ABSTRACT
Numerous animal and plant viruses are transmitted by arthropod vectors in a persistent, circulative manner. Tomato yellow leaf curl virus (TYLCV) is transmitted by the sweet potato whitefly Bemisia tabaci. We report here that infection with Rickettsia spp., a facultative endosymbiont of whiteflies, altered TYLCV-B. tabaci interactions. A B. tabaci strain infected with Rickettsia acquired more TYLCV from infected plants, retained the virus longer, and exhibited nearly double the transmission efficiency compared to an uninfected B. tabaci strain with the same genetic background. Temporal and spatial antagonistic relationships were discovered between Rickettsia and TYLCV within the whitefly. In different time course experiments, the levels of virus and Rickettsia within the insect were inversely correlated. Fluorescence in situ hybridization analysis of Rickettsia-infected midguts provided evidence for niche exclusion between Rickettsia and TYLCV. In particular, high levels of the bacterium in the midgut resulted in higher virus concentrations in the filter chamber, a favored site for virus translocation along the transmission pathway, whereas low levels of Rickettsia in the midgut resulted in an even distribution of the virus. Taken together, these results indicate that Rickettsia, by infecting the midgut, increases TYLCV transmission efficacy, adding further insights into the complex association between persistent plant viruses, their insect vectors, and microorganism tenants that reside within these insects.
IMPORTANCE Interest in bacterial endosymbionts in arthropods and many aspects of their host biology in agricultural and human health systems has been increasing. A recent and relevant studied example is the influence of Wolbachia on dengue virus transmission by mosquitoes. In parallel with our recently studied whitefly-Rickettsia-TYLCV system, other studies have shown that dengue virus levels in the mosquito vector are inversely correlated with bacterial load. Our work here presents evidence of unifying principles between vectors of plant and animal viruses in a role for endosymbionts in manipulating vector biology and pathogen transmission. Our results demonstrate the influence of an interesting and prominent bacterial endosymbiont in Bemisia tabaci in TYLCV transmission, a worldwide disease infecting tomatoes. Besides its agricultural importance, this system provides interesting insights into Bemisia interaction with these newly discovered endosymbionts.
INTRODUCTION
Tomato yellow leaf curl virus (TYLCV) is a monopartite single-stranded DNA (ssDNA) plant-infecting begomovirus in the family Geminiviridae. The whitefly vector, Bemisia tabaci, transmits TYLCV in a persistent, circulative manner. Circulative viruses movement throughout the body of the insect and interact with insect gut, salivary, and hemolymph tissues. The viral genome is packaged in a nonenveloped capsid, and the capsid is the sole vehicle for genome movement within the insect vector. Some data show that TYLCV possesses some level of vector pathogenicity (1), even though the majority of begomoviruses do not replicate in the vector and are not passed to offspring. TYLCV has been shown to be the exception to the latter rule (2) and has recently been shown to manipulate vector feeding behavior (3), which makes it a useful virus for investigating the complex interactions between the virus and the insect vector.
Circulative transmission of begomoviruses involves virion ingestion and internalization by the vector. Virions must be transported across gut cellular membranes, occupy a temporary residence in the hemolymph, and then enter the salivary system (4–6). Begomovirus transmission requires a minimal access period (AAP) of 1 to 3 h on the infected plant, a minimal latent period of 6 to 12 h during which the virus is not initially transmissible but becomes so once “acquired” (e.g., following salivary gland entry), and a minimal 30- to 60-min inoculation access period (IAP) on the host plant. Once acquired, virions are transmissible for the life of the whitefly (4, 7). The current model for the transmission pathway is based on the aphid-transmitted polero- and luteoviruses (luteovirids), in vivo transmission parameters, anatomical studies, and transmission electron microscopy localization (4–6, 8–10). In aphids and whiteflies, the gut membrane is thought to regulate transmission competency (8, 11), whereas the salivary tissues regulate the specificity of vector-virus interactions. In this model, virions are transported into the cytoplasm in vesicles that fuse with the basal plasma membrane, releasing particles between the membrane and the basal lamina (5, 6, 10, 12). The organ of specificity for luteovirids is the accessory salivary gland (13); however, labeled TYLCV and Squash leaf curl virus have been observed only in the primary salivary glands of B. tabaci (14).
