previous reports that demonstrate that the pRB-E2F1 complex formed through their alternate interaction is a poor regulator of cell cycle E2F transcriptional targets (13, 21, 22). Importantly, regulation of a TA-p73 reporter, a uniquely E2F1-responsive proapoptotic target gene (16), is similar between wild type and ΔG-pRB (Fig. 2B). Taken together with the in vitro binding assays above, the ΔG substitutions dramatically reduce the affinity of pRB for the E2F transactivation domain and prevent transcriptional repression of canonical E2F-responsive genes, even when overexpressed.
FIG 2.

Regulation of E2F transcriptional activity by ΔG-pRB. (A) The E2F4B-luciferase construct containing four tandem E2F recognition sites was cotransfected with E2F1/DP1, E2F2/DP1, or E2F3/DP1. Where indicated, wild-type pRB or ΔG-pRB was transfected to assess the ability of each to regulate E2F. (B) A reporter construct containing the TA-p73 promoter was transfected with E2F1/DP1 and the indicated pRB expression vectors to assess regulation of E2F1. Error bars indicate 1 standard deviation from the mean (n = 3). An asterisk indicates a statistically significant difference from E2F/DP1 transfection alone (t test, P < 0.05).
Introduction of an E2F-binding-deficient allele of Rb1 into the endogenous murine locus.
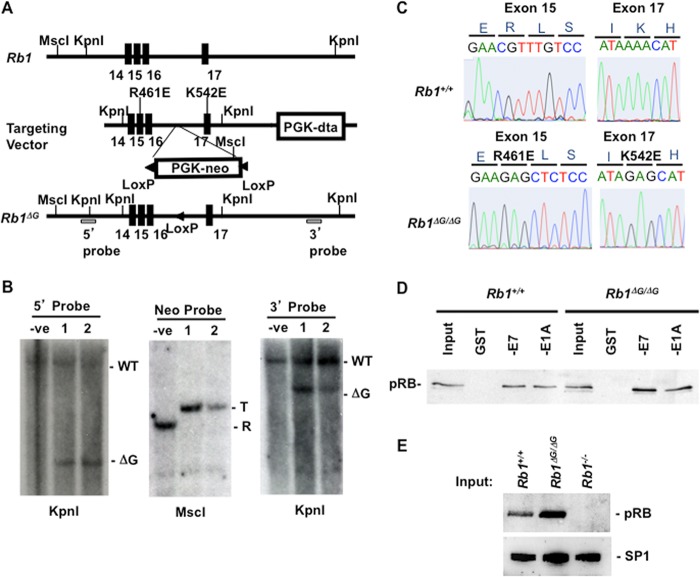
Our next goal was to introduce the R467E and K548E substitutions (R461E and K542E in murine numbering) into the Rb1 gene and create an endogenous Rb1ΔG mutant. Figure 3A contains a diagram of the Rb1 locus, the targeting vector used to introduce the ΔG mutations, along with relevant restriction enzyme cut sites that were used to map homologous recombination. Southern blotting assays were performed using 5′ and 3′ external probes, as well as the neomycin resistance gene, for two candidate clones (Fig. 3B). The smaller KpnI fragments found in both targeted clones are indicative of homologous recombination at both ends of the targeting vector. The single band in the MscI-digested, Neor gene-probed lane indicated that the targeting vector was integrated only once. These clones were expanded and used to produce chimeric male mice that were bred to B6; FvB EIIa-cre transgenic animals to establish germ line transmission, excise the selectable marker, and create the gene structure shown at the bottom of Fig. 3A. Success in creating the Rb1ΔG allele was assessed by breeding to homozygosity and sequencing exons 15 and 17 (Fig. 3C), which demonstrated that the relevant codons were successfully changed to encode glutamate. Furthermore, GST-E7 and GST-E1A pulldowns from Rb1+/+ and Rb1ΔG/ΔG fibroblasts were used to confirm that ΔG-pRB in these extracts was capable of binding to viral oncoproteins (Fig. 3D). Lastly, we examined ΔG-pRB expression by Western blotting in comparison with wild-type and knockout MEFs (Fig. 3E). This revealed a slight increase in ΔG-pRB expression relative to the wild-type control. Taken together, these experiments reveal that the Rb1ΔG allele stably expresses pRB and that ΔG-pRB is capable of binding viral proteins through its pocket domain, suggesting that it is correctly folded.
FIG 3.
Gene targeting of the murine Rb1ΔG allele. (A) The region of the Rb1 gene targeted in this study is shown on top. The locations of exons are indicated by black rectangles, and relevant KpnI locations are shown. The targeting vector is shown in the middle with the locations of selectable markers (and additional KpnI and MscI cut sites), as well as newly introduced restriction sites and the mutations in exons 15 and 17 (amino acid positions are described using murine numbering). The bottom diagram shows the Rb1ΔG allele after homologous recombination and removal of the PGK-Neo cassette by breeding to EIIa-cre transgenic mice. The locations of 5′ and 3′ probes used in Southern blotting are shown. (B) Genomic DNA from two candidate clones was digested with the indicated restriction enzymes and analyzed by Southern blotting to demonstrate homologous recombination and single integration. T, targeted; R, random integration. (C) DNA was extracted from Rb1ΔG/ΔG embryos, and exons 15 and 17 were PCR amplified and sequenced. Electropherograms of the relevant regions are shown for wild type and Rb1ΔG/ΔG (using murine codon numbering). (D) Extracts were prepared from wild-type and Rb1ΔG/ΔG MEFs, and GST, GST-E7, or GST-E1A was added to precipitate pRB. The amount of precipitated pRB was determined by Western blotting. (E) Nuclear extracts from cells of the indicated genotypes were analyzed by SDS-PAGE and Western blotting for pRB and SP1.
