ABSTRACT
Human herpesvirus 6 (HHV-6) is widely spread in the human population and has been associated with several neuroinflammatory diseases, including multiple sclerosis. To develop a small-animal model of HHV-6 infection, we analyzed the susceptibility of several lines of transgenic mice expressing human CD46, identified as a receptor for HHV-6. We showed that HHV-6A (GS) infection results in the expression of viral transcripts in primary brain glial cultures from CD46-expressing mice, while HHV-6B (Z29) infection was inefficient. HHV-6A DNA persisted for up to 9 months in the brain of CD46-expressing mice but not in the nontransgenic littermates, whereas HHV-6B DNA levels decreased rapidly after infection in all mice. Persistence in the brain was observed with infectious but not heat-inactivated HHV-6A. Immunohistological studies revealed the presence of infiltrating lymphocytes in periventricular areas of the brain of HHV-6A-infected mice. Furthermore, HHV-6A stimulated the production of a panel of proinflammatory chemokines in primary brain glial cultures, including CCL2, CCL5, and CXCL10, and induced the expression of CCL5 in the brains of HHV-6A-infected mice. HHV-6A-induced production of chemokines in the primary glial cultures was dependent on the stimulation of toll-like receptor 9 (TLR9). Finally, HHV-6A induced signaling through human TLR9 as well, extending observations from the murine model to human infection. Altogether, this study presents a first murine model for HHV-6A-induced brain infection and suggests a role for TLR9 in the HHV-6A-initiated production of proinflammatory chemokines in the brain, opening novel perspectives for the study of virus-associated neuropathology.
IMPORTANCE HHV-6 infection has been related to neuroinflammatory diseases; however, the lack of a suitable small-animal infection model has considerably hampered further studies of HHV-6-induced neuropathogenesis. In this study, we have characterized a new model for HHV-6 infection in mice expressing the human CD46 protein. Infection of CD46 transgenic mice with HHV-6A resulted in long-term persistence of viral DNA in the brains of infected animals and was followed by lymphocyte infiltration and upregulation of the CCL5 chemokine in the absence of clinical signs of disease. The secretion of a panel of chemokines was increased after infection in primary murine brain glial cultures, and the HHV-6-induced chemokine expression was inhibited when TLR9 signaling was blocked. These results describe the first murine model for HHV-6A-induced brain infection and suggest the importance of the TLR9 pathway in HHV-6A-initiated neuroinflammation.
INTRODUCTION
Human herpesvirus (HHV)-6A and -6B are two closely related yet distinct betaherpesviruses. They share more than 90% nucleotide sequence identity but differ in several genetic features and clinical manifestations (1, 2). HHV-6B has long been identified as the etiological agent for exanthem subitum, a common infant febrile illness (3), while primary infection with HHV-6A has not yet been conclusively associated with any specific disease. Although the presence of DNA and RNA sequences from HHV-6A and HHV-6B has been described in the central nervous system (CNS) of healthy individuals (4–8), several studies have correlated HHV-6 infection with neuroinflammatory diseases. In immunosuppressed patients, HHV-6A and -6B often reactivate and are able to provoke neurological complications, such as severe encephalitis or meningitis (9). In addition, both viruses have been proposed to play a role in the pathogenesis of the autoimmune neurological disease multiple sclerosis (10–13). However, the mechanisms explaining how HHV-6 could be involved in such diseases remain to be elucidated.
The human transmembrane protein CD46 was identified as the receptor for HHV-6 entry into host cells (14). This complement regulatory protein is also the receptor for several other pathogens, such as measles virus and Neisseria (15–17). Alternative splicing mechanisms lead to the expression of different isoforms of the protein, which can be placed into two groups according to their cytoplasmic tail, CD46-cyt1 (short) or CD46-cyt2 (long), which exhibit different immunoregulatory properties (18). CD46 expression in humans is ubiquitous (19); therefore, it provides a wide range of potentially susceptible cell types for HHV-6 infection. Although lymphocytes are known as the main target cells, several CNS cell types, including astrocytes and oligodendrocytes, have been successfully infected by HHV-6A and, with lower efficiency, by HHV-6B (20–27). Recently, CD134 (OX40), a member of the tumor necrosis factor (TNF) receptor superfamily that was expressed on activated T lymphocytes, has been identified as a receptor molecule for HHV-6B (28).
Like most herpesviruses, HHV-6 can establish long-term latency in its hosts using immunomodulatory mechanisms to evade the immune system. HHV-6 is indeed known to infect T cells very efficiently and to reduce their proliferation (29–31), and some viral proteins have been shown to inhibit signaling pathways involved in immune responses (32–34). However, HHV-6 is also able to promote inflammation by inducing the development of a Th1 phenotype in T cells (35, 36), enhancing the cytotoxicity of NK cells (37), and by increasing the production of proinflammatory cytokines and chemokines in different cell types (38, 39). In particular, HHV-6A was shown to induce the expression of CCL5 (RANTES) in human astrocytes, endothelial cells, and tonsilar cells (40–42). Interestingly, HHV-6 was found to establish latency in the host by integrating into human chromosomes and allowing the virus to be transmitted in the germ line in some individuals (43), which so far is unique among human herpesviruses.
Animal models are of critical importance to better understand the pathogenesis of HHV-6 infections; nevertheless, only a few are currently available. Shortly after the discovery of HHV-6, Yalcin et al. described an asymptomatic infection of cynomolgus macaques and African green monkeys with an HHV-6B strain associated with a rise in HHV-6-specific IgG levels (44). HHV-6A infection was performed later in pig-tailed macaques and led to moderate symptoms, antibody (Ab) response, and plasma viremia (45). Recently, a marmoset model was used to study HHV-6A and -6B infection and showed that HHV-6A, but not HHV-6B, is able to provoke neurological symptoms (46). However, monkey experiments are often limited, as they imply severe ethical constraints and elevated costs. Therefore, the development of a more suitable small-animal model would be of high interest for the study of HHV-6 pathogenesis and of HHV-6-related neuroinflammation in particular. Mice were initially described to be resistant to HHV-6 infection (47), possibly due to the testis-restricted expression and low sequence homology of the murine CD46 homologue (48). Humanized mice, including SCID mice transplanted with human fetal liver and thymus (49) and Rag2−/−γc−/− mice reconstituted with cord blood-derived human hematopoietic stem cells (50), have been used to study HHV-6 infection. However, in these models, mice were used only as an in vivo environment to study the infection of human cells or tissues, and the infection of murine tissues was not described. In this study, we characterized a new model for HHV-6 infection in mice expressing the human CD46 protein. Infection of these mice with HHV-6A but not HHV-6B resulted in the persistence of viral DNA for up to 9 months in the brains of infected animals. HHV-6A infection was followed by lymphocyte infiltration and CCL5 upregulation, although clinical signs of disease were not observed. The secretion of a panel of chemokines was increased after infection in primary brain glial cultures generated from CD46 transgenic mice as well, and the HHV-6-induced chemokine expression was inhibited when TLR9 signaling was blocked. These results describe the first murine model for HHV-6A brain infection and support the hypothesis that HHV-6A can initiate neuroinflammation via the TLR9 pathway.
MATERIALS AND METHODS
Cell lines and viruses.
Murine lymphoid B-cell M12 and L fibroblast lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS). M12 and L cells stably expressing CD46 Cyt2 isoforms (M12-CD46 and L-CD46) were described previously (17). HSB2 and MOLT3 human T cell lines were cultured in complete RPMI medium supplemented with 10% FCS. Human embryonic kidney (HEK) cells stably expressing the human TLR9 (hTLR9) gene and the luciferase reporter gene were kindly provided by Kate Fitzgerald (UMASS, Worcester, MA) and cultured in complete DMEM supplemented with 10% FCS.
HHV-6A (GS strain) and HHV-6B (Z29 strain) were kindly provided by L. Naesens (Belgium) and propagated in HSB2 and MOLT3 cells, respectively. For virus production, cells were infected at a multiplicity of infection (MOI) of 0.005 for 1.5 h at 37°C. At maximum cytopathic effects (CPE), cells were centrifuged, resuspended in a 10-fold lower volume of fresh RPMI supplemented with 20% of FCS, aliquoted, and stored at −80°C. For cell-free virus production, infected cell suspensions at maximum CPE were frozen at −80°C. After 3 freeze-thaw cycles, virus suspensions were clarified by centrifugation (20 min, 4,000 × g, 4°C) and loaded on a 15% sucrose layer. Viruses were then pelleted by ultracentrifugation (2 h, 28,500 rpm; SW32Ti rotor) and resuspended in cold FCS. Noninfected cell suspensions were processed under identical conditions and were used for mock infections. Experimental HHV-6 infection of nonadherent cells, M12 and M12-CD46, was performed in suspension for 1.5 h at 37°C at an MOI of 1. Cells were then pelleted and resuspended in fresh medium for further culture.
Virus titers were determined by immunofluorescence. HSB2 or MOLT3 cells were cultured in 96-well plates and infected in quadruplicate with serial dilutions of viral stock. After 5 days of culture, cells were harvested on 10-well slides, fixed in cold acetone, and stained with mouse anti-HHV-6 p41 primary antibody (Ab; Santa Cruz Biotechnology) and Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (Molecular Probes). Titers were calculated in 50% tissue culture infective doses per ml (TCID50/ml).
Animals and infection protocols.
Transgenic mice ubiquitously expressing the cyt1 (CD46-cyt1 mice) or cyt2 (CD46-cyt2 mice) isoform of CD46 in the C57BL/6J background (18), mice transgenic for the whole genomic sequence and the promoter of CD46 expressing both cyt1 and cyt2 cytoplasmic tails (CD46ge mice) (51), and CD46-cyt2 mice crossed into the interferon type I (IFN-I) receptor knock out (IFNARKO) background (52) were bred and used in the animal facility PBES of ENS Lyon, France.
Four- to 7-week-old mice were anesthetized with a solution of ketamine (Pharma) and xylazine (Rompun) and then received an intracranial (i.c.) injection of 50 μl of HHV-6A or -6B (105 TCID50 per mouse), UV-inactivated virus (30 min at 254 nm), or heat-inactivated virus (15 min at 80°C) in the right hemisphere. Control mice were injected with mock solution (described above). An additional intraperitoneal (i.p.) injection of 2 × 106 HHV-6A- or HHV-6B-infected HSB2 or MOLT3 cells, respectively, was performed 1 week after i.c. injection. Animals were sacrificed, and brains, spleens, and blood were collected at several time points after infection. Brains were immediately frozen at −80°C for further analysis. Splenocytes were isolated by mechanical disruption followed by red blood cell lysis before nucleic acid extraction. For the analysis of chemokine expression and histology, mice received only i.c. injection with purified virus and were perfused with PBS before brain sampling.
All animals were handled in strict accordance with good animal practice as defined by the French national charter on the ethics of animal experimentation. Animal work was approved by the regional ethical committee (Comité Régional d'Ethique pour l'Experimentation Animal de la Région Rhône-Alpes; CREEA) and by the Regional Ethical Committee (CECCAPP). Experiments were performed in the PBES (Plateau de Biologie Expérimentale de la Souris), ENS-Lyon, in Lyon, France.
Enrichment of primary splenic T lymphocytes.
