ABSTRACT
Emerging and zoonotic pathogens pose continuing threats to human health and ongoing challenges to diagnostics. As nucleic acid tests are playing increasingly prominent roles in diagnostics, the genetic characterization of molecularly uncharacterized agents is expected to significantly enhance detection and surveillance capabilities. We report the identification of two previously unrecognized members of the family Orthomyxoviridae, which includes the influenza viruses and the tick-transmitted Thogoto and Dhori viruses. We provide morphological, serologic, and genetic evidence that Upolu virus (UPOV) from Australia and Aransas Bay virus (ABV) from North America, both previously considered potential bunyaviruses based on electron microscopy and physicochemical features, are orthomyxoviruses instead. Their genomes show up to 68% nucleotide sequence identity to Thogoto virus (segment 2; ∼74% at the amino acid level) and a more distant relationship to Dhori virus, the two prototype viruses of the recognized species of the genus Thogotovirus. Despite sequence similarity, the coding potentials of UPOV and ABV differed from that of Thogoto virus, instead being like that of Dhori virus. Our findings suggest that the tick-transmitted viruses UPOV and ABV represent geographically distinct viruses in the genus Thogotovirus of the family Orthomyxoviridae that do not fit in the two currently recognized species of this genus.
IMPORTANCE Upolu virus (UPOV) and Aransas Bay virus (ABV) are shown to be orthomyxoviruses instead of bunyaviruses, as previously thought. Genetic characterization and adequate classification of agents are paramount in this molecular age to devise appropriate surveillance and diagnostics. Although more closely related to Thogoto virus by sequence, UPOV and ABV differ in their coding potentials by lacking a proposed pathogenicity factor. In this respect, they are similar to Dhori virus, which, despite the lack of a pathogenicity factor, can cause disease. These findings enable further studies into the evolution and pathogenicity of orthomyxoviruses.
INTRODUCTION
Upolu virus (UPOV) strain C5581, an enveloped spherical virus with a diameter of approximately 100 nm, was isolated in 1966 from adult Ornithodoros capensis ticks that infested a sooty tern (Onychoprion fuscatus [protonym, Sterna fuscata]) colony on Upolu Cay, a small atoll of the Great Barrier Reef, Australia (1). No serologic relationship of UPOV to other viruses was demonstrated until 1975, when three antigenically related isolates of Aransas Bay virus (ABV) were obtained from ticks of the same species complex collected from seabird nests on islands off the southern Texas coast (2). UPOV and ABV were considered to form a distinct antigenic group.
UPOV and ABV do not propagate in mosquitoes but replicate in mammalian cell cultures (African green monkey kidney [Vero], baby hamster kidney [BHK], Madin-Darby canine kidney [MDCK], and human embryonic kidney 293 [HEK 293] cells) (2, 3). An incompatibility of tick-derived arboviruses with mosquito physiology has also been observed for other related tick-associated viruses such as Quaranfil and Johnston Atoll viruses (4, 5). Based on physicochemical and morphological features reported for UPOV, the viruses of the Upolu virus serogroup (UPOV and ABV) were tentatively placed into the family Bunyaviridae as two species not assigned to one of the genera of this family of enveloped, negative-sense, single-stranded RNA viruses with tripartite genomes (3, 6). Here, we report data clearly demonstrating that UPOV and ABV are orthomyxoviruses.
The family Orthomyxoviridae includes the influenza viruses in the genera Influenzavirus A, Influenzavirus B, and Influenzavirus C; Infectious salmon anemia virus (ISAV) in the genus Isavirus; and the tick-transmitted viruses Thogoto virus (THOV) and Dhori virus (DHOV) in the genus Thogotovirus (6). In addition, several not yet formally classified viruses related to known orthomyxoviruses were recently described (7, 8). The genomes of orthomyxoviruses consist of 6 segments (thogotoviruses) to 8 segments (influenza viruses) of negative-sense, single-stranded RNA (9). Replication and transcription take place in the cell nucleus, where the viral polymerase complex, consisting of polymerase basic subunit 1 (PB1), polymerase basic subunit 2 (PB2), and the polymerase acidic subunit (PA), synthesizes negative-strand, viral genomic RNA (vRNA), positive-strand RNA (cRNA) complementary to vRNA, and capped polyadenylated mRNAs that are shorter than vRNA and cRNA (10, 11). A function in cap binding and mRNA synthesis has been assigned to PB2 (12–18), RNA chain elongation has been assigned to PB1 (19–21), and cRNA and vRNA synthesis as well as cap cleavage have been assigned to PA, possibly regulated by phosphorylation (22–26). Whereas the three polymerase subunits are encoded by the three largest genome segments in all orthomyxoviruses, coding assignments for the smaller segments differ between genera. In the tick-transmitted thogotoviruses, the fourth largest segment codes for a surface glycoprotein (GP) with a distant relationship to that of baculoviruses (27, 28), segment 5 encodes the nucleoprotein (NP), and segment 6 encodes the matrix protein (M) and in some species also encodes an elongated accessory M-long (ML) protein that interferes with the host innate immune response (9).
We present data that demonstrate genetic as well as serologic relationships of UPOV and ABV to the thogotoviruses. The morphology of UPOV and ABV is compatible with that of orthomyxoviruses; serologically, they cross-react with THOV, and the complete genome sequences determined for both viruses are more closely related to the sequence of THOV than to that of DHOV, but both viruses have coding repertoires similar to that of DHOV and not that of THOV. Analyses of the increasing sequence diversity of thogotovirus genomes have begun to delineate highly conserved protein domains that may point to novel therapeutic targets of orthomyxoviruses.
MATERIALS AND METHODS
Viruses.
Virus stocks of Upolu virus (UPOV) strain C5581 (1) and Aransas Bay virus (ABV) strain RML65660-8 (2) were obtained from the World Reference Center for Emerging Viruses and Arboviruses collection at the University of Texas Medical Branch, Galveston, TX. Total RNA was extracted with Tri reagent (MRC, Cincinnati, OH) from 250 μl of virus stock, suspended in 35 μl nuclease-free water, and stored at −80°C.
Transmission electron microscopy.
Vero E6 cells infected with UPOV or ABV were fixed for 1 h in a mixture of 2.5% paraformaldehyde and 0.1% glutaraldehyde in 0.05 M cacodylate (pH 7.3), to which 0.03% picric acid and 0.03% CaCl2 were added. Fixed monolayers were washed with 0.1 M cacodylate, cells were scraped, and pelleted cells were postfixed with 1% OsO4 in 0.1 M cacodylate for 1 h. Cells were washed with distilled water and finally stained en block with 2% aqueous uranyl acetate for 20 min at 60°C. Preparations were dehydrated in ethanol, processed through propylene oxide, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on a Leica EM UC7 ultramicrotome (Leica Microsystems, Buffalo Grove, IL), stained with lead citrate, and examined with a Philips 201 transmission electron microscope at 60 kV.
Serologic tests.
Viral antigens used in serologic tests were not inactivated and were prepared by sucrose/acetone extraction of BHK cells, hamster liver, or newborn mouse liver (29) infected with the respective viruses. Mouse hyperimmune ascites fluids served as antibody preparations. Four intraperitoneal injections of antigen (10% homogenates of infected newborn mouse brain or liver in phosphate-buffered saline [PBS]) mixed with Freund's complete adjuvant were given at weekly intervals; thereafter, mice were inoculated with sarcoma cells, and immune ascitic fluid was collected. Complement fixation (CF) tests were performed in a microtiter plate format by incubation at 4°C overnight in the presence of 2 U guinea pig complement (30, 31). On a scale from 0 (complete hemolysis) to 4+ (no hemolysis), CF titers were scored as the highest antibody/antigen titer that gave a 3+/4+ fixation of complement; antibody titers of ≥1:8 were rated positive. Hemagglutination inhibition (HI) tests were also done in microtiter plates (31, 32). Nonspecific hemagglutinin inhibitors were removed by acetone extraction, sera were rehydrated in 0.05 M borate–0.12 M NaCl (pH 9), and naturally occurring agglutinins were adsorbed to male goose erythrocytes (29). HI was assessed with 4 units of antigen extracted (8.5% sucrose [pH 5.75]–acetone) from ABV- or UPOV-infected BHK cells, THOV-infected hamster liver, or DHOV-infected mouse liver and tested against 2-fold serial dilutions of pretreated serum beginning at a dilution of 1:10 and male goose erythrocytes. Animal work was performed under an IACUC-approved protocol at the University of Texas Medical Branch.