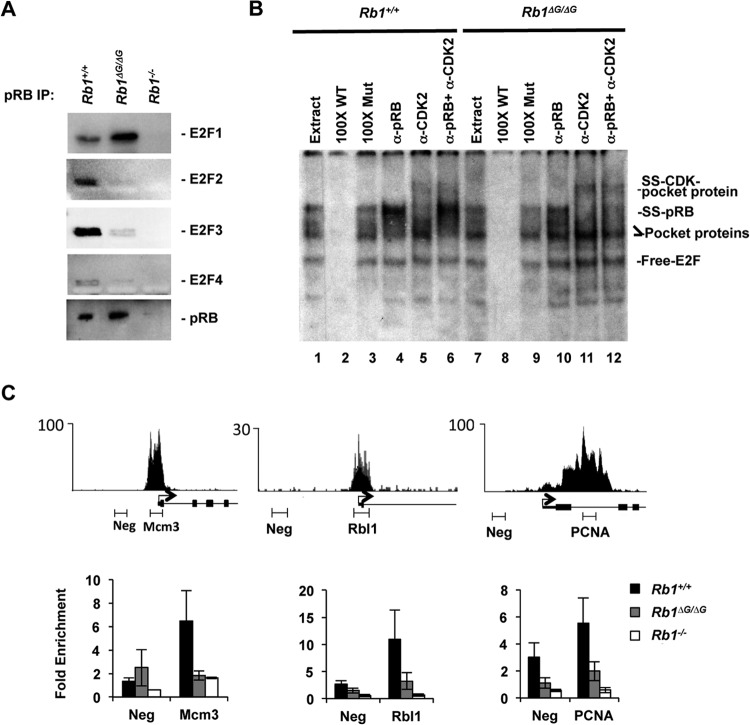
The properties of ΔG-pRB were investigated further in Rb1ΔG/ΔG cells by immunoprecipitation and Western blotting (Fig. 4A). Anti-pRB precipitates were blotted for E2F1, E2F2, E2F3, and E2F4, and in this analysis, E2F2, E2F3, and E2F4 levels were greatly reduced but E2F1 levels were considerably increased (Fig. 4A). This suggests that E2F1's interaction with pRB at its alternative interaction site may be competitive with pRB interactions with E2Fs through the general interaction site that we disrupted by mutation. Because the specific pRB-E2F1 complex prefers to bind to sequences such as those in the TA-p73 promoter and not the consensus E2F sites found in cell cycle genes, we used DNA binding specificity to identify the type of pRB-E2F1 complexes present in Rb1ΔG/ΔG extracts. To investigate the configuration of pRB-E2F1 complexes that form in Rb1ΔG/ΔG cells, we took advantage of the fact that the pRB-E2F1 specific interaction has low affinity for consensus E2F binding elements by EMSA (Fig. 4B). This analysis revealed that pRB-E2F complexes can readily be detected in wild-type extracts using an antibody supershift for pRB (Fig. 4B; compare lanes 3 and 4). However, the presence of other E2F-containing complexes that migrate to the same position obscures this complex, and they can be shifted by adding anti-Cdk2 antibodies (Fig. 4B; compare lanes 3 and 5). Gel shifts of Rb1ΔG/ΔG extracts demonstrate that pRB-E2F complexes are undetectable, even when anti-Cdk2 antibodies are used to shift other complexes away from this position in the gel (Fig. 4B; compare lanes 6 and 12). Based on this analysis, pRB-E2F complexes that are competent to repress cell cycle target genes appear to be absent in Rb1ΔG/ΔG cells.
FIG 4.
Loss of ΔG-pRB binding at E2F-responsive promoters. MEFs were induced to exit the cell cycle by serum withdrawal, and pRB's interaction with E2Fs was investigated. (A) Anti-pRB antibodies were used to precipitate pRB, and associated E2F transcription factors were detected by Western blotting. (B) EMSAs were performed to compare the abundance of all pRB-E2F-containing complexes in Rb1+/+ and Rb1ΔG/ΔG nuclear extracts. The migration positions of free E2F, E2Fs bound to RB family proteins (labeled as pocket protein), antibody-supershifted pRB-E2F complexes (SS-pRB), and antibody-supershifted complexes containing p107/p130–E2F-cyclin-CDK (SS-CDK-pocket protein) are all indicated to the right. Cold competitor probes (100× WT and 100× Mut), as well as the antibodies used to shift complexes, are listed on top. (C) Sequence read peaks from ChIP sequence analysis of human pRB are shown for Mcm3, Rbl1, and Pcna on top. The locations of negative-control and proximal promoter PCR amplicons used in this study are shown along with the transcriptional start site and exons. ChIP quantitative PCR analysis was undertaken for pRB at the indicated promoters. All ChIP enrichment values are scaled relative to a neutral genome location, the GAPDH promoter. Error bars indicate the standard errors.
Given the importance of disrupting endogenous pRB-E2F complexes in the interpretation of phenotypes in Rb1ΔG/ΔG mice, we also investigated this question by chromatin immunoprecipitation (ChIP). The presence of pRB-E2F complexes was quantitatively assessed by ChIP in which pRB was precipitated, and known E2F-responsive promoter regions were PCR amplified. ChIP sequence tracks for human pRB (32), at three E2F target genes that were used as a guide to design positive- and negative-control PCR amplicons to determine if ΔG-pRB can associate with these promoters (Fig. 4C, top). This experiment demonstrates that PCR amplification of sequences within the peak of pRB occupancy can readily detect wild-type pRB at these promoters, whereas it is reduced or missing at the negative-control location (Fig. 4C, bottom; compare black bars). PCR amplification of ΔG-pRB precipitates demonstrates that occupancies are similar between the peak and negative-control amplicons (Fig. 4C, bottom; compare gray bars), and these are generally equivalent to background levels defined by ChIP from Rb1−/− cells (Fig. 4C, bottom; compare white bars).
The characterization of pRB from Rb1ΔG/ΔG cells reveals that it has a specific defect in E2F interactions. Presumably because pRB can autoregulate itself through repression of E2F transcription factors (37), pRB is mildly increased in Rb1ΔG/ΔG cells. Despite its overexpression, and increased association with E2F1, pRB-E2F complexes that are capable of interaction with cell cycle E2F promoter elements are undetectable in Rb1ΔG/ΔG MEFs.
Normal cell cycle progression in asynchronous Rb1ΔG/ΔG MEFs.
Given the biochemical defect in pRB-E2F interactions described above, we sought to understand the functional consequences of this defect in transcriptional and cell cycle control. Asynchronously proliferating MEFs were pulse-labeled with BrdU and processed for flow cytometry to examine cell cycle phases in Rb1ΔG/ΔG cells. As shown in Fig. 5A, Rb1ΔG/ΔG MEFs have a cell cycle phase distribution very similar to that of the wild type, and this is clearly different from Rb1−/− cells that are characterized by lower G1, higher S-phase, and higher G2/M levels (38). We also carried out growth curves to see if the proliferation rate of Rb1ΔG/ΔG cells differs from those of knockout or wild-type controls. As shown in Fig. 5B, the quantity of cells increased similarly between genotypes over a 5-day period, suggesting that proliferation rates were similar across all genotypes. It is known that Rb1−/− MEFs enter S phase prematurely under these growth conditions, and this is detectable by smaller cell size in G1 (38). Figure 5C demonstrates that the average forward scatter size measurements are similar between wild-type and Rb1ΔG/ΔG cells but statistically different from that of Rb1−/− cells. Taken together with cell cycle phase proportions in Fig. 5A and similar proliferative rates in Fig. 5B, this suggests that cell cycle phase lengths, particularly G1, are unchanged between wild-type and Rb1ΔG/ΔG MEFs. Lastly, we also quantitated 8N cells between these genotypes of MEFs as a surrogate marker for endoreduplication. Again, Rb1ΔG/ΔG fibroblasts are similar to those of the wild type, whereas Rb1−/− cells have elevated levels of 8N cells (Fig. 5D). Taken together, these analyses demonstrate the surprising finding that impairing E2F transcriptional repression by pRB has little effect on cell cycle progression.