Spleen cells from CD46ge mice or nontransgenic littermates were enriched in T lymphocytes first by adherence of monocytes for 3 h at 37°C. Supernatants were then harvested, and B lymphocytes were depleted by magnetic cell sorting (MACS) using anti-mouse IgG microbeads (Miltenyi Biotec). Separation was performed according to the manufacturer's instructions using an AutoMACS separator. After separation, enrichment in T lymphocytes was checked by flow cytometry, and the percentage of CD3+ cells in the cell suspensions was higher than 70%. Before infection with HHV-6, cells were activated with concanavalin A (ConA) (3 μg/ml) at 37°C overnight.
Primary brain glial cell cultures.
Brains were extracted from newborn CD46 transgenic or wild-type mice (3 to 4 days old), washed in DMEM, homogenized by passing through a sterile syringe and needle (19 gauge), and cultured as described previously (53). Briefly, 12-well plates were precoated overnight with poly-d-lysine (50 μg/ml; Sigma), and cells were cultured in 1 ml of DMEM supplemented with 20% FCS for the first 48 h. Further culture was performed using 10% FCS, and medium was renewed three times per week. Cell cultures usually appeared confluent after 6 to 8 days and contained astrocytes (∼90%), oligodendrocytes, and microglial cells. Cultures were infected for 2 h at 37°C with 5 × 105 TCID50 per well of HHV-6A, HHV-6B, or UV-irradiated virus or were mock infected in a final volume of 0.5 ml, and fresh medium was then added. TLR9 blocking treatments were performed using ODN 2088 (Invivogen) at 10 μg/ml. At 1, 2, or 5 days postinfection, culture supernatants were collected and cells were washed twice with PBS before total RNA or genomic DNA extraction.
Nucleic acids extraction and quantitative PCR (qPCR).
Genomic DNA was extracted from both cells and brain tissue using Nucleospin tissue columns (Macherey-Nagel), and total RNAs were extracted using the RNeasy kit (Qiagen) including DNase I treatment. RNAs were additionally treated with Turbo DNase (Ambion) to exclude any contamination with genomic DNA. Retrotranscription was performed on 0.5 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). Products were diluted (1/10), and 5 μl of the dilution was used for DNA amplification.
Quantitative PCRs on genomic DNA were performed using 50 ng of DNA template and a set of primers specific for the HHV-6 gene U41 (Table 1). For normalization of murine and human samples, genomic DNA was run in parallel with murine β-actin primers or human β-globin primers, respectively (Table 1).
TABLE 1.
HHV-6 specific primers used in qPCR analyses
| Target genea | Sequence |
Amplicon length (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| Genomic DNA templates | |||
| U41 | 5′-GTCATAGACCGGAGCATCGT-3′ | 5′-TGAGGTGATGAGGGATAGGG-3′ | 196 |
| Murine β-actin | 5′-GGTACTAACAATGGCTCGTGTGAC-3′ | 5′-TCAGGGCAGGTGAAACTGTATGG-3′ | 123 |
| Human β-globin | 5′-CCCTTGGACCCAGAGGTTCT-3′ | 5′-CGAGCACTTTCTTGCCATGA-3′ | 101 |
| cDNA templates | |||
| U79 (E) | 5′-AACGACGAAGACAAGCAACCG-3′ | 5′-TGTTATGCCATCCTCGTGACTTTG-3′ | 123 |
| U86 (IE2, LAT) | 5′-ACCCACAGACAATGCACATCC-3′ | 5′-TGGGCTGTAGGAGTTGATTCG-3′ | 106 |
| U90 (IE1, LAT) | 5′-TGGAAACACAGACCCATCAGAC-3′ | 5′-TCTGGAGAAGGAGTGCATGGAT-3′ | 141 |
| U94 (IE, LAT) | 5′-CGCCCCTGATTTCCGTTGTG-3′ | 5′-CCTGCAAAGTGGTACGCTCAAG-3′ | 121 |
| U100 (L) | 5′-AACTGAGGAACTGATGGAAACC-3′ | 5′-GCTTACGATGGCAGGATCTATG-3′ | 109 |
| Murine CCL5 | 5′-GCACCTGCCTCACCATATGG-3′ | 5′-AGCACTTGCTGCTGGTGTAG-3′ | 112 |
| Murine CCL2 | 5′-ACCAGCACCAGCCAACTC-3′ | 5′-CAGAAGCATGACAGGGACC-3′ | 75 |
| Murine CXCL10 | 5′-GCAACTGCATCCATATCG-3′ | 5′-GACATCTCTGCTCATCATTC-3′ | 135 |
| Murine GAPDH | 5′-GCATGGCCTTCCGTGTCC-3′ | 5′-TGTCATCATACTTGGCAGGTTTCT-3′ | 84 |
| Human GAPDH | 5′-CACCCACTCCTCCACCTTTGAC-3′ | 5′-GTCCACCACCCTGTTGCTGTAG-3′ | 112 |
E, early; IE, immediate early; L, late; LAT, latency-associated transcript.
Quantitative PCRs on cDNA templates were performed with primers specific to the viral genes U79, U94, and U100 (Table 1). As transcripts of U79 (early transcript) and U100 (late transcript) genes are extensively spliced, primers were designed to span exon/exon junctions in order to discriminate between cDNA and potential genomic DNA contamination.
The expression of the genes coding for murine CCL2, CCL5, and CXCL10 were analyzed using primers presented in Table 1. Glyceraldehyde 3′-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene for viral mRNA quantification in both human and murine samples (Table 1). The PCR efficiency of each set of primers was determined using the standard curves obtained by performing real-time PCR on 10-fold dilutions of PCR products. These dilutions of PCR products were then used as standard references and included in each PCR run in order to standardize the PCR run with respect to template integrity, sample loading, and inter-PCR variations.
All real-time PCRs were performed in duplicate using the Platinum Sybr green qPCR supermix-UDG (Invitrogen). Samples were run in an ABI Prism 7000 SDS, and results were analyzed using ABI Prism 7000 SDS software. Levels of viral DNA were normalized to the number of cells in each sample by comparison to the β-actin gene and expressed as U41 copies per copy of β-actin. Calculations were done using the 2ΔΔCT model according to MIQE guidelines (54, 55), and the expression levels of all mRNAs analyzed were normalized to that of GAPDH.
Cytokine assay.
Supernatants from HHV-6A or mock-infected primary brain glial cell cultures from wild-type or CD46-transgenic mice were collected 48 h postinfection and analyzed with a Proteome Profiler antibody array for mouse cytokines (R&D Systems), which can detect 40 different murine cytokines. Each supernatant was mixed and incubated with a cocktail of biotinylated detection antibodies and blotted onto a nitrocellulose membrane previously spotted in duplicate with capture antibodies according to the manufacturer's instructions. Revelation was performed by chemiluminescence using horseradish peroxidase (HRP)-coupled streptavidin. Data were analyzed using the software Quantity One and expressed as mean pixel density.
Antibody detection.
Ninety-six-well plates were coated overnight at 4°C with UV-irradiated purified HHV-6A virions diluted in carbonate/bicarbonate buffer (pH 9.6). Wells were washed and blocked for 1 h at room temperature with PBS supplemented with 3% bovine serum albumin (PBS-BSA 3%). Tenfold dilutions of serum were added to each well in duplicate and incubated for 2 h at 37°C. HHV-6-specific Abs were revealed using HRP-conjugated anti-murine IgG Ab (Promega). Standard curves, obtained by serial dilution of positive serum from mice immunized with HHV-6 antigens in complete Freund adjuvant, were run for each experiment. One arbitrary unit (AU) was attributed to the 10-fold dilution of the control serum. After washing, coloration was observed by adding 200 μl/well of orthophenylenediamine solution (Sigma) after incubation for 30 min at 37°C. Absorbance was measured using a VersaMax microplate reader (Molecular Devices). Data were analyzed with Softmax Pro 5.3 software using a standard curve and expressed as AU.
Immunofluorescence.
For immunofluorescence staining, primary brain cells were seeded on coverslips previously coated with poly-d-lysine (50 μg/ml; Sigma). After infection, cells were washed with PBS, fixed in cold acetone for 10 min, and rehydrated in PBS overnight. Aspecific binding sites were blocked with PBS-BSA 3% before incubation with mouse anti-HHV-6 p41 Ab (9A5D12; Santa Cruz) diluted 1/100 or mouse anti-HHV-6 gp116 Ab (6A5G3; HHV-6 foundation) diluted 1/100, as well as rabbit anti-glial fibrillary acidic protein (GFAP) Ab diluted 1/750 (Dako), for 1.5 h at 37°C. The specific labeling was revealed using Alexa Fluor 555-coupled anti-rabbit and Alexa Fluor 488-coupled anti-mouse (Molecular Probes) secondary Abs for 45 min at 37°C. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (1/1,000). Slides were observed by confocal microscopy using an LSM710 spectral microscope (Zeiss).
For immunohistofluorescence analysis, sections of 7 μm were cut from frozen brains, fixed in 90% cold acetone for 10 min, and rehydrated in PBS for 15 min. Sections were then incubated for 30 min with Fc receptor blocker agent (Innovex Biosciences). Aspecific binding sites were blocked with PBS-BSA 3% before incubation with rabbit polyclonal anti-CD3 Ab (Dako), rat monoclonal anti-CD19 Ab (Serotec), or rat anti-mouse F4/80 Ab (specific for murine monocytes and macrophages) at 4°C overnight. The specific labeling was revealed using Alexa Fluor 488-coupled anti-rabbit and Alexa Fluor 546-coupled anti-rat secondary Abs (Molecular Probes) for 45 min at 37°C. Cell nuclei were stained with DAPI, and slides were observed using an Axiovert 200M microscope (Zeiss). Quantification of fluorescence staining was done on 2 sections from 4 to 8 different mice in each group. All sections were processed under identical conditions during CD3 staining, and images of the left and right periventricular areas were acquired using the same exposure time to allow proper comparison between slides. Image J software was then used for image analysis to determine CD3 signal density. Briefly, the same arbitrary threshold for positive fluorescence was applied to all images, and signal density was calculated as a ratio between the area of positive fluorescence (above the threshold) and the total area analyzed.
Luciferase assay.
Stably transfected HEK 293 cells expressing both human TLR9 and the luciferase reporter gene under the control of the pELAM promoter, which contains critical NF-κB-binding sites, were seeded in 96-well plates at a density of 4 × 104 cells per well and stimulated with purified HHV-6A, HHV-6B, or UV-inactivated virus at an MOI of 0.5, 1, and 2 in triplicate. Nonstimulated, mock-stimulated, and CpG-stimulated controls were added, and an additional control with TLR9 antagonist ODN 2088 (Invivogen) treatment was performed during HHV-6A stimulation. Cell lysates were assayed for luciferase activity using a luciferase reporter assay system (Promega). For each experiment, luminescence intensity values were compared to those of the nonstimulated controls.
Statistical analyses.
Data were analyzed for statistical significance using the Mann-Whitney nonparametric U test, and Student's t test and results were considered significant when the P value was below 0.05.
RESULTS
Infection of CD46-transfected murine lymphocyte lines with HHV-6.