Unbiased high-throughput sequencing, reverse transcription-PCR, and rapid amplification of cDNA ends.
Genomic sequences were generated by applying a combination of unbiased high-throughput sequencing (UHTS), subsequent consensus reverse transcription-PCR (RT-PCR), and rapid amplification of cDNA ends (RACE) assays. Aliquots of total RNA extracts (0.5 μg) were treated with DNase I (DNA-free; Ambion, Austin, TX, USA) prior to reverse transcription with Superscript III (Invitrogen, Carlsbad, CA, USA) with random octamer primers linked to an arbitrary, defined 17-mer primer sequence. The cDNA was RNase H treated and randomly amplified by PCR with AmpliTaq (Applied Biosystems, Foster City, CA, USA) and a primer mix including the octamer-linked 17-mer-sequence primer in combination with the defined 17-mer-sequence primer in a 1:9 ratio (33). Amplification products of >70 bp were purified (MinElute; Qiagen, Hilden, Germany) and ligated to linkers for sequencing on a GS-FLX sequencer (454 Life Sciences, Branford, CT, USA) (34). Sequence reads were stripped of primer sequences and highly repetitive elements and then clustered and assembled into contiguous fragments (contigs) for comparison to data in the GenBank database by using the Basic Local Alignment Search Tool (blast) (35) at the nucleotide (blastn) and deduced amino acid (blastx) levels.
Various specific primer sets for the validation of draft genome sequences were designed based on the UHTS data as well as sequences of THOV, DHOV, and another related orthomyxovirus, Batken virus (BKNV) (primer sequences are available upon request). Gaps between contigs were filled, and the completed draft genomes were resequenced by overlapping PCR products. Reactions included routinely 1 μl random hexamer-primed cDNA (Superscript II; Invitrogen), primers at 0.2 mM, and Platinum Taq DNA polymerase (Invitrogen). Products were purified (QIAquick PCR purification kit; Qiagen) and directly dideoxy sequenced on both strands (Genewiz, South Plainfield, NJ, USA). Genomic termini were characterized by using RACE kits (Invitrogen). For 5′-RACE, first-strand cDNA was synthesized from total RNA by using custom gene-specific primer 1 (GSP1) and Superscript III. After purification using SNAP columns, a homopolymeric tail was added with terminal deoxynucleotidyl transferase (TdT; Invitrogen) and dCTP followed by PCR amplification using Platinum Taq DNA polymerase (Invitrogen) and nested primer GSP2 combined with the 5′-RACE deoxyinosine-containing anchor primer. Depending on the choice of GSP1 and GSP2, the 5′ ends of genomic RNA (corresponding to the 3′ end of antigenomic RNA) or the antigenomic RNA were determined. Products were cloned into the pCR-TOPO vector (Invitrogen). Transcriptional termination sites were mapped by 3′-RACE employing the poly(A) tail of the (shorter) mRNA transcripts for cDNA priming with an Invitrogen oligo(dT) adaptor primer. Thereafter, cDNA was amplified by PCR using a primer complementary to the introduced adaptor sequence and a custom sequence-specific primer.
PCRs to assess splicing events were performed with forward primer p1 (5′-GCT AAT CGG GTG GAT GGA TG for UPOV and 5′-GCT GAT CGG GTG GAT GGA C for Jos virus [JOSV; an orthomyxovirus related to THOV]) (7) and two reverse primers, p2 (5′-GGC CGC TTT TTT TTT TTT TTT TTT ATT AAA AT for UPOV and 5′-ATG CGG CCG CTT TTT TTT TTT TTT TTT TAA CAC C for JOSV) and p3 (5′-ccg ccA GAG ATA TCA AGG CA for UPOV and 5′-gcc gcc AGA GAA ATC AAG GCA for JOSV; lowercase indicates nonviral bases added to raise primer binding temperature). Nucleic acid extracts for amplification were generated from crude cell homogenates (cellular RNA) or nuclease-treated (8 ng/μl RNase A [Ambion] for 15 min at room temperature and 0.3 U/μl Benzonase nuclease [Qiagen] and 0.06 U/μl Turbo DNase [Ambion] for 45 min at room temperature, followed by 8 ng/μl RNase A and 0.08 U/μl RNase H [Invitrogen] for 2 h at 37°C) cell culture supernatants (genomic RNA) obtained from virus-infected HEK 293 cells harvested at 72 h postinfection. PCR products were analyzed by agarose gel electrophoresis and visualization by GelGreen staining (Biotium, Hayward, CA, USA).
Sequence analyses.
Sequence assembly and analysis were done by employing programs of the Wisconsin GCG Package (version 10.3; Accelrys Inc., San Diego, CA), MEGA 5 (36), Geneious 5.5 (37), and NewblerAssembler 2.4. Identities of nucleotide and amino acid sequences were calculated with the Needleman-Wunsch algorithm, applying an EBLOSUM62 substitution matrix (gap open/extension penalties of 12/2 for nucleotide and 6/1 for amino acid alignments; EMBOSS [38]) and a Perl script to parse the results for all comparisons. Topology and targeting predictions were obtained by using SignalP, NetNGlyc, and TMHMM (http://www.cbs.dtu.dk/services); Phobius (http://phobius.sbc.su.se); and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) (39, 40). Multiple-sequence alignments were generated with CLUSTAL (41), and programs implemented in MEGA and Geneious software were applied for phylogenetic analyses.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the segments of Upolu virus and of Aransas Bay virus are KC506156 to KC506161 and KC506162 to KC506167, respectively.
RESULTS
Recognition of UPOV and ABV as orthomyxoviruses.
The failure to obtain amplification products from nucleic acids of UPOV or ABV by reverse transcription-PCR (RT-PCR) using a panel of degenerate bunyaviral consensus primers led us to pursue UHTS. Sequence libraries were prepared from total RNA extracted from an ABV stock. Sequencing on the Roche GS-FLX platform yielded 94,835 reads with a mean length of 222 bases (range, 29 to 382 bases) that generated contiguous sequence assemblies (contigs) with homology to THOV in regions corresponding to approximately 30% to 80% of the six THOV genome segments (segment 1, ∼70%; segment 2, ∼60%; segment 3, ∼30%; segment 4, ∼40%; segment 5, ∼70%; segment 6, ∼80%). Continuous coding sequences for UPOV and ABV were subsequently generated through consensus RT-PCR using primers representing the ABV contigs as well as sequences of THOV, DHOV, and the related orthomyxovirus BKNV (42, 43). Rapid amplification of cDNA ends (RACE) was applied to determine 5′ and 3′ genomic and 3′ mRNA termini (Tables 1 and 2).
TABLE 1.
Properties of UPOV and ABV genome segmentsa
| Virus | Segment | Segment length (nt) | 5′ UTR length (nt) | ORF length (aa) | 3′ UTR length (nt) | Predicted molecular mass (kDa) | pI | FLUAV/THOV homolog |
|---|---|---|---|---|---|---|---|---|
| UPOV | 1 | 2,385 | 27 | 774 | 36 | 89.4 | 9.0 | PB2 |
| ABV | 2,384 | 27 | 774 | 35 | 89.1 | 9.0 | ||
| UPOV | 2 | 2,245 | 45 | 716 | 52 | 81.3 | 7.5 | PB1 |
| ABV | 2,246 | 45 | 716 | 53 | 81.4 | 8.0 | ||
| UPOV | 3 | 1,984 | 35 | 629 | 62 | 72.5 | 5.7 | PA |
| ABV | 1,984 | 35 | 629 | 62 | 72.5 | 5.7 | ||
| UPOV | 4 | 1,635 | 23 | 524 | 40 | 59.1 | 8.7 | GP |
| ABV | 1,630 | 23 | 521 | 44 | 59.1 | 6.4 | ||
| UPOV | 5 | 1,542 | 30 | 470 | 102 | 53.2 | 9.0 | NP |
| ABV | 1,544 | 32 | 470 | 102 | 53.1 | 9.1 | ||
| UPOV | 6 | 973 | 32 | 271 | 128 | 30.2 | 6.6 | M |
| ABV | 983 | 32 | 271 | 138 | 30.2 | 6.6 |
nt, nucleotides; aa, amino acids.