FIG 5.

Normal cell cycle progression in asynchronous Rb1ΔG/ΔG MEFs. (A) Asynchronously proliferating cell cultures were pulse-labeled and stained for BrdU incorporation along with total DNA using propidium iodide. The proportion of cells in each respective cell cycle phase was determined by flow cytometry. (B) Growth curve for Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells over a 5-day period of proliferation. (C and D) Mean forward scatter (C) and 8N DNA content (D) from cultures of asynchronously grown Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells. Error bars indicate 1 standard deviation from the mean (n = 3). An asterisk represents a statistically significant difference from the wild-type control (t test, P < 0.05).
Discrete defects in E2F transcriptional control in Rb1ΔG/ΔG fibroblasts in cell cycle arrest.
The retinoblastoma protein is perhaps best known for its role in mediating negative growth signals and arresting the cell cycle (39). For this reason, we investigated a number of cell cycle exit scenarios to determine the effects of the Rb1ΔG/ΔG genotype on proliferative control. In response to serum deprivation for 60 h, we discovered that known E2F transcriptional targets of pRB fail to be repressed in Rb1ΔG/ΔG cells compared to wild-type levels (Fig. 6A). Similar levels of gene expression were found in Rb1−/− cells following the same treatment. In addition, we also utilized BrdU labeling and flow cytometry to investigate proliferative control at the same time point following serum withdrawal, and this revealed that Rb1ΔG/ΔG fibroblasts respond equivalently to the wild type in their ability to exit the cell cycle (Fig. 6B). Conversely, Rb1−/− cells are defective for cell cycle withdrawal under these conditions (Fig. 6B). Since this cell cycle exit scenario reveals an instance in which Rb1ΔG/ΔG cells resemble a wild-type cell cycle arrest, we further investigated E2F regulation under these conditions. Expression levels of known E2F target genes were compared between wild-type, Rb1ΔG/ΔG, and Rb1−/− cells by microarray (Fig. 6C). Log2 ratios were generated comparing Rb1ΔG/ΔG expression levels relative to that of the wild type, as well as ratios of Rb1−/− expression levels to that of the wild type. Gene expression changes are clustered based on similarity, and this reveals that Rb1ΔG/ΔG cells are mostly defective in repressing E2F targets that function in DNA replication (Fig. 6C). Interestingly, this class of E2F target genes is known to be direct regulatory targets of pRB in senescence (32). Not surprisingly, since Rb1−/− cells fail to arrest under these serum deprivation conditions, most E2F targets display increased expression relative to the wild type in this microarray experiment (Fig. 6C). The implication of these experiments is that ΔG-pRB may have the ability to arrest the cell cycle independently of E2F repression at cell cycle genes.
FIG 6.
Discrete defects in E2F transcriptional control in Rb1ΔG/ΔG MEFs. (A) The relative expression levels of six pRB-regulated E2F cell cycle target genes are shown. Each transcript was quantified from serum-starved Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− fibroblasts with the relative level of message present in wild type scaled to 1. (B) Serum-starved cell cultures were pulse-labeled and stained for BrdU incorporation along with total DNA using propidium iodide. The proportion of cells in each respective cell cycle phase was determined by flow cytometry. (C) Microarray analysis was performed on serum-starved Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells, and E2F target gene expression is shown. Log ratios of expression of Rb1ΔG/ΔG or Rb1−/− relative to that of wild-type control are shown as a heat map, and genes were clustered based on similarity of expression. The genotypes of each lane are shown above. Categories of E2F target genes are shown to the right. (D) Serum-starved cells of the indicated genotypes were stimulated to reenter the cell cycle with 10% FBS. Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells were pulse-labeled with BrdU and harvested at the indicated time points. All graphs represent at least 3 individual experiments, and error bars indicate 1 standard deviation from the mean. An asterisk represents a statistically significant difference from the wild-type control (t test, P < 0.05).
Much of our knowledge of pRB-E2F control of transcription and their response to cyclin/CDK regulation comes from serum starvation and restimulation experiments. Under these circumstances, Rb1−/− and p107−/−; p130−/− double-knockout cells display accelerated progression through G1 and premature expression of E2F target genes (38, 40). We subjected wild-type, Rb1ΔG/ΔG, and Rb1−/− cells to serum stimulation and monitored their progress through G1 and into S phase by BrdU labeling and flow cytometry analysis. This experiment revealed that Rb1ΔG/ΔG cells progress rapidly through G1 in response to serum, and they reach peak BrdU incorporation at the same time as Rb1−/− cells. In this respect, Rb1ΔG/ΔG MEFs very much resemble knockout cells, and this suggests a context where pRB-dependent repression of E2F transcription is key to regulating cell cycle progression.
In addition to serum withdrawal, we also tested if ΔG-pRB could be activated through inhibition of cyclin D-associated kinases and gamma irradiation as a means to assess if it could arrest the cell cycle in response to these signals. Ectopic expression of p16Ink4a in proliferating wild-type, Rb1ΔG/ΔG, and Rb1−/− cells was used to induce a G1 arrest, and BrdU labeling and flow cytometry were used to measure DNA replication 3 days later (Fig. 7A). These data demonstrate that Rb1ΔG/ΔG cells exhibit a reduction in BrdU incorporation similar to that of the wild type, whereas Rb1−/− MEFs continue to proliferate. At the same time point that these cultures were analyzed for cell cycle progression, we also extracted RNA and measured relative expression levels of E2F target genes. These experiments reveal a modest but similar reduction in E2F target gene expression in Rb1+/+ and Rb1ΔG/ΔG MEFs (Fig. 7B). Surprisingly, a similar reduction in expression was observed for Ccne1 and Tyms in Rb1−/− cells (Fig. 7B). We also carried out PI-BrdU analysis and E2F gene expression profiling of Rb1 wild-type, mutant, and knockout MEFs in response to gamma irradiation (Fig. 7C and D). These experiments again revealed that Rb1ΔG/ΔG cells were fully capable of arresting proliferation in response to DNA breaks. Furthermore, investigation of E2F target gene expression again revealed that expression in Rb1ΔG/ΔG cells was reduced as much as it was in the wild type, and Rb1−/− cells also reduced expression of E2F targets even though they did not arrest.
