ABSTRACT
Tick-borne encephalitis virus (TBEV) and Omsk hemorrhagic fever virus (OHFV) are highly pathogenic tick-borne flaviviruses; TBEV causes neurological disease in humans, while OHFV causes a disease typically identified with hemorrhagic fever. Although TBEV and OHFV are closely related genetically, the viral determinants responsible for these distinct disease phenotypes have not been identified. In this study, chimeric viruses incorporating components of TBEV and OHFV were generated using infectious clone technology, and their pathological characteristics were analyzed in a mouse model to identify virus-specific determinants of disease. We found that only four amino acids near the C terminus of the NS5 protein were primarily responsible for the development of neurological disease. Mutation of these four amino acids had no effect on viral replication or histopathological features, including inflammatory responses, in mice. These findings suggest a critical role for NS5 in stimulating neuronal dysfunction and degeneration following TBEV infection and provide new insights into the molecular mechanisms underlying the pathogenesis of tick-borne flaviviruses.
IMPORTANCE Tick-borne encephalitis virus (TBEV) and Omsk hemorrhagic fever virus (OHFV) belong to the tick-borne encephalitis serocomplex, genus Flavivirus, family Flaviviridae. Although TBEV causes neurological disease in humans while OHFV causes a disease typically identified with hemorrhagic fever. In this study, we investigated the viral determinants responsible for the different disease phenotypes using reverse genetics technology. We identified a cluster of only four amino acids in nonstructural protein 5 primarily involved in the development of neurological disease in a mouse model. Moreover, the effect of these four amino acids was independent of viral replication property and did not affect the formation of virus-induced lesions in the brain directly. These data suggest that these amino acids may be involved in the induction of neuronal dysfunction and degeneration in virus-infected neurons, ultimately leading to the neurological disease phenotype. These findings provide new insight into the molecular mechanisms of tick-borne flavivirus pathogenesis.
INTRODUCTION
Tick-borne encephalitis virus (TBEV) and Omsk hemorrhagic fever virus (OHFV) belong to the tick-borne encephalitis (TBE) serocomplex, genus Flavivirus, family Flaviviridae, which includes TBEV, Powasson virus, Langat virus, Louping ill virus, OHFV, Alkhurma virus (ALKV), and Kyasanur Forest disease virus (KFDV) (1). Although the majority of TBE complex viruses cause encephalitis, OHFV, ALKV, and KFDV are known to cause hemorrhagic disease.
TBE is endemic in Europe, Russia, and Far-East Asia, where about 10,000 cases are reported annually. TBEV can be divided into three subtypes: the Far-Eastern subtype, known as Russian spring summer encephalitis virus; the European subtype, known as Central European encephalitis subtype; and the Siberian subtype (2). In human patients, TBEV produces febrile illness, characterized by flu-like symptoms, followed by neurological symptoms, including febrile headache, visual changes, paralysis, seizures, and coma (3). Among TBEV subtypes, the Far-Eastern subtype is particularly virulent, with mortality rates ranging from 20 to 60%. TBE therefore represents a significant threat to public health in regions where TBEV is endemic.
OHFV is endemic in a localized region near the Omsk Oblast of southwestern Siberia (4). Human OHFV infection results in clinical symptoms quite different from those caused by TBEV, characterized by high continuous fever, headache, muscle pain, dehydration, and often a distinct hemorrhagic syndrome, including visceral hemorrhages of the nose, gums, uterus, and lungs. Unlike ALKV and KFDV, OHFV infections are rarely associated with neurological symptoms or sequelae (1).
Mice have been used as a reliable model with which to study disease progression after TBEV or OHFV infection. In our previous studies, we showed that virus-infected mice develop clinical signs and pathology similar to those seen in humans (5, 6). TBEV-infected mice experience severe encephalitis resulting in paralysis, ranging from hind limb paresis to complete paralysis, while OHFV-infected mice exhibit viscerotropic disease with limited signs of neurological disease despite multiplication of the virus in the brain.
The flavivirus genome consists of a positive-polarity, single-stranded RNA of ∼11 kb, which encodes three structural proteins (the core [C], premembrane [prM], and envelope [E] proteins) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), within a single open reading frame (7). The 5′ and 3′ untranslated regions predict secondary structures that are implicated in viral replication, translation, and packaging of the genome (8, 9). Although TBEV and OHFV exhibit >90% amino acid sequence identity (Table 1), the viral determinants responsible for disease-specific phenotypes have not been identified.
TABLE 1.
Amino acid differences between TBEV (strain Oshima 5–10) and OHFV (strain Guriev)
| Protein | Length (no. of amino acids) | No. of amino acid differences | Identity (%) |
|---|---|---|---|
| C | 96 | 16 | 83.3 |
| prM | 184 | 31 | 83.2 |
| E | 496 | 39 | 92.1 |
| NS1 | 352 | 36 | 90.0 |
| NS2A | 230 | 41 | 82.1 |
| NS2B | 131 | 15 | 88.5 |
| NS3 | 621 | 50 | 91.9 |
| NS4A | 149 | 18 | 87.9 |
| NS4B | 252 | 23 | 90.9 |
| NS5 | 904 | 74 | 91.8 |
| Total | 3415 | 343 | 90.0 |
Recently, we constructed infectious cDNA clones of both TBEV and OHFV (10–12). In the present study, we used these cDNA clones to generate TBEV-OHFV chimeric viruses with the specific objective of identifying viral determinants critical for the development of neurological disease in mice. Using this technology, we were able to identify a four-amino-acid region near the C terminus of the viral RNA-dependent RNA polymerase (RdRp) that is critical for neuropathogenesis. This discovery will allow us to further characterize specific virus-host cell interactions responsible for the development of severe disease.
MATERIALS AND METHODS
Cells.
BHK-21 cells were grown at 37°C in Eagle minimum essential medium (E-MEM) supplemented with 8% fetal calf serum (FCS) and l-glutamine. Mouse neuroblastoma NA cells (kindly provided by M. Sugiyama and N. Ito of Gifu University) were maintained in E-MEM supplemented with 10% FCS. PC12 cells were maintained at 37°C in RPMI 1640 supplemented with 10% horse serum and 5% FCS.
Viruses.
Recombinant TBEV (Oshima 5-10 strain) and OHFV (Guriev strain) were recovered from infectious cDNA clones of the respective viruses (TBEV-pt and OHF-pt, respectively) as reported previously (10, 11). To prepare the infectious cDNA clones of chimeric viruses (Fig. 1), cDNA fragments were synthesized by standard fusion-PCR and subcloned into TBEV-pt and OHF-pt in a stepwise manner.
FIG 1.
Neurological disease in mice infected with chimeric viruses. The coding regions for OHFV and TBEV proteins are shown in white and black, respectively. The percentages of mice exhibiting signs of neurological disease are shown on the right. (A) Chimeric viruses were constructed by replacement of the coding region for each viral protein. (B) The coding region of OHFV nonstructural protein NS5 was partially replaced with that of TBEV; nucleotide positions and restriction enzyme sites used for this replacement are indicated. (C) The amino acids at positions 879, 880, 881, and/or 891 of NS5 were substituted. Normal and boldface letters indicate the amino acid sequences of OHFV and TBEV, respectively.
Infectious RNA was transcribed from the infectious cDNA clones using mMESSAGE mMACHINE SP6 or T7 kits (Ambion, Austin, TX) and transfected into BHK-21 cells using TransIT-mRNA (Mirus Bio LLC, Madison, WI), as described previously (13). At 3 days postinfection (dpi), viral particles were collected from culture supernatants. Stocks of all viruses were propagated in BHK-21 cells. Successful recombination of recovered viruses was confirmed by sequencing of reverse transcription-PCR (RT-PCR) fragments.
All experiments using recombinant viruses were performed according to the Law Concerning the Conservation and Sustainable Use of Biological Diversity through Regulations on the Use of Living Modified Organisms of Japan, a Japanese law ensuring compliance with the Cartagena Protocol on Biosafety. All experiments using live viruses were performed in a BSL-3 facility.
Virus titration.
For viral titrations, monolayers of BHK-21 cells prepared in multiwell plates were incubated with serial dilutions of virus for 1 h, overlaid with E-MEM containing 2% FCS and 1.5% carboxymethyl cellulose (Sigma-Aldrich, St. Louis, MO), and incubated for 5 days. After incubation, the cells were fixed and stained with 0.25% crystal violet in 10% buffered formalin. Plaques were counted and expressed as PFU/ml.
Growth curves of the recombinant viruses.
BHK-21 cells were infected with each recombinant virus at a multiplicity of infection (MOI) of 0.01. After virus adsorption for 1 h, the inocula were removed. Cells were then washed with phosphate-buffered saline (PBS) and incubated in E-MEM containing 2% FCS. Media were harvested at 24, 48, and 72 h postinfection and stored at −80°C until titration.