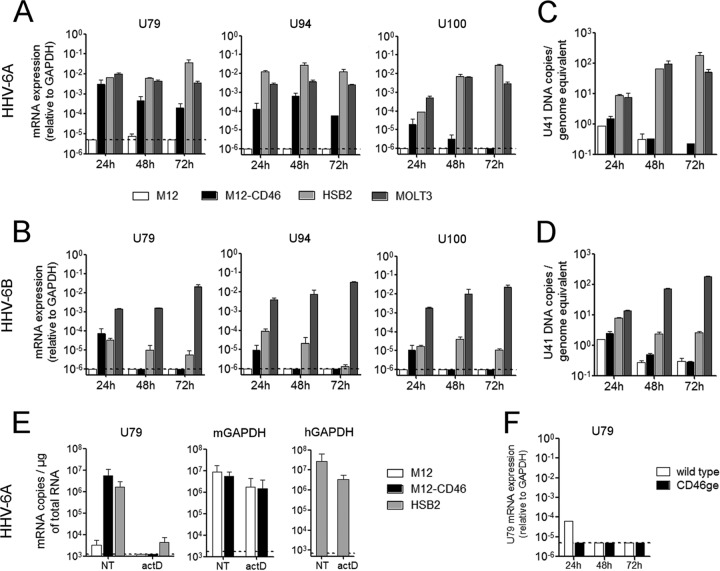
To analyze the infection of murine cells by HHV-6, we initially compared the permissiveness of the murine B-lymphoblastic cell line M12, stably expressing human CD46 (M12-CD46), to that of two human T-lymphoid cell lines, HSB2 and MOLT3, which allow productive infection by HHV-6A and -6B, respectively. Cells were infected with either HHV-6A or HHV-6B at an MOI of 1. While infection led to important CPE in the human cells, no morphological changes were observed in either M12 or M12-CD46 cells. In addition, we monitored the production of viral mRNA by reverse transcription-qPCR (RT-qPCR) during 3 days postinfection (p.i.) (Fig. 1). After infection with HHV-6A (Fig. 1A), the expression of the early gene U79 and the immediate-early gene U94 was detected only in the M12-CD46 cells and not in the parental M12 cells. The levels of expression at 24 h were comparable to those observed in human HSB2 and MOLT3 cells but slowly decreased with time, probably due to cell multiplication and dilution of the transcripts. Similarly, expression of the late transcript U100 was also detected in M12-CD46 cells only, although the level of expression was lower. Levels of all three HHV-6 transcripts were analyzed at later time points up to 5 days p.i. and continued to decrease following cell division (data not shown). In the case of HHV-6B infection, viral RNA was found at lower levels in M12-CD46 cells at 24 h p.i. only and was not detectable afterwards in murine cells (Fig. 1B). In human lymphocyte lines infected with HHV-6B, expression of all three transcripts highly increased in the MOLT3 cell line, which is classically used for the production of the HHV-6B Z29 strain, while it decreased with time in the other human cell line, HSB2, which is used for the production of HHV-6A (strain GS). Finally, HHV-6A infection of murine L fibroblasts and L-CD46 cells gave results very similar to those observed with M12 and M12-CD46 cells, while HHV-6B infection was inefficient in all murine lines tested (data not shown).
FIG 1.
HHV-6 infection of murine and human lymphoid cells. Murine lymphoid M12 cells, stably expressing the human protein CD46 (M12-CD46), parental M12 cell line, and human T cell lines HSB2 and MOLT3, were infected with HHV-6A (A and C) or HHV-6B (B and D) at an MOI of 1. (A and B) mRNA levels of the viral genes U79 (early), U94 (immediate early), and U100 (late) were determined using RT-qPCR and expressed relative to that of murine GAPDH (M12 and M12-CD46) or human GAPDH (HSB2 and MOLT3). (C and D) Genomic DNA levels of the viral gene U41 were determined using qPCR, and means and standard deviations from two independent experiments are presented. (E) Before infection with HHV-6A, M12, M12-CD46, and HSB2 cells were treated with actinomycin D (actD) at 5 μg/ml for 1 h or left untreated (NT). Total RNA was extracted at 8 h postinfection, and U79 mRNA levels were quantified by RT-qPCR. Results are presented as the number of copies per μg of total RNA and are representative of two independent experiments. (F) Primary murine T lymphocytes were enriched from spleens of wild-type (open bars) or CD46ge (closed bars) mice, ConA activated, and infected with HHV-6A at an MOI of 1. mRNA expression of U79 was quantified relative to that of murine GAPDH by RT-qPCR at different time points postinfection. Dotted lines represent the limit of detection of the qPCR system.
We then analyzed the levels of HHV-6 DNA in infected cells. As shown in Fig. 1C and D, genomic DNA levels increased rapidly in the productively infected human cell lines, whereas they decreased in murine cells. To determine whether the HHV-6A mRNA detected in M12-CD46 cells is produced following infection by the murine cells or represents a carryover from the initial virus inoculum, we pretreated cells with actinomycin D for 1 h before infection to block new RNA synthesis and quantified the expression of the early gene U79 8 h p.i. As shown in Fig. 1E, this treatment completely blocked the production of HHV-6A mRNA in both murine and human cells, while it only moderately decreased the level of the abundantly expressed housekeeping gene GAPDH mRNA in these cells, demonstrating that U79 is transcribed by HHV-6A-infected M12-CD46 cells. Finally, as HHV-6 primary targets in humans are T lymphocytes, we used ConA-activated CD3+ lymphocytes from spleens of CD46 transgenic mice to infect them using the same experimental conditions as those for cell lines described above. However, infection with either HHV-6A (Fig. 1F) or HHV-6B (not shown) did not allow the detection of viral transcripts.
Altogether, these results suggest that HHV-6A but not HHV-6B could enter CD46-expressing murine B cell line and L fibroblasts and start RNA transcription, although later stages of productive infection and cytopathic effect were not observed. However, in vitro HHV-6A infection of primary CD46+ T lymphocytes was not efficient.
Infection of murine primary brain glial cultures with HHV-6.
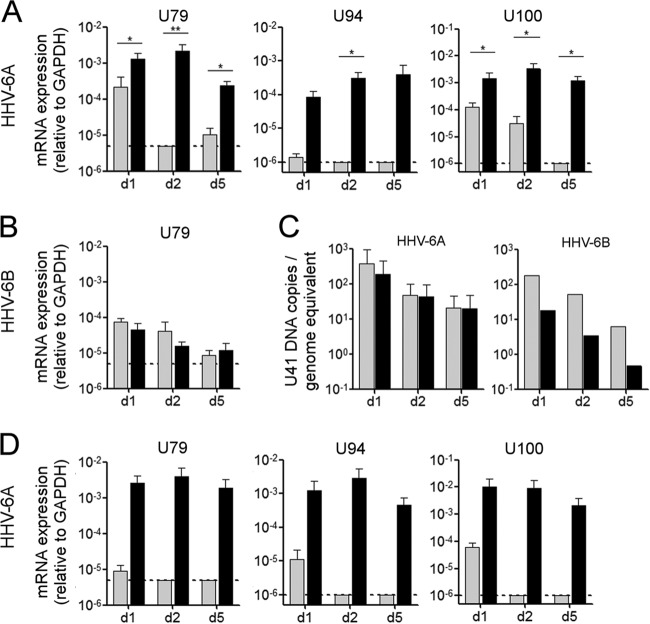
As HHV-6 is known to be neurotropic in humans and to infect both human astrocytes and oligodendrocytes (22–25), we next analyzed the permissiveness of primary brain glial cultures, which mostly contain astrocytes and, to a lesser extent, oligodendrocytes and microglial cells (data not shown), to HHV-6 infection. These cultures were obtained from either CD46-cyt1 mice, expressing the cyt1 isoform of CD46, or wild-type littermate controls (Fig. 2). After HHV-6A infection, expression of U79, U94, and U100 genes was significantly higher in CD46-expressing cells than in wild-type cells (Fig. 2A) and was maintained for up to 7 days (not shown). Basal levels of transcripts were also detected in CD46-negative cells but decreased rapidly with time. On the contrary, only a basal level of expression of U79 (Fig. 2B), U94, or U100 (not shown) was observed after HHV-6B infection, and no difference was found between cells from CD46-cyt1 mice and cells from wild-type mice. This suggested that HHV-6A, but not HHV-6B, could infect CD46-expressing murine glial cells. The presence of viral DNA in the cultures then was analyzed by qPCR (Fig. 2C), and for both HHV-6A and HHV-6B, infection resulted in a decrease in viral DNA loads in all murine cultures analyzed (Fig. 2C), showing that HHV-6A infection of CD46-expressing murine glial cells is nonproductive. Moreover, CPE in terms of syncytium formation were not observed during 2 weeks p.i. under these experimental conditions. Similar results were obtained with cultures from other mouse lines expressing CD46-cyt2 or both isoforms of CD46 (CD46-cyt2 mice and CD46ge mice) (not shown), indicating that infection of murine glial cells is independent from the CD46 isoform expressed. As IFN-I is important for the control of viral infections in the early stages (52, 56), we prepared primary brain cultures from either mice deficient for the expression of IFN-I receptor (IFNARKO mice) or CD46-cyt2 mice crossed into the IFNARKO background. Surprisingly, HHV-6A infection resulted in similar patterns of production of U79, U94, and U100 mRNA (Fig. 2D) and in the absence of formation of syncytia (not shown), suggesting that limitation in the progression toward later stages of infection in murine cells is independent of IFN type I signaling.
FIG 2.
HHV-6 infection of murine primary glial brain cultures. (A to C) Primary glial cultures were generated from the brains of wild-type mice (gray bars) or CD46-cyt1 mice (black bars) and infected with HHV-6A or HHV-6B at an MOI of 0.5. Levels of mRNA expression of the viral gene U79 (A and B) and U94 and U100 genes (A) were determined at 1, 2, and 5 days p.i. using RT-qPCR. (C) Viral DNA levels were determined by qPCR using genomic DNA. Means and standard deviations from 2 to 5 independent experiments are plotted. (D) HHV-6A infection was performed with cultures generated from IFNARKO mice (gray bars) and IFNRKO × CD46-cyt2 mice (black bars), and the mRNA levels of U79, U94, and U100 were analyzed. Dotted lines represent the limit of detection of the quantitative PCR system.
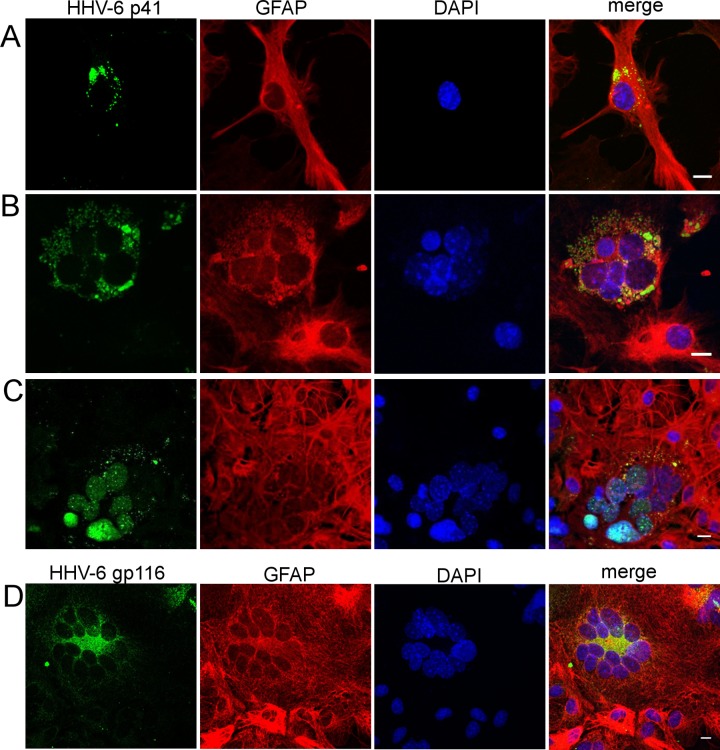
Several studies on human glial cells have suggested that cocultures with productively infected lymphocytes increase the efficiency of the infection (22, 27, 40). To analyze whether infection also could be amplified in murine cells during continuous contact with cells producing infectious virus, primary brain cultures were overlaid with HHV-6A-infected HSB2 cells. The confocal microscopy approach allowed us to analyze staining for viral proteins in GFAP+ murine cells, thereby excluding any potential residual infected human lymphocytes. Indeed, production of HHV-6 p41 early antigen (Fig. 3A to C) as well as gp116 glycoprotein (Fig. 3D) was observed in CD46+ cultures 7 days after the establishment of coculture with cytoplasmic/perinuclear (Fig. 3A, B, and D) and nuclear (Fig. 3C) localization. Furthermore, the formation of syncytia was observed after 2 to 3 days of coculture and increased with time, and syncytium-forming cells were positive for the astrocyte marker GFAP and viral antigens p41 and gp116. The expression of viral proteins and formation of syncytia was observed with nontransgenic cultures as well, although at lower levels (data not shown), suggesting that fusion of murine glial cells during coculture occurs with lower efficiency in the absence of CD46. These results suggested that astrocytes can be infected by HHV-6A and form syncytia when they are in contact with infected human lymphocytes, indicating that HHV-6A infection could progress in primary murine cells if adequate conditions are provided.