Circulative plant virus transmission is a very specific and regulated process controlled by both insect and virus proteins (15–18), with the crucial participation of a third player—bacterial endosymbionts. Evidence points to the direct involvement of whitefly bacterial endosymbiont proteins in begomovirus-whitefly circulative transmission (4, 7). Like other phloem feeders, B. tabaci harbors a diverse array of endosymbionts, including the primary endosymbiont Portiera aleyrodidarum (19), and several other facultative secondary symbionts, including Rickettsia, Hamiltonella, Wolbachia, Arsenophonus, Cardinium, and Fritschea (19, 20). Nearly all secondary symbionts in B. tabaci colocalize with the primary endosymbiont inside the bacteriocytes, a specialized organ for housing bacterial endosymbionts, ensuring their vertical transmission. Several in vitro and in vivo assays have shown that a chaperone GroEL protein produced by Hamiltonella in B. tabaci B biotype interacts with the TYLCV coat protein (CP), while other GroELs produced by other secondary endosymbionts in both B and Q biotypes do not interact with the CP. The release of virions protected by GroEL occurs adjacent to the primary salivary glands (21). Only Rickettsia localizes outside the bacteriocytes, appears in most of the body cavity, and infects the majority of the internal organs, excluding the bacteriocytes (22, 23). Rickettsia infects B. tabaci's digestive, salivary, and reproductive organs and cells and reaches exceptionally high concentrations within the host (22). These recent findings raised the possibility that the unique localization patterns and infection of pivotal organs in the insect host may significantly influence the insect's biology. Indeed, one study has shown that Rickettsia-infected B. tabaci females, compared to noninfected insects with the same genetic background, exhibit increased fecundity, a greater rate of survival, and host reproduction manipulation via the production of a higher proportion of daughters (24), attributes that may also significantly impact virus transmission.
In the present study, we investigated the impact of Rickettsia spp. on B. tabaci-TYCLV interactions. The availability of sibling Rickettsia-infected and noninfected populations enabled us to appraise interactions between TYLCV, Rickettsia, and B. tabaci. We measured the effects of these putative tritrophic interactions on virus acquisition, retention, and transmission, as well as effects on TYLCV and Rickettsia distribution within the insect. By comparing these interactions using a virus host and a nonhost plant over time, we tested the dynamics of the triple interaction and explored how the virus and/or infected plant can affect the vector and thereby significantly impact virus plant-to-plant spread and, hence, disease epidemiology.
MATERIALS AND METHODS
Insects and plant growth conditions.
Rickettsia-free (Rick−) and Rickettsia-infected (Rick+) B-biotype B. tabaci populations were reared on cotton seedlings (Gossypium hirsutum L. cv. Acala) maintained inside insect-proof cages and growth rooms under standard conditions of 25°C ± 2°C, 60% relative humidity, and a 14-h light/10-h dark photoperiod. Viruliferous Rick− and Rick+ B biotype B. tabaci populations were reared on tomato seedlings, maintained inside insect-proof cages in growth rooms as previously described (25). A healthy tomato seedling 4 to 5 weeks of age was added once a month, while older plants were removed from the cage at different times.
Whitefly biotype identification.
Identification of the B biotype was based on microsatellite markers (25). The two strains of B. tabaci biotype B used in the present study were established by selection of isofemale strains as previously described (25). The original B biotype population used to establish the isofemale strains was collected from the Ayalon Valley in Israel (31°52′3″N, 34°57′34″E) in 1987 and cultured under laboratory conditions. This population tested positive for Portiera and Hamiltonella. The Rickettsia-positive and -negative strains used in the present study are referred to as Rick+ and Rick− strains, respectively.
Whitefly DNA extraction and PCR amplification.
Two methods of DNA isolation were used to confirm the presence of Rickettsia or TYLCV: one for single insects and one for pools of insects. To isolate DNA from single insects for PCR, 20 adult B. tabaci were individually homogenized in lysis buffer as previously described (22, 25). The lysate was then incubated at 65°C for 15 min, followed by incubation at 95°C for 10 min. DNA extraction from pools of whiteflies was done using an extraction buffer containing 10 mM Tris-HCl, 1.4 M NaCl, 2 mM EDTA, 2% CTAB (cetyltrimethylammonium bromide), and 0.2% β-mercaptoethanol. A total of 45 to 50 whiteflies were collected per sample, homogenized in the extraction buffer, and incubated at 37°C for 45 min. Samples were centrifuged for 55 min at 16,300 × g, and an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added. Samples were centrifuged again for 5 min at 16,300 × g, and the upper phase was transferred to a new tube. One volume of chloroform was added, followed by another centrifugation. The upper phase was collected, and nucleic acids were precipitated using 0.2 volumes of 5 M sodium chloride (NaCl) and 1 volume of isopropanol at 4°C overnight. After 12 h, the DNA was pelleted using centrifugation, and the impurities were removed from the pellets using 70% ethanol. DNA for PCR was dissolved in 40 μl of double-distilled H2O (ddH2O). DNA for real-time qPCR was dissolved in 30 μl of buffer that included 70 mM HEPES (pH 8.4) and 8 mM NaOH (pH 9).