Pathogenicity of the recombinant viruses in mice.
Five- to six-week-old BALB/c mice (Japan SLC, Inc., Shizuoka, Japan) were challenged with 10,000 PFU of each virus subcutaneously. The physical conditions of the mice were observed, and the body weights were measured daily. Simple neurological assessments, including landing tests, balance tests, and grasping tests, were performed as described previously (5).
For analysis of viral distribution in tissues, three to four mice were sacrificed on 1, 3, 5, 7, 9, and 11 dpi, and sera, brains, and spleens were collected after perfusion with cold PBS. Organs were individually weighed, homogenized, and prepared as 10% (wt/vol) suspensions in PBS supplemented with 10% FCS. Suspensions were then clarified by centrifugation (4,000 rpm for 5 min at 4°C), and the supernatants were titrated.
The animal experiments were performed in accordance with the recommendations in the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology. The experimental protocols were approved by the Animal Care and Use Committee of the Hokkaido University (approval 09-0071/11-0065).
Scoring.
Mice were scored based on the severity of neurological signs. Signs of paralysis and loss of balance were typically associated with viral infection and were scored as 0 (absent), 1 (present), or 2 (severe). Mice were also weighed at each scoring session, with mice exhibiting >10% loss of body weight defined as sick. Scoring for paralysis was assigned as follows: 0, normal; 1, dragging limbs or inversion of dorsum pedis; and 2, complete paralysis and no spontaneous movement. Scoring for loss of balance was assigned as follows: 0, normal; 1, leaning of head or trunk posture to one side; and 2, inability to retain posture and falling to one side or a circling movement to one side. Total scores were quantified and were expressed as means ± the standard errors of the mean. Neurological disease was defined as a total score of >1.0.
Histopathology and immunohistochemistry.
The brains of infected mice were collected and fixed in 10% neutral phosphate-buffered formalin at 7 dpi or at the terminal stages of infection for each virus. Paraffin embedded brains were cut into 4-μm-thick sections, stained with hematoxylin and eosin, and examined by light microscopy. For the detection of viral antigens, the sections were incubated with rabbit polyclonal antibody against flavivirus E protein (14) and stained using the streptavidin-biotin-immunoperoxidase complex method (Histofine SAB-PO kit; Nichirei, Tokyo, Japan). Sections were counterstained with Mayer's hematoxylin. For the detection of activated caspase-3, rabbit polyclonal anti-cleaved caspase 3 (1:500; Cell Signaling Technology, Beverly, MA) was used.
TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay.
For the detection of DNA breaks in neuronal cells, sections were incubated with 0.02 mg of proteinase K (Sigma)/ml and treated with a methanol solution containing 3% H2O2 to block endogenous peroxidase reactivity. Sections were then incubated with 0.5 μM terminal deoxynucleotidyltransferase (TdT; Invitrogen, Carlsbad, CA) and 10 μM biotin-16-dUTP (Roche, Penzberg, Germany) in TdT buffer for 90 min at 37°C, followed by peroxidase-conjugated streptavidin. Sections were then counterstained with methyl green.
Quantitative RT-PCR of inflammatory cytokines.
Total RNA was extracted from the brains of infected mice using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. First-strand cDNA was synthesized from 0.4 μg of total RNA using Moloney murine leukemia virus reverse transcriptase (Life Technologies, Carlsbad, CA) and oligo(dT) primers. To quantify cytokine mRNA levels, real-time PCR was performed using a Kapa SYBR Fast qPCR kit (Kapa Biosystems, Woburn, MA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The primer sets used here are listed in Table 2. Relative quantification of gene expression was normalized against GAPDH. Expression levels are represented as the level of gene expression relative to uninfected controls.
TABLE 2.
Primer sets used in the quantitative RT-PCR
| Primer | Orientation | Sequence (5′–3′) |
|---|---|---|
| TNF-α | Sense | CAAATGGCCTCCCTCTCATC |
| Antisense | CTCCAGCTGCTCCTCCACTT | |
| IL-1β | Sense | CCTTCCAGGATGAGGACATGA |
| Antisense | CAGCACGAGGCTTTTTTGTTG | |
| IL-6 | Sense | GGGACTGATGCTGGTGACAA |
| Antisense | TCCACGATTTCCCAGAGAACA | |
| GAPDH | Sense | GCACCACCACTGCTTAGCC |
| Antisense | GGATGCAGGGATGATGTTCTG |
Measurement of the neurite length of PC12 cells.
PC12 cells were seeded onto collagen-coated eight-well chamber slides (Matsunami Glass Industries, Ltd., Kishiwada, Japan). Cells were infected with virus at a multiplicity of infection (MOI) of 10 and, at 24 h postinfection, were treated with 150 ng of rat 2.5S nerve growth factor (NGF; Becton Dickinson, Franklin Lakes, NJ)/ml in RPMI 1640 supplemented with 1% horse serum. After 72 h of treatment, the cells were fixed in 4% (wt/vol) paraformaldehyde for 20 min at 37°C and observed by BZ-9000 (Keyence, Osaka, Japan). Cells bearing neurites equivalent in length to the cell body diameter were scored using BZ-2 Analyser software (Keyence). A total of 80 cells acquired from three independent experiments were quantified.
Statistical analysis.
Data are expressed as means ± the standard errors. The Tukey-Kramer test was used to determine statistical significance of differences in the mean values of neurological scores (see Table 3), virus titers at each time point (see Fig. 2, 3, 4, and 7), inflammatory cytokines (see Fig. 6), and neurite outgrowth (see Fig. 7). The Kaplan-Meier survival curves and the log-rank test were used to evaluate the survival of infected mice (Table 3).
TABLE 3.
Pathogenicity of chimeric viruses in a mouse modela
| Chimeric virus | No. of mice | Morbidity (%) | Mean time to onset of disease (days) ± SDb | Mortality (%)c | Mean survival time (days) ± SDb | Neurological disease (%)d | Neurological scoree |
|---|---|---|---|---|---|---|---|
| OHFV-pt | 32 | 96.9 (31/32) | 9.7 ± 2.6 | 96.9 (31/32) | 11.9 ± 3.3 | 6.4 (2/31) | 0.11 ± 0.07 |
| TBEV-pt | 31 | 100 (31/31) | 9.0 ± 1.4 | 67.7 (21/31) | 14.2 ± 3.6 | 81.0 (17/21) | 1.71 ± 0.27** |
| OHFV/TBE-ME | 15 | 93.3 (14/15) | 10.1 ± 2.2 | 93.3 (14/15) | 12.2 ± 2.9 | 14.2 (2/14) | 0.27 ± 0.18 |
| TBEV/OHF-ME | 15 | 100 (15/15) | 8 ± 0.93 | 100 (15/15) | 10.3 ± 1.5 | 93.3 (14/15) | 2.27 ± 0.25** |
| OHFV/TBE-C | 12 | 100 (12/12) | 7.3 ± 0.87 | 83.3 (10/12) | 9.1 ± 2.0 | 10 (1/10) | 0.17 ± 0.17 |
| OHFV/TBE-NS1 | 12 | 75 (9/12) | 10.2 ± 3.2 | 75 (9/12) | 14.5 ± 5.7 | 11.1 (1/9) | 0.08 ± 0.08 |
| OHFV/TBE-NS2A | 12 | 100 (12/12) | 9.8 ± 2.6 | 100 (12/12) | 13.1 ± 3.5 | 16.7 (2/12) | 0.33 ± 0.22 |
| OHFV/TBE-NS2B | 12 | 91.7 (11/12) | 10.3 ± 3 | 83.3 (10/12) | 13.4 ± 3.7 | 18.2 (2/11) | 0.42 ± 0.23 |
| OHFV/TBE-NS3 | 12 | 91.7 (11/12) | 10.5 ± 4.0 | 91.7 (11/12) | 13.5 ± 4.1 | 45.5 (5/11) | 1 ± 0.37 |
| OHFV/TBE-NS4AB | 12 | 91.7 (11/12) | 10.7 ± 3. | 91.7 (11/12) | 13.7 ± 3.1 | 9.1 (1/11) | 0.17 ± 0.17 |
| OHFV/TBE-NS5 | 18 | 100 (18/18) | 8.6 ± 2.1 | 100 (18/18) | 11.6 ± 3.1 | 88.9 (16/18) | 2.17 ± 0.31** |
| OHFV/TBE-Mtase | 12 | 100 (12/12) | 10.6 ± 3.8 | 100 (12/12) | 13.3 ± 4.1 | 16.7 (2/12) | 0.25 ± 0.18 |
| OHFV/TBE-RdRp | 18 | 88.9 (16/18) | 11.2 ± 3.7 | 88.9 (16/18) | 14.1 ± 4.0 | 75 (12/16) | 1.78 ± 0.38** |
| OHFV/TBE-8458-9488 | 10 | 90 (9/10) | 11.3 ± 4.2 | 90 (9/10) | 13.7 ± 4.3 | 11.1 (1/9) | 0.4 ± 0.27 |
| OHFV/TBE-9488-10295 | 10 | 20 (2/10) | 9.0 ± 1.4 | 20 (2/10)* | 13.5 ± 2.1 | 0 (0/2) | 0 |
| OHFV/NS5 879KFK891D | 27 | 96.2 (26/27) | 9 ± 3.7 | 96.2 (26/27) | 11.4 ± 4.3 | 80.8 (21/26) | 1.65 ± 0.22** |
| OHFV/NS5 879KFK | 10 | 100 (10/10) | 8.1 ± 1.5 | 100 (10/10) | 11.4 ± 2.5 | 20 (2/10) | 0.5 ± 0.34 |
| OHFV/NS5 891D | 10 | 100 (10/10) | 9.1 ± 2.4 | 90 (10/10) | 13 ± 3.2 | 22.2 (2/9) | 0.4 ± 0.27 |
| TBEV/NS5 879RYS891E | 15 | 100 (15/15) | 9.3 ± 1.4 | 66.7 (10/15) | 14 ± 2.8 | 30 (3/10) | 0.33 ± 0.19 |
For the morbidity and mortality columns, the number of mice affected/the number of mice tested is indicated in parentheses.