FIG 3.
Production of HHV-6 proteins in primary murine glial brain cultures overlaid with HHV-6A-infected HSB2 lymphocytes. Primary murine brain glial cells generated from CD46-transgenic (CD46-cyt1) mice were cocultured with HHV-6A-infected HSB2 cells. Seven days after the establishment of coculture, supernatants as well as nonadherent cells, were removed, and adherent cells were fixed and analyzed for the presence of viral antigens by confocal microscopy. Cells were stained with antibodies against HHV-6 protein (green) p41 (A to C) or gp116 (D) or with glial fibrillary acidic protein (GFAP) antibody (red), and cell nuclei were stained with DAPI (blue). With p41 antibody, both cytoplasmic (A and B) and nuclear (C) staining was observed. Scale bar, 10 μm.
Infection of CD46 transgenic mice with HHV-6.
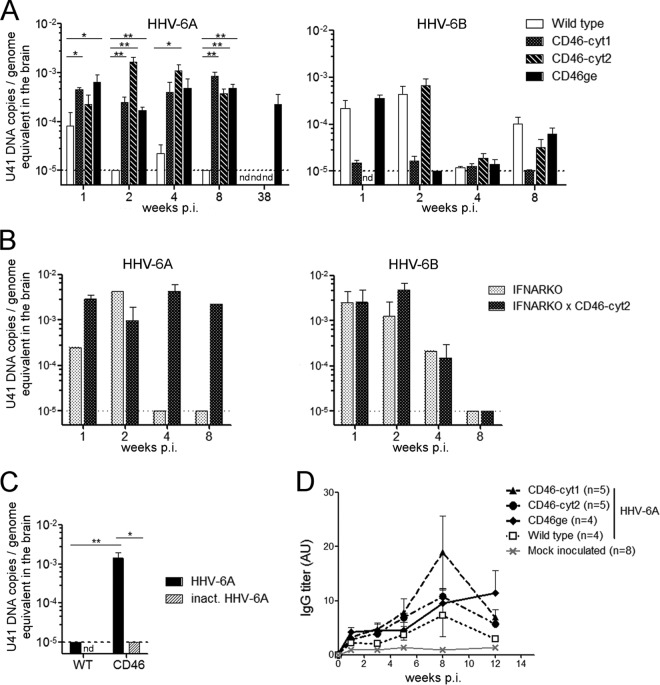
CD46 cytoplasmic tails, CD46-cyt1 (short) and CD46-cyt2 (long), were shown to exhibit opposite roles in the control of inflammatory response, with cyt1 having anti-inflammatory activities and cyt2 rather proinflammatory effects (18). Although most of the cells express both types of CD46 cytoplasmic tails, cyt2 is preferentially expressed in the human brain (57). Therefore, transgenic mice expressing either one or the other CD46 isoform in the brain may have differential susceptibility to the infection and to potential viral neuroinflammatory effects. Thus, we analyzed the susceptibility of several lines of CD46 transgenic mice expressing different cytoplasmic domains of CD46 or being crossed into the IFNARKO background to infection with both HHV-6A and HHV-6B (Fig. 4). In order to target both brain tissue and the periphery, mice were first inoculated i.c. with 105 TCID50 of HHV-6A or HHV-6B and 1 week later received i.p. injection. As HHV-6A and HHV-6B are known to be highly cell-associated viruses (2), we used HHV-6A-infected HSB2 or HHV-6B-infected MOLT3, respectively, as carrier cells for the i.p. injection to increase viral load and stimulate a secondary immune response in mice after a peripheral viral challenge. Mice were monitored for 8 weeks; however, visible clinical signs potentially related to the infection were not observed. Brains and spleens were taken at different time periods and analyzed by qPCR for the presence of HHV-6 DNA and mRNA. Interestingly, following HHV-6A infection, viral DNA was detected in the brains of all CD46 transgenic mouse lines in significantly larger amounts than in wild-type mice, and they persisted for up to 38 weeks p.i. in 3 out of 4 infected CD46 transgenic mice, whereas the levels decreased below the limit of detection after 2 weeks p.i. in the brain of wild-type mice. Viral RNA, however, was not detected in organs from infected mice, including several latency-associated transcripts, U86 (IE2), U90 (IE1), and U94, tested 1 month after infection (data not shown).
FIG 4.
In vivo infection with HHV-6A or HHV-6B in different CD46-transgenic mouse lines. (A) CD46-cyt1, CD46-cyt2, or CD46ge mice or wild type littermates received i.c. injection of HHV-6A (4 to 9 mice/group) or HHV-6B (4 to 9 mice/group), followed 1 week later by i.p. injection of HHV-6A-infected HSB2 cells or HHV-6B-infected MOLT3 cells. (B) The same type of infection was performed with IFNARKO and IFNARKO × CD46-cyt2 mice with either HHV-6A or HHV-6B (1 to 4 mice/group). Brains were collected at different time points after infection, and levels of U41 DNA were determined by qPCR. Data were normalized to genome equivalents using the murine cellular gene β-actin. (C) Control experiment carried out using CD46 transgenic mice (CD46-cyt1 and CD46ge) and nontransgenic littermates (5 to 6 mice/group), which received i.c. injection of either HHV-6A or heat-inactivated (inact.) HHV-6A (nd, not done). Brains were collected 3 weeks after i.c. injection. (A to C) Increase in viral loads compared to wild-type control mice were analyzed using the Mann-Whitney test (*, P ≤ 0.05; **, P ≤ 0.01). Dotted lines represent the limit of detection of the quantitative PCR method. (D) Plasma from CD46-cyt1, CD46-cyt2, and CD46ge mice and nontransgenic littermates, inoculated with HHV-6A or mock infection solution, were collected before and after virus administration every 2 weeks for 12 weeks. HHV-6-specific IgG was detected by ELISA, using purified virions for coating, and expressed in arbitrary units (AU). Means and standard errors are plotted.
In agreement with the results obtained in vitro, HHV-6B infection did not result in significant augmentation of HHV-6 DNA in CD46 transgenic brains compared to nontransgenic brains and was observed principally by 2 weeks after infection (Fig. 4B). Interestingly, neither HHV-6A nor HHV-6B infection resulted in important changes in DNA levels in the spleen of CD46 transgenic mice, where low levels were detected only occasionally in both transgenic and wild-type mice (data not shown). Furthermore, similar to what was observed with primary brain cultures, the absence of a functional IFN type I system did not significantly change the pattern of HHV-6A infection in the brain, although HHV-6B infection persisted longer in the IFNARKO background without any visible difference between CD46 transgenic and nontransgenic mice (Fig. 4B). As genetic background was found to play a role in the susceptibility of mice to herpesvirus infection (58), CD46 transgenic mice crossed into the BALB/c background were analyzed as well. The HHV-6A infection resulted in increased DNA levels in the brain, similar to what was observed in the other CD46 lines, in the C57BL/6 genetic background (data not shown), without any clinical sign of disease. Altogether, these results underline the necessity of CD46 expression for the persistence of HHV-6A DNA in the brain of infected mice irrespectively of the expressed CD46 isoform and IFN type I signaling. Furthermore, to test whether an infectious virus is required for the persistence of viral DNA in the brain of CD46 transgenic mice, we performed an additional control experiment using inactivated virus (Fig. 4C). As UV irradiation was found to prevent DNA quantification by the real-time PCR approach, heat inactivation (80°C, 15 min), which preserves integrity of viral DNA (data not shown), was used. DNA from infectious virus, but not from heat-inactivated virus, was observed in CD46 transgenic mice 3 weeks after i.c. injection (Fig. 4C), confirming that viral infection leads to DNA persistence in the murine brain.
We then analyzed the production of HHV-6-specific antibodies in the serum of infected mice using a specific enzyme-linked immunosorbent assay (ELISA). Blood was taken at different time periods p.i., and antibody titers were monitored up to 3 months after infection. As shown in Fig. 4D, virus-specific antibodies were regularly detected in all infected mice. The levels of HHV-6-specific IgG were generally higher in CD46 transgenic than in wild-type mice, with a peak of production observed at 8 weeks p.i. These results demonstrate the development of an HHV-6-specific humoral immune response in infected animals and prompted us to further analyze a potential involvement of the cellular component of the immune response and the presence of infiltrating lymphocytes in the brain of infected mice.
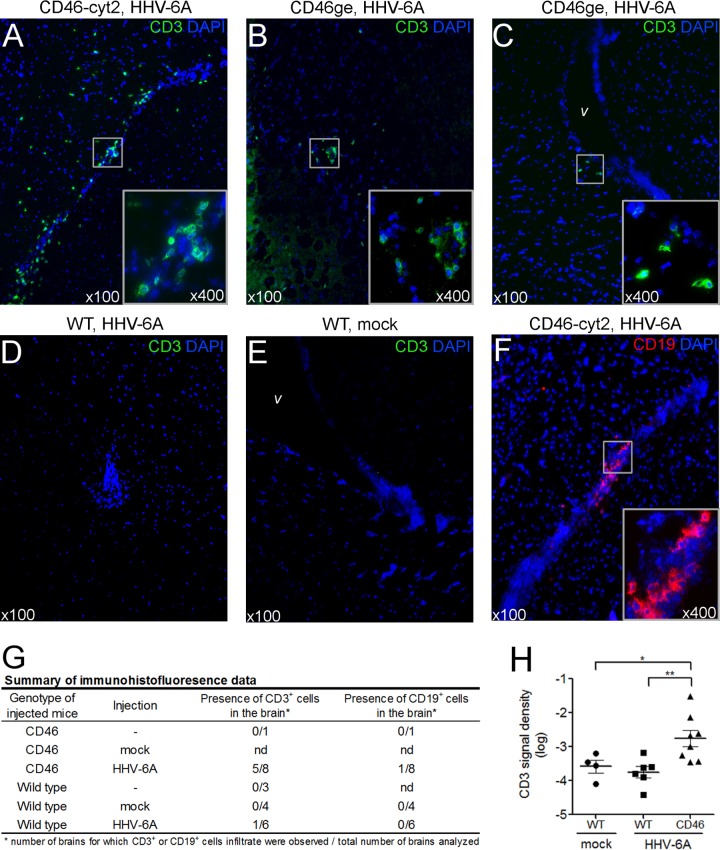
To study whether HHV-6A could induce any neuropathology, we performed immunohistofluorescence analyses on brain sections. CD46 transgenic and wild-type mice were injected i.c. with 105 TCID50 of purified HHV-6A. Three weeks after infection, mice were perfused and brains were analyzed for the presence of CD3+ T lymphocytes, CD19+ B lymphocytes (Fig. 5), and F4/80+ macrophages (data not shown). CD3+ infiltrates were regularly found, mostly in periventricular areas, in infected CD46 transgenic mice, as shown in Fig. 5A to C, but they were absent or detected at a very low frequency in infected wild-type and noninfected mice (Fig. 5D to G). Additionally, CD3 signal was quantified using ImageJ software on images from the right lateral ventricle region and was found to be significantly increased in the HHV-6A-infected CD46 transgenic group compared to the wild-type and noninfected control groups (Fig. 5H). Similar results were obtained with images from the left lateral ventricle area (not shown), indicating that infiltration is not restricted to the side of injection. CD19+ infiltrate was found only occasionally in CD46 transgenic infected mice (Fig. 5F). Although F4/80+ cells were more frequent in the brain of infected CD46 transgenic mice, F4/80 staining was observed in both infected and noninfected brains (data not shown), probably due to the presence of activated resident microglial cells which also express F4/80. These results suggested that HHV-6A infection results in lymphocyte infiltration in the brain and encouraged us to further investigate the mechanisms which may be involved in that recruitment.
FIG 5.
Lymphocyte infiltration in the brain of HHV-6A-infected mice. CD46-cyt2 mice (A and F), CD46ge mice (B and C), and wild-type littermates (D and E) received a single i.c. injection of purified HHV-6A or mock infection solution (E) in the right brain hemisphere. Three weeks after injection, mice were perfused with PBS and brains were collected and frozen. Coronal brain sections were fixed and analyzed by immunofluorescence using CD3 (green) (A to E) or CD19-specific (red) (F) antibodies. Cell nuclei were stained with DAPI (blue). Sections were observed at a magnification of ×100, and infiltrates are presented in inserts at higher magnification (×400). Images of the left (A and C to F) or right (B) lateral ventricle (v) areas are presented. (G) Summary of immunohistofluorescence observations. Brain slides from wild-type or CD46-cyt1, CD46-cyt2, and CD46ge mice (grouped as CD46), injected with HHV-6A, mock infection solution, or left uninjected, were analyzed for the presence of CD3+ and CD19+ cells. Data are presented as the number of brains for which CD3+ or CD19+ infiltrating cells was observed out of the total number of brains analyzed. nd, not done. (H) Two sections from each brain presented in panel G were used for quantification of CD3 fluorescent signal density. Images from the right lateral ventricle region were analyzed, and CD3 signal density (fluorescent area out of the total area analyzed) was determined using Image J software. Each point corresponds to the average value of 2 sections analyzed from one brain. Means ± standard errors for each group are presented, and statistical analysis was performed using Mann-Whitney test.
Modulation of chemokine secretion in murine primary brain glial cells.
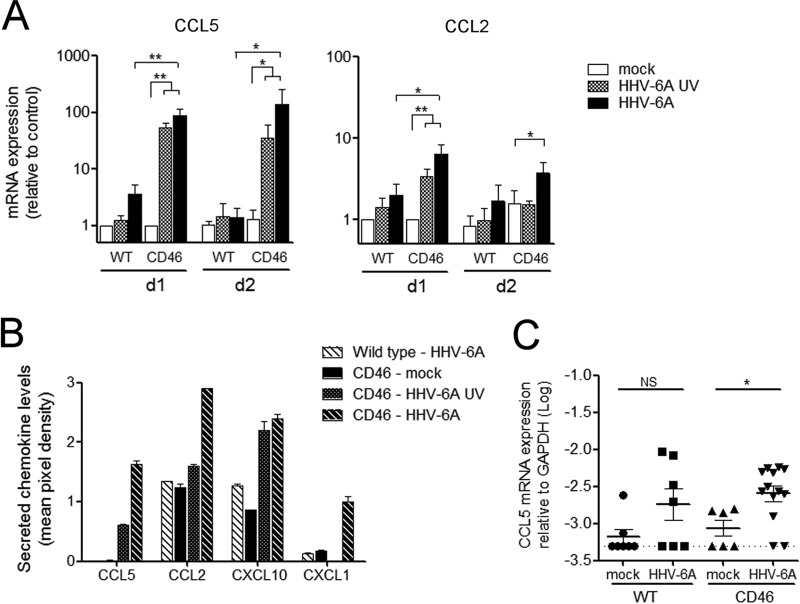
As chemokines play a critical role in the recruitment of inflammatory cells, we next analyzed whether HHV-6A infection could modulate the production of different chemokines in primary murine brain glial cell cultures. Cells were infected with HHV-6A and analyzed initially for the production of two chemokines, CCL2 and CCL5, known for their chemoattractant properties on monocytes/macrophages and lymphocytes and their involvement in neuroinflammatory processes. HHV-6A infection of primary brain cultures resulted in an important increase of both CCL2 and CCL5 mRNA production at 24 h p.i. (Fig. 6A). Interestingly, UV-irradiated virus increased the chemokine production as well, although to a lesser extent than infectious virus. In addition, HHV-6A did not require IFN type I signaling to induce CCL5 production, as demonstrated in primary cultures derived from IFNARKO mice (data not shown). To obtain a more global view of chemokines affected by HHV-6A infection in brain cells, we performed an analysis of the production of 40 different cytokines using a Proteome Profiler array (Fig. 6B). This approach allowed us to confirm the secretion of CCL2 and CCL5 at the protein level in the supernatant of primary brain cultures 48 h p.i. and also revealed the production of other chemokines, including CXCL10 and CXCL1. Finally, we analyzed the production of CCL5 mRNA in HHV-6A-infected murine brains (Fig. 6C). Three weeks after i.c. injection, the level of CCL5 transcripts was significantly increased in the brains of CD46 transgenic mice compared to that of noninfected controls. Some increase in CCL5 expression was observed in infected wild-type mice compared to control mice, although the difference was not significant. Contrary to what was observed in vitro, no statistical difference was observed between infected wild-type and CD46 transgenic mice, potentially due to the kinetic of infection. CCL5 was analyzed at 3 weeks p.i. to avoid any bias related to i.c. injection. However, this could be too late to observe differences between wild-type and CD46 mice in vivo, as the highest increase of CCL5 expression was seen at early time points in vitro. Altogether, these results suggest that HHV-6A infection induces the production of a panel of chemokines and that the mechanisms of this induction involve both infectious and noninfectious components present in UV-inactivated HHV-6 particles.
FIG 6.
HHV-6A increases chemokine production in primary brain glial cells from CD46 transgenic mice. (A and B) Primary cultures generated from CD46 transgenic mice (CD46-cyt1 and CD46-cyt2) or wild-type mice were inoculated with either HHV-6A, UV-irradiated virus (HHV-6A UV), or mock solution. (A) At 24 and 48 h postinfection, total RNA was extracted and analyzed for the expression of CCL5 and CCL2 by RT-qPCR. CCL5 and CCL2 mRNA levels were normalized using GAPDH, and ratios relative to basal levels of mock-infected controls at 24 h postinfection are presented as means and standard errors of four (WT) or six (CD46) independent experiments. Statistical analyses were performed using the Mann-Whitney test. (B) Culture supernatants from one representative experiment were collected at 48 h postinfection, and the secretion of 40 cytokines was analyzed in duplicate using a Proteome Profiler antibody array. Results for chemokines for which consistent variations were observed are presented as mean pixel density and standard deviations from duplicates. (C) CD46 transgenic mice (CD46-cyt1 and CD46-cyt2) and wild-type mice received a single i.c. injection of purified HHV-6A or mock solution. CCL5 mRNA expression in the brain was analyzed 3 weeks after injection. Statistical analyses were performed using Student's t test (*, P ≤ 0.05; **, P ≤ 0.01).
HHV-6A-induced chemokine production requires TLR9 signaling.
HHV-6-induced chemokine upregulation was not dependent on the CD46 isoform expressed, as no difference was observed in CCL5 or CCL2 synthesis in primary brain cultures from CD46-cyt1, CD46-cyt2, or CD46ge mice (data not shown). Moreover, engagement of CD46 using anti-CD46 antibody did not modulate CCL5 expression (data not shown), indicating that HHV-6A-induced chemokine activation does not require signaling via CD46.
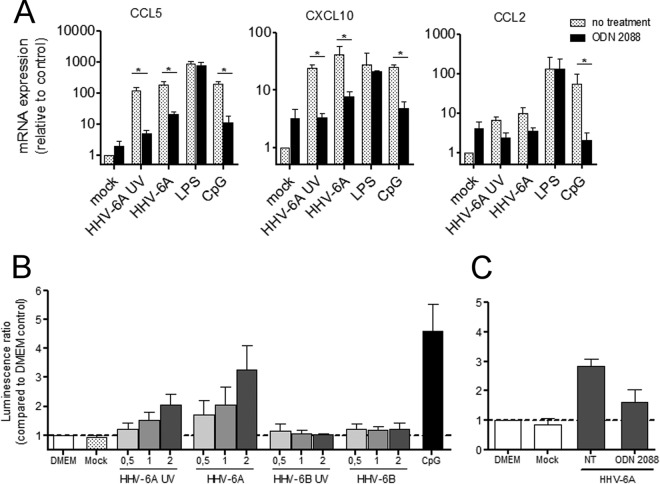
HHV-6B was previously shown to impair TLR signaling in human dendritic cells (DCs) (59) and to induce the production of IFN-λ1 through toll-like receptor 9 (TLR9) in plasmacytoid DCs (pDCs) (60), while HHV-6A could upregulate TLR9 expression in CD4+ T lymphocytes (61). As murine astrocytes and microglia express TLR9 (62–64), we next analyzed whether HHV-6A-induced secretion of proinflammatory chemokines requires TLR9. Primary murine brain cultures were treated with the TLR9 antagonist ligand ODN 2088 and infected with HHV-6A or UV-inactivated HHV-6A (Fig. 7A). Treatment with TLR9 antagonist significantly decreased HHV-6A-induced as well as CpG (natural TLR9 ligand)-induced production of CCL5 and CXCL10. A decrease in CCL2 production was also observed, although it was not as prominent as that with the other two chemokines. Furthermore, the ODN 2088 pretreatment did not affect lipopolysaccharide (LPS)-induced chemokine increase, which requires the engagement of TLR4, demonstrating its specificity of action in this experimental system.
FIG 7.
Role of TLR9 signaling in the induction of chemokine secretion by HHV-6A. (A) Primary cultures of CD46-transgenic mice were inoculated with either UV-inactivated or infectious HHV-6A or mock solution. Control stimulations with LPS and CpG were performed in parallel. During stimulation, cells were treated with the TLR9 antagonist ligand ODN 2088 (black bars) or left untreated (gray bars). CCL5, CXCL10, and CCL2 mRNA expression was analyzed 24 h after stimulation. Means and standard deviations from 4 independent experiments are presented. (B) HEK cells stably expressing human TLR9 and the luciferase reporter construct were stimulated with UV-inactivated or infectious HHV-6A and HHV-6B at an MOI of 0.5, 1, or 2 in triplicate. Nonstimulated (DMEM), mock-stimulated, and CpG-stimulated controls were analyzed in parallel. Means and standard errors of the means from three independent experiments are presented. (C) hTLR9-HEK reporter cells were treated with the TLR9 antagonist ODN 2088 or left untreated and then were stimulated with HHV-6A. Luminescence values are expressed relative to the nonstimulated DMEM control. Means and standard deviations of triplicate values from one representative of three independent experiments are plotted (*, P ≤ 0.05; Mann-Whitney test).
Finally, to verify whether HHV-6A-induced stimulation of TLR9 is restricted to the murine system or is extended to the human TLR9 as well, we analyzed whether HHV-6 could interact with human TLR9, using HEK cells stably expressing both human TLR9 and the luciferase reporter gene, under the control of NF-κB-inducible promoter. While purified HHV-6A induced luciferase expression in a dose-dependent fashion, purified HHV-6B did not have any effect (Fig. 7B). Both infectious and UV-treated HHV-6A were able to interact with TLR9, although the effect of UV-treated HHV-6A was weaker, and ODN 2088 inhibited the stimulatory effect (Fig. 7C). Altogether, these results strongly suggest that HHV-6A but not HHV-6B could interact with TLR9 and induce consecutively proinflammatory chemokine responses.
DISCUSSION
A growing number of studies have linked HHV-6 infection to different neurological diseases, such as multiple sclerosis, chronic fatigue syndrome, and epilepsy (65, 66). However, host factors that contribute to protective and pathological immune responses to HHV-6 remain undefined. Thus, the development of small-animal models is of great interest for the study of HHV-6 immunopathogenesis and the analysis of its involvement in neuropathology. This report presents the first murine model for HHV-6A-induced neuropathogenesis and highlights significant differences between HHV-6A and HHV-6B infection. Our in vitro results with both murine lymphoid cell lines and primary brain cell cultures suggest that CD46 expression on murine cells can mediate HHV-6A entry in these cells, in agreement with previous studies (14, 67). Although we did not observe production of viral particles, de novo synthesis of several viral transcripts was detected, including the U79 gene, which is considered to be involved in viral DNA replication (68). Thus, this finding suggests the start of a replication cycle in murine cells after HHV-6A exposure. The expression of the U94 gene, described as a latency-associated gene (69, 70), was observed concomitantly with other early and late genes, which is reminiscent of productive viral infection. However, the absence of consecutive DNA replication in the analyzed cell types suggests abortive or nonproductive infection, possibly due to either the presence of mouse-specific restriction factors or the absence of some human intracellular factors needed for a productive infection. Nevertheless, HHV-6 infection in human cells is not always productive, and an absence of cytopathic effects was reported when primary human astrocytes were infected with cell-free virus (40). Therefore, the nonproductive infection observed in these studies may be related to intrinsic characteristics of the infected cell type or potential necessity for prior virus adaptation to murine cells. Finally, when primary murine brain cultures were overlaid with HHV-6A-infected human lymphocytes continuously releasing large quantities of virus, formation of syncytia and production of viral proteins was observed in the murine astrocytes, similar to what has been observed with human astrocytes in an analogous infection environment (40), suggesting permissiveness of murine neural cells to HHV-6A infection when adequate conditions are provided.
In vivo, we observed long-term persistence of HHV-6A DNA in CD46 transgenic mice, which is in contrast to wild-type mice. This strongly suggests that HHV-6A is able to infect murine CNS in a CD46-dependent manner and to establish a persistent infection without being cleared by the immune system. Absence of detection of viral RNA or protein expression does not contradict it, as the infection in the murine brain may be restricted to some areas or cell types; thus, low levels of RNA could be below the level of detection in the whole-brain extracts that we analyzed here. Moreover, only a few genes were analyzed among the numerous HHV-6 genes which could be expressed. Therefore, we cannot exclude that some other nontested RNA transcripts, potentially different from those known to be associated with latency in human cells, are expressed during HHV-6A infection in the murine brain and participate in the persistence of HHV-6A. Viral RNA and proteins are very rarely detected in the brain of healthy people harboring HHV-6A DNA without HHV-6-related pathology (7, 8, 12, 71). In humans, HHV-6 infection of tissue in vivo seems to be much less efficient than that in vitro. In the Rag2−/−γc−/− murine model, in which mice were humanized with cord blood-derived human hematopoietic stem cells, only HHV-6A DNA and hardly detectable green fluorescent protein (GFP) expression were observed in the blood (50). Likewise, asymptomatic HHV-6 infection in our model goes along with the absence of virally encoded transcripts and viral antigens. Other models for HHV-6A infection developed in monkeys exhibited very few clinical signs (45). The recent study by Leibovitch et al. described neurological signs, including motor weakness and sensory deficits (46). Although we did not observe any clear limb weakness in mice, some mild neurological symptoms might have been easily missed in this murine model.
IFN signaling was shown to be important in numerous neurotropic viral infections, including measles (72), vesicular stomatitis virus (52), and henipavirus (53). Moreover, HHV-6A was shown to be sensitive to IFN-I, contrary to HHV-6B, as it is unable to block IFN-I signaling in infected cells (73). Surprisingly, the susceptibility to HHV-6 infection was not increased in CD46 transgenic mice deficient in IFN-I receptor, suggesting that IFN-I signaling does not play a critical role in the control of HHV-6A infection in mice. It may be possible that an adaptive immune system efficiently controls HHV-6 infection, along with the regular production of HHV-6-specific antibodies we have observed in mice. As the absence of virus-specific humoral response was related to increased peripheral viral spreading in the marmoset model (46) and HHV-6 reactivation is most often observed in immunosuppressed people (9), we postulate that mice deficient in components of the adaptive immune system are more susceptible to HHV-6A infection.
The expression of proinflammatory chemokines is critical in the generation of neuroinflammation, as they are key elements for leukocyte recruitment and their passage through the blood-brain barrier. CCL5 (RANTES) was shown to be induced by HHV-6A in human astrocytes, endothelial cells, and tonsilar cells (40–42). Our results in the mouse model are consistent with these data, as HHV-6A also strongly upregulated CCL5 in primary brain cultures as well as other proinflammatory chemokines, including CCL2, CXCL10, and CXCL1. This suggests that the presence of HHV-6A in the CNS, even in the absence of a productive infection, triggers the recruitment of several types of immune cells expressing these receptors, such as neutrophils, monocytes/macrophages, and lymphocytes. Indeed, we observed the presence of infiltrating T and B lymphocytes in several CD46 transgenic mice, which further confirms the ability of HHV-6A to induce leukocyte trafficking to the brain. Recently, Hashimoto's thyroiditis has been shown to be associated with HHV-6A infection (74), and it is possible that the abundant lymphocyte infiltrates observed in this disease can be induced by HHV-6A-stimulated chemokine production, leading to further triggering of the autoimmune response.
Chemokine upregulation shown in this study could be achieved with both infectious and inactivated HHV-6A, and our results revealed a critical role for TLR9 in the induction of chemokines. Murine as well as human TLR9 was indeed shown to be expressed in microglial cells and astrocytes (62–64, 75, 76). Both of these cell types may be responsible for the increase in chemokine secretion in mixed glial cultures. In the murine system, chemokine activation requires both CD46 expression on the surface and signaling through TLR9, which suggests that CD46 mediates the capture and internalization of HHV-6A viral particles, allowing access of viral DNA to the endosomal compartment containing TLR9. Indeed, CD46 was shown to efficiently internalize opsonized bacteria (77) and to transport measles virus into the endosome/lysosome compartment for efficient major histocompatibility complex class II-restricted presentation in both human (78) and murine cells (79). Furthermore, our results demonstrate that in addition to the murine TLR9, HHV-6A could stimulate human TLR9, underlining the significance of the murine model for studying this aspect of HHV-6A pathogenesis. TLR9 was also shown to be an essential modulator of an autoimmune process in an animal model of multiple sclerosis (80). These results suggest the potential of HHV-6A to induce neuroinflammation via TLR9 and may have important clinical importance, as TLR9 inhibitors could be used to decrease the production of proinflammatory chemokines and restrain the CNS pathogenesis.
Although CD46 cytoplasmic tails cyt1 and cyt2 have significant signaling functions (81–83) and were shown to exhibit opposite roles in the control inflammatory response (18), we did not find any difference in the susceptibility of CD46 cyt1 and cyt2 transgenic mice to HHV-6 infection or in chemokine production from these transgenic lines. These results confirm a predominant role for the CD46 ectodomain in the infection and are in agreement with the available data (67, 84), excluding the importance of the differential CD46 signaling in HHV-6A persistence in the brain. However, in contrast to HHV-6A infection, CD46 expression did not seem to play any role in the infection by HHV-6B, either in vitro or in vivo, suggesting that CD46 alone could not allow HHV-6B infection in murine cells. Furthermore, HHV-6B did not trigger TRL9 signaling in this system, whereas it was shown to induce IFN-λ1 via binding to TLR9 in human pDCs (60). We could postulate that, like in the murine cells, HHV-6B is not able to enter human HEK cells despite CD46 expression and could not reach the intracellular compartment where TLR9 is located. This suggests the presence of another receptor for HHV-6B on human pDCs not expressed on HEK cells. Together with the results obtained with the murine cells, these data cast serious doubts on the role of CD46 in HHV-6B infection and support previous studies suggesting differential roles for CD46 in HHV-6A and HHV-6B entry (67, 85) and a probable role of the other recently discovered human cell membrane protein, CD134, as a specific HHV-6B entry receptor (28).
Altogether, our results suggest that HHV-6A infection in CD46 transgenic mice can mimic some aspects of human brain infection and help in better understanding virus-induced neuropathogenesis. Our data support the existence of a relationship between HHV-6A residence in the brain and neuroinflammatory processes and are in agreement with recent results obtained with the marmoset model showing a link between HHV-6A infection and neuropathology (46). Furthermore, this study provides evidence for the long-term persistence of HHV-6A DNA in the brain of infected mice. As HHV-6A was showed to reside in the human brain as well (4–7), the CD46 transgenic murine model also could provide a useful tool to study the mechanisms involved in HHV-6A persistence in the CNS. The availability of a mouse model is of high interest in this field, as it offers a number of genetic tools for the analysis of the role of different host components in HHV-6 infection and may open new perspectives in the study of antiviral immune responses in the brain and of the host factors potentially involved in virus-induced neuroinflammation.
ACKNOWLEDGMENTS
This work was supported by grants from INSERM and ARSEP. J.R. was supported by a doctoral fellowship from the French Ministry of Research. J.F.J. was the recipient of a fellowship from ARSEP and Finovi.