The samples were tested for the presence of Rickettsia by PCR using two Rickettsia-specific primers for amplification of the 16S rRNA gene fragment: Rb-F (5′-GCTCAGAACGAACGCTATC-3′) and Rb-R (5′-GAAGGAAAGCATCTCTGC-3′) (25). PCRs (20 μl) were carried out using 2 μl of template DNA lysate, 20 pmol of each primer, 10 mM deoxynucleoside triphosphates, 1× DreamTaq buffer, and 1 U of DreamTaq DNA polymerase (Fermentas). PCR-amplified products were visualized on a 1% agarose gel containing ethidium bromide.
qPCR analysis.
To verify the exact concentrations of TYLCV and Rickettsia within the whiteflies, a quantitative PCR (qPCR) approach was used. The primer pair used for the amplification of TYLCV was previously described (4). The Rickettsia gene gltA was used for quantification with the following primers: forward, 5′-ATGACTAATGGCAATAATAA-3′; and reverse, 5′-CATAACCAGTGTAAAGCTG-3′. Actin forward primer 5′-TCTTCCAGCCATCCTTCTTG-3′, reverse primer 5′-CGGTGATTTCCTTCTGCATT-3′, and an expected product size of 81 bp were used with each sample as a normalization gene for verifying equal concentrations of whitefly genomic DNA. Amplifications were performed using 1× Absolute Blue qPCR SYBR green ROX Mix (Thermo Scientific) and 5 pmol of each primer. To ensure the reproducibility of the measurements and data, the expression of each gene was tested in triplicate in each of three biologically independent experiments. A biological replicate consisted of a pool of 45 to 50 whiteflies. The cycling conditions were as follows: 15 min of activation at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 58°C, and 30 s at 72°C. A melting ramp from 72 to 95°C was used, with a 1°C rise at each step and a 5-s wait between steps. The channel source was 470 nm, and the detector was set at 510 nm. A Rotor-Gene 6000 machine (Corbett Robotics Pty, Ltd., Brisbane, Australia) and the accompanying software were used for qPCR data normalization and quantification.
Plant DNA extraction and PCR amplification.
To confirm TYLCV infection, young leaves were collected from plants homogenized in buffer (100 mM Tris [pH 8], 50 mM EDTA [pH 8.5], 500 mM NaCl, 10 mM β-mercaptoethanol). A 40-μl portion of 20% sodium dodecyl sulfate (SDS) was added, and the mixture was then incubated at 65°C for 10 min. After incubation, 200 μl of 5 M potassium acetate was added, and the solution was placed in 4°C for at least 20 min. Samples were centrifuged at room temperature at 20,000 × g for 20 min. A total of 600 μl of the supernatant was removed, and another 480 μl of isopropanol was added. The solution was placed in −20°C for 30 min to precipitate the DNA. After 30 min, the samples were centrifuged for 15 min in room temperature at 15,600 × g. The supernatant was removed, and the tubes were washed with 98% ethanol. DNA was dissolved in 40 μl of ddH2O.
The samples were tested for the presence of TYLCV by PCR using the following TYLCV-specific primers to amplify the coat protein gene fragment: C473 (5′-AGTCACGGGCCCTTACA-3′) and V61 (5′-ATACTTGGACACCTAATGGC-3′) (4). The reaction was carried out in a volume of 50 μl containing 300 μg of template DNA, 20 pmol of each primer, 10 mM deoxynucleoside triphosphates, 1× DreamTaq buffer, and 1 U of DreamTaq DNA polymerase (Fermentas). PCR-amplified products were visualized following electrophoresis in a 1% agarose gel containing ethidium bromide.
TYLCV transmission experiments.
To calculate the TYLCV transmission efficiencies of the whitefly populations, Rick− and Rick+ B biotype B. tabaci whiteflies were provided a 48-h AAP on a TYLCV-infected tomato plant. Whiteflies were provided with a 7-day IAP on 4-week-old, healthy tomato plants. After an additional 2 weeks, young leaves were collected from the plants for DNA extraction and PCR for verification of the TYLCV infection. A total of 60 plants were tested for each strain.
FISH.