FIG 7.
Rb1ΔG/ΔG MEFs are able to exit the cell cycle in response to p16 expression or DNA damage. (A) Fibroblast cells of the indicated genotypes were transduced with control or p16INK4A-expressing retroviruses. Following drug selection, cells were pulse-labeled with BrdU and the percentage of positive cells was determined by flow cytometry. All graphs represent at least 3 individual experiments, and error bars indicate 1 standard deviation from the mean. An asterisk represents a statistically significant difference from the wild-type control (t test, P < 0.05). (B) Expression of E2F target genes was measured at the same time point as was BrdU labeling in panel A. Fold repression of six pRB-dependent E2F cell cycle target genes is shown for Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells. Measurements are expressed as a log2 ratio of p16-expressing cells over empty vector for each respective gene target and genotype of cells. All graphs represent at least 3 individual experiments, and error bars indicate 1 standard deviation from the mean. (C) Fibroblast cells of the indicated genotypes were treated with 15 Gy of radiation or allowed to grow asynchronously. Forty-eight hours following exposure, cells were pulse-labeled with BrdU and the percentage of positive cells was determined by flow cytometry. Statistical analysis is as reported for panel A. (D) Following irradiation, the expression of E2F target genes was measured in Rb1+/+, Rb1ΔG/ΔG, and Rb1−/− cells. Gene expression measurements were made as in panel B.
In summary, Rb1ΔG/ΔG MEFs undertake relatively normal progression through the cell cycle during asynchronous proliferation. In response to serum deprivation, ectopic p16 expression, and gamma irradiation, Rb1ΔG/ΔG cells again display wild-type levels of cell cycle arrest activity. Importantly, Rb1ΔG/ΔG cells exhibit an acceleration through G1 in response to serum stimulation that very much resembles the defect found in Rb1−/− cells. From this perspective, Rb1ΔG/ΔG MEFs have defects in cell cycle control that are consistent with restraining cell cycle entry rather than facilitating cell cycle exit. During some situations of cell cycle arrest, E2F target gene expression in wild-type, Rb1ΔG/ΔG, and Rb1−/− cells was decreased. While this was surprising, it should be noted that p107 and p130 are also capable of repressing E2F target genes and that these pocket proteins would be expected to be dephosphorylated and active in cells that successfully arrest cell cycle progression. The implications of these experiments for linking E2F transcriptional control and cell cycle progression will be discussed later.
Cell cycle control in development and homeostasis of Rb1ΔG/ΔG mice.
In addition to the cell cycle defects observed in Rb1−/− fibroblasts, Rb1 knockout mice also have defects in development that lead to embryonic lethality beginning at 13.5 days of gestation (E13.5) (41–43). We investigated Rb1ΔG/ΔG mutants to characterize the role of E2F transcriptional repression in these developmental contexts. Live Rb1ΔG/ΔG mice were obtained and are indistinguishable from wild-type littermates on a gross anatomical level (Fig. 8A and B). We also examined embryos at distinct developmental stages, and newborns, to characterize the viability of Rb1ΔG/ΔG mice. As shown in Table 1, Rb1ΔG/ΔG embryos were obtained at the expected Mendelian ratios up until birth. After birth, approximately half of the Rb1ΔG/ΔG animals die. Based on these observations, we focused our investigation at E18.5 to search for developmental defects. Figure 8C shows hematoxylin and eosin (H&E) staining as well as BrdU immunohistochemistry on sections from a number of major organs and tissues. We observed that approximately half of the Rb1ΔG/ΔG animals display an atrophy phenotype in their skeletal muscle without accompanying increases in proliferation (Fig. 8C and D). Most notably, we observed this phenotype in the diaphragm, which may explain newborns that were observed struggling to breathe. Analysis of dying and surviving newborns at postnatal day 0.5 (P0.5) revealed that defective skeletal muscle correlates with poor survival (Fig. 9A). This prompted us to investigate muscle development in Rb1ΔG/ΔG mice and cells. We isolated RNA from skeletal muscle and analyzed E2F target gene expression. As shown in Fig. 9B, there were few differences between wild-type muscle, histologically normal muscle from Rb1ΔG/ΔG mice, and atrophied muscle from Rb1ΔG/ΔG animals. This further suggests that the muscle defects observed in some Rb1ΔG/ΔG mice were not caused by aberrant proliferation or loss of transcription of these E2F cell cycle target genes. However, to investigate the question of cell cycle exit in Rb1ΔG/ΔG muscle development in a separate context where we can better detect proliferation, we generated myotubes in culture. Interestingly, differentiation of wild-type, Rb1ΔG/ΔG, or Rb1−/− MEFs into myotubes, followed by restimulation with serum, indicated that only the Rb1−/− controls can be induced to incorporate BrdU (Fig. 9C and D). This suggests that muscle defects in Rb1ΔG/ΔG cells are very unlikely to be caused by cell cycle arrest deficiency, and our gene expression profiling suggests that E2F target expression levels do not correlate with this phenotype.
FIG 8.
Viability of Rb1ΔG/ΔG mutant mice and proliferative control during early development. (A) Photographs of 1-year-old wild-type and Rb1ΔG/ΔG mice. (B) Photographs of newborn pups at P0.5. The wild-type animal and the Rb1ΔG/ΔG mutant were both viable. (C) Embryos were isolated at E18.5 and fixed in formalin, and serial sections were cut and stained with either hematoxylin and eosin or BrdU to assess tissue architecture and proliferation. Representative images of major organs, including heart, lung, diaphragm, skeletal muscle, intestine, and kidney, from Rb1+/+ and Rb1ΔG/ΔG mice are included. In total, 9 embryos from each genotype were examined and stained for BrdU incorporation. (D) Quantification of BrdU-positive cells in the indicated tissue types. Error bars indicate 1 standard deviation from the mean (n = 4). Bars, 20 μm.
TABLE 1.
Early development of Rb1ΔG/ΔG micea
| Genotype | No. of offspring at time point: |
|||
|---|---|---|---|---|
| E13.5 | E18.5 | P0.5 | P14 | |
| Rb1+/+ | 11 (16) | 24 (23) | 15 (15) | 163 (127) |
| Rb1ΔG/+ | 34 (32) | 46 (46) | 29 (31) | 291 (254) |
| Rb1ΔG/ΔG | 19 (16) | 22 (23) | 9 live; 9 dead (15) | 54 (127) |
| Total | 64 | 92 | 62 | 508 |
Intercrosses between Rb1ΔG/+ mice were used to determine the frequency of wild-type, heterozygous, and Rb1ΔG/ΔG animals. Numbers in parentheses represent the expected values based on Mendelian predictions.