There were no statistically significant differences in the average onset of disease and the survival time in each group.
The survival of the mice was analyzed by the Kaplan-Meier method. *, Significant difference from OHFV-pt (P < 0.001).
That is, the percentage of mice showing neurological symptoms before death. The number of mice showing neurological symptoms before death/the number of dead mice is indicated in parentheses.
That is, the neurological scores for the severity of neurological signs were quantified as described in Materials and Methods. **, Significant difference from the score of OHFV-pt (P < 0.01).
FIG 2.
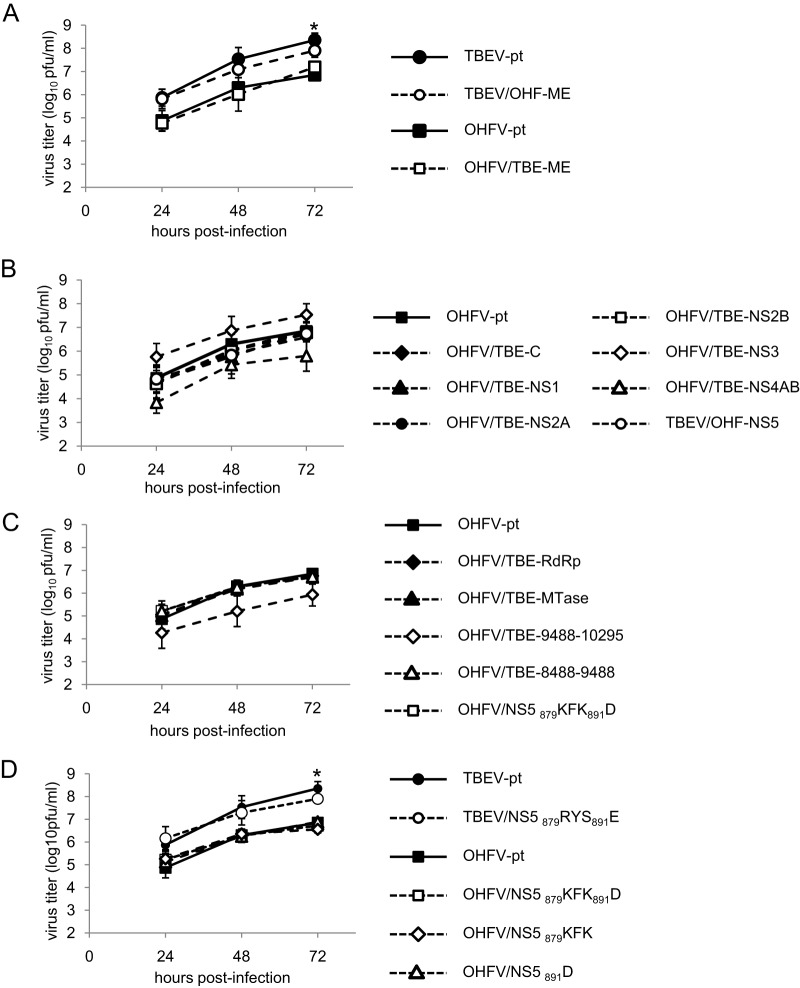
Growth curves of chimeric viruses. A Monolayer of BHK-21 cells was infected with wild-type and chimeric viruses at an MOI of 0.01. Media were harvested at each time point, and virus titers were determined by plaque assay in BHK-21 cells. The chimeric viruses showed no significant differences from parental virus at any time point examined. (A) Chimeric viruses with replacement of viral envelope proteins (prM/E). *, Significant difference between TBEV-pt and OHFV-pt and between TBEV-pt and OHFV/TBE-ME (P < 0.05). (B) Chimeric OHFV with replacement of the C or each NS protein. (C) Chimeric OHFV with partial replacement in the NS5 protein. (D) Chimeric viruses with substitutions in the KFK-D motif. *, Significant difference between TBEV-pt and all chimeric OHFV (P < 0.05).
FIG 3.
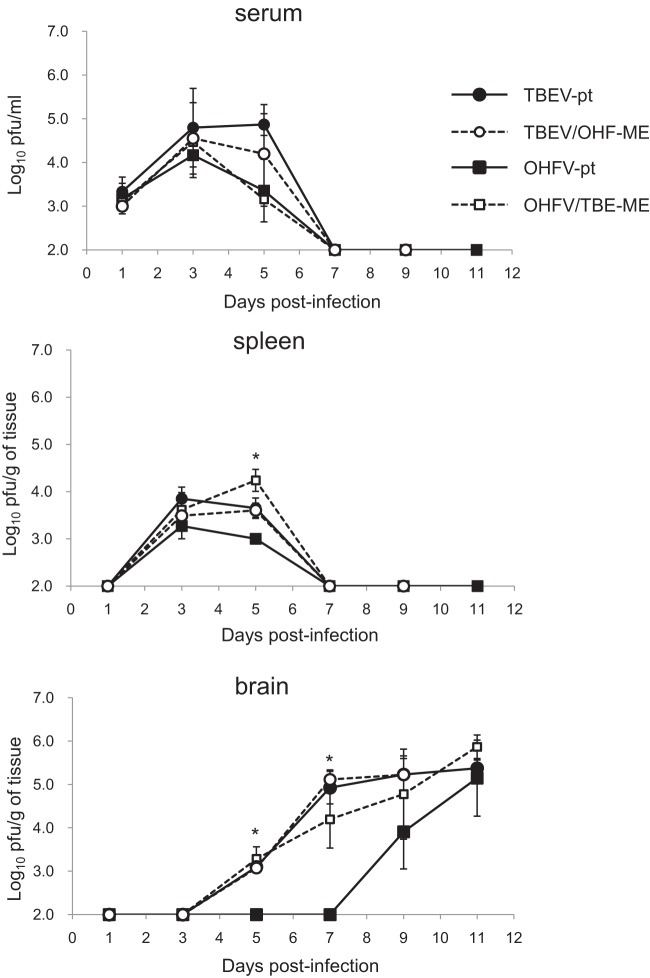
Multiplication in organs of chimeric viruses following replacement of the envelope proteins. Mice were infected with 10,000 PFU of each virus (TBEV-pt, TBEV/OHF-ME, OHFV-pt, and OHFV/TBE-ME). Virus titers in the serum, spleen, and brain were determined by plaque assays on the days indicated. Error bars represent the standard deviations (n = 4). By 11 dpi, all mice inoculated with TBEV/OHF-ME had died. *, Significant difference between TBEV-pt and OHFV-pt (P < 0.05). No significant differences were observed between TBEV-pt, TBEV/OHF-ME, and OHF/TBE/ME.
FIG 4.
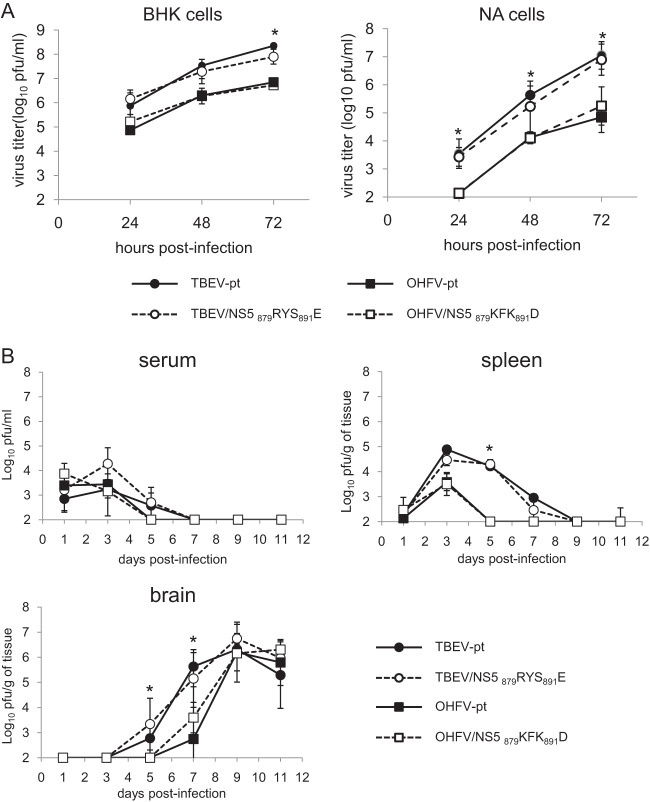
Growth properties of chimeric viruses containing NS5 amino acid substitutions. (A) Monolayers of BHK-21 or NA cells were infected with wild-type and chimeric viruses at an MOI of 0.01. Media were harvested at each time point, and virus titers were determined by plaque assay in BHK-21 cells. (B) Mice were infected with 10,000 PFU of each virus. Virus titers in the serum, spleen, and brain were determined by plaque assays on the days indicated. Error bars represent the standard deviations (n = 3). *, Significant difference between TBEV-pt and OHFV-pt and between TBEV-pt and OHFV/NS5 879KFK891D (P < 0.05). No significant differences were observed between TBEV-pt and TBEV/NS5 879RYS891E or between OHFV-pt and OHFV/NS5 879KFK891D at any time point.
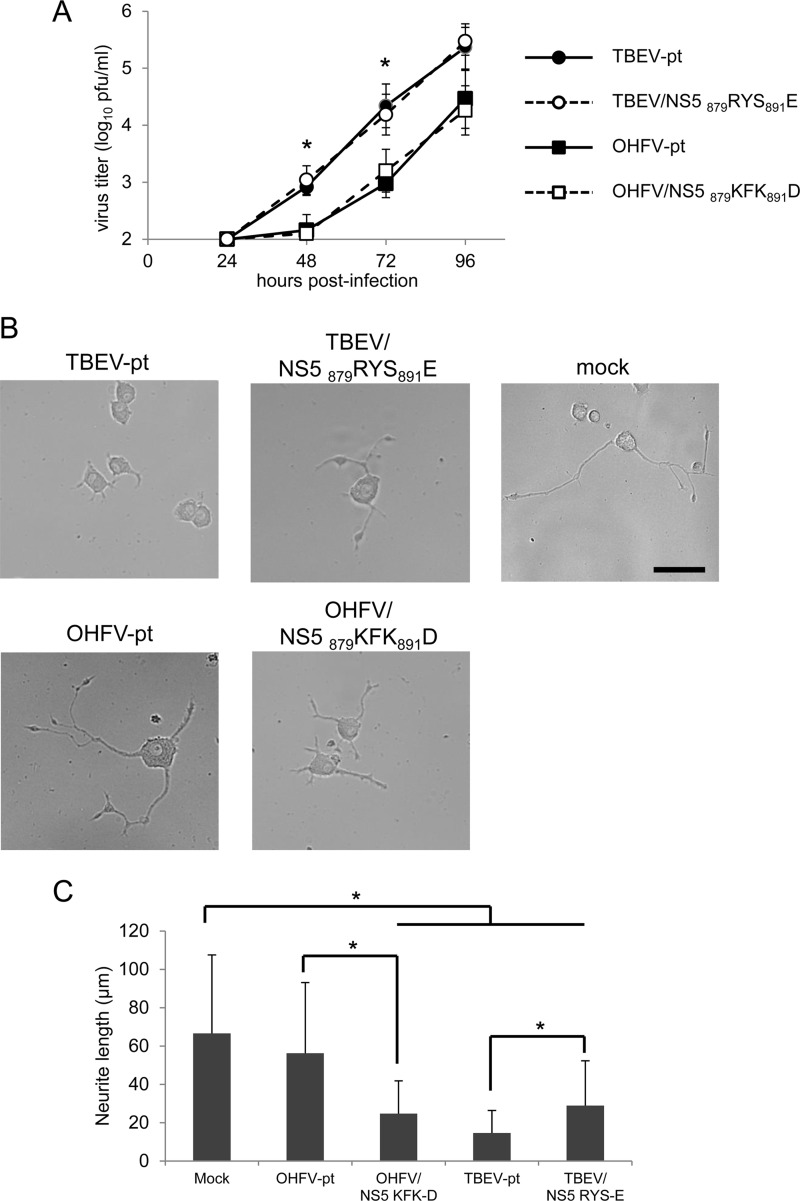
FIG 7.
Neurite formation by PC12 cells infected with chimeric viruses containing NS5 amino acid substitutions. PC12 cells were infected with TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D at an MOI of 10 and treated with 150 ng of NGF/ml at 24 h postinfection. (A) Media were harvested at each time point, and virus titers were determined by plaque assay in BHK-21 cells. *, Significant difference between TBEV-pt and OHFV-pt and between TBEV-pt and OHFV/NS5 879KFK891D (P < 0.05). No significant differences were observed between TBEV-pt and TBEV/NS5 879RYS891E or between OHFV-pt and OHFV/NS5 879KFK891D at any time point. (B) Typical images of the cells 72 h after NGF treatment are shown. Scale bar, 50 μm. (C) The average of neurite length was quantified 72 h after NGF treatment. *, Significant difference (P < 0.01).
FIG 6.
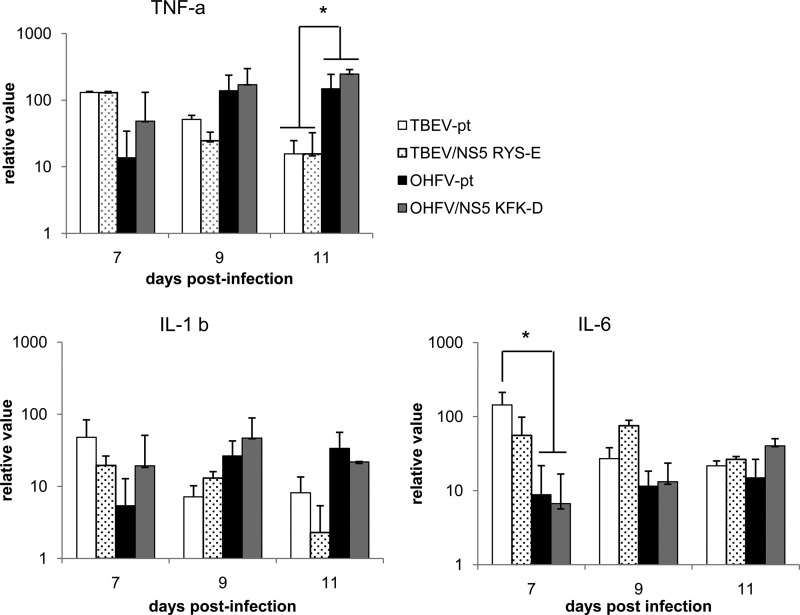
Expression of inflammatory cytokines in the brains of mice infected with chimeric viruses. Mice were inoculated with 10,000 PFU of TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D, and the expression of inflammatory cytokines in the brain was measured by real-time PCR. The levels of TNF-α, IL-1β, and IL-6 mRNA expression were measured at the time points indicated and normalized against GAPDH. Expression levels are shown relative to uninfected controls. *, Significant difference (P < 0.05).
RESULTS
Effects of viral structural protein replacement on pathogenicity.
The viral structural proteins, prM and E, have been shown to play important roles in the tissue tropism and neuropathogenesis of flaviviruses (15–17). To assess whether these proteins are responsible for the differences in pathogenicity between TBEV and OHFV, the chimeric infectious clones TBEV/OHF-ME and OHFV/TBE-ME were generated by replacing the prM and E genes of each virus with those from the other species (Fig. 1A). Viable chimeric viruses were recovered from cells transfected with synthetic mRNA derived from the plasmid template and sequencing of each progeny virus confirmed their chimeric composition. Basic replication characteristics were investigated in BHK-21 cells (Fig. 2A). Although TBEV grew more rapidly than OHFV (P < 0.05), similar growth curves were obtained between parental viruses and the virus with replacement of the prM and E genes (no significant differences), indicating that prM and E did not affect viral replication properties in cultured cells.
The pathogenicities of the chimeric viruses were evaluated in BALB/c mice using nonchimeric wild-type viruses as controls (i.e., TBEV-pt and OHFV-pt). Mortality rates of >70% were seen in each group, with no significant differences in onset of disease and survival time between groups (Table 3); however, animals could be readily divided into two groups based on clear differences in disease phenotype. Mice infected with chimeric viruses containing the TBEV NS protein genes (i.e., TBEV-pt or TBEV/OHF-ME) began to show general signs, such as hunched posture, ruffled fur, and general malaise at 7 to 9 dpi. The majority of mice that succumbed to infection with either TBEV-pt or TBEV/OHF-ME (81.0 and 93.3%, respectively, Table 3) showed typical indications of neurological illness such as loss of balance, paresis, hind-limb paralysis, or tremor in the final stage of disease, similar to the observations with other neurotropic flaviviruses. In contrast, OHFV-pt- or OHFV/TBE-ME-infected mice exhibited general signs of illness (hunched posture, ruffled fur, and general malaise), but the majority of animals exhibited mild or no signs of neurological illness (OHFV-pt, 6.4%; OHFV/TBE-ME, 14.2%). In semiquantitative neurological assessments, the animals showed little or no indication of a physical inability to perform the assessment tests. In the final stages of infection, OHFV-pt- and OHFV/TBE-ME-infected mice exhibited obvious signs of weakness and were unable to complete the assessments, but they attempted to perform the required tasks. The neurological scores for severity in mice infected with either TBEV-pt or TBEV/OHF-ME were significantly higher than those in mice infected with OHFV-pt- or OHFV/TBE-ME (P < 0.01). Taken together, these results indicated that the structural proteins prM and E are not responsible for the differences in disease phenotype elicited by TBEV and OHFV.
Major organs (i.e., spleen, liver, lung, and brain) and serum were harvested at various time points to determine virus titers in mice infected with parental and chimeric viruses. Virus was initially detected between 1 and 3 dpi, with virus titers peaking between 3 and 5 dpi in serum and spleen (Fig. 3). No significant differences in virus titer were observed in the serum or peripheral organs of mice infected with parental or chimeric viruses.
Similar virus titers were observed in the brains of mice infected with TBEV-pt and TBEV/OHF-ME, with virus first detected at 5 dpi, and peaking at 7 dpi. In contrast, virus was not detected in the brains of OHFV-pt-infected mice until 9 dpi, with virus titers reaching levels similar to TBEV-pt or TBEV/OHF-ME-infected mice at 11 dpi.
Despite differences in disease phenotype, viral replication in OHFV/TBE-ME-infected mice was similar to that seen in mice infected with TBEV-pt or TBEV/OHF-ME. These data indicate that the replacement of viral prM and E proteins from OHFV with those of TBEV increased the neuroinvasiveness of the virus but did not directly affect disease phenotype.
The NS3 and NS5 genes determine neurological disease caused by TBEV in mice.
To identify specific genetic determinants of disease phenotypes in TBEV and OHFV, we constructed chimeric OHFV viruses with the C protein or each of the NS protein genes replaced with the equivalent gene from TBEV (Fig. 1A). Viable chimeric viruses were recovered and basic replication characteristics were investigated in BHK-21 cells. While growth of OHFV-TBE-NS3 was elevated slightly, there were no significant differences between parental OHFV and each chimeric virus (Fig. 2B).
The pathogenicity of each chimeric virus was examined in a mouse model (Table 3). Significant lethality was seen for each of the eight chimeric viruses, with most mice exhibiting general signs of disease (i.e., ruffled fur, decreased activity, and weight loss). There were no significant differences in disease onset or survival times between groups. Fewer than 20% of mice infected with OHFV/TBE-C, OHFV/TBE-NS1, OHFV/TBE-NS2A, OHFV/TBE-NS2B, or OHFV/TBE-NS4AB showed evidence of neurological disease. However, approximately half (45.5%) of the mice infected with OHFV/TBE-NS3 exhibited clear neurological signs, including loss of balance, paresis, and hind-limb paralysis. Even higher rates of neurological disease were seen in mice infected with OHFV/TBE-NS5, with 88.9% of mice exhibiting significant neurological signs. These data suggested that the NS5 protein of TBEV was critical for development of severe neurological disease in mice, with the NS3 protein of TBEV also affecting the disease phenotype to some extent.
To further delineate specific regions within the NS5 protein involved in development of severe neurological disease, we constructed OHFV chimeras in which the NS5 protein was partially substituted with that of TBEV (Fig. 1B). Flavivirus NS5 consists of two principle domains, the methyltransferase (MTase) domain located on the N-terminal side of the protein and the RNA-dependent RNA polymerase (RdRp) domain on the C-terminal side (18, 19). We therefore constructed chimeric OHFV viruses replacing either the MTase or RdRp domains of NS5 with that of TBEV. Each chimeric virus showed replication properties similar to those of the parental OHFV in BHK cells (Fig. 2C). As shown in Fig. 1B and Table 3, replacement of the RdRp domain substantially increased the frequency (75%) at which mice developed signs of neurological disease, while replacement of the MTase domain had only a minimal impact compared to the parental OHFV (16.7% versus 6.4%, respectively) (Fig. 1 and Table 3).
The RdRp domain was further divided into three regions, and each region of OHFV was substituted with that of TBEV (Fig. 1B). As shown in Table 3, replacement of the N terminus of the RdRp domain (nucleotides [nt] 8458 to 9488) had no impact on the development of neurological signs in infected mice (11.1%). Replacement of the middle region of the RdRp domain (nt 9488 to 10295) markedly reduced the virulence, with the majority of infected mice exhibiting no signs of disease and surviving; animals that did eventually succumb to infection exhibited no neurological signs prior to death. This low morbidity may have been related to the lower replication efficacy compared to parental OHFV observed in BHK-21 cells (Fig. 2C). Replacement of the C terminus of the RdRp domain (nt 10295 to 10377) resulted in a frequency of severe neurological signs similar to the parental TBEV (80.8% versus 81%, respectively). This result was surprising given that there are only four amino acid differences between TBEV and OHFV in this region: at amino acids 879 to 881 (Lys/Phe/Lys in TBEV versus Arg/Tyr/Ser in OHFV) and at amino acid 891 (Asp in TBEV versus Glu in OHFV).
To determine the effects of the four amino acid differences on the pathogenicity of TBEV and OHFV, we made a series of chimeric viruses substituting the 879-881 triplet and residue 891 either individually or in tandem (Fig. 1C). Insertion of the TBEV amino acid triplet 879-881 or residue 891 into the NS5 protein into OHFV led to only modest increases in the rate of neurological signs compared to the OHFV parental strain (20 and 22.2%, respectively, versus 6.3%; Fig. 1C and Table 3). However, substitution of all four amino acids (that is, amino acids 879 to 881 and amino acid 891) resulted in a significant increase in the frequency of neurological signs, similar to that seen with TBEV (Fig. 1C and Table 3).
To further demonstrate the critical roles of these four residues in the development of severe neurological disease, we constructed a recombinant TBEV in which the four amino acids in the C terminus of NS5 were substituted with those of OHFV. Fewer mice infected with the chimeric TBEV showed neurological signs compared to those infected with the parental TBEV. However, as shown in Table 3, the four amino acid substitutions did not impact the morbidity, mortality, or survival curves of TBEV and OHFV. These results indicated that the combined motif (designated as the KFK-D motif), including amino acids Lys879/Phe880/Lys881 and Asp891 in the C terminus of NS5, is a critical determinant of neurological disease in mice infected with TBEV.
Effects of the four amino acid substitutions in NS5 on viral characteristics.
To determine the relationship between neurological disease development and viral replication, the effects of the KFK-D motif on viral growth characteristics were investigated both in vitro and in vivo. BHK or mouse neuroblastoma NA cells were infected with TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D at an MOI of 0.01. Virus was harvested 24 to 72 h postinfection and quantified by a plaque assay. As shown in Fig. 2D and 4A, although TBEV grew more rapidly than OHFV (P < 0.05), similar growth curves were obtained in both BHK and NA cells between parental viruses and the virus with four amino acid substitutions in NS5 (no significant differences), indicating that these mutations did not affect viral replication properties in cultured cells.
Viral loads in serum, spleen, and brain of infected mice were compared between mice inoculated with TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D (Fig. 4B). Transient viremia and multiplication in the spleen were observed in mice infected with each virus. Detection of virus in the brain was delayed in OHFV-infected mice compared to those infected with TBEV (P < 0.05). However, the four amino acid substitutions in NS5 did not affect viral multiplication in any of these organs (no significant differences). These results indicated that the differences seen in the neuropathogenesis of OHFV and TBEV are not due to alterations in viral replication.
To determine the relationship between central nervous system (CNS) pathology and neurological disease, histopathological features of mice were examined following infection with either TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D. At 7 dpi, nonsuppurative encephalitis with mild perivascular cuffing and meningitis was observed in the brains of some mice, but there were no significant differences between groups (data not shown).