Fluorescence in situ hybridization (FISH) was performed as previously described (20). Briefly, specimens were fixed overnight in Carnoy's fixative (chloroform-ethanol-glacial acetic acid, 6:3:1 [vol/vol/vol]), decolorized in 6% H2O2 in ethanol for 2 h, washed in 100% ethanol, and hybridized overnight in hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% SDS, 30% formamide) containing 10 pmol of fluorescent probes per ml. Dissected midguts were fixed for 5 min in Carnoy's fixative and hybridized overnight. For specific targeting of Portiera and Rickettsia, the probes BTP1-Cy3 (5′-Cy3-TGTCAGTGTCAGCCCAGAAG-3′) and Rb1-Cy5 (5′-Cy5-TCCACGTCGCCGTCTTGC-3′), respectively, were used. For specific targeting of TYLCV, the probe Cy5-5′-GGAACATCAGGGCTTCGATA-3′, which is complementary to a sequence in TYLCV coat protein gene, was used. Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole) at 0.1 mg ml−1. The stained samples were mounted whole in hybridization buffer and viewed under an IX81 Olympus FluoView500 confocal microscope. Optical sections (0.7 to 1.0 μm thick) were prepared from each specimen. The specificity of detection was confirmed by using no-probe and Rickettsia-free whitefly controls.
Statistical analyses.
Student's t test was performed to compare the results from experiments using two treatments. In experiments with more than two treatments, a one-way analysis of variance, followed by a Tukey-Kramer honest significant difference (HSD) test, was performed. For all statistical analyses, JMP4 (SAS Institute, Inc.) was used. In all of the figures, different letters between treatments indicate significant differences between treatments (P < 0.05).
RESULTS AND DISCUSSION
Rickettsia-infected and noninfected B biotype isofemale strains differ in their TYLCV transmission efficacy.
Two isofemale strains, one Rickettsia infected (Rick+) and the other Rickettsia noninfected (Rick−), were established from an inbred B biotype strain (∼300 generations) as previously described (25) and were reared under laboratory conditions. Each individual from the infected strain contained Rickettsia, whereas none originating from the Rickettsia-free strain were infected. Both strains harbored Portiera, the primary endosymbiont, and the secondary endosymbiont, Hamiltonella (data not shown). To investigate whether the Rick+ and Rick− strains show differences in TYLCV transmission efficiency, three different independent transmission experiments of TYLCV to healthy tomato were conducted, using a single whitefly per plant. All insects were given a 48-h AAP on TYLCV-infected plants. Although the transmission experiments conducted with the Rick− strain resulted in 41% of the plantlets being infected with TYLCV, 79% of the plantlets were infected by the Rick+ strain (Fig. 1). These results suggest that the presence of Rickettsia, the only symbiont that differs between the two tested populations with the same genetic background, influences TYLCV transmission.
FIG 1.
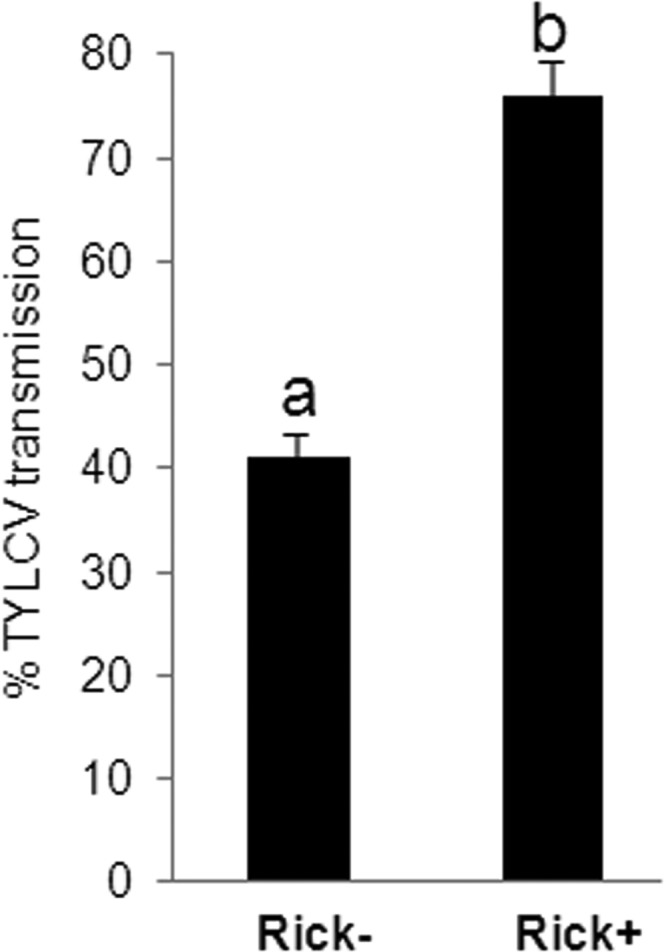
TYLCV transmission experiments by Rickettsia-infected and uninfected strains of the B biotype of B. tabaci. Rick+ and Rick− strains harbor Hamiltonella, an additional secondary endosymbiont, and Portiera, the primary endosymbiont of whiteflies. The Rick+ strain exhibits nearly twice the transmission efficiency as the Rick− strain, controlling for genetic background using inbred, isofemale lines. A total of 20 plants per transmission experiments were used. Altogether, 60 plants were examined for each population.