FIG 9.
Muscle degeneration in Rb1ΔG/ΔG embryos. (A) H&E staining of tissue sections from newborn diaphragms of the indicated genotypes. The leftmost Rb1ΔG/ΔG sample is from a viable newborn, and the rightmost Rb1ΔG/ΔG sample is from a newborn that was found struggling to breathe and died shortly after birth. Analogous sections from embryos isolated at E18.5 with either normal or atrophied diaphragm (and other skeletal muscle) are presented. (B) The relative expression levels of six pRB-dependent E2F cell cycle target genes are shown. Each transcript was quantified from RNA isolated from dissected quadriceps muscle from E18.5 mice. The relative level of message present in the wild type was scaled to 1, and the Rb1ΔG/ΔG embryos were stratified into normal and atrophied based upon H&E staining of skeletal muscle. Error bars indicate 1 standard deviation from the mean (n = 3). Statistically significant differences are indicated by an asterisk (t test, P > 0.05). (C) Wild-type, Rb1ΔG/ΔG, and Rb1−/− MEFs were infected with a MyoD-expressing retrovirus and induced to differentiate into myotubes. Upon differentiation, cells were stimulated with 15% FBS and labeled with BrdU for 24 h. Cells were fixed and stained for myosin heavy chain (MHC), BrdU incorporation, and DNA as shown. The arrow indicates an MHC- and BrdU-positive cell. (D) The frequency of double-positive BrdU- and MHC-stained cells, as well as single-positive cells that stained only for BrdU, was determined. Graphs indicate the averages, error bars represent 1 standard deviation, and an asterisk indicates a statistically significant difference from the wild-type control (t test, P < 0.05). Bars, 20 μm.
In addition to the developmental stage where some Rb1ΔG/ΔG animals fail to survive, we also searched for examples of defective proliferative control and E2F gene derepression in adult mice. Previously, we have demonstrated that transforming growth factor β (TGF-β)-induced arrest of mammary epithelium requires E2F repression (44). Accordingly, we investigated the histology of mammary ductal epithelium and determined its state in 6- to 8-week-old females (Fig. 10A and B). Mutant ducts were hyperplastic, as characterized by additional layers of epithelium, and these were more frequent in Rb1ΔG/ΔG females than in controls (Fig. 10B). From this perspective, loss of E2F repression by ΔG-pRB leads to excessive proliferation but does not compromise the ability of Rb1ΔG/ΔG females to nurse their pups. Interestingly, numerous other tissues display normal histology with little evidence of hyperplasia. Figures 10C, E, and G show normal histology of Rb1ΔG/ΔG lungs, cardiac muscle, and brain, respectively. We have also examined skeletal muscle, kidney, liver, and intestines with similar results (Fig. 10I and data not shown). Measurement of pRB-E2F target transcript levels revealed that upregulation of at least some of these genes is detectable in these tissues (Fig. 10D, F, and H). We also investigated cell cycle arrest and E2F target gene expression in columnar epithelial cells from the surface of intestinal villi, as they are known to require pRB function for cell cycle arrest. Unlike most other tissue isolates that are a mix of cell types, they can be isolated to allow gene expression to be assessed specifically within this cell type (45). Rb1ΔG/ΔG intestinal epithelium retained normal tissue structure, characterized by a single layer of polarized epithelial cells (Fig. 10I), despite derepression of E2F transcriptional targets in these cells (Fig. 10J). BrdU labeling and staining of intestines further revealed that DNA synthesis was absent in epithelial cells of Rb1ΔG/ΔG and wild-type villi, but conditional deletion of Rb1 resulted in BrdU labeling of more than 10% of epithelial cells (Fig. 10K and L).
FIG 10.
Limited hyperplasia in tissues of Rb1ΔG/ΔG mice. (A) Sections of mammary ducts from 8-week-old wild-type and Rb1ΔG/ΔG mice stained with H&E. Arrows indicate cell layers in the lumen of the duct. (B) Quantification of the proportions of hyperplastic (hp) ducts found in wild-type and Rb1ΔG/ΔG mammary glands. Ducts three or more cells thick were scored as hyperplastic. Proportions were compared by χ2 test (*, significant difference, P < 0.05). (C) H&E staining of lung tissue from 8-week-old mice. (D) Relative mRNA levels for cyclin E1 (Ccne1), cyclin A2 (Ccna2), p107 (Rbl1), thymidylate synthase (Tyms), Pcna, and Mcm3 were also determined in wild-type and Rb1ΔG/ΔG lung tissue from 8-week-old mice. Wild-type expression levels are scaled to 1. (E and F) H&E staining of cardiac muscle from 6- to 8-week-old mice is shown along with expression analysis of E2F transcriptional targets. (G and H) H&E staining of brain tissue from 8-week-old mice. Accompanying analysis of E2F transcriptional targets from this tissue is shown to the right. (I) H&E staining of crypts and villi from the small intestines of 8-week-old mice. (J) The relative expression level of six E2F cell cycle target genes in mRNA prepared from isolated villi is shown. (K) Eight-week-old mice of the indicated genotypes were injected with BrdU 2 h prior to sacrifice. Tissue sections from intestines were stained for BrdU incorporation (red) and DNA (blue). (L) The frequency of BrdU-positive nuclei in columnar epithelial cells of villi is shown. Each graph represents at least 3 individual experiments, and error bars indicate 1 standard deviation from the mean. An asterisk represents a statistically significant difference from the wild-type control (t test, P < 0.05). Bars, 20 μm (5 μm in panel C).
Taken together, results of histological analysis of developing Rb1ΔG/ΔG mice demonstrate that loss of E2F repression by ΔG-pRB is largely tolerated during developmental proliferative control events. Most tissues tested demonstrate derepression of some E2F targets with few examples of hyperplasia, and the strongest developmental phenotype, partially penetrant muscle atrophy, appears not to be proliferation related.
Rb1ΔG/ΔG mice do not develop spontaneous tumors.
Possession of one null allele of Rb1 predisposes mice to develop pituitary tumors within the first year of life (46, 47). Chimeric mice containing Rb1−/− cells, or conditionally deleted for Rb1 in the pituitary, succumb to pituitary tumors in the first 4 months of life (48, 49). Consequently, we followed a cohort of wild-type and Rb1ΔG/ΔG mice to investigate the incidence of spontaneous tumor formation in these animals (Fig. 11A). Wild-type and Rb1ΔG/ΔG animals were tumor free beyond 1.5 years of life; in contrast, Rb1+/− control mice developed pituitary tumors with a mean survival of 400 days. This suggests that loss of E2F repression by pRB alone is not sufficient to predispose these mice to cancer.