TABLE 2.
Sequence conservation at the termini of genome segmentsa
| Segment | 3′ terminus (genomic orientation) | 5′ terminus (genomic orientation) |
|---|---|---|
| UPOV | ||
| 1 (PB2) | 3′-UCG UUA UCG UUC GUC AAAA GUA | 5′-AGA GAU AUC AAA GCA G UUU UUU |
| 2 (PB1) | UCG UUA UCG UCC GUC AAAA GUU | AGA GAU AUC AAG GCA G UUU UUU |
| 3 (PA) | UCG UUA UCG UUC GUC AAAA GUU | AGA GAA AUC AAA GCA G UUU UUU |
| 4 (HP) | UCG UUA UUG UCC GUC AAAA GUU | AGA GAA AUC AAG GCA G UUU UUU |
| 5 (NP) | UCG UUU UCG UCC GUC AAAA GUU | AGA GAC AUC AAG GCA G UUU UUC |
| 6 (M) | UCG UUA UUG UCC GUC AACA GAU | AGA GAU AUC AAG GCA G UUU UUU |
| ABV | ||
| 1 (PB2) | 3′-UCG UUA UCG UUC GUC AAAG UGA | 5′-AGA GAU AUC AAA GCA G UUU UUU |
| 2 (PB1) | UCG UUA UCG UCC GUC AAAA AGU | AGA GAU AUC AAG GCA G UUU UUU |
| 3 (PA) | UCG UUA UCG UUC GUC AAAA GUU | AGA GAA AUC AAA GCA G UUU UUC |
| 4 (HP) | UCG UUU UCG UCC GUC AAAA GUU | AGA GAA AUC AAG GCA G UUU UUU |
| 5 (NP) | UCG UUU UCG UCC GUC AAAA GUU | AGA GAU AUC AAG GCA G UUU UUU |
| 6 (M) | UCG UUA UCG UCC GUC AAAGAAU | AGA GAU AUC AAG GCA G UUU UUU |
| THOV | consensus, UCG UUU UUG UYC GYB WVCW KKK | consensus, AGA GAW AUC AAR GCR S UUU UUU |
| 1 (PB2) | 3′-UCG UUU UUG UUC GCU ACCU GUC | 5′-AGA GAA AUC AAG GCG A UUU UUC |
| 2 (PB1) | UCG UUU UUG UCC GCG AGGU UUG | AGA GAA AUC AAG GCG C UUU UUU |
| 3 (PA) | UCG UUU UUG UUC GUG AACU GUA | AGA GAA AUG AAA GCA C UUU UUU |
| 4 (HP) | UCG UUU UUG UUC GUC UACA AGG | NGA GAU AUC AAA GCA G UUU UUU |
| 5 (NP) | UCG UUU UUG UCC GUC AGUU UUA | AGA GAA AUC AAG GCA G UUU UUU |
| 6 (M) | UCACCU UUG UCC GUC ACCU CUA | AGA GAA AUC AAG GCA G UUU UUU |
| DHOV | ||
| 1 (PB2) | 3′-UCG UUU UUG UUC GUC AAAU CUG | 5′-AGA GAA AUC AAA GCA G UUU UUC |
| 2 (PB1) | UCG UUU UUG UUC GUC AACU GUC | AGA GAU AUC AAA GCA G UUU UUU |
| 3 (PA) | UCG UUU UUG UUC GUC AAUG GUG | AGA GAA AUC AAA GCA G UUU UUU |
| 4 (HP) | UCG UUU UUG UUC GUC AAUGCUA | AGA GAA AUC AAA GCA G UUU UUC |
| 5 (NP) | UCG UUA UUG UUC GUC AAAGCUU | AGA GAU AUC AAA GCA G UUU UUU |
| 6 (M) | UCG UUA UUG UUC GUC AUGA UCU | AGA GAA AUC AAA GCA G UUU UUU |
| JOSV | ||
| 1 (PB2) | 3′-UCG UUU UUG UUC NUC AAAA GUU | 5′-AGA GAA AUC AAA GCA G UUU UUU |
| 2 (PB1) | UCG UUU UUG UCC GUC AAAG GGU | NA |
| 3 (PA) | NA | guuucccaguaggucuc AGA GAU AUC AAG GCA G UUU UUU |
| 4 (HP) | UCG UUU UUC UCC UCA AAAA CCU | AGA GAA AUC AAG GCA G UUU UUU |
| 5 (NP) | UCG UUU UUC UCC UGU ACCU CGA | NA |
| 6 (M) | g UCACCU UUG UCC GUC AAAA GCU | AGA GAA AUC AAG GCA G UUU UUU |
Morphology of UPOV and ABV virions.
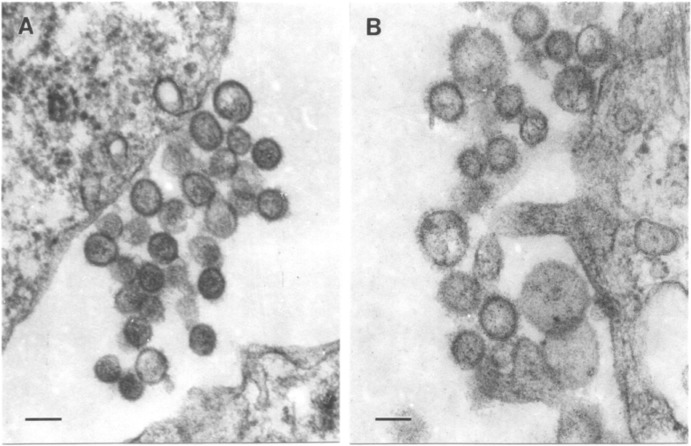
Transmission electron microscopy of ultrathin sections showed UPOV and ABV virions in clusters at the cell surface of infected Vero E6 cells (Fig. 1). Virions of UPOV were either round, with diameters of 75 to 95 nm, or slightly oval, with sizes ranging from 75 by 85 nm to 105 by 120 nm (Fig. 1A). Virions of ABV were more polymorphic and partly larger, ranging from 75 by 85 nm up to 105 by 130 and 120 by 140 nm (Fig. 1B).
FIG 1.
Ultrastructure of Upolu virus (A) and Aransas Bay virus (B) in infected Vero E6 cell cultures. Bar = 100 nm.
Genetic and serologic characterization of UPOV and ABV.