We thank T. Seya for CD46ge transgenic mice; L. Naesens, H. Agut, P. Giraudon, A. Epstein, D. Gerlier, and the HHV-6 Foundation for their helpful suggestions or insights and/or providing the reagents for this study; C. Genain for stimulating discussions during the initial stages of the project; N. Goutagny and K. Fitzgerald for HEK-TLR9 cell;, and I. Grosjean from the CelluloNet facility, SFR Biosciences Gerland-Lyon Sud (UMS344/US8), and A. Ruiz, M. Chalons, B. Piasecka, C. Sellin, and members of the CIRI group Immunobiology of Viral Infections for their help in the achievement of this study.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, Diluca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Höllsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 6 November 2013. Classification of HHV-6A and HHV-6B as distinct viruses. Arch. Virol. 10.1007/s00705-013-1902-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun DK, Dominguez G, Pellett PE. 1997. Human herpesvirus 6. Clin. Microbiol. Rev. 10:521–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067 [DOI] [PubMed] [Google Scholar]
- 4.Luppi M, Barozzi P, Maiorana A, Marasca R, Torelli G. 1994. Human herpesvirus 6 infection in normal human brain tissue. J. Infect. Dis. 169:943–944. 10.1093/infdis/169.4.943 [DOI] [PubMed] [Google Scholar]
- 5.Chan PK, Ng HK, Hui M, Ip M, Cheung JL, Cheng AF. 1999. Presence of human herpesviruses 6, 7, and 8 DNA sequences in normal brain tissue. J. Med. Virol. 59:491–495. [DOI] [PubMed] [Google Scholar]
- 6.Chan PK, Ng HK, Hui M, Cheng AF. 2001. Prevalence and distribution of human herpesvirus 6 variants A and B in adult human brain. J. Med. Virol. 64:42–46. 10.1002/jmv.1015 [DOI] [PubMed] [Google Scholar]
- 7.Cuomo L, Trivedi P, Cardillo MR, Gagliardi FM, Vecchione A, Caruso R, Calogero A, Frati L, Faggioni A, Ragona G. 2001. Human herpesvirus 6 infection in neoplastic and normal brain tissue. J. Med. Virol. 63:45–51. [DOI] [PubMed] [Google Scholar]
- 8.Opsahl ML, Kennedy PGE. 2005. Early and late HHV-6 gene transcripts in multiple sclerosis lesions and normal appearing white matter. Brain 128:516–527. 10.1093/brain/awh390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerr DM. 2006. Human herpesvirus 6 and central nervous system disease in hematopoietic cell transplantation. J. Clin. Virol. 37(Suppl 1):S52–S56. 10.1016/S1386-6532(06)70012-9 [DOI] [PubMed] [Google Scholar]
- 10.Soldan SS, Berti R, Salem N, Secchiero P, Flamand L, Calabresi PA, Brennan MB, Maloni HW, McFarland HF, Lin HC, Patnaik M, Jacobson S. 1997. Association of human herpes virus 6 (HHV-6) with multiple sclerosis: increased IgM response to HHV-6 early antigen and detection of serum HHV-6 DNA. Nat. Med. 3:1394–1397. 10.1038/nm1297-1394 [DOI] [PubMed] [Google Scholar]
- 11.Chapenko S, Millers A, Nora Z, Logina I, Kukaine R, Murovska M. 2003. Correlation between HHV-6 reactivation and multiple sclerosis disease activity. J. Med. Virol. 69:111–117. 10.1002/jmv.10258 [DOI] [PubMed] [Google Scholar]
- 12.Challoner PB, Smith KT, Parker JD, MacLeod DL, Coulter SN, Rose TM, Schultz ER, Bennett JL, Garber RL, Chang M. 1995. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 92:7440–7444. 10.1073/pnas.92.16.7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman AD, Mock DJ, Powers JM, Baker JV, Blumberg BM. 2003. Human herpesvirus 6 genome and antigen in acute multiple sclerosis lesions. J. Infect. Dis. 187:1365–1376. 10.1086/368172 [DOI] [PubMed] [Google Scholar]
- 14.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827. 10.1016/S0092-8674(00)81678-5 [DOI] [PubMed] [Google Scholar]
- 15.Källström H, Liszewski MK, Atkinson JP, Jonsson AB. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639–647. 10.1046/j.1365-2958.1997.4841857.x [DOI] [PubMed] [Google Scholar]
- 16.Dörig RE, Marcil A, Chopra A, Richardson CD. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295–305. 10.1016/0092-8674(93)80071-L [DOI] [PubMed] [Google Scholar]
- 17.Naniche D, Varior-Krishnan G, Cervoni F, Wild TF, Rossi B, Rabourdin-Combe C, Gerlier D. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marie JC, Astier AL, Rivailler P, Rabourdin-Combe C, Wild TF, Horvat B. 2002. Linking innate and acquired immunity: divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nat. Immunol. 3:659–666 [DOI] [PubMed] [Google Scholar]
- 19.Johnstone RW, Loveland BE. 1993. Identification and quantification of complement regulator CD46 on normal human tissues. Immunology 79:341–347 [PMC free article] [PubMed] [Google Scholar]
- 20.Gardell JL, Dazin P, Islar J, Menge T, Genain CP, Lalive PH. 2006. Apoptotic effects of human herpesvirus-6A on glia and neurons as potential triggers for central nervous system autoimmunity. J. Clin. Virol. 37(Suppl 1):S11–S16. 10.1016/S1386-6532(06)70005-1 [DOI] [PubMed] [Google Scholar]
- 21.Ahlqvist J, Fotheringham J, Akhyani N, Yao K, Fogdell-Hahn A, Jacobson S. 2005. Differential tropism of human herpesvirus 6 (HHV-6) variants and induction of latency by HHV-6A in oligodendrocytes. J. Neurovirol. 11:384–394. 10.1080/13550280591002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albright AV, Lavi E, Black JB, Goldberg S, O'Connor MJ, González-Scarano F. 1998. The effect of human herpesvirus-6 (HHV-6) on cultured human neural cells: oligodendrocytes and microglia. J. Neurovirol. 4:486–494. 10.3109/13550289809113493 [DOI] [PubMed] [Google Scholar]
- 23.Dietrich J, Blumberg BM, Roshal M, Baker JV, Hurley SD, Mayer-Pröschel M, Mock DJ. 2004. Infection with an endemic human herpesvirus disrupts critical glial precursor cell properties. J. Neurosci. 24:4875–4883. 10.1523/JNEUROSCI.5584-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donati D, Martinelli E, Cassiani-Ingoni R, Ahlqvist J, Hou J, Major EO, Jacobson S. 2005. Variant-specific tropism of human herpesvirus 6 in human astrocytes. J. Virol. 79:9439–9448. 10.1128/JVI.79.15.9439-9448.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Filippis L, Foglieni C, Silva S, Vescovi AL, Lusso P, Malnati MS. 2006. Differentiated human neural stem cells: a new ex vivo model to study HHV-6 infection of the central nervous system. J. Clin. Virol. 37(Suppl 1):S27–S32. 10.1016/S1386-6532(06)70008-7 [DOI] [PubMed] [Google Scholar]
- 26.Gu B, Zhang G-F, Li L-Y, Zhou F, Feng D-J, Ding C-L, Chi J, Zhang C, Guo D-D, Wang J-F, Zhou H, Yao K, Hu W-X. 2011. Human herpesvirus 6A induces apoptosis of primary human fetal astrocytes via both caspase-dependent and -independent pathways. Virol. J. 8:530. 10.1186/1743-422X-8-530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, McCarthy M, Zhou Y, Chandran B, Wood C. 1996. Infection of primary human fetal astrocytes by human herpesvirus 6. J. Virol. 70:1296–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, Yamanishi K, Mori Y. 2013. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc. Natl. Acad. Sci. U. S. A. 110:9096–9099. 10.1073/pnas.1305187110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flamand L, Gosselin J, Stefanescu I, Ablashi D, Menezes J. 1995. Immunosuppressive effect of human herpesvirus 6 on T-cell functions: suppression of interleukin-2 synthesis and cell proliferation. Blood 85:1263–1271 [PubMed] [Google Scholar]
- 30.Li L, Gu B, Zhou F, Chi J, Wang F, Peng G, Xie F, Qing J, Feng D, Lu S, Yao K. 2011. Human herpesvirus 6 suppresses T cell proliferation through induction of cell cycle arrest in infected cells in the G2/M phase. J. Virol. 85:6774–6783. 10.1128/JVI.02577-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Øster B, Bundgaard B, Höllsberg P. 2005. Human herpesvirus 6B induces cell cycle arrest concomitant with p53 phosphorylation and accumulation in T cells. J. Virol. 79:1961–1965. 10.1128/JVI.79.3.1961-1965.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. 2007. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J. Virol. 81:5737–5748. 10.1128/JVI.02443-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kofod-Olsen E, Ross-Hansen K, Schleimann MH, Jensen DK, Møller JML, Bundgaard B, Mikkelsen JG, Höllsberg P. 2012. U20 is responsible for human herpesvirus 6B inhibition of tumor necrosis factor receptor-dependent signaling and apoptosis. J. Virol. 86:11483–11492. 10.1128/JVI.00847-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne RS, Mattick C, Nicholson L, Devaraj P, Alcami A, Gompels UA. 2000. RANTES binding and down-regulation by a novel human herpesvirus-6 beta chemokine receptor. J. Immunol. 164:2396–2404 http://www.jimmunol.org/content/164/5/2396 [DOI] [PubMed] [Google Scholar]
- 35.Mayne M, Cheadle C, Soldan SS, Cermelli C, Yamano Y, Akhyani N, Nagel JE, Taub DD, Becker KG, Jacobson S. 2001. Gene expression profile of herpesvirus-infected T cells obtained using immunomicroarrays: induction of proinflammatory mechanisms. J. Virol. 75:11641–11650. 10.1128/JVI.75.23.11641-11650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikuta H, Nakane A, Lu H, Taguchi Y, Minagawa T, Matsumoto S. 1990. Interferon induction by human herpesvirus 6 in human mononuclear cells. J. Infect. Dis. 162:35–38. 10.1093/infdis/162.1.35 [DOI] [PubMed] [Google Scholar]
- 37.Flamand L, Stefanescu I, Menezes J. 1996. Human herpesvirus-6 enhances natural killer cell cytotoxicity via IL-15. J. Clin. Investig. 97:1373–1381. 10.1172/JCI118557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arena A, Liberto MC, Capozza AB, Focà A. 1997. Productive HHV-6 infection in differentiated U937 cells: role of TNF alpha in regulation of HHV-6. New Microbiol. 20:13–20 [PubMed] [Google Scholar]
- 39.Inagi R, Guntapong R, Nakao M, Ishino Y, Kawanishi K, Isegawa Y, Yamanishi K. 1996. Human herpesvirus 6 induces IL-8 gene expression in human hepatoma cell line, Hep G2. J. Med. Virol. 49:34–40. [DOI] [PubMed] [Google Scholar]
- 40.Meeuwsen S, Persoon-Deen C, Bsibsi M, Bajramovic JJ, Ravid R, De Bolle L, van Noort JM. 2005. Modulation of the cytokine network in human adult astrocytes by human herpesvirus-6A. J. Neuroimmunol. 164:37–47. 10.1016/j.jneuroim.2005.03.013 [DOI] [PubMed] [Google Scholar]
- 41.Caruso A, Rotola A, Comar M, Favilli F, Galvan M, Tosetti M, Campello C, Caselli E, Alessandri G, Grassi M, Garrafa E, Cassai E, Di Luca D. 2002. HHV-6 infects human aortic and heart microvascular endothelial cells, increasing their ability to secrete proinflammatory chemokines. J. Med. Virol. 67:528–533. 10.1002/jmv.10133 [DOI] [PubMed] [Google Scholar]