Previous studies have compared the transmission capabilities of B. tabaci populations; however, these have focused on differences between different biotypes, primarily the B and Q biotypes. The B and Q biotypes from Spain were shown to transmit TYLCV, and the Q biotype was found to be slightly more efficient than the B biotype (26). Experiments with different B biotype populations from Israel have shown high levels of TYLCV transmission with variations in the transmission ability of individual populations. Gottlieb et al. demonstrated the involvement of Hamiltonella, an additional secondary endosymbiont harbored only in the B biotype in Israel, in the transmission of TYLCV (21). Interestingly, contrary to the Q biotype from Israel, which does not harbor Hamiltonella, the Spanish Q population was found to harbor Hamiltonella. This later study showed that a GroEL protein produced by Hamiltonella is an important factor in facilitating TYLCV transmission (21). These populations were all infected with Rickettsia. The influence of Rickettsia in TYLCV transmission was not tested because the critical biological tools, e.g., isofemale lines that differentially harbor Rickettsia, were not available for these populations. Biotype B whitefly populations that harbor Rickettsia transmit TYLCV with a significantly higher efficiency (Fig. 1), and this supports the hypothesis that the presence of this secondary endosymbiont within the whitefly significantly influences the transmission of the virus.
Rickettsia-infected B. tabaci B biotype individuals acquire more TYLCV than noninfected individuals.
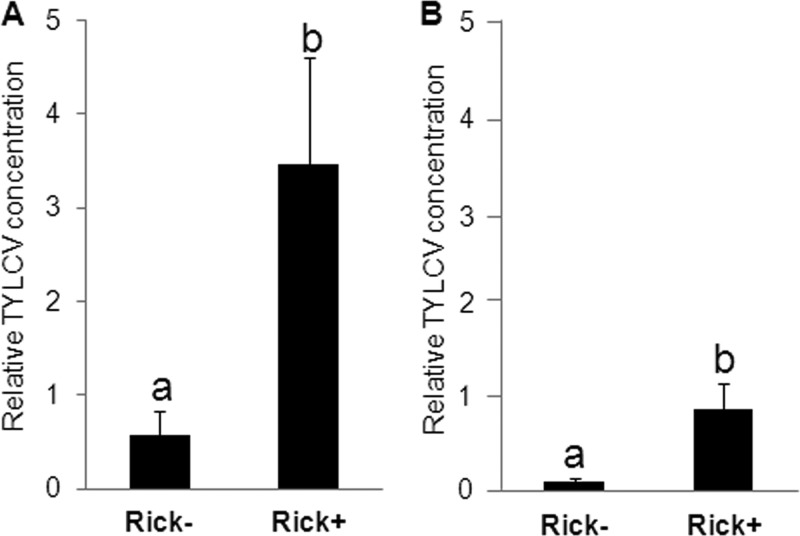
One hypothesis to explain the increase in the transmission efficiency in the Rick+ strain is that this strain acquires more TYLCV than does a Rick− strain. For the aphid-borne viruses in the family Luteoviridae, differences in virus acquisition among aphid biotypes and genotypes impact the transmission efficiency of the tested virus strain (27–30). To test this hypothesis, the amounts of TYLCV were measured, using a qPCR approach, in Rick+ and Rick− strains that were continuously reared on TYLCV-infected tomato plants. The results showed ∼6-fold significantly more TYLCV DNA in the Rick+ strain than in the Rick− strain (Fig. 2A). To further investigate whether the increased accumulation of TYLCV in Rick+ whiteflies occurred during the time frame of acquisition for circulative viruses, age-synchronized 4-day-old adult whiteflies from both strains were used in a TYLCV acquisition experiment for 48 h, after which the TYLCV amounts were quantified using the same qPCR approach. Approximately 8-fold more TYLCV was detected in the Rick+ strain, suggesting that the Rick+ strain acquires more TYLCV (Fig. 2B). If the presence of Rickettsia alters the ability of whiteflies to acquire TYLCV, these data show that changes in the insect are expressed within the time needed for efficient acquisition and transmission of circulative viruses and/or are sustained in the whitefly populations continuously exposed to infected plants.
FIG 2.
Quantification of acquisition of TYLCV in Rick+ and Rick− whitefly strains using a qPCR approach. The quantification was performed relative to actin, a housekeeping gene that did not change over time under these conditions. Whiteflies were collected from a colony continuously reared on TYLCV-infected tomato plants (A) or reared on TYLCV-infected tomato plants for 48 h (B).
Difference in TYLCV dynamics in Rickettsia-infected and noninfected strains.