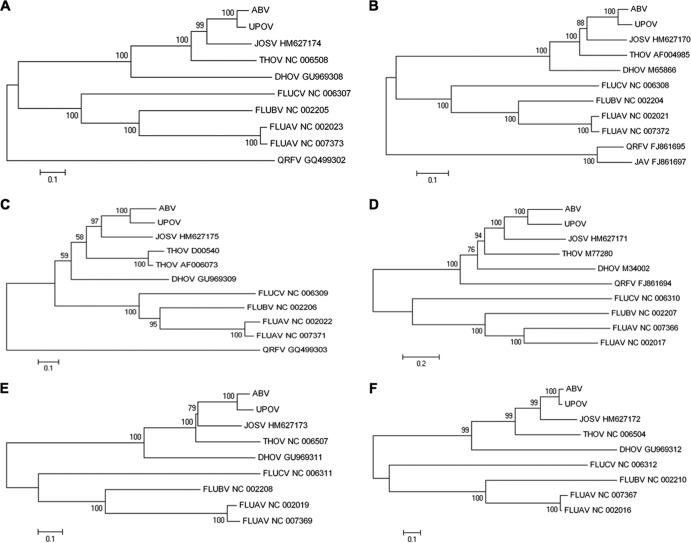
UPOV and ABV display terminal sequences that are semicomplementary and conserved among the six segments and the two viruses (Table 2). Overall, the termini of each segment adhere to consensus sequences determined for THOV (3′-UCG UUU UUG UU/CC GU/CC/G/U and 5′-AGA GAA/U AUC AAG/A GCA/G G/C UUU UUU), although specific differences are evident at the 3′ terminus in position 6 (“A,” similar to DHOV segments 5 and 6), position 8 (“C,” similar to influenza viruses), and positions 16 to 19 (conserved AAA/CA/G, similar to Jos virus [JOSV]) (7) as well as in position 6 of the 5′ terminus of UPOV segment 5 and ABV segment 3 (“C”). In THOV and JOSV, the 3′-terminal sequence of segment 6 differs from that of all other segments. No specific difference in the 3′-terminal sequence of segment 6 was found for UPOV and ABV, similar to DHOV. Analogous to influenza virus, the formation of a forked terminal panhandle has been shown to be essential for promoter function in THOV although with potential differences in the intrastrand base pairing of vRNA and cRNA “hook” structures (44–47). Compared to this, the changes in the terminal sequences of UPOV and ABV are located either at the unpaired fork region (3′/5′ position 6) (Fig. 2), with no compensating base change at the opposite terminus, or in the paired panhandle region, with compensating mutations at the opposite terminus (3′ position 11/5′ position 12; genomic orientation). In addition, 3′ C8 (genomic orientation) allows for a second paired base of a potential 3′ hook in several of the segments, and potential wobbling between the intrastrand pairing of 3′ C2/G9-5′ G2/C9 and the interstrand pairing of 3′ C2/5′ G2-3′G9/5′ C9 may provide options for “breathing” of the structure (Fig. 2). Termination of mRNA transcripts occurred at a conserved oligo(U)5-6 signal located 17 nucleotides (nt) from the 5′ end of vRNA templates, as indicated by RACE with oligo(dT) priming. The level of coding sequence similarity between individual segments of UPOV and ABV, and to corresponding segments of other orthomyxoviruses, was variable (Table 3). Phylogenetic analysis indicated that the evolutionary relationship for all segments is consistently closest between UPOV and ABV and that both viruses are more closely related to the recently characterized JOSV and THOV than to DHOV or the influenza viruses (Fig. 3).
FIG 2.
Potential base pairing of UPOV and ABV segment terminal bases.
TABLE 3.
Percent sequence identitiesa
| Sequence and virus | % sequence identity |
|||||
|---|---|---|---|---|---|---|
| UPOV | ABV | JOSV | THOV | DHOV | FLUAV | |
| PB2 (aa)/S1 (nt) | ||||||
| UPOV | 78.3 | 67.9 | 61.7 | 54.2 | 46.6 | |
| ABV | 92.1 | 68.1 | 62.1 | 54.4 | 47.8 | |
| JOSV | 71.1 | 71.3 | 62.4 | 53.5 | 49.7 | |
| THOV | 61.2 | 60.9 | 61.0 | 54.2 | 48.0 | |
| DHOV | 36.8 | 36.6 | 36.3 | 36.6 | 46.5 | |
| FLUAV | 22.5 | 21.9 | 20.7 | 22.3 | 22.7 | |
| PB1 (aa)/S2 (nt) | ||||||
| UPOV | 78.8 | 67.8 | 68.1 | 63.5 | 51.5 | |
| ABV | 92.7 | 68.1 | 68.2 | 62.9 | 51.2 | |
| JOSV | 75.1 | 76.1 | 66.2 | 61.5 | 50.1 | |
| THOV | 73.8 | 73.6 | 71.9 | 63.7 | 51.6 | |
| DHOV | 62.3 | 62.1 | 60.6 | 61.9 | 51.5 | |
| FLUAV | 28.4 | 28.8 | 29.3 | 30.3 | 32.1 | |
| PA (aa)/S3 (nt) | ||||||
| UPOV | 79.0 | 63.1 | 59.5 | 55.4 | 48.1 | |
| ABV | 86.0 | 63.8 | 60.7 | 55.5 | 48.5 | |
| JOSV | 63.1 | 64.8 | 58.7 | 53.7 | 43.0 | |
| THOV | 46.5 | 46.4 | 45.5 | 55.0 | 47.1 | |
| DHOV | 39.4 | 39.1 | 40.6 | 35.1 | 49.5 | |
| FLUAV | 22.0 | 23.5 | 23.2 | 22.4 | 24.3 | |
| GP (aa)/S4 (nt) | ||||||
| UPOV | 69.3 | 60.5 | 57.1 | 53.9 | 49.8 | |
| ABV | 67.1 | 59.5 | 55.7 | 53.2 | 50.0 | |
| JOSV | 52.9 | 51.9 | 55.5 | 54.6 | 44.8 | |
| THOV | 42.4 | 43.6 | 42.9 | 53.6 | 47.0 | |
| DHOV | 36.3 | 32.6 | 33.3 | 35.3 | 46.2 | |
| FLUAV | 22.0 | 19.3 | 21.6 | 21.1 | 19.4 | |
| NP (aa)/S5 (nt) | ||||||
| UPOV | 79.9 | 57.4 | 58.8 | 55.9 | 39.1 | |
| ABV | 89.1 | 59.2 | 59.6 | 56.7 | 48.1 | |
| JOSV | 64.3 | 65.2 | 63.6 | 52.7 | 42.1 | |
| THOV | 59.1 | 59.9 | 63.2 | 55.2 | 49.0 | |
| DHOV | 40.7 | 41.3 | 43.6 | 42.1 | 49.5 | |
| FLUAV | 22.4 | 23.0 | 21.2 | 21.0 | 23.0 | |
| M (aa)/S6 (nt) | ||||||
| UPOV | 81.9 | 64.4 | 60.0 | 50.3 | 49.7 | |
| ABV | 95.9 | 64.7 | 59.2 | 50.7 | 50.2 | |
| JOSV | 71.2 | 70.5 | 61.7 | 51.1 | 49.1 | |
| THOV | 45.4 | 43.5 | 48.7 | 51.3 | 48.0 | |
| DHOV | 27.9 | 28.3 | 28.5 | 23.4 | 47.4 | |
| FLUAV | 18.1 | 16.3 | 19.7 | 18.3 | 23.3 | |
Pairwise sequence identities between Upolu virus (UPOV), Aransas Bay virus (ABV), Jos virus (JOSV), Thogoto virus (THOV), Dhori virus (DHOV), and influenza A virus (FLUAV) at the nucleotide (nt) level (values above the diagonal) and at the amino acid (aa) level (values below the diagonal). S1, segment 1.
FIG 3.
Phylogenetic analysis of deduced amino acid sequences of UPOV and ABV in comparison to those of other selected orthomyxoviruses, indicated by their GenBank accession number and abbreviation (UPOV, Upolu virus; ABV, Aransas Bay virus; JOSV, Jos virus; THOV, Thogoto virus; DHOV, Dhori virus; FLUCV, influenza C virus; FLUBV, influenza B virus; FLUAV, influenza A virus; QRFV, Quaranfil virus; JAV, Johnston Atoll virus). Neighbor-joining trees were constructed under a Jukes-Cantor model, running 1,000 pseudoreplicates; bootstrap values of >50% are indicated at the respective nodes; and scale bars indicate substitutions per site. (A) PB2 (segment 1); (B) PB1 (segment 2); (C) PA (segment 3); (D) GP (segment 4); (E) NP (segment 5); (F) M (segment 6).
The largest segments of UPOV and ABV show sequence homology to orthomyxoviral PB2 gene sequences (Pfam accession number PF00604; “Flu_PB2” [http://pfam.sanger.ac.uk/]) (Tables 1 and 3). Although the PB2 sequence is the least conserved among orthomyxoviral polymerase subunits, UPOV and ABV sequences closely match those of JOSV and THOV, with DHOV being more distantly related, particularly in the C-terminal portion. Only a few amino acid motifs (D89LG, R149KPV, W225LP, and I314CRVALG in UPOV) are conserved with respect to influenza viruses outside an N-terminal motif (F40 to L56 in UPOV) that is recognizable throughout influenza and tick-transmitted viruses and located in a region that is implicated in PB1 binding in influenza A virus (FLUAV) (48). Only limited conservation was noted for the cap-binding domain defined in FLUAV, although the secondary structure of the N-terminal part and aromatic residues corresponding to FLUAV F330, F363, and F404 (but not F323/F325) are maintained, as also previously reported for THOV (18, 49). Consistent with the nuclear replication of orthomyxoviruses, a nuclear localization signal (NLS) (K745RRX11KRPRR), resembling the bipartite NLS identified in FLUAV (K736RKRX12KRIR) (50–52), is present. However, mutational analysis of THOV did not support a functional NLS role for its homologous K753RRR motif (53).