- 42.Grivel J-C, Santoro F, Chen S, Fagá G, Malnati MS, Ito Y, Margolis L, Lusso P. 2003. Pathogenic effects of human herpesvirus 6 in human lymphoid tissue ex vivo. J. Virol. 77:8280–8289. 10.1128/JVI.77.15.8280-8289.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbuckle JH, Medveczky PG. 2011. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect. 13:731–741. 10.1016/j.micinf.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yalcin S, Mukai T, Kondo K, Ami Y, Okawa T, Kojima A, Kurata T, Yamanishi K. 1992. Experimental infection of cynomolgus and African green monkeys with human herpesvirus 6. J. Gen. Virol. 73(Part 7):1673–1677. 10.1099/0022-1317-73-7-1673 [DOI] [PubMed] [Google Scholar]
- 45.Lusso P, Crowley RW, Malnati MS, Di Serio C, Ponzoni M, Biancotto A, Markham PD, Gallo RC. 2007. Human herpesvirus 6A accelerates AIDS progression in macaques. Proc. Natl. Acad. Sci. U. S. A. 104:5067–5072. 10.1073/pnas.0700929104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leibovitch E, Wohler JE, Cummings Macri SM, Motanic K, Harberts E, Gaitán MI, Maggi P, Ellis M, Westmoreland S, Silva A, Reich DS, Jacobson S. 2013. Novel marmoset (Callithrix jacchus) model of human herpesvirus 6A and 6B infections: immunologic, virologic and radiologic characterization. PLoS Pathog. 9:e1003138. 10.1371/journal.ppat.1003138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lusso P. 1996. Human herpesvirus 6 (HHV-6). Antiviral Res. 31:1–21. 10.1016/0166-3542(96)00949-7 [DOI] [PubMed] [Google Scholar]
- 48.Tsujimura A, Shida K, Kitamura M, Nomura M, Takeda J, Tanaka H, Matsumoto M, Matsumiya K, Okuyama A, Nishimune Y, Okabe M, Seya T. 1998. Molecular cloning of a murine homologue of membrane cofactor protein (CD46): preferential expression in testicular germ cells. Biochem. J. 330(Part 1):163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gobbi A, Stoddart CA, Malnati MS, Locatelli G, Santoro F, Abbey NW, Bare C, Linquist-Stepps V, Moreno MB, Herndier BG, Lusso P, McCune JM. 1999. Human herpesvirus 6 (HHV-6) causes severe thymocyte depletion in SCID-hu Thy/Liv mice. J. Exp. Med. 189:1953–1960. 10.1084/jem.189.12.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanner A, Carlson SA, Nukui M, Murphy EA, Berges BK. 2013. Human herpesvirus 6A infection and immunopathogenesis in humanized Rag2−/− γc−/− mice. J. Virol. 87:12020–12028. 10.1128/JVI.01556-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shingai M, Inoue N, Okuno T, Okabe M, Akazawa T, Miyamoto Y, Ayata M, Honda K, Kurita-Taniguchi M, Matsumoto M, Ogura H, Taniguchi T, Seya T. 2005. Wild-type measles virus infection in human CD46/CD150-transgenic mice: CD11c-positive dendritic cells establish systemic viral infection. J. Immunol. 175:3252–3261 http://www.jimmunol.org/content/175/5/3252 [DOI] [PubMed] [Google Scholar]
- 52.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- 53.Dhondt KP, Mathieu C, Chalons M, Reynaud JM, Vallve A, Raoul H, Horvat B. 2013. Type I interferon signaling protects mice from lethal henipavirus infection. J. Infect. Dis. 207:142–151. 10.1093/infdis/jis653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 56.Katze MG, He Y, Gale M., Jr 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675–687. 10.1038/nri888 [DOI] [PubMed] [Google Scholar]
- 57.Johnstone RW, Russell SM, Loveland BE, McKenzie IF. 1993. Polymorphic expression of CD46 protein isoforms due to tissue-specific RNA splicing. Mol. Immunol. 30:1231–1241. 10.1016/0161-5890(93)90038-D [DOI] [PubMed] [Google Scholar]
- 58.Lopez C. 1975. Genetics of natural resistance to herpesvirus infections in mice. Nature 258:152–153. 10.1038/258152a0 [DOI] [PubMed] [Google Scholar]
- 59.Murakami Y, Tanimoto K, Fujiwara H, An J, Suemori K, Ochi T, Hasegawa H, Yasukawa M. 2010. Human herpesvirus 6 infection impairs Toll-like receptor signaling. Virol. J. 7:91. 10.1186/1743-422X-7-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nordström I, Eriksson K. 2012. HHV-6B induces IFN-Lambda1 responses in cord plasmacytoid dendritic cells through TLR9. PLoS One 7:e38683. 10.1371/journal.pone.0038683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi J, Wang F, Li L, Feng D, Qin J, Xie F, Zhou F, Chen Y, Wang J, Yao K. 2012. The role of MAPK in CD4(+) T cells toll-like receptor 9-mediated signaling following HHV-6 infection. Virology 422:92–98. 10.1016/j.virol.2011.09.026 [DOI] [PubMed] [Google Scholar]
- 62.Bowman CC, Rasley A, Tranguch SL, Marriott I. 2003. Cultured astrocytes express toll-like receptors for bacterial products. Glia 43:281–291. 10.1002/glia.10256 [DOI] [PubMed] [Google Scholar]
- 63.McKimmie CS, Fazakerley JK. 2005. In response to pathogens, glial cells dynamically and differentially regulate Toll-like receptor gene expression. J. Neuroimmunol. 169:116–125. 10.1016/j.jneuroim.2005.08.006 [DOI] [PubMed] [Google Scholar]
- 64.El-Hage N, Podhaizer EM, Sturgill J, Hauser KF. 2011. Toll-like receptor expression and activation in astroglia: differential regulation by HIV-1 Tat, gp120, and morphine. Immunol. Investig. 40:498–522. 10.3109/08820139.2011.561904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao K, Crawford JR, Komaroff AL, Ablashi DV, Jacobson S. 2010. Review part 2: human herpesvirus-6 in central nervous system diseases. J. Med. Virol. 82:1669–1678. 10.1002/jmv.21861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynaud JM, Horvat B. 2013. Human herpesvirus 6 and neuroinflammation. ISRN Virol. 2013:11. 10.3389/fmicb.2013.00174 [DOI] [Google Scholar]
- 67.Mori Y, Seya T, Huang HL, Akkapaiboon P, Dhepakson P, Yamanishi K. 2002. Human herpesvirus 6 variant A but not variant B induces fusion from without in a variety of human cells through a human herpesvirus 6 entry receptor, CD46. J. Virol. 76:6750–6761. 10.1128/JVI.76.13.6750-6761.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniguchi T, Shimamoto T, Isegawa Y, Kondo K, Yamanishi K. 2000. Structure of transcripts and proteins encoded by U79-80 of human herpesvirus 6 and its subcellular localization in infected cells. Virology 271:307–320. 10.1006/viro.2000.0326 [DOI] [PubMed] [Google Scholar]
- 69.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. 1998. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. U. S. A. 95:13911–13916. 10.1073/pnas.95.23.13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caselli E, Bracci A, Galvan M, Boni M, Rotola A, Bergamini C, Cermelli C, Dal Monte P, Gompels UA, Cassai E, Di Luca D. 2006. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology 346:402–414. 10.1016/j.virol.2005.11.018 [DOI] [PubMed] [Google Scholar]
- 71.Chi J, Gu B, Zhang C, Peng G, Zhou F, Chen Y, Zhang G, Guo Y, Guo D, Qin J, Wang J, Li L, Wang F, Liu G, Xie F, Feng D, Zhou H, Huang X, Lu S, Liu Y, Hu W, Yao K. 2012. Human herpesvirus 6 latent infection in patients with glioma. J. Infect. Dis. 206:1394–1398. 10.1093/infdis/jis513 [DOI] [PubMed] [Google Scholar]
- 72.Mrkic B, Pavlovic J, Rülicke T, Volpe P, Buchholz CJ, Hourcade D, Atkinson JP, Aguzzi A, Cattaneo R. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaworska J, Gravel A, Flamand L. 2010. Divergent susceptibilities of human herpesvirus 6 variants to type I interferons. Proc. Natl. Acad. Sci. U. S. A. 107:8369–8374. 10.1073/pnas.0909951107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caselli E, Zatelli MC, Rizzo R, Benedetti S, Martorelli D, Trasforini G, Cassai E, Degli Uberti EC, Di Luca D, Dolcetti R. 2012. Virologic and immunologic evidence supporting an association between HHV-6 and Hashimoto's thyroiditis. PLoS Pathog. 8:e1002951. 10.1371/journal.ppat.1002951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, Shapiro A, Antel JP. 2005. TLR signaling tailors innate immune responses in human microglia and astrocytes. J. Immunol. 175:4320–4330 [DOI] [PubMed] [Google Scholar]
- 76.Carpentier PA, Begolka WS, Olson JK, Elhofy A, Karpus WJ, Miller SD. 2005. Differential activation of astrocytes by innate and adaptive immune stimuli. Glia 49:360–374. 10.1002/glia.20117 [DOI] [PubMed] [Google Scholar]
- 77.Li K, Feito MJ, Sacks SH, Sheerin NS. 2006. CD46 (membrane cofactor protein) acts as a human epithelial cell receptor for internalization of opsonized uropathogenic Escherichia coli. J. Immunol. 177:2543–2551 [DOI] [PubMed] [Google Scholar]
- 78.Gerlier D, Trescol-Biémont MC, Varior-Krishnan G, Naniche D, Fugier-Vivier I, Rabourdin-Combe C. 1994. Efficient MHC class II-restricted presentation of measles virus to T cells relies on its targeting to its cellular receptor human CD46 and involves an endosomal pathway. Cell Biol. Int. 18:315–320. 10.1006/cbir.1994.1080 [DOI] [PubMed] [Google Scholar]
- 79.Rivailler P, Trescol-Biémont MC, Gimenez C, Rabourdin-Combe C, Horvat B. 1998. Enhanced MHC class II-restricted presentation of measles virus (MV) hemagglutinin in transgenic mice expressing human MV receptor CD46. Eur. J. Immunol. 28:1301–1314. [DOI] [PubMed] [Google Scholar]
- 80.Prinz M, Garbe F, Schmidt H, Mildner A, Gutcher I, Wolter K, Piesche M, Schroers R, Weiss E, Kirschning CJ, Rochford CDP, Brück W, Becher B. 2006. Innate immunity mediated by TLR9 modulates pathogenicity in an animal model of multiple sclerosis. J. Clin. Investig. 116:456–464. 10.1172/JCI26078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang G, Liszewski MK, Chan AC, Atkinson JP. 2000. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J. Immunol. 164:1839–1846 http://www.jimmunol.org/content/164/4/1839 [DOI] [PubMed] [Google Scholar]
- 82.Zaffran Y, Destaing O, Roux A, Ory S, Nheu T, Jurdic P, Rabourdin-Combe C, Astier AL. 2001. CD46/CD3 costimulation induces morphological changes of human T cells and activation of Vav, Rac, and extracellular signal-regulated kinase mitogen-activated protein kinase. J. Immunol. 167:6780–6785 http://www.jimmunol.org/content/167/12/6780 [DOI] [PubMed] [Google Scholar]
- 83.Ludford-Menting MJ, Thomas SJ, Crimeen B, Harris LJ, Loveland BE, Bills M, Ellis S, Russell SM. 2002. A functional interaction between CD46 and DLG4: a role for DLG4 in epithelial polarization. J. Biol. Chem. 277:4477–4484. 10.1074/jbc.M108479200 [DOI] [PubMed] [Google Scholar]
- 84.Greenstone HL, Santoro F, Lusso P, Berger EA. 2002. Human herpesvirus 6 and measles virus employ distinct cd46 domains for receptor function. J. Biol. Chem. 277:39112–39118. 10.1074/jbc.M206488200 [DOI] [PubMed] [Google Scholar]
- 85.Mori Y, Akkapaiboon P, Yonemoto S, Koike M, Takemoto M, Sadaoka T, Sasamoto Y, Konishi S, Uchiyama Y, Yamanishi K. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J. Virol. 78:4609–4616. 10.1128/JVI.78.9.4609-4616.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]