Abstract
RIG-I-like receptors (RLRs) play important roles in the host defense to numerous viral pathogens. Since they were discovered, much light has been shed on the molecular details of how these cytoplasmic viral RNA receptors sense viral infection and orchestrate antiviral innate immunity. Intriguingly, in addition to viral RNA binding, a series of posttranslational modifications (PTMs) is required for the rapid activation of RLRs and, inversely, for the prevention of aberrant innate immune signaling. Recent discoveries have shown that viruses manipulate the PTMs of RLRs to escape innate immune detection. This article highlights some of these recent findings in this fast-evolving field.
RIG-I-LIKE RECEPTOR (RLR)-MEDIATED SENSING OF VIRAL INFECTION
For the detection of viral pathogens, mammalian cells are equipped with a sophisticated immune surveillance apparatus comprised of a defined repertoire of molecular sensors, classically termed pattern-recognition receptors (PRRs). Upon recognition of pathogen-associated molecular patterns (PAMPs), PRRs initiate downstream signaling that results in the gene expression of antiviral molecules and many cytokines, including type I interferons (alpha/beta interferons [IFN-α/β]). Through the upregulation of interferon-stimulated genes (ISGs), IFNs then induce an antiviral state in both infected and uninfected cells, as well as tailor adaptive immune responses.
At least three main classes of PRRs have been implicated in the detection of viral nucleic acid: (i) Toll-like receptors (TLRs), which sense incoming virions in endolysosomes by binding to viral RNA (TLR3 and -7/8) or CpG-containing DNA (TLR9), (ii) the recently identified, structurally diverse group of viral DNA sensors, including cGAS, IFI16, and DAI, and (iii) RIG-I-like receptors (RLRs), identified in 2004, that are essential for the detection of viral RNA in the cytoplasm of most cell types.
RLRs, comprising RIG-I, MDA5, and LGP2, are characterized by a conserved domain structure, consisting of a central DExD/H-box helicase domain and a C-terminal domain (CTD), both of which are responsible for binding viral RNA. In addition, RIG-I and MDA5 harbor two N-terminal caspase activation and recruitment domains (CARDs) which, upon virus sensing, initiate downstream signaling, leading to type I IFN gene expression. In contrast, LGP2 lacks the CARD signaling module and has been shown to exert a regulatory role in RLR signaling; its precise action, however, is yet to be defined (reviewed in reference 1).
Virus replication studies revealed that RIG-I confers resistance to many negative-sense RNA viruses, including orthomyxoviruses, rhabdoviruses, bunyaviruses, and paramyxoviruses as well as the positive-strand hepatitis C virus (HCV); in contrast, MDA5 was shown to primarily detect members of the Picornaviridae and Caliciviridae families. Despite these early studies suggesting that RIG-I and MDA5 detect mainly nonoverlapping subsets of viral pathogens, there is new evidence that numerous viruses, including dengue virus, West Nile virus (WNV), reoviruses, and several paramyxoviruses (e.g., measles virus and Sendai virus [SeV]), are sensed by both RIG-I and MDA5 (reviewed in reference 2). Furthermore, studies using synthetic or purified viral RNA revealed important molecular signatures that are required for RLR activation. It is now well established that a 5′triphosphate (5′ppp) moiety, present in the genomic RNA of many viruses, in concert with short blunt-end double-stranded RNA (dsRNA) stretches, such as “panhandle” structures, are critical for RIG-I's ability to discriminate non-self from self RNA. The sequence composition of the RNA ligand also seems to play a role in RIG-I activation: for example, poly(U/UC) motifs found in the genomic RNA of HCV were shown to stimulate RIG-I when combined with a 5′ppp group. In addition to the detection of RNA viruses, RIG-I also has been shown to contribute to the detection of DNA viruses, such as Epstein-Barr virus, by recognizing 5′ppp-containing small RNA species generated through transcription of viral DNA by RNA polymerase III. In contrast to RIG-I agonists, the characteristics of the RNA ligands sensed by MDA5 are largely unknown. The current view is that MDA5 recognizes long dsRNA organized in web-like structures, as found in picornavirus-infected cells (2).
Despite these important insights into the distinct viral RNA structures that can trigger RLR activation, the important question of what the physiological ligand during an actual viral infection is has just begun to be elucidated. Next-generation sequencing of viral RNA complexed with RIG-I in cells infected with influenza A virus (IAV) or SeV confirmed that short 5′ppp-containing viral RNAs produced during replication are physiological ligands for RIG-I (2). In addition to sensing viral replication products, can RIG-I also recognize virion RNA that is tightly packed with viral proteins? This question was recently answered by Weber et al., who showed that the nucleoprotein-encapsidated 5′ppp-RNA of incoming virions triggers RIG-I activation immediately after entry into the cell (3). Together, these studies indicate that during probably most viral infections, multiple RNA species—internalized with the virion and produced during viral replication—distinctly trigger RIG-I and/or MDA5 activation, likely at different time points during infection. In support of this model, distinct viral RNA products generated during WNV infection were shown to sequentially stimulate RIG-I and MDA5 activation: RIG-I early during infection, and MDA5 at later time points (4). More-detailed studies are needed to identify the authentic RNA ligands for RIG-I and MDA5 and to define the contributions and dynamics of action of these two sensors during other viral infections.
INTERPLAY OF UBIQUITINATION AND PHOSPHORYLATION REGULATES RLR SIGNALING
Upon viral RNA recognition by the CTD and helicase, RIG-I and MDA5 initiate antiviral signaling by interacting through their CARDs with the CARD of the adaptor protein MAVS (also called IPS-1, VISA, or CARDIF). MAVS contains a transmembrane domain that anchors it to both mitochondria and peroxisomes. Intriguingly, from these two organelles, MAVS was shown to induce a biphasic antiviral response (5). From peroxisomes, MAVS activates the transcription factors IFN regulatory factor 1 (IRF1) and IRF3, leading to the rapid expression of a select group of antiviral genes and triggering an immediate, IFN-independent antiviral response. At later time points during infection, mitochondrion-localized MAVS induces IRF3/7 activation to trigger IFN-α/β gene expression and the subsequent upregulation of ISGs via IFN-α/β receptor signaling.
Over the past few years, substantial progress has been made toward understanding the molecular details of how RNA binding to RIG-I and MDA5 results in their CARD-dependent downstream signaling. It is now well established that the signal-transducing activities of RIG-I and MDA5 are tightly regulated by a combinatorial posttranslational modification (PTM) code, with ubiquitination and phosphorylation being the best characterized PTMs. The first evidence of RIG-I regulation by PTMs was provided by mass spectrometry analysis of purified RIG-I CARDs, identifying covalent Lys63-linked polyubiquitination attached to Lys172 (and five other lysines) located in the CARD2 of RIG-I (6). Intriguingly, this ubiquitin mark does not lead to RIG-I degradation but instead is critical for RIG-I activation by facilitating binding to MAVS and thereby IFN induction (Fig. 1). Mechanistically, Lys63-ubiquitin chains are believed to serve as a scaffold for RIG-I oligomerization and subsequent MAVS binding. The same study also identified the enzyme responsible for RIG-I ubiquitination, TRIM25, a RING-dependent ubiquitin E3 ligase belonging to the tripartite motif (TRIM) protein family, comprised of more than 80 members in humans. Viral replication studies in TRIM25 knockdown and knockout cells demonstrated that TRIM25 is required for effective IFN induction and RIG-I-dependent restriction of multiple RNA viruses, including IAV, paramyxoviruses, and vesicular stomatitis virus (6). More recently, RIG-I was shown to bind noncovalent Lys63-ubiquitin chains in vitro, and it was suggested that free ubiquitin binding is important for RIG-I signaling (7); however, the physiological role of noncovalent ubiquitin binding in RIG-I activation in infected cells warrants investigation. The progress on RIG-I activation by ubiquitination was further advanced by the discovery of a second ubiquitin E3 ligase, called Riplet or REUL, that induces Lys63-linked ubiquitination at the CTD of RIG-I (8). How these two Lys63-ubiquitin marks—at the CARDs and CTD—induce RIG-I signaling has just recently been unveiled. Riplet-mediated ubiquitination of the CTD, together with viral RNA binding, is believed to induce a conformational change in RIG-I that exposes the N-terminal CARDs, thus promoting TRIM25 binding to CARD1 and the subsequent ubiquitination of Lys172 in CARD2 (9). TRIM25 itself is tightly regulated by Lys48-linked ubiquitination/deubiquitination, determining the protein's half-life in infected cells (10). Conflicting data have been reported about Lys63-linked ubiquitination of MDA5, leaving it unclear whether MDA5 requires Lys63-ubiquitin chains for antiviral signal transduction.
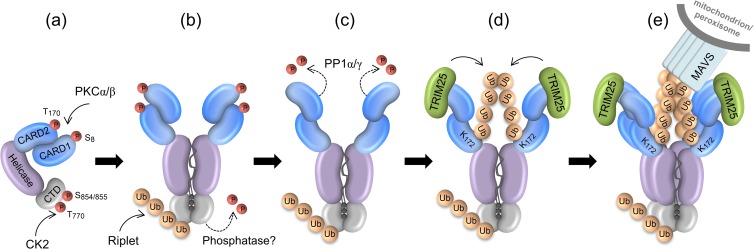
FIG 1.
Current model of RLR regulation (depicted for RIG-I). (a) RIG-I (and MDA5) are kept inactive in uninfected cells by two mechanisms: a constitutive phosphorylation of their CARDs (Ser8 and Thr170 in RIG-I; Ser88 in MDA5) and the CTD (Thr770 and Ser854/855 in RIG-I) and a closed conformation. (b) Viral RNA binding to the CTD/helicase, together with CTD dephosphorylation and Lys63-linked ubiquitination by Riplet, induces RIG-I dimerization and a conformational change, exposing the N-terminal CARDs. (c) The exposed CARDs recruit the phosphatases PP1α/γ, which dephosphorylate Ser8 and Thr170 in RIG-I (and Ser88 in MDA5). Dephosphorylation possibly induces a structural rearrangement within the tandem CARD, which (d) allows TRIM25 to bind to CARD1 and to induce Lys63-linked ubiquitination of Lys172 in CARD2. (e) The ubiquitin-bound CARDs facilitate RIG-I oligomerization and binding to MAVS, ultimately inducing antiviral signaling. Ub, ubiquitin; P, phosphorylation.
As aberrant or premature immune signaling may be harmful to the host, effective control mechanisms are required to prevent RLR activation in uninfected cells. A recent series of studies indicated that RIG-I and MDA5 are kept inactive by at least two mechanisms in uninfected cells: a constitutive phosphorylation of specific Ser/Thr residues in the CARDs and CTD, and a closed conformation in which binding of the helicase to the CARDs prevents downstream signaling. Conventional protein kinase C alpha/beta (PKC-α/β) and casein kinase II (CK2) were shown to be responsible for RIG-I CARD and CTD phosphorylation, respectively (11, 12). The kinase responsible for MDA5 CARD phosphorylation is still unknown. Biochemical studies using phospho-specific antibodies against the identified phosphorylation sites in RIG-I and MDA5 revealed that upon virus infection, both sensors are rapidly dephosphorylated, resulting in immune signaling. A phosphatome RNA interference (RNAi) screen identified the phosphatases PP1α and PP1γ (PP1α/γ) as being responsible for RIG-I and MDA5 dephosphorylation (13). PP1α and PP1γ, but not the isoenzyme PP1β, are recruited to the exposed CARDs, leading to their dephosphorylation. Mechanistically, dephosphorylation likely induces a conformational change within the tandem CARD, facilitating binding of TRIM25 to the CARDs and ubiquitination-dependent MAVS interaction. Studies are under way to investigate the precise details of how the phosphatases PP1α/γ are activated in response to viral infection and what determines their substrate specificities toward RLRs.
MANIPULATION OF RLR'S SIGNALING ACTIVITIES BY VIRAL PATHOGENS
Coevolution with their hosts enabled successful viral pathogens to manipulate and shape innate immune responses for their own benefit. Many different viral strategies for RLR evasion have been identified, including modification of the 5′ppp signature moiety in the viral genome and inhibition of key signaling molecules downstream of RLRs, such as MAVS. Recent studies provided evidence that viruses manipulate critical PTMs of RLRs to escape innate immunity. Several viruses have been shown to specifically modulate the Lys63-linked ubiquitination of RIG-I through targeting of the E3 ligases TRIM25 and Riplet. IAV, using its NS1 protein, targets TRIM25 through a direct interaction with its coil-coiled domain (CCD) (14). Mechanistically, NS1 binding to the CCD prevents TRIM25 from self-assembling into its oligomeric, enzymatically active form, thereby suppressing RIG-I CARD ubiquitination. Interestingly, the NS1 proteins of some strains of IAV also target Riplet, thereby blocking ubiquitination of RIG-I at the CTD (15). Moreover, it has been reported recently that the NS3-4A protease of HCV targets Riplet, but not TRIM25, for cleavage (9). Thus, HCV NS3-4A blunts RIG-I signaling at two distinct steps: by cleaving MAVS, as previously reported, and by cleaving Riplet. Furthermore, Kaposi's sarcoma-associated herpesvirus, arteriviruses, and nairoviruses encode viral deubiquitinating enzymes to actively remove Lys63-ubiquitin chains from the RIG-I CARDs, thereby suppressing downstream signaling (16, 17). As phosphorylation/dephosphorylation of RLRs is critical for their immune signaling ability, it is conceivable that viruses have also evolved means of manipulating the RLR phosphorylation state. To keep RLRs in the phosphorylated, inactive state, viruses may either directly induce RLR phosphorylation or block their dephosphorylation by PP1α/γ. Indeed, while encephalomyocarditis virus and poly(I·C)-RNA efficiently triggered RLR dephosphorylation, indicative of their activation, some members of the Paramyxoviridae family did not induce RLR dephosphorylation, indicating that these viruses manipulate the RLR phosphorylation state to escape immune detection (M. E. Davis, M. K. Wang, L. J. Rennick, F. Full, S. Gableske, A. W. Mesman, S. I. Gringhuis, T. B. H. Geijtenbeek, W. P. Duprex, and M. U. Gack, submitted for publication).
CONCLUSION
The recent discovery of specific PTM marks that determine the signaling “on” or “off” state of RIG-I and MDA5 may greatly facilitate research investigating RLR activation during viral infection and pathological conditions, such as autoimmune disease. New insights into the host regulatory mechanisms of RLR signaling may stimulate drug development designed to either boost antiviral signaling or dampen it in situations where the RLR response has gone awry. Furthermore, the discovery of novel virus-host interactions to escape the RLR response may open up novel therapeutic avenues for infectious diseases.
ACKNOWLEDGMENTS
I apologize to all whose work could not be cited due to space constraints.
Current research in the Gack laboratory is supported by grants from the NIH (AI087846, AI097699, and AI104415), the Armenise-Harvard Foundation, and the Alexander and Margaret Stewart Trust Foundation.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlee M. 2013. Master sensors of pathogenic RNA–RIG-I like receptors. Immunobiology 218:1322–1335. 10.1016/j.imbio.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. 10.1016/j.chom.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J. Virol. 87:11416–11425. 10.1128/JVI.01488-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141:668–681. 10.1016/j.cell.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- 7.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330. 10.1016/j.cell.2010.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807–817. 10.1074/jbc.M804259200 [DOI] [PubMed] [Google Scholar]
- 9.Oshiumi H, Miyashita M, Matsumoto M, Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 9:e1003533. 10.1371/journal.ppat.1003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. 2014. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 7:ra3. 10.1126/scisignal.2004577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maharaj NP, Wies E, Stoll A, Gack MU. 2012. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J. Virol. 86:1358–1371. 10.1128/JVI.06543-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Ren H, Liu Y, Teeling JL, Gu J. 2011. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 85:1036–1047. 10.1128/JVI.01734-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38:437–449. 10.1016/j.immuni.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. 10.1016/j.chom.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 8:e1003059. 10.1371/journal.ppat.1003059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, Ou JH, Jung JU. 2011. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899–10904. 10.1128/JVI.00690-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, Garcia-Sastre A, Snijder EJ, Kikkert M. 2012. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 86:773–785. 10.1128/JVI.06277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]