FIG 11.

Rb1ΔG/ΔG mutant mice have normal life expectancy. (A) Mice of the indicated genotypes were allowed to age to determine their life span and natural demise. Kaplan-Meier plots depict tumor-free survival of the indicated genotypes. Tick marks indicate the ages at which individual mice were analyzed anatomically and histologically for abnormalities. Wild-type (n = 40) and Rb1ΔG/ΔG (n = 42) mice are significantly different from Rb1+/− (n = 9) mice (log rank test, P < 0.05). (B) Photographs of pituitary glands from wild-type, Rb1ΔG/ΔG, and Rb1+/− mice are representative of findings for animals between 8 and 12 months of age. Arrows identify the locations of pituitary glands. Bars, 1 cm. (C) H&E staining of a tissue section from the pituitary gland of a 2-year-old Rb1ΔG/ΔG mutant. The different lobes of the pituitary are labeled: A, anterior lobe; I, intermediate lobe; P, posterior lobe. Bar, 1 mm.
While Rb1ΔG/ΔG mutants did not succumb to pituitary or other cancers, we searched for evidence of neoplastic lesions in these mice (Fig. 11B). Pituitary adenocarcinomas are known to be slowly progressing tumors, so we searched for evidence of hyperplasia in 8- to 11-month-old animals, as well as in aged Rb1ΔG/ΔG mice. However, gross morphology of Rb1ΔG/ΔG pituitaries was indistinguishable from that of wild-type controls (Fig. 11B). Furthermore, H&E staining of pituitaries failed to reveal aberrant proliferation even in aged Rb1ΔG/ΔG mutants (Fig. 11C). Lastly, it has been demonstrated that pituitary tumorigenesis can be suppressed in Rb1+/− mice if the intermediate lobe fails to properly develop (50). We note that the intermediate lobe is present in Rb1ΔG/ΔG animals and shows normal histology (Fig. 11C, marked by “I”).
Throughout this report, we have provided evidence that pRB's specific interaction with E2F1 represents a separate biochemical function that is not related to the control of cell cycle E2F target genes. However, we decided to challenge this interpretation by crossing Rb1ΔG/ΔG mutants with E2f1−/− mice to see if compound mutant mice or cells have more severe phenotypes that do Rb1ΔG/ΔG mice alone. We found that compound mutant mice displayed a reduction in survival similar to that of Rb1ΔG/ΔG mutants based on the genotype of P14 pups (Fig. 12A and Table 1). In addition, preparation of compound mutant fibroblasts and subjection of them to serum withdrawal growth arrest failed to reveal a defect in this cell cycle exit paradigm (Fig. 12B). Lastly, we followed small cohorts of E2f1−/− and Rb1ΔG/ΔG; E2f1−/− mice over a 450-day period and failed to observe spontaneous tumor formation in compound mutant mice (Fig. 12C). Based on these further analyses of pRB-E2F1 function, we conclude that the specific pRB-E2F1 interaction is unlikely to replace general E2F interactions to control cell cycle E2F target genes.
FIG 12.

E2F1 loss does not affect cell cycle control, viability, or cancer susceptibility in Rb1ΔG/ΔG mice. (A) Genotypes of offspring at P14 from a cross of compound heterozygote Rb1ΔG/+; E2f1+/− mice. The expected number of mice based on Mendelian ratios is presented in parentheses. (B) Serum-starved cell cultures from compound mutant fibroblasts of the indicated genotypes were pulse-labeled and stained for BrdU incorporation along with total DNA using propidium iodide. The proportion of cells in each respective cell cycle phase was determined by flow cytometry. (C) Mice of the indicated genotypes were allowed to age to determine their life span and natural demise. Kaplan-Meier plots depict tumor-free survival of the indicated genotypes. Tick marks indicate individual mice that were analyzed anatomically and histologically for abnormalities.
Collectively, our analysis of cancer incidence in Rb1ΔG/ΔG mice suggests that loss of E2F transcriptional repression is insufficient to cause tumor formation and has limited effects on cell cycle control. Our data suggest that tumor suppression by pRB is likely to be a more complex process than this individual biochemical function.
DISCUSSION
The strongest phenotype observed in Rb1ΔG/ΔG mice is partially penetrant muscle degeneration. Previous work has suggested that pRB plays a crucial role in regulating autophagy and that in its absence muscle cells catabolize themselves, leading to an apoptosis-independent death (51). Importantly, inhibition of autophagy rescues this phenotype even in the absence of pRB function (51). Intriguingly, Araki et al. have recently demonstrated that loss of nuclear localization by pRB interferes with sarcomere structure in skeletal muscle, leading to a nonapoptotic cell death (52). This may offer a nontranscriptional explanation of our phenotype, as loss of E2F binding likely reduces pRB anchorage in the nucleus. It is not clear if both of these mechanisms or others are contributing to the observed defect in Rb1ΔG/ΔG mice, but our results further strengthen the role for pRB in muscle development. We expect that the partial penetrance of this muscle phenotype is due to genetic modifiers. Crossing homozygous mutants leads to full viability of offspring (data not shown), indicating that this trait could be bred out of our colony. We note that there are other examples of genetic background effects on pRB- and E2F-related phenotypes such as the role of E2F3 in proliferation (53), the impact of E2F1 on tumorigenesis (54), and the effects of p107 and p130 on proliferative control in development (55, 56).
Interactions between pRB and E2Fs are commonly considered to be the mechanism that regulates the transition between the G1 and S phases of the cell cycle (1). Our data demonstrate that the Rb1ΔG mutation disrupts this regulatory interaction, but mice carrying this allele tolerate its effects and display few defects in cell cycle control. The strongest defect in proliferative control that we observed in Rb1ΔG/ΔG mutants was in restraining cell cycle entry following serum stimulation. This is intriguing because a number of key cell cycle regulators, D- and E-type cyclins, are dispensable for asynchronous proliferation and knockout embryos develop almost to term (57, 58). Strikingly, knockout MEFs deficient for all D- or E-type cyclins are impaired for cell cycle reentry following serum starvation. This suggests that the cellular response to serum stimulation may be fundamentally different from cell cycle exit paradigms as cyclin-CDK and now pRB-E2F functions are essential in coordinating cell cycle reentry in response to growth factor signaling.