The sequences of UPOV and ABV segments 2 correspond to orthomyxoviral PB1 sequences (Pfam accession number PF00602; “Flu_PB1” [http://pfam.sanger.ac.uk/]) (Tables 1 and 3) and show conservation of the polymerase motifs pre-A, A, B, C, D, and E (20, 54–56). Conservation was also noted for amino acids maintained between THOV, DHOV, and influenza viruses in the second half of the N-terminal domain involved in PA binding in FLUAV (Y22-Y47 in UPOV) (57–59) and a downstream motif present throughout the orthomyxoviruses (L118-T124 in UPOV). PB1s of UPOV and ABV have a rather neutral pI (Table 1), more similar to PB1 of THOV than to that of influenza viruses. No conservation was obvious in the region of the FLUAV bipartite NLS (60), as is also the case for THOV and DHOV.
Segments 3 of UPOV and ABV encode a PA-like protein (Pfam accession number PF00603; “Flu_PA” [http://pfam.sanger.ac.uk/]) (Tables 1 and 3). The endonuclease motif PDXn(D/E) described previously for FLUAV (24, 25) corresponds to a P96HX16D motif in UPOV and ABV that is not surrounded by additional characteristic primary or secondary sequence conservation as reported for FLUAV. An elevated level of conservation was noted for the C-terminal part of the sequence (around Q426-F452 in UPOV), which has been implicated in interactions with PB1 in FLUAV (61).
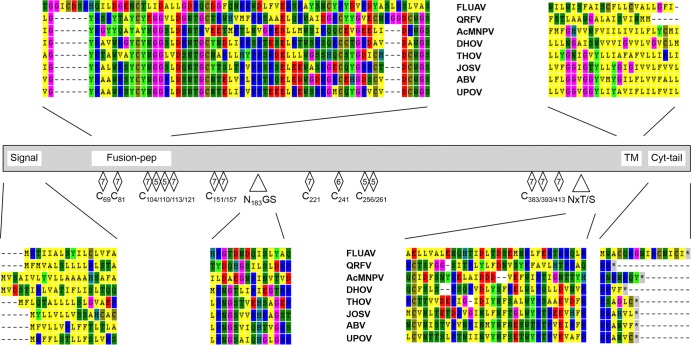
The putative glycoproteins (GPs) of UPOV and ABV are encoded by segment 4 (Tables 1 and 3). Instead of showing conservation with respect to influenza virus-like orthomyxoviral GPs, the overall structures of UPOV and ABV GPs are similar to those of the corresponding proteins of THOV and the “baculovirus gp64 envelope glycoprotein family” (Pfam accession number PF03273 [http://pfam.sanger.ac.uk/]) (27, 28), including conservation of glycosylation sites around positions 183 and 415/428 of UPOV (Fig. 4). Primary sequence conservation was observed for the N-terminal region containing a potential fusion peptide cleavage site (V59GY-WGS116 in UPOV, homologous to the A61GY-WGS118 sequence proposed for THOV [28]) and for the motifs W155RCGV, upstream of the only strictly conserved glycosylation site, N183GS; S351LSKIDERLIG, S391NC, D401GRW, and G444VIEDEEGWNF. There are significant differences in the cytoplasmic tail regions of GPs of the various orthomyxovirus species (Fig. 4). Serologic analyses by hemagglutination inhibition (HI) tests indicate limited cross-reactivity between UPOV, ABV, and THOV (Table 4). Interestingly, antigenic relatedness was greater between ABV and THOV than between UPOV and THOV or UPOV and ABV, pointing to sequence areas divergent between UPOV and ABV as potentially being involved in HI epitopes (possibly including I43-E55, W98-C110, L122-K134, K171-V175, C225-H235, L364-K371, W404-I424, and, particularly, L264-H306, which includes indel regions, in UPOV).
FIG 4.
Schematic of glycoprotein alignment including the tick-borne orthomyxoviruses Upolu virus (UPOV), Aransas Bay virus (ABV), Jos virus (JOSV), Thogoto virus (THOV), Dhori virus (DHOV), and Quaranfil virus (QRFV) as well as influenza A virus (FLUAV) and the insect Autographa californica multicapsid polyhedrosis virus (AcMNPV), showing the signal peptide (Signal); motifs of a potential fusion peptide cleavage site proposed for THOV (Fusion-pep); cysteine (C) residues conserved in all orthomyxoviruses or in the tick-borne orthomyxoviruses and AcMNPV  , in tick-borne viruses and AcMNPV except DHOV
, in tick-borne viruses and AcMNPV except DHOV  , or in thogoto- and dhoriviruses or in thogotoviruses and AcMNPV
, or in thogoto- and dhoriviruses or in thogotoviruses and AcMNPV  ; conserved glycosylation sites surrounding position 183 (N183GS/N183GT; N197VT in AcMNPV) and position 415/428 (NxT/S, including N415/412/410XT/S in UPOV, ABV, and JOSV; N428/427/423/416XT/S in UPOV, ABV, JOSV, and THOV; N378NT in THOV; N396HS in DHOV; N422VS in QRFV; and N384NS/N426TT in AcMNPV); the trans-membrane anchor (TM); and amino acids of the cytoplasmic tail region (Cyt-tail).
; conserved glycosylation sites surrounding position 183 (N183GS/N183GT; N197VT in AcMNPV) and position 415/428 (NxT/S, including N415/412/410XT/S in UPOV, ABV, and JOSV; N428/427/423/416XT/S in UPOV, ABV, JOSV, and THOV; N378NT in THOV; N396HS in DHOV; N422VS in QRFV; and N384NS/N426TT in AcMNPV); the trans-membrane anchor (TM); and amino acids of the cytoplasmic tail region (Cyt-tail).
TABLE 4.
Serologic analyses
| Serologic analysis | Titer |
||||
|---|---|---|---|---|---|
| ABV | UPOV | Araguari virus | DHOV | THOV | |
| Hemagglutination inhibition | |||||
| Antibody | Antigen (4 U)a |
||||
| ABV | 1,280 | 40 | ND | <10 | 160 |
| UPOV | 160 | 320 | ND | <10 | 40 |
| Araguari virus | <10 | <10 | 640 | <10 | 10 |
| DHOV | <10 | <10 | <10 | 2,560 | 10 |
| THOV | 1,280 | 40 | 10 | 80 | 5,120 |
| Complement fixation | |||||
| Antigen | Antibodyb |
||||
| ABV | ≥64/≥8 | <8/<8 | <8/<8 | <8/<8 | 32/≥8 |
| UPOV | ≥64/≥1 | ≥64/≥1 | <8/<8 | <8/<8 | <8/<8 |
| Araguari virus | <8/<8 | <8/<8 | ≥64/≥8 | <8/<8 | <8/<8 |
| DHOV | <8/<8 | <8/<8 | <8/<8 | ≥64/≥8 | <8/<8 |
| THOV | 32/≥8 | <8/<8 | <8/<8 | <8/<8 | ≥64/≥8 |
Reciprocal of the serum dilution giving complete inhibition of agglutination with 4 units of antigen. ND, not done.
Reciprocal of the serum dilution/optimal antigen dilution resulting in fixation of complement (2 units of guinea pig complement).
The nucleoprotein (NP) of orthomyxoviruses represents the main type-specific antigen recognized in complement fixation (CF) tests (Table 4) and has been widely used to assess phylogenetic relationships. The open reading frame (ORF) encoded by segments 5 of UPOV and ABV is conserved with respect to “influenza virus nucleoprotein” (Pfam accession number PF00506 [http://pfam.sanger.ac.uk/]) (Tables 1 and 3). Although only a low level of conservation was observed for the characterized N-terminal NLS in the NP of FLUAV (62, 63), which is also the case for THOV and DHOV, greater conservation was noted for the second half of a region that has been proposed for RNA interactions in FLUAV (L134, V137, L139, T143, I147, Q150K, V160, A168, G170, I173, R176, and G186 in UPOV) (64, 65). Conservation was also evident in the previously characterized internal NP regions 2 to 5 (66). This includes a sequence in region 4 that corresponds to a proposed nuclear accumulation motif of FLUAV (S329AGEDLGLLS in UPOV) (67, 68) and a motif in region 5 that is similar to a C-terminal bipartite NLS motif found in THOV and JOSV (K388RX9KGKR in UPOV) (7) but not in DHOV. The internal bipartite NLS characterized for THOV and FLUAV is conserved (K195RX9KTKR in UPOV) (69).
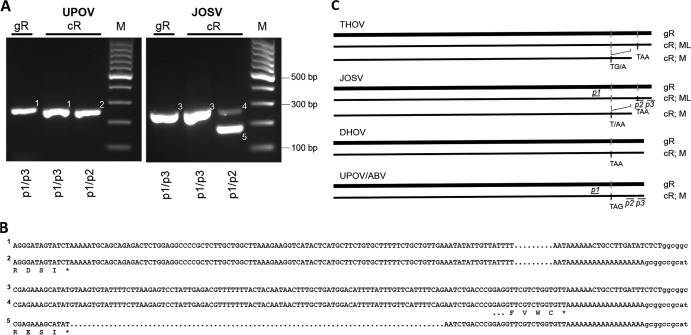
Segments 6 of UPOV and ABV show no homology to entries in the protein family database. The nucleotide sequences align only with the segment 6 sequence of JOSV and the C-terminal quarter of that of THOV but not with that of DHOV or the influenza viruses (Tables 1 and 3). Limited conservation with respect to DHOV was discernible at the deduced amino acid level for a short motif (A249KGVSYQVL in UPOV) and strictly conserved amino acids E175, N181T, E212, Y224D, G232, E236, and I240 located in the C-terminal region that has been proposed for the matrix protein (M) of THOV to inhibit viral polymerase activity (70). Segments 6 of UPOV, ABV, and DHOV have longer untranslated regions (UTRs) than those of THOV and JOSV (DHOV, 121 nt; UPOV and ABV, 128 and 138 nt, respectively). PCR analyses of genomic and mRNA preparations indicated that only a single-size segment 6 mRNA transcript was generated by UPOV, whereas two differently sized mRNA transcripts were generated by JOSV (Fig. 5) (7). This correlates with different coding strategies used by the viruses. Whereas segment 6 of DHOV codes only for an M protein (71) that terminates in a position analogous to that of the putative M ORFs of UPOV and ABV, THOV and JOSV are known to generate two products through splicing (72); ML is generated from nonspliced transcripts, resulting in a UTR of 20 nt (73), while M is generated from a spliced transcript by the creation of a stop codon at the splice junction, which is located in a position corresponding to the stop codons for M in UPOV, ABV, and DHOV (Fig. 5C). Of note, the level of sequence conservation between UPOV and ABV is highest for segment 6, and this segment's sequence is also one of the closest to those of JOSV and THOV (Table 3), despite the observed differences in coding potential.
FIG 5.
Segment 6 coding strategies. (A) RNA extracts obtained from HEK 293 cells infected with Upolu virus (UPOV) or Jos virus (JOSV) (cellular RNA [cR]) or from DNase- and RNase-treated supernatants (genomic RNA [gR]). cDNA was amplified with primers located upstream of a potential splice region (p1) and downstream at an mRNA polyadenylation signal (p2) or at the segment terminal sequence (p3). M indicates molecular size markers. Only a single-size amplification product was observed with the UPOV template (bands 1 and 2), whereas differently sized products were generated with the JOSV template (bands 3 to 5). (B) Relevant sequences obtained from the respective bands shown in panel A. (C) Schematic of segment 6 coding strategies of Thogoto virus (THOV) and JOSV, and of Dhori virus (DHOV), Aransas Bay virus (ABV), and UPOV, indicating locations of primers p1, p2, and p3; ML or M ORF termination codons (ochre, opal, and amber); and splice sites.
DISCUSSION
Analyses of the genome sequences of UPOV from Australia and ABV from North America show that they are up to ∼75% identical at the amino acid level (∼68% identical at the nucleotide level) (Table 3) to viruses in the family Orthomyxoviridae. The genetic distances of these viruses are shortest with respect to JOSV and THOV, ranging from approximately 76% amino acid and 68% nucleotide (PB1) to 52% amino acid and 60% nucleotide (GP) identity with JOSV and from approximately 74% amino acid and 68% nucleotide (PB1) to 43% amino acid and 56% nucleotide (GP) identity with THOV. However, the coding strategy of segments 6 of UPOV and ABV differs from that of JOSV and THOV and is similar to that of DHOV. Differences in the commonly conserved segment termini are also compatible with a significant evolutionary distance of UPOV and ABV from the species Thogoto virus. The species Dhori virus includes two viruses, DHOV and BKNV, which share approximately 97% and 90% amino acid (87% and 80% nucleotide) identity among their available partial NP and GP sequences, respectively. In comparison, DHOV and THOV share only between 42% and 35% amino acid (55% and 54% nucleotide) identity for their NP and GP sequences, respectively. This is also reflected by the serological reaction between the viruses. Whereas DHOV and BKNV cross-react, DHOV and THOV are antigenically distinct. This provided the basis for the inclusion of BKNV together with DHOV in a single species, Dhori virus, separate from the species Thogoto virus (6, 43). Both UPOV and ABV are antigenically closer to THOV than to DHOV by HI tests, whereas by CF tests there are differences in their cross-reactivities to THOV and to each other when ABV antigen/UPOV antibody is tested. These serologic results, combined with the <60% amino acid (<60% nucleotide) sequence identity of their NP or GP sequences to that of THOV or DHOV and the observed differences in coding capacity, suggest that UPOV and ABV should be considered separate species within the genus Thogotovirus, distinct from the species Dhori virus and Thogoto virus. In addition, amino acid sequence identities between UPOV and ABV of as little as 86% (78% nucleotide identity), and even less for the immunoreactive GP, combined with the serologic differences observed between them, may justify their classification as two separate species.
Due to their distinct structures, the GPs of the tick-infecting orthomyxoviruses have been classified as class 3 penetrenes, distinct from the class 2 penetrenes in alphaviruses and flaviviruses and the class 1 penetrenes in the influenza viruses (28). Furthermore, it has been hypothesized, based on sequence homologies, that GPs of viruses in the genus Thogotovirus may have been derived from a common ancestor with insect baculoviruses (27, 28). Thus, the tick-infecting orthomyxoviruses represent an evolutionary lineage distinct from that of the influenza viruses, and an ancestral relationship of either orthomyxoviral line to the other is not apparent from available data (Fig. 3). Nonetheless, the tick-adapted orthomyxoviral GPs are compatible with mammalian receptors, as exemplified by previous reports of human THOV and DHOV infections. In central Africa and regions of southern Europe, THOV has also been isolated from various ruminant species (61, 74). The geographic distribution of DHOV includes primarily India and eastern Russia but also East Africa, Egypt, and other Mediterranean countries, where serologic data indicate circulation in ruminants as well as waterfowl (61, 75–78). Migratory waterfowl are also reservoirs of influenza A viruses (78, 79). Cases of natural human infection have been reported for THOV from Africa (77), and accidental laboratory infections with DHOV indicate that this virus can also act as a human pathogen (76), despite the lack of an ML protein (71, 73). UPOV and ABV productively infect BHK, Vero, or HEK 293 cells and are lethal to newborn mice after intracerebral inoculation (1, 2, 61), suggesting that mammalian pathogenicity is also conceivable for UPOV and ABV.
In FLUAV, reassortment of genome segments is a well-known phenomenon that leads to sudden genetic shifts that can result in dramatic changes in pathogenicity. Reassortment in arthropod and vertebrate hosts has also been demonstrated for THOV in experimental settings (80, 81). The dissemination of genetically related tick-transmitted orthomyxoviruses over large distances by migratory birds (74) may support genome segment reassortment culminating in the emergence of novel genotypes with altered pathogenicity and host range. Indeed, the recent implication of other tick-borne orthomyxoviruses of the proposed genus Quarjavirus in human febrile illness (82) and the discovery of variants with high bird pathogenicity (83, 84) reinforce the need for comprehensive surveillance and characterization of this growing group of viruses to closely monitor their potential as emerging pathogens.
ACKNOWLEDGMENTS
We thank Alla Tashmukhamedova and Aaloki Shah at the Center of Infection and Immunity for expert technical assistance and Meera Bhat at the Center of Infection and Immunity for project management.