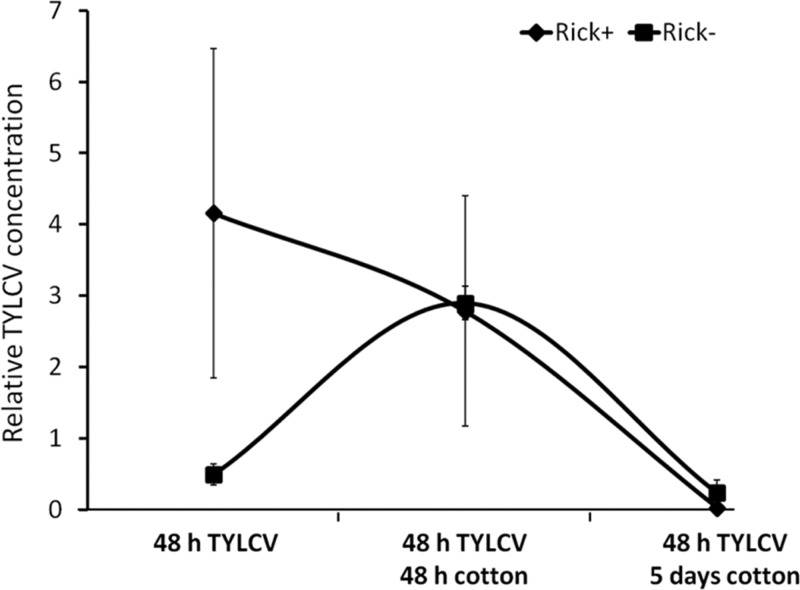
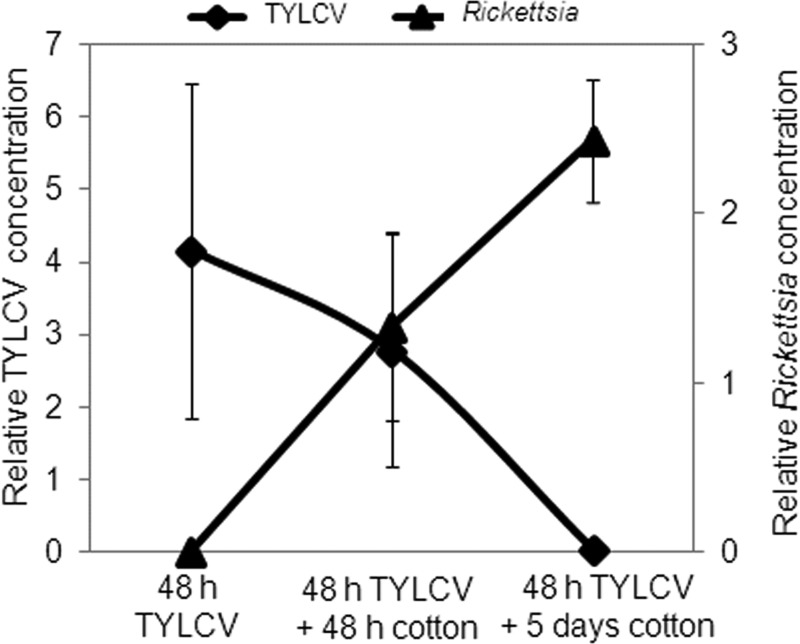
The results presented in Fig. 1 and 2 show that the Rick+ strain transmits TYLCV more efficiently than the Rick− strain and that the Rick+ strain acquired more virus than did Rick− insects. Together, these data may indicate that increased TYLCV acquisition in the Rick+ populations may be responsible for the higher transmission efficiency. However, an alternative hypothesis could be that the virus is retained longer in the Rick+ strain. An enhanced retention ability in the whitefly vector may result in higher titers of viable virus particles available for transmission. To test this hypothesis, synchronized adult whiteflies aged up to 4 days from both the Rick+ and the Rick− strains were provided with a 48-h AAP on TYLCV-infected tomato plants and then transferred to cotton, a host for B. tabaci and a nonhost for TYLCV, for various time periods. Transferring insects to a nonhost plant for the virus would enable us to measure viral DNA acquired during the AAP. Since TYLCV is thought to be nonpropagative, enhanced virus retention can be measured after transfer of whiteflies to a nonhost plant of the virus. Insects were divided into three cohorts: one collected off the plants (referred to as 48 h on TYLCV-infected tomato plants), the second transferred to cotton and collected after an additional 48 h on cotton (referred to as 48 h on TYLCV and 48 h on cotton), and the third transferred to cotton and collected after 5 days on cotton (referred to as 48 h on TYLCV and 5 days on cotton). Each cohort from the two strains was used to quantify TYLCV using qPCR. The Rick+ strain contained on average ∼8-fold significantly more TYLCV than the Rick− strain after the initial AAP. Strikingly, after 48 h on cotton, the amounts of TYLCV in the Rick− strain increased by ∼6-fold relative to the level immediately after the 48-h AAP on TYLCV-infected tomato (Fig. 3). In contrast, the virus amounts gradually decreased over time in the Rick+ strain. After 5 days on cotton, the virus levels in both strains were at the lower limit of detection (Fig. 3).