We suggest that the data in this report should be considered carefully. While our biochemical measurements of pRB-E2F interaction and regulation indicate a strong loss of function in the ΔG-pRB protein, it is important to separate its effects in isolation from the broader role of E2F transcriptional control in cell proliferation. For example, p16 expression and gamma irradiation experiments indicate that Rb1ΔG/ΔG cells can arrest; however, E2F transcript levels drop even at E2F target genes, where we show that pRB no longer localizes. In these arrest assays, p16 and gamma irradiation inhibit cyclin-CDK activity and the other pocket proteins become active and can block E2F transcription. Importantly, it is known that p107/p130 and E2F4/E2F5 are required to arrest in response to p16 (59, 60). For this reason, we think that our data do not indicate that the E2F transcriptional regulatory network is unnecessary for proliferative control but rather that it retains some function independent of pRB. This is reinforced by the reduction in E2F gene expression that takes place in Rb1−/− cells even though they do not arrest proliferation in response to p16 or gamma irradiation. Lastly, in 1998 Dyson (61) described the lack of E2F regulation data in cell cycle exit paradigms such as these as a “quirk” of the pRB-E2F literature, and we suggest that our study adds valuable new information in this area.
E2F gene expression levels in Rb1ΔG/ΔG embryonic and adult tissues are upregulated at a modest level, and there is no evidence of increased proliferation in these tissues. Conversely, conditional deletion of Rb1 in intestinal epithelia as reported by Chong et al. displays approximately 20- to 40-fold-increased expression of E2F target genes (45). In considering these Rb1 knockout data, it is important to remember that loss of Rb1 in these cells is accompanied by proliferation. In quiescent cells, p130 is dephosphorylated and active for repression of E2F target genes (40, 62). Furthermore, when cells are stimulated to proliferate, E2Fs and Myc transcribe E2F genes, further leading to amplification of E2F transcription levels (61). For this reason, it is difficult to examine the expression data in proliferating knockout cells and quiescent Rb1ΔG/ΔG mutant cells and determine the level of expression that is necessary to advance the cell cycle. As with our cell cycle exit experiments, these measurements also reinforce the idea that loss of pRB-E2F transcriptional control does not necessarily stimulate advancement of the cell cycle or fully activate the E2F transcriptional program.
Based on data from this study, we propose the models in Fig. 13. Figure 13A shows the RB pathway in cell cycle regulatory control as it is widely accepted, in which pRB control of E2F transcription is central to the regulation of entry into S phase. Figure 13B depicts cell cycle regulation in the absence of pRB repression of E2Fs in which alternate pathways allow control of S-phase entry even in the absence of physical control of E2Fs by pRB (14). Mechanisms in which pRB function influences cell cycle control outside E2F transcription, but before the commitment to DNA replication, may prove to be important means to influence cell cycle progression.
FIG 13.

Model for cell cycle control in Rb1ΔG/ΔG mutant cells. (A) Cell cycle regulation by the RB pathway in normal cells. pRB restricts E2F transcription to prevent Cdk2 activation, negative feedback to pRB, and cell cycle advancement. (B) ΔG-pRB is responsive to p16 activation and, in the absence of regulation of E2F transcription factors, retains the ability to prevent cell cycle entry.
ACKNOWLEDGMENTS
We thank numerous colleagues for advice and reagents. We specifically thank Kristen Kernohan and Nathalie Bérubé for their assistance with developing and analyzing our ChIP experiments.
M.J.C. is the recipient of a CIHR M.D./Ph.D. studentship. M.J.C., M.J.T., S.T., and S.M.F. are/were members of the CIHR-Strategic Training Program in Cancer Research and Technology Transfer. F.A.D. is the Wolfe Senior Fellow in Tumor Suppressor Genes. This work was funded by an operating grant from the CIHR (MOP-89765) to F.A.D. and a grant from the NIH (RO1 CA98956) to J.M.P. and G.L.
Footnotes
Published ahead of print 24 March 2014
REFERENCES
- 1.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100:57–70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 2.Massague J. 2004. G1 cell-cycle control and cancer. Nature 432:298–306. 10.1038/nature03094 [DOI] [PubMed] [Google Scholar]
- 3.Classon M, Harlow E. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910–917. 10.1038/nrc950 [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, McCormick F. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103–112. 10.1016/S1535-6108(02)00102-2 [DOI] [PubMed] [Google Scholar]
- 5.DeCaprio JA. 2009. How the Rb tumor suppressor structure and function was revealed by the study of adenovirus and SV40. Virology 384:274–284. 10.1016/j.virol.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 6.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 7.Chellappan S, Kraus KB, Kroger B, Munger K, Howley PM, Phelps WC, Nevins JR. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and the human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. U. S. A. 89:4549–4553. 10.1073/pnas.89.10.4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583–7591. 10.1128/JVI.75.16.7583-7591.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiebert SW. 1993. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol. Cell. Biol. 13:3384–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. 1992. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 6:177–185. 10.1101/gad.6.2.177 [DOI] [PubMed] [Google Scholar]
- 11.Qin X-Q, Livingston DM, Ewen M, Sellers WR, Arany Z, Kaelin WG. 1995. The transcription factor E2F-1 is a downstream target of RB action. Mol. Cell. Biol. 15:742–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin XQ, Chittenden T, Livingston DM, Kaelin WG., Jr 1992. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 6:953–964. 10.1101/gad.6.6.953 [DOI] [PubMed] [Google Scholar]
- 13.Dick FA, Dyson N. 2003. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 12:639–649. 10.1016/S1097-2765(03)00344-7 [DOI] [PubMed] [Google Scholar]
- 14.Dick FA, Rubin SM. 2013. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 14:297–306. 10.1038/nrm3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA. 2009. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15:184–194. 10.1016/j.ccr.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti A, Screpanti I, Balsano C, Alesse E, Gulino A, Levrero M. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5:552–558. 10.1038/ncb998 [DOI] [PubMed] [Google Scholar]
- 17.Avni D, Yang Martelli HF, Hofmann F, ElShamy WM, Ganesan S, Scully R, Livingston DM. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12:735–746. 10.1016/S1097-2765(03)00355-1 [DOI] [PubMed] [Google Scholar]
- 18.Mendoza-Maldonado R, Paolinelli R, Galbiati L, Giadrossi S, Giacca M. 2010. Interaction of the retinoblastoma protein with Orc1 and its recruitment to human origins of DNA replication. PLoS One 5:e13720. 10.1371/journal.pone.0013720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ. 2003. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 22:1445–1460. 10.1038/sj.onc.1206264 [DOI] [PubMed] [Google Scholar]
- 20.Chau BN, Pan CW, Wang JY. 2006. Separation of anti-proliferation and anti-apoptotic functions of retinoblastoma protein through targeted mutations of its A/B domain. PLoS One 1:e82. 10.1371/journal.pone.0000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cecchini MJ, Dick FA. 2011. The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem. J. 434:297–308. 10.1042/BJ20101210 [DOI] [PubMed] [Google Scholar]
- 22.Julian LM, Palander O, Seifried LA, Foster JE, Dick FA. 2008. Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene 27:1572–1579. 10.1038/sj.onc.1210803 [DOI] [PubMed] [Google Scholar]
- 23.Carnevale J, Palander O, Seifried LA, Dick FA. 2012. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol. Cell. Biol. 32:900–912. 10.1128/MCB.06286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calbo J, Parreno M, Sotillo E, Yong T, Mazo A, Garriga J, Grana X. 2002. G1 cyclin/cyclin-dependent kinase-coordinated phosphorylation of endogenous pocket proteins differentially regulates their interactions with E2F4 and E2F1 and gene expression. J. Biol. Chem. 277:50263–50274. 10.1074/jbc.M209181200 [DOI] [PubMed] [Google Scholar]
- 25.Cecchini MJ, Amiri M, Dick FA. 2012. Analysis of cell cycle position in mammalian cells. J. Vis. Exp. 2012(59):3491. 10.3791/3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifried LA, Talluri S, Cecchini M, Julian LM, Mymryk JS, Dick FA. 2008. pRB-E2F1 complexes are resistant to adenovirus E1A-mediated disruption. J. Virol. 82:4511–4520. 10.1128/JVI.02713-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field SJ, Tsai F-Y, Kuo F, Zubiaga AM, Kaelin JWG, Livingston DM, Orkin SH, Greenberg ME. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549–561. 10.1016/S0092-8674(00)81255-6 [DOI] [PubMed] [Google Scholar]
- 28.Dick FA, Sailhamer E, Dyson NJ. 2000. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715–3727. 10.1128/MCB.20.10.3715-3727.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392–8396. 10.1073/pnas.90.18.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novitch BG, Mulligan GJ, Jacks T, Lassar AB. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441–456. 10.1083/jcb.135.2.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talluri S, Isaac CE, Ahmad M, Henley SA, Francis SM, Martens AL, Bremner R, Dick FA. 2010. A G1 checkpoint mediated by the retinoblastoma protein that is dispensable in terminal differentiation but essential for senescence. Mol. Cell. Biol. 30:948–960. 10.1128/MCB.01168-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17:376–387. 10.1016/j.ccr.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 36.Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM, Pipas JM. 2005. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. J. Virol. 79:7492–7502. 10.1128/JVI.79.12.7492-7502.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan B, Chang CY, Jones D, Lee WH. 1994. The transcription factor E2F-1 mediates the autoregulation of RB gene expression. Mol. Cell. Biol. 14:299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 16:2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 40.Hurford RK, Jr, Cobrinik D, Lee MH, Dyson N. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447–1463. 10.1101/gad.11.11.1447 [DOI] [PubMed] [Google Scholar]
- 41.Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. 1992. Requirement for a functional Rb-1 gene in murine development. Nature 359:328–330. 10.1038/359328a0 [DOI] [PubMed] [Google Scholar]
- 42.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. 1992. Effects of an Rb mutation in the mouse. Nature 359:295–300. 10.1038/359295a0 [DOI] [PubMed] [Google Scholar]
- 43.Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288–294. 10.1038/359288a0 [DOI] [PubMed] [Google Scholar]
- 44.Francis SM, Bergsied J, Isaac CE, Coschi CH, Martens AL, Hojilla CV, Chakrabarti S, Dimattia GE, Khoka R, Wang JY, Dick FA. 2009. A functional connection between pRB and transforming growth factor beta in growth inhibition and mammary gland development. Mol. Cell. Biol. 29:4455–4466. 10.1128/MCB.00473-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. 2009. E2f1–3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462:930–934. 10.1038/nature08677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison DJ, Hooper ML, Armstrong JF, Clarke AR. 1995. Effects of heterozygosity for the Rb-1t19neo allele in the mouse. Oncogene 10:1615–1620 [PubMed] [Google Scholar]
- 47.Hu N, Gutsmann A, Herbert DC, Bradley A, Lee W-H, Lee EY. 1994. Heterozygous Rb-1 delta 20/+ mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene 9:1021–1027 [PubMed] [Google Scholar]
- 48.Vooijs M, van der Valk M, te Riele H, Berns A. 1998. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene 17:1–12. 10.1038/sj.onc.1202169 [DOI] [PubMed] [Google Scholar]
- 49.Williams BO, Schmitt EM, Remington L, Bronson RT, Albert DM, Weinbert RA, Jacks T. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, Zhu L. 2010. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nat. Genet. 42:83–88. 10.1038/ng.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciavarra G, Zacksenhaus E. 2010. Rescue of myogenic defects in Rb-deficient cells by inhibition of autophagy or by hypoxia-induced glycolytic shift. J. Cell Biol. 191:291–301. 10.1083/jcb.201005067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araki K, Kawauchi K, Hirata H, Yamamoto M, Taya Y. 2013. Cytoplasmic translocation of the retinoblastoma protein disrupts sarcomeric organization. eLife 2:e01228. 10.7554/eLife.01228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690–703 [PMC free article] [PubMed] [Google Scholar]
- 54.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. 1998. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat. Genet. 18:360–364. 10.1038/ng0498-360 [DOI] [PubMed] [Google Scholar]
- 55.LeCouter JE. 1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125:4669–4679 [DOI] [PubMed] [Google Scholar]
- 56.LeCouter JE, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. 1998. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol. Cell. Biol. 18:7455–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. 2003. Cyclin E ablation in the mouse. Cell 114:431–443. 10.1016/S0092-8674(03)00645-7 [DOI] [PubMed] [Google Scholar]
- 58.Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, Akashi K, Sicinski P. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118:477–491. 10.1016/j.cell.2004.07.025 [DOI] [PubMed] [Google Scholar]
- 59.Bruce JL, Hurford RKJ, Classon M, Koh J, Dyson N. 2000. Requirements for cell cycle arrest by p16INK4a. Mol. Cell 6:737–742. 10.1016/S1097-2765(00)00072-1 [DOI] [PubMed] [Google Scholar]
- 60.Gaubatz S, Lindeman GJ, Jakoi L, Nevins JR, Livingston DM, Rempel RE. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729–735. 10.1016/S1097-2765(00)00071-X [DOI] [PubMed] [Google Scholar]
- 61.Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245–2262. 10.1101/gad.12.15.2245 [DOI] [PubMed] [Google Scholar]
- 62.Moberg K, Starz MA, Lees JA. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceno C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ. 2003. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc. Natl. Acad. Sci. U. S. A. 100:2363–2368. 10.1073/pnas.0436813100 [DOI] [PMC free article] [PubMed] [Google Scholar]