This work was supported by National Institutes of Health award AI57158 (Northeast Biodefense Center—Lipkin); National Institutes of Health contract HHSN2722010000401/HHSN27200004/D04; the U.S. Department of Defense; and the U.S. Agency for International Development (USAID) Emerging Pandemic Threats (EPT) Program, PREDICT project, under terms of cooperative agreement number GHN-A-OO-09-00010-00.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Doherty RL, Whitehead RH, Wetters EJ. 1968. Isolation of viruses from Ornithodoros capensis Neumann from a tern colony on the Great Barrier Reef, North Queensland. Aust. J. Sci. 31:363–364 [Google Scholar]
- 2.Yunker CE, Clifford CM, Keirans JE, Thomas LA, Rice RCA. 1979. Aransas Bay virus, a new arbovirus of the Upolu serogroup from Ornithodoros capensis (Acari: Argasidae) in costal Texas. J. Med. Entomol. 16:453–460 [Google Scholar]
- 3.El Mekki AA, Nieuwenhuysen P, van der Groen G, Pattyn SR. 1981. Characterization of some ungrouped viruses. Trans. R. Soc. Trop. Med. Hyg. 75:799–806. 10.1016/0035-9203(81)90416-8 [DOI] [PubMed] [Google Scholar]
- 4.Carley JG, Standfast HA, Kay BH. 1973. Multiplication of viruses isolated from arthopods [sic] and vertebrates in Australia in experimentally infected mosquitoes. J. Med. Entomol. 10:244–249 [DOI] [PubMed] [Google Scholar]
- 5.Hurlbut HS, Thomas JI. 1960. The experimental host range of the arthropod-borne animal viruses in arthropods. Virology 12:391–407. 10.1016/0042-6822(60)90162-8 [DOI] [PubMed] [Google Scholar]
- 6.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 7.Bussetti AV, Palacios G, Travassos da Rosa A, Savji N, Jain K, Guzman H, Hutchison S, Popov VL, Tesh RB, Lipkin WI. 2012. Genomic and antigenic characterization of Jos virus. J. Gen. Virol. 93:293–298. 10.1099/vir.0.035121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Silva EV, Da Rosa AP, Nunes MR, Diniz JA, Tesh RB, Cruz AC, Vieira CM, Vasconcelos PF. 2005. Araguari virus, a new member of the family Orthomyxoviridae: serologic, ultrastructural, and molecular characterization. Am. J. Trop. Med. Hyg. 73:1050–1058 http://www.ajtmh.org/content/73/6/1050.long [PubMed] [Google Scholar]
- 9.Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 10.Bouloy M, Plotch SJ, Krug RM. 1978. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 75:4886–4890. 10.1073/pnas.75.10.4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krug RM, Broni BA, Bouloy M. 1979. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 18:329–334. 10.1016/0092-8674(79)90052-7 [DOI] [PubMed] [Google Scholar]
- 12.Ulmanen I, Broni BA, Krug RM. 1981. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 78:7355–7359. 10.1073/pnas.78.12.7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaas D, Patzelt E, Kuechler E. 1982. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10:4803–4812. 10.1093/nar/10.15.4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa Y, Kimura N, Toyoda T, Mizumoto K, Ishihama A, Oda K, Nakada S. 1995. The RNA polymerase PB2 subunit is not required for replication of the influenza virus genome but is involved in capped mRNA synthesis. J. Virol. 69:728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L, Galarza JM, Summers DF. 1996. Recombinant-baculovirus-expressed PB2 subunit of the influenza A virus RNA polymerase binds cap groups as an isolated subunit. Virus Res. 42:1–9. 10.1016/0168-1702(96)01289-0 [DOI] [PubMed] [Google Scholar]
- 16.Honda A, Mizumoto K, Ishihama A. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4:475–485. 10.1046/j.1365-2443.1999.00275.x [DOI] [PubMed] [Google Scholar]
- 17.Li ML, Rao P, Krug RM. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078–2086. 10.1093/emboj/20.8.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500–506. 10.1038/nsmb.1421 [DOI] [PubMed] [Google Scholar]
- 19.Braam J, Ulmanen I, Krug RM. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609–618 [DOI] [PubMed] [Google Scholar]
- 20.Biswas SK, Nayak DP. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi M, Toyoda T, Ishihama A. 1996. Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch. Virol. 141:525–539. 10.1007/BF01718315 [DOI] [PubMed] [Google Scholar]
- 22.Sanz-Ezquerro JJ, Fernandez Santaren J, Sierra T, Aragon T, Ortega J, Ortin J, Smith GL, Nieto A. 1998. The PA influenza virus polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79(Part 3):471–478 [DOI] [PubMed] [Google Scholar]
- 23.Huarte M, Falcon A, Nakaya Y, Ortin J, Garcia-Sastre A, Nieto A. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007–6013. 10.1128/JVI.77.10.6007-6013.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. 10.1038/nature07745 [DOI] [PubMed] [Google Scholar]
- 25.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913. 10.1038/nature07720 [DOI] [PubMed] [Google Scholar]
- 26.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989–9001. 10.1128/JVI.76.18.8989-9001.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse MA, Marriott AC, Nuttall PA. 1992. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology 186:640–646. 10.1016/0042-6822(92)90030-S [DOI] [PubMed] [Google Scholar]
- 28.Garry CE, Garry RF. 2008. Proteomics computational analyses suggest that baculovirus GP64 superfamily proteins are class III penetrenes. Virol. J. 5:28. 10.1186/1743-422X-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke DH, Casals J. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561–573 [DOI] [PubMed] [Google Scholar]
- 30.Beaty BJ, Calisher CH, Shope RE. 1989. Arboviruses, p 797–855 In Schmidt NJ, Emmons RW. (ed), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 6th ed. American Public Health Association, Washington, DC [Google Scholar]
- 31.Beaty BJ, Calisher CH, Shope RE. 1995. Arboviruses, p 189–212 In Lennette EH, Lennette DA, Lennette ET. (ed), Diagnostic procedures for viral, rickettsial and chlamydial infections, 7th ed. American Public Health Association, Washington, DC [Google Scholar]
- 32.Shope RE. 1963. The use of a micro hemagglutination-inhibition test to follow antibody response after arthropod-borne virus infection in a community of forest animals. An. Microbiol. 11(Part A):167–169 [Google Scholar]
- 33.Quan PL, Palacios G, Jabado OJ, Conlan S, Hirschberg DL, Pozo F, Jack PJ, Cisterna D, Renwick N, Hui J, Drysdale A, Amos-Ritchie R, Baumeister E, Savy V, Lager KM, Richt JA, Boyle DB, Garcia-Sastre A, Casas I, Perez-Brena P, Briese T, Lipkin WI. 2007. Detection of respiratory viruses and subtype identification of influenza A viruses by GreeneChipResp oligonucleotide microarray. J. Clin. Microbiol. 45:2359–2364. 10.1128/JCM.00737-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2011. Geneious v5.5. http://www.geneious.com/ [Google Scholar]
- 38.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 39.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 40.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 42.Lvov DK, Karas FR, Tsyrkin YM, Vargina SG, Timofeev EM, Osipova NZ, Veselovskaya OV, Grebenyuk YI, Gromashevski VL, Fomina KB. 1974. Batken virus, a new arbovirus isolated from ticks and mosquitoes in Kirghiz S.S.R. Arch. Gesamte Virusforsch. 44:70–73. 10.1007/BF01242183 [DOI] [PubMed] [Google Scholar]
- 43.Frese M, Weeber M, Weber F, Speth V, Haller O. 1997. Mx1 sensitivity: Batken virus is an orthomyxovirus closely related to Dhori virus. J. Gen. Virol. 78(Part 10):2453–2458 [DOI] [PubMed] [Google Scholar]
- 44.Weber F, Haller O, Kochs G. 1997. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch. Virol. 142:1029–1033. 10.1007/s007050050138 [DOI] [PubMed] [Google Scholar]
- 45.Leahy MB, Dessens JT, Nuttall PA. 1997. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J. Virol. 71:8352–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leahy MB, Dessens JT, Pritlove DC, Nuttall PA. 1998. The Thogoto orthomyxovirus cRNA promoter functions as a panhandle but does not stimulate cap snatching in vitro. J. Gen. Virol. 79(Part 3):457–460 [DOI] [PubMed] [Google Scholar]
- 47.Tchatalbachev S, Flick R, Hobom G. 2001. The packaging signal of influenza viral RNA molecules. RNA 7:979–989. 10.1017/S1355838201002424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perales B, de la Luna S, Palacios I, Ortin J. 1996. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70:1678–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fechter P, Mingay L, Sharps J, Chambers A, Fodor E, Brownlee GG. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381–20388. 10.1074/jbc.M300130200 [DOI] [PubMed] [Google Scholar]
- 50.Mukaigawa J, Nayak DP. 1991. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J. Virol. 65:245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14:229–233. 10.1038/nsmb1212 [DOI] [PubMed] [Google Scholar]
- 52.Resa-Infante P, Jorba N, Zamarreno N, Fernandez Y, Juarez S, Ortin J. 2008. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3:e3904. 10.1371/journal.pone.0003904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber F, Gruber S, Haller O, Kochs G. 1999. PB2 polymerase subunit of Thogoto virus (Orthomyxoviridae family). Arch. Virol. 144:1601–1609. 10.1007/s007050050613 [DOI] [PubMed] [Google Scholar]
- 54.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin DA, Roychoudhury S, Palese P, Clay WC, Fuller FJ. 1991. Evolutionary relatedness of the predicted gene product of RNA segment 2 of the tick-borne Dhori virus and the PB1 polymerase gene of influenza viruses. Virology 182:1–7. 10.1016/0042-6822(91)90641-N [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller R, Poch O, Delarue M, Bishop DH, Bouloy M. 1994. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 75(Part 6):1345–1352 [DOI] [PubMed] [Google Scholar]
- 57.Perez DR, Donis RO. 1995. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 69:6932–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez S, Zurcher T, Ortin J. 1996. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24:4456–4463. 10.1093/nar/24.22.4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leahy MB, Dessens JT, Weber F, Kochs G, Nuttall PA. 1997. The fourth genus in the Orthomyxoviridae: sequence analyses of two Thogoto virus polymerase proteins and comparison with influenza viruses. Virus Res. 50:215–224. 10.1016/S0168-1702(97)00072-5 [DOI] [PubMed] [Google Scholar]
- 60.Nath ST, Nayak DP. 1990. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol. Cell. Biol. 10:4139–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zurcher T, de la Luna S, Sanz-Ezquerro JJ, Nieto A, Ortin J. 1996. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77(Part 8):1745–1749 [DOI] [PubMed] [Google Scholar]
- 62.Wang P, Palese P, O'Neill RE. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71:9690–9700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi M, Toyoda T, Adyshev DM, Azuma Y, Ishihama A. 1994. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J. Virol. 68:8433–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albo C, Valencia A, Portela A. 1995. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J. Virol. 69:3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuller FJ, Freedman-Faulstich EZ, Barnes JA. 1987. Complete nucleotide sequence of the tick-borne, orthomyxo-like Dhori/Indian/1313/61 virus nucleoprotein gene. Virology 160:81–87. 10.1016/0042-6822(87)90047-X [DOI] [PubMed] [Google Scholar]
- 67.Davey J, Dimmock NJ, Colman A. 1985. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell 40:667–675. 10.1016/0092-8674(85)90215-6 [DOI] [PubMed] [Google Scholar]
- 68.Weber F, Haller O, Kochs G. 1996. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J. Virol. 70:8361–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber F, Kochs G, Gruber S, Haller O. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250:9–18. 10.1006/viro.1998.9329 [DOI] [PubMed] [Google Scholar]
- 70.Hagmaier K, Gelderblom HR, Kochs G. 2004. Functional comparison of the two gene products of Thogoto virus segment 6. J. Gen. Virol. 85:3699–3708. 10.1099/vir.0.80300-0 [DOI] [PubMed] [Google Scholar]
- 71.Clay WC, Fuller FJ. 1992. Nucleotide sequence of the tick-borne orthomyxo-like Dhori/India/1313/61 virus membrane protein gene. J. Gen. Virol. 73(Part 10):2609–2616 [DOI] [PubMed] [Google Scholar]
- 72.Kochs G, Weber F, Gruber S, Delvendahl A, Leitz C, Haller O. 2000. Thogoto virus matrix protein is encoded by a spliced mRNA. J. Virol. 74:10785–10789. 10.1128/JVI.74.22.10785-10789.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagmaier K, Jennings S, Buse J, Weber F, Kochs G. 2003. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J. Virol. 77:2747–2752. 10.1128/JVI.77.4.2747-2752.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Calisher CH, Karabatsos N, Filipe AR. 1987. Antigenic uniformity of topotype strains of Thogoto virus from Africa, Europe, and Asia. Am. J. Trop. Med. Hyg. 37:670–673 [DOI] [PubMed] [Google Scholar]
- 75.Sang R, Onyango C, Gachoya J, Mabinda E, Konongoi S, Ofula V, Dunster L, Okoth F, Coldren R, Tesh R, da Rossa AT, Finkbeiner S, Wang D, Crabtree M, Miller B. 2006. Tickborne arbovirus surveillance in market livestock, Nairobi, Kenya. Emerg. Infect. Dis. 12:1074–1080. 10.3201/eid1207.060253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaidamovich SY, Butenko AM, Leschinskaya HV. 2000. Human laboratory acquired arbo-, arena-, and hantavirus infections. J. Am. Biol. Safety Assoc. 5:5–11 http://www.absa.org/abj/abj/000501Gaidamovich.pdf [Google Scholar]
- 77.Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, Akinkugbe FM, David-West TS, Kemp GE. 1975. Arthropod-borne viral infections of man in Nigeria, 1964-1970. Ann. Trop. Med. Parasitol. 69:49–64 [DOI] [PubMed] [Google Scholar]
- 78.Iashkulov KB, Shchelkanov M, L'Vov SS, Dzhambinov SD, Galkina IV, Fediakina IT, Bushkieva B, Morozova TN, Kireev DE, Akanina DS, Litvin KE, Usachev EV, Prilipov AG, Grebennikova TV, Gromashevskii VL, Iamnikova SS, Zaberezhnyi AD, L'Vov KD. 2008. Isolation of influenza virus A (Orthomyxoviridae, Influenza A virus), Dhori virus (Orthomyxoviridae, Thogotovirus), and Newcastle's disease virus (Paromyxoviridae, Avulavirus) on the Malyi Zhemchuzhnyi Island in the north-western area of the Caspian Sea. Vopr. Virusol. 53:34–38 (In Russian.) [PubMed] [Google Scholar]
- 79.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones LD, Davies CR, Green BM, Nuttall PA. 1987. Reassortment of Thogoto virus (a tick-borne influenza-like virus) in a vertebrate host. J. Gen. Virol. 68(Part 5):1299–1306 [DOI] [PubMed] [Google Scholar]
- 81.Davies CR, Jones LD, Green BM, Nuttall PA. 1987. In vivo reassortment of Thogoto virus (a tick-borne influenza-like virus) following oral infection of Rhipicephalus appendiculatus ticks. J. Gen. Virol. 68(Part 9):2331–2338 [DOI] [PubMed] [Google Scholar]
- 82.Presti RM, Zhao G, Beatty WL, Mihindukulasuriya KA, da Rosa AP, Popov VL, Tesh RB, Virgin HW, Wang D. 2009. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 83:11599–11606. 10.1128/JVI.00677-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mickely R. 2012. Investigating the newly described Wellfleet Bay virus. Carrier 4(1):4 http://www.aphis.usda.gov/wildlife_damage/nwdp/Carrier/pdfs/The%20Carrier%20Vol%204%20Iss%201.pdf [Google Scholar]
- 84.Kessell A, Hyatt A, Lehmann D, Shan S, Crameri S, Holmes C, Marsh G, Williams C, Tachedjian M, Yu M, Bingham J, Payne J, Lowther S, Wang J, Wang LF, Smith I. 2012. Cygnet River virus, a novel orthomyxovirus from ducks, Australia. Emerg. Infect. Dis. 18:2044–2046. 10.3201/eid1812.120500 [DOI] [PMC free article] [PubMed] [Google Scholar]