FIG 3.
Quantification of TYLCV retention in Rick+ and Rick− strains using qPCR. A Whitefly population was reared on TYLCV-infected tomato plants for a 48-h AAP, and the virus concentration was quantified in one-third of the whiteflies. Two-thirds of the remaining insects were used for additional quantification of the virus after an additional 48 h and 5 days on cotton plants.
Collectively, the results shown in Fig. 1 to 3 suggest significant relationships between the presence of TYLCV and Rickettsia in the B. tabaci B biotype. One hypothesis to explain these results is directly related to differences in feeding behaviors between Rick+ and Rick− strains. The larger amounts of TYLCV in the Rick+ strain might be a result of differential acquisition of phloem sap, in which the virus is located. To test this hypothesis, additional experiments using an electrical penetrating graph technique, in which the general feeding behaviors can be directly measured, may confirm or rebut this hypothesis (31). However, an alternative and non-mutually exclusive hypothesis involves more complex, direct or indirect, interactions between Rickettsia and the virus. The difference in TYLCV amounts between Rick+ and Rick− strains after transfer to a nonhost plant (Fig. 3) offers hints on virus-bacterium or tritrophic virus-bacterium-whitefly interactions. The rise in the virus levels in the Rick− strain indicates some replication of the virus in the absence of Rickettsia that is rapidly shut down, resulting in undetectable virus after 5 days on a TYLCV nonhost plant (Fig. 3). If the replication occurs in the Rick− strain, the bacterium in the Rick+ strain might have a role in directly or indirectly suppressing replication.
Rickettsia has been shown to influence several biological characteristics of the whitefly (20, 22–25, 32, 33). These studies suggest that this bacterium may constitutively activate the expression of stress- and immunity-related genes. The activation of such genes can be advantageous for the whitefly under stress conditions in a way that makes the insect become prepared for stress conditions, as previously shown for whitefly resistance to heat stress upon infection with Rickettsia (25). The presence of TYLCV in the whitefly has been shown to induce negative effects in the whitefly, such as reducing its fecundity and fertility (34) and altering its general gene expression (35). Such negative effects may induce stress-related genes that in turn suppress or alter the initial replication of TYLCV in the Rick+ strain.
Dynamics of Rickettsia, Hamiltonella, and Portiera levels upon TYLCV acquisition and retention.