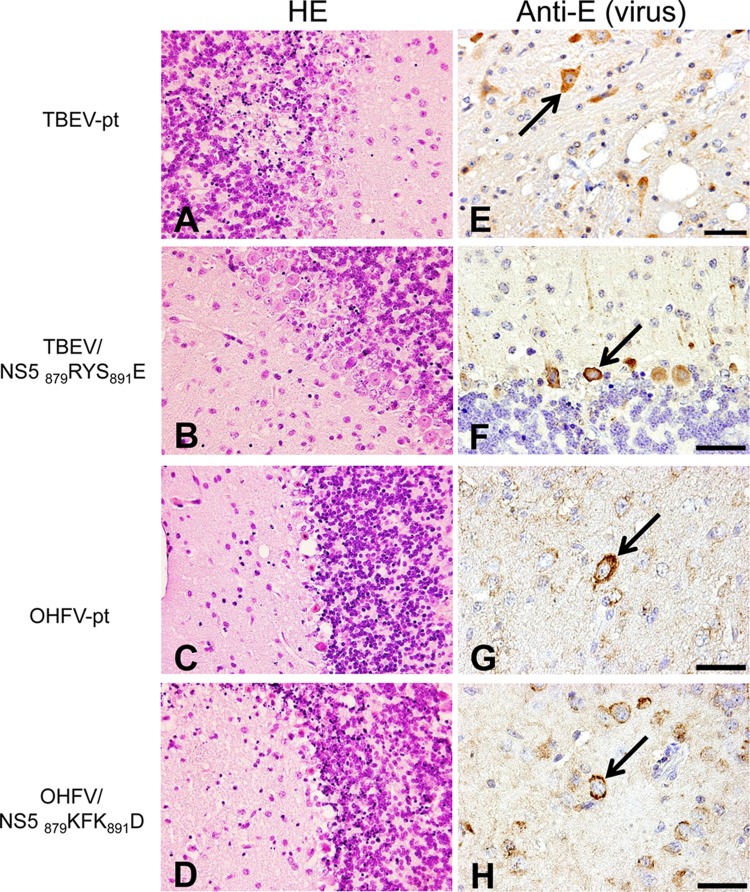
Next, dying mice exhibiting obvious signs of severe illness, including the inability to stand or move, total paralysis, and/or weight loss of >30%, were sacrificed for histopathological examination at the terminal phase of disease (8 to 14 dpi). Mild to severe nonsuppurative encephalitis, including neuronal degeneration, activation of microglial cells, and infiltration of mononuclear cells in the perivascular area, was observed throughout the cerebral cortex, cerebellum, and brain stem (Fig. 5A to D). Pathological lesions accompanied by vacuolation, nuclear pyknosis of neuronal cells and ischemic changes (necrosis) of neurons were prominent in the brains of animals infected with each virus. Using immunohistochemistry, viral antigens were diffusely detected in all groups in the cytoplasm of neurons of the cerebral cortex, hippocampus, and brain stem, as well as in the Purkinje cells and granule cells of the cerebellum (Fig. 5E to H). By utilizing TUNEL assays and immunohistochemistry for active caspase-3, apoptotic cells were identified primarily as cerebral neurons and granule cells in cerebellum (data not shown). Although virus-induced encephalitis was confirmed in all groups, no clear differences in histopathological features or virus distribution were observed between parental and chimeric viruses containing the four amino acid KFK-D motif.
FIG 5.
Histopathological features of the brains of mice infected with chimeric viruses containing NS5 amino acid substitutions. Mice were inoculated with 10,000 PFU of TBEV-pt (A and E), TBEV/NS5 879RYS891E (B and F), OHFV-pt (C and G), or OHFV/NS5 879KFK891D (D and H), and sacrificed at terminal stages of disease (7 to 13 dpi). Brain histopathology consisted of marked nonsuppurative encephalitis in all mice. Severe tissue damage with degeneration and necrosis of Purkinje cells, and pyknosis of granule cells in the cerebellum (A to D; original magnification, ×400). Viral antigens (arrows) were detected in the cytoplasm of various neuronal cells in the brains of virus-infected mice (E to H; scale bars, 50 μm).
The inflammatory response following infection with parental or chimeric viruses was also assessed by measuring expression levels of inflammatory cytokines in brains of infected mice at 7 to 11 dpi. Although levels of inflammatory cytokines tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), and IL-6 increased after infection with each virus, no significant differences which could be correlated with substitutions in NS5 were observed (Fig. 6). Taken together, these results suggest that differences in neurological disease induced by KFK-D motif substitutions were the result of factors other than direct lesions of the brain caused by viral cytopathic effects and inflammatory responses.
A previous study by Wigerius et al. suggested that NS5 was involved in the attenuation of neurite outgrowth using PC12 cells derived from rat pheochromocytoma (20). PC12 cells resemble neurons in many respects, and when grown in the presence of NGF, they differentiate into a neuronal phenotype by developing neurites, becoming electrically excitable and increasing the synthesis of various neurotransmitters (21). We investigated the effects of the KFK-D motif of TBEV on neurite formation and development using PC12 cells.
PC12 cells were infected with TBEV-pt, TBEV/NS5 879RYS891E, OHFV-pt, or OHFV/NS5 879KFK891D and were then treated with NGF. As shown in Fig. 7A, although TBEV grew faster than OHFV (P < 0.05), similar growth curves were obtained between parental viruses and those with four amino acid substitutions in NS5 (no significant differences), as observed in BHK and NA cells. During the experiments, infected cells remained viable up to at least 7 dpi. However, significant differences were observed in neurite outgrowth in infected cells. Neurite length in cells infected with viruses with the KFK-D motif was significantly shorter than that in cells infected with viruses without the KFK-D motif (Fig. 7B and C). These results indicated that the KFK-D motif of TBEV is involved in the attenuation of neurite outgrowth.
DISCUSSION
In this study, we utilized infectious clones of two viruses with different disease phenotypes to determine the genetic determinants of neurological disease. We identified four amino acids in the C-terminal region of TBEV nonstructural protein NS5 that were critical determinants of neurological disease but did not affect viral replication or histopathological features.
Replacement of the prM and E proteins of OHFV with the equivalent proteins from TBEV increased viral neuroinvasiveness. In several reports, amino acid changes in the E protein have been shown to affect the neuroinvasiveness of tick-borne flaviviruses (15, 17, 22, 23), although the detailed mechanism of viral entry into the CNS remains unclear. There are a total of 39 amino acid differences between TBEV strain Oshima and OHFV strain Guriev, but none has been previously reported to be involved in viral neuroinvasiveness. The identification of amino acids responsible for facilitating the rapid neuroinvasiveness of TBEV could lead to new insights into viral pathogenesis and help to clarify the mechanism by which the virus gains entry into the CNS. TBEV/OHF-ME, carrying the prM and E genes of OHFV and the other genes for TBEV, entered the brain earlier than OHFV. Thus, the regions other than the prM and E proteins are also responsible for the neuroinvasiveness of TBEV. Several reports indicated amino acid changes in the NS proteins affect the neuroinvasiveness of tick-borne flaviviruses (24, 25) and mosquito-borne flaviviruses (26–30). Multiplication of TBEV/OHF-ME in the brain was similar to that of parental TBEV. Regions other than the prM and E proteins may compensate for the effects of replacement of the prM and E proteins on entry and multiplication in the brain because parental TBEV is highly neuroinvasive.
Although recombinant chimeric OHFV incorporating the TBEV prM and E proteins multiplied in the brain similar to TBEV, this failed to translate into neurological disease. Therefore, factors other than viral multiplication in the brain are involved in the induction of neurological disease, and these factors are distinct from prM and E.
Four amino acids in the C terminus of the NS5 were identified as critical determinants of neurological disease following TBEV infection in mice. The NS5 protein of flaviviruses is a multifunctional protein containing an N-terminal MTase domain and a C-terminal RdRp domain (18, 19), separated by an interdomain region with nuclear localization sequences (31, 32). NS5 has also been shown to have interferon antagonist activity in several flavivirus studies (33–36). The four amino acids identified in the present study are located in the C-terminal region of the RdRp domain. Several studies have shown that amino acid changes in the RdRp domain affect flavivirus genome replication and disease development (10, 37). However, the KFK-D motif described here did not affect viral multiplication either in vitro or in vivo. In addition, no significant differences in histopathological features, such as inflammatory response or viral antigen distribution, were observed in animals infected with OHFV chimeras incorporating the KFK-D motif. Therefore, the differential disease phenotype induced by making KFK-D motif substitutions in OHFV was not a result of alterations in the viral replication properties of NS5 or of the induction of viral lesions in the brain directly. Instead, these effects appear to be the result of other factors such as induction of neuronal dysfunction and/or degeneration in virus-infected neurons, resulting in the neurological disease phenotype in mice.
Neuronal dysfunction and degeneration have been associated with a number of neurotropic viral infections. It has been suggested that neuronal dysfunction, rather than neuronal death, is likely responsible for the severe neurological symptoms caused by rabies virus, since the neuropathological findings are relatively mild. Rabies infection induces electrophysiological alterations, including effects on ion channels and neurotransmission, which may be the cause of functional impairment (38). Alterations in synaptic function have been also reported in borna disease virus and human immunodeficiency virus (HIV) infections (39, 40), whereas axonal degeneration is instrumental in the development of neuronal dysfunction during herpesvirus and HIV infections (41–44). Despite these clear associations between neurotropic viruses and neuronal dysfunction, the majority of research examining neurological diseases caused by tick-borne flaviviruses has focused on virus-induced cytopathic effects or immunopathogenic responses. We showed here that the KFK-D motif was involved in the arrest of neurite outgrowth in PC12 cells. Impaired neurite outgrowth has been linked to various neurological disorders, and it may be involved in the development of neurological disease by TBEV infection. Further studies focusing on the biological activity of the KFK-D motif of NS5 and its role in neuronal dysfunction, including neurite differentiation, may provide new insight into the molecular mechanisms of the pathogenicity of neurological disease following tick-borne flavivirus infection.
As shown in Fig. 8, the KFK-D motif is found on the lateral surface of the thumb domain of flavivirus RdRp using the crystal structure of the West Nile virus RdRp domain as a template (PDB code 2HFZ). Similar results were obtained using that of Japanese encephalitis virus as a template (PDB code 4K6M). It is possible that individual amino acids play important roles in the interaction with unidentified host factors within neurons. The tick-borne flavivirus NS5 has been shown to possess a C-terminal PDZ binding motif (PBM), Ser/Ile/Ile (45), thereby facilitating binding to a variety of PDZ domain-containing proteins (46). PDZ domains are protein interaction modules that are often found in multidomain scaffolding proteins. PDZ-containing scaffolds assemble specific proteins into large molecular complexes involved in maintaining cell polarity and regulation of synaptic plasticity and synaptic vesicle dynamics (47–49). In a previous study of TBEV, NS5 opposed neuronal differentiation by binding to the PDZ domain protein Scribble (20). It was reported that a second anchorage site upstream of the C-terminal PBM supported the interaction between the PDZ domain protein and the PBM (50). Therefore, it is possible that the substitutions of the four amino acids in the C terminus of NS5 might affect the interaction between NS5 and host proteins, such as PDZ domain-containing proteins, resulting in neuronal dysfunction and degeneration, through alteration of synaptic plasticity and axonal degeneration.
FIG 8.
Amino acids involved in the neurological disease caused by tick-borne flaviviruses. A three-dimensional model of the tick-borne flavivirus RdRp domain in NS5 was constructed based on the crystal structure of West Nile virus polymerase (PDB code 2HFZ). The structure of tick-borne flavivirus polymerase is shown in ribbon (upper) and surface (lower) representations. Amino acid positions 879, 880, 881, and 891 are colored blue.
Interestingly, the amino acids 879 to 881 Lys/Phe/Lys and 891 Asp are highly conserved among TBEV (Fig. 9). Eighty percent of TBEV strains encode these residues, with residues 880 Phe and 891 Asp conserved across all strains. In contrast, all reported OHFVs encode 879-881 Arg/Tyr/Ser and 891 Glu. Residues 879-880 Arg/Tyr and 891 Glu are also conserved in the other hemorrhagic tick-borne flaviviruses, KFDV and ALKV, despite their phylogenetic distance from OHFV. This conservation of the KFK-D motif among TBEV strains supports the suggestion that this motif is important in the development of neurological disease of TBE and that the conserved amino acids in hemorrhagic tick-borne flaviviruses play a role in the development of hemorrhagic disease. The amino acid differences between these viruses may also be the result of adaptive evolution within a particular tick species leading to the selection of different virus variants, since tick-borne flaviviruses are maintained predominantly in ticks (51).
FIG 9.
Alignment of C-terminal amino acids (841 to 900 in TBEV) of NS5 for flaviviruses. The KFK-D motifs of TBEV are shown in boldface type and are shaded.
In conclusion, this study provides the first description of critical viral genetic factors important for the different disease manifestations of tick-borne flaviviruses. Four amino acids near the C terminus of the viral NS5 were shown to be critical for the development of neurological disease in TBEV infection in mice. Mutation of these amino acids did not directly affect viral replication or histopathological features, including inflammatory responses, suggesting that neuronal dysfunction and degeneration are involved in neurological disease manifestations. These insights may provide important information for identifying the mechanisms of the pathogenesis of tick-borne flaviviruses.
ACKNOWLEDGMENTS
This study was supported by Grants-in-Aid for Scientific Research (24780293, 22780268, and 21405035) and the Global COE Program from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan and Health Sciences Grants for Research on Emerging and Re-emerging Infectious Disease from the Ministry of Health, Labor, and Welfare of Japan. M.R.H. is supported as a subcontractor with Battelle Memorial Institute under its prime contract with the National Institute of Allergy and Infectious Diseases (HHSN272200200016I).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Burke DS, Monath TP. 2001. Flaviviruses, p 1043–1125 In Knipe DM, Howley PM. (ed), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 2.Gritsun TS, Lashkevich VA, Gould EA. 2003. Tick-borne encephalitis. Antivir. Res. 57:129–146. 10.1016/S0166-3542(02)00206-1 [DOI] [PubMed] [Google Scholar]
- 3.Lindquist L, Vapalahti O. 2008. Tick-borne encephalitis. Lancet 371:1861–1871. 10.1016/S0140-6736(08)60800-4 [DOI] [PubMed] [Google Scholar]
- 4.Ruzek D, Yakimenko VV, Karan LS, Tkachev SE. 2010. Omsk haemorrhagic fever. Lancet 376:2104–2113. 10.1016/S0140-6736(10)61120-8 [DOI] [PubMed] [Google Scholar]
- 5.Holbrook MR, Aronson JF, Campbell GA, Jones S, Feldmann H, Barrett AD. 2005. An animal model for the tickborne flavivirus: Omsk hemorrhagic fever virus. J. Infect. Dis. 191:100–108. 10.1086/426397 [DOI] [PubMed] [Google Scholar]
- 6.Tigabu B, Juelich T, Bertrand J, Holbrook MR. 2009. Clinical evaluation of highly pathogenic tick-borne flavivirus infection in the mouse model. J. Med. Virol. 81:1261–1269. 10.1002/jmv.21524 [DOI] [PubMed] [Google Scholar]
- 7.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649–688. 10.1146/annurev.mi.44.100190.003245 [DOI] [PubMed] [Google Scholar]
- 8.Gritsun TS, Venugopal K, Zanotto PM, Mikhailov MV, Sall AA, Holmes EC, Polkinghorne I, Frolova TV, Pogodina VV, Lashkevich VA, Gould EA. 1997. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′ UTRs. Virus Res. 49:27–39. 10.1016/S0168-1702(97)01451-2 [DOI] [PubMed] [Google Scholar]
- 9.Proutski V, Gould EA, Holmes EC. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194–1202. 10.1093/nar/25.6.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshii K, Igarashi M, Ito K, Kariwa H, Holbrook MR, Takashima I. 2011. Construction of an infectious cDNA clone for Omsk hemorrhagic fever virus, and characterization of mutations in NS2A and NS5. Virus Res. 155:61–68. 10.1016/j.virusres.2010.08.023 [DOI] [PubMed] [Google Scholar]
- 11.Hayasaka D, Gritsun TS, Yoshii K, Ueki T, Goto A, Mizutani T, Kariwa H, Iwasaki T, Gould EA, Takashima I. 2004. Amino acid changes responsible for attenuation of virus neurovirulence in an infectious cDNA clone of the Oshima strain of tick-borne encephalitis virus. J. Gen. Virol. 85:1007–1018. 10.1099/vir.0.19668-0 [DOI] [PubMed] [Google Scholar]
- 12.Takano A, Yoshii K, Omori-Urabe Y, Yokozawa K, Kariwa H, Takashima I. 2011. Construction of a replicon and an infectious cDNA clone of the Sofjin strain of the Far-Eastern subtype of tick-borne encephalitis virus. Arch. Virol. 156:1931–1941. 10.1007/s00705-011-1066-0 [DOI] [PubMed] [Google Scholar]
- 13.Yoshii K, Holbrook MR. 2009. Subgenomic replicon and virus-like particles of Omsk hemorrhagic fever virus. Arch. Virol. 154:573–580. 10.1007/s00705-009-0345-5 [DOI] [PubMed] [Google Scholar]
- 14.Yoshii K, Konno A, Goto A, Nio J, Obara M, Ueki T, Hayasaka D, Mizutani T, Kariwa H, Takashima I. 2004. Single point mutation in tick-borne encephalitis virus prM protein induces a reduction of virus particle secretion. J. Gen. Virol. 85:3049–3058. 10.1099/vir.0.80169-0 [DOI] [PubMed] [Google Scholar]
- 15.Goto A, Hayasaka D, Yoshii K, Mizutani T, Kariwa H, Takashima I. 2003. A BHK-21 cell culture-adapted tick-borne encephalitis virus mutant is attenuated for neuroinvasiveness. Vaccine 21:4043–4051. 10.1016/S0264-410X(03)00269-X [DOI] [PubMed] [Google Scholar]
- 16.Mandl CW, Allison SL, Holzmann H, Meixner T, Heinz FX. 2000. Attenuation of tick-borne encephalitis virus by structure-based site-specific mutagenesis of a putative flavivirus receptor binding site. J. Virol. 74:9601–9609. 10.1128/JVI.74.20.9601-9609.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rumyantsev AA, Murphy BR, Pletnev AG. 2006. A tick-borne Langat virus mutant that is temperature sensitive and host range restricted in neuroblastoma cells and lacks neuroinvasiveness for immunodeficient mice. J. Virol. 80:1427–1439. 10.1128/JVI.80.3.1427-1439.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin EV. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74(Pt 4):733–740. 10.1099/0022-1317-74-4-733 [DOI] [PubMed] [Google Scholar]
- 19.Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726–733. 10.1126/science.4023707 [DOI] [PubMed] [Google Scholar]
- 20.Wigerius M, Melik W, Elvang A, Johansson M. 2010. Rac1 and Scribble are targets for the arrest of neurite outgrowth by TBE virus NS5. Mol. Cell. Neurosci. 44:260–271. 10.1016/j.mcn.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 21.Greene LA, Tischler AS. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U. S. A. 73:2424–2428. 10.1073/pnas.73.7.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurrelbrink RJ, McMinn PC. 2003. Molecular determinants of virulence: the structural and functional basis for flavivirus attenuation. Adv. Virus Res. 60:1–42. 10.1016/S0065-3527(03)60001-1 [DOI] [PubMed] [Google Scholar]
- 23.Kozlovskaya LI, Osolodkin DI, Shevtsova AS, Romanova L, Rogova YV, Dzhivanian TI, Lyapustin VN, Pivanova GP, Gmyl AP, Palyulin VA, Karganova GG. 2010. GAG-binding variants of tick-borne encephalitis virus. Virology 398:262–272. 10.1016/j.virol.2009.12.012 [DOI] [PubMed] [Google Scholar]
- 24.Kentaro Y, Yamazaki S, Mottate K, Nagata N, Seto T, Sanada T, Sakai M, Kariwa H, Takashima I. 2013. Genetic and biological characterization of tick-borne encephalitis virus isolated from wild rodents in southern Hokkaido, Japan, in 2008. Vector-Borne Zoonotic Dis. 13:406–414. 10.1089/vbz.2012.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruzek D, Gritsun TS, Forrester NL, Gould EA, Kopecky J, Golovchenko M, Rudenko N, Grubhoffer L. 2008. Mutations in the NS2B and NS3 genes affect mouse neuroinvasiveness of a Western European field strain of tick-borne encephalitis virus. Virology 374:249–255. 10.1016/j.virol.2008.01.010 [DOI] [PubMed] [Google Scholar]
- 26.Whiteman MC, Wicker JA, Kinney RM, Huang CY, Solomon T, Barrett AD. 2011. Multiple amino acid changes at the first glycosylation motif in NS1 protein of West Nile virus are necessary for complete attenuation for mouse neuroinvasiveness. Vaccine 29:9702–9710. 10.1016/j.vaccine.2011.09.036 [DOI] [PubMed] [Google Scholar]
- 27.Wicker JA, Whiteman MC, Beasley DW, Davis CT, McGee CE, Lee JC, Higgs S, Kinney RM, Huang CY, Barrett AD. 2012. Mutational analysis of the West Nile virus NS4B protein. Virology 426:22–33. 10.1016/j.virol.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, Hall R, Khromykh AA. 2010. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 84:1641–1647. 10.1128/JVI.01979-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396–2404. 10.1128/JVI.80.5.2396-2404.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engel AR, Rumyantsev AA, Maximova OA, Speicher JM, Heiss B, Murphy BR, Pletnev AG. 2010. The neurovirulence and neuroinvasiveness of chimeric tick-borne encephalitis/dengue virus can be attenuated by introducing defined mutations into the envelope and NS5 protein genes and the 3′ non-coding region of the genome. Virology 405:243–252. 10.1016/j.virol.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. 2002. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J. Biol. Chem. 277:36399–36407. 10.1074/jbc.M204977200 [DOI] [PubMed] [Google Scholar]
- 32.Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. 1999. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 257:731–737. 10.1006/bbrc.1999.0370 [DOI] [PubMed] [Google Scholar]
- 33.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503–3515. 10.1128/JVI.01161-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908–5918. 10.1128/JVI.02714-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828–12839. 10.1128/JVI.79.20.12828-12839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200:1261–1270. 10.1086/605847 [DOI] [PubMed] [Google Scholar]
- 37.Van Slyke GA, Ciota AT, Willsey GG, Jaeger J, Shi PY, Kramer LD. 2012. Point mutations in the West Nile virus (Flaviviridae; Flavivirus) RNA-dependent RNA polymerase alter viral fitness in a host-dependent manner in vitro and in vivo. Virology 25:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu ZF, Jackson AC. 2005. Neuronal dysfunction and death in rabies virus infection. J. Neurovirol. 11:101–106. 10.1080/13550280590900445 [DOI] [PubMed] [Google Scholar]
- 39.Volmer R, Monnet C, Gonzalez-Dunia D. 2006. Borna disease virus blocks potentiation of presynaptic activity through inhibition of protein kinase C signaling. PLoS Pathog. 2:e19. 10.1371/journal.ppat.0020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volmer R, Prat CM, Le Masson G, Garenne A, Gonzalez-Dunia D. 2007. Borna disease virus infection impairs synaptic plasticity. J. Virol. 81:8833–8837. 10.1128/JVI.00612-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Regge N, Nauwynck HJ, Geenen K, Krummenacher C, Cohen GH, Eisenberg RJ, Mettenleiter TC, Favoreel HW. 2006. Alpha-herpesvirus glycoprotein D interaction with sensory neurons triggers formation of varicosities that serve as virus exit sites. J. Cell Biol. 174:267–275. 10.1083/jcb.200510156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soffer D, Martin JR. 1989. Axonal degeneration and regeneration in sensory roots in a genital herpes model. Acta Neuropathol. 77:605–611. 10.1007/BF00687888 [DOI] [PubMed] [Google Scholar]
- 43.Michaud J, Fajardo R, Charron G, Sauvageau A, Berrada F, Ramla D, Dilhuydy H, Robitaille Y, Kessous-Elbaz A. 2001. Neuropathology of NFHgp160 transgenic mice expressing HIV-1 Env protein in neurons. J. Neuropathol. Exp. Neurol. 60:574–587 [DOI] [PubMed] [Google Scholar]
- 44.Robinson B, Li Z, Nath A. 2007. Nucleoside reverse transcriptase inhibitors and human immunodeficiency virus proteins cause axonal injury in human dorsal root ganglia cultures. J. Neurovirol. 13:160–167. 10.1080/13550280701200102 [DOI] [PubMed] [Google Scholar]
- 45.Werme K, Wigerius M, Johansson M. 2008. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell. Microbiol. 10:696–712. 10.1111/j.1462-5822.2007.01076.x [DOI] [PubMed] [Google Scholar]
- 46.Melik W, Ellencrona K, Wigerius M, Hedstrom C, Elvang A, Johansson M. 2012. Two PDZ binding motifs within NS5 have roles in Tick-borne encephalitis virus replication. Virus Res. 169:54–62. 10.1016/j.virusres.2012.07.001 [DOI] [PubMed] [Google Scholar]
- 47.Hung AY, Sheng M. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277:5699–5702. 10.1074/jbc.R100065200 [DOI] [PubMed] [Google Scholar]
- 48.Roche JP, Packard MC, Moeckel-Cole S, Budnik V. 2002. Regulation of synaptic plasticity and synaptic vesicle dynamics by the PDZ protein Scribble. J. Neurosci. 22:6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim E, Sheng M. 2004. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5:771–781. 10.1038/nrn1517 [DOI] [PubMed] [Google Scholar]
- 50.Terrien E, Chaffotte A, Lafage M, Khan Z, Prehaud C, Cordier F, Simenel C, Delepierre M, Buc H, Lafon M, Wolff N. 2012. Interference with the PTEN-MAST2 Interaction by a Viral Protein Leads to Cellular Relocalization of PTEN. Sci. Signal. 5:ra58. [DOI] [PubMed] [Google Scholar]
- 51.Gould EA, de Lamballerie X, Zanotto PM, Holmes EC. 2001. Evolution, epidemiology, and dispersal of flaviviruses revealed by molecular phylogenies. Adv. Virus Res. 57:71–103. 10.1016/S0065-3527(01)57001-3 [DOI] [PubMed] [Google Scholar]