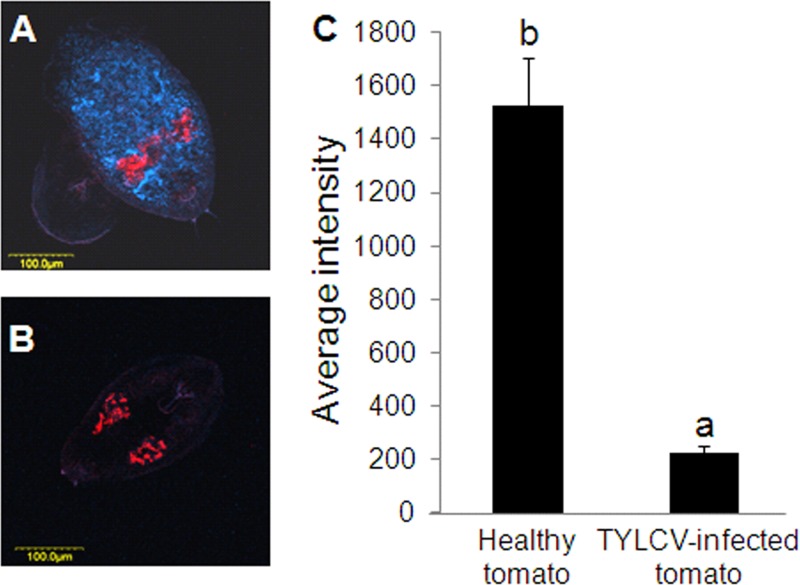
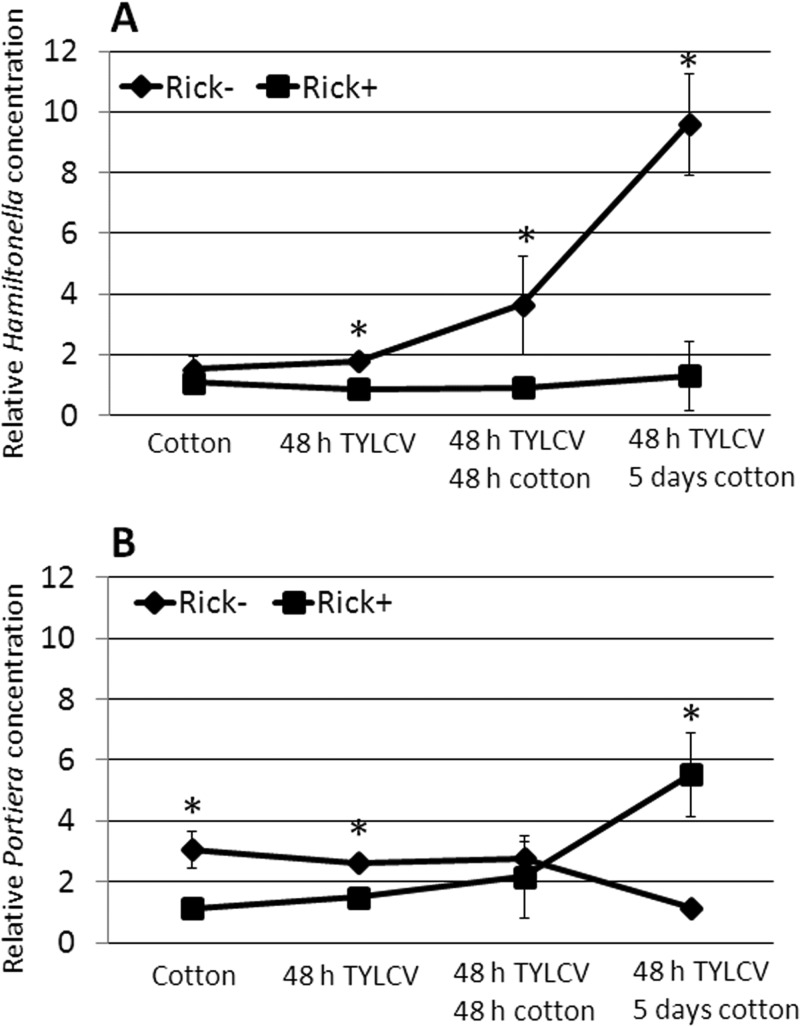
Our data show that TYLCV levels and transmission by whiteflies are influenced by Rickettsia infection and that the phenotype may be partially mediated by insect proteins, such as HSP70 and other stress-related proteins, that respond to the presence of the virus at the molecular level. The high levels of the virus in the Rick+ strain may be due to better feeding; however, they may also be influenced by the amounts of Rickettsia or possibly other endosymbionts in the Rick+ strain. Bacterial load within whiteflies can change upon feeding on infected plants or upon the virus presence. Large amounts of virus may compete with Rickettsia and other bacterial endosymbionts for access to whitefly resources or introduce additional indirect effects, which may in turn influence Rickettsia levels. To test this hypothesis, FISH analysis on B. tabaci B biotype nymphs that were reread on TYLCV-infected tomato plants was performed and compared to nymphs reared on uninfected plants. Although the levels of Rickettsia were exceptionally high in nymphs reared on healthy tomato plants (Fig. 4A), the levels of the bacterium in nymphs that were reared on TYLCV-infected tomato plants were low (Fig. 4B). The fluorescence intensity in both treatments showed highly significant differences (Fig. 4C). Interestingly, the levels of Hamiltonella and Portiera, the primary endosymbiont, fluctuated across the different time points after TYLCV acquisition and retention (Fig. 5). Hamiltonella, a bacterial endosymbiont previously implicated in TYLCV transmission (17), did not change across the different time points in Rick+ strain; however, it gradually increased in the Rick− strain (Fig. 5A). Expression of the Hamiltonella GroEL protein was previously implicated in TYLCV transmission via a direct physical interaction with TYLCV virions. One hypothesis is that Hamiltonella GroEL is functioning differentially in Rick+ and Rick− strains. The levels of Portiera slightly fluctuated across the different time points in the two strains; however, they did not significantly differ by 48 h after TYLCV acquisition (Fig. 5B), suggesting that Portiera is likely not to play a role in the different transmission abilities of the two strains. A previous study further demonstrated that Portiera GroEL, unlike the one encoded by Hamiltonella, did not interact with TYLCV (17).
FIG 4.
Fluorescence in situ hybridization (FISH) on whitefly nymphs infected with Rickettsia reared on healthy tomato plants (A) or on TYLCV-infected tomato plants (B). (C) Fluorescence quantification of 10 Rick+ nymphs reared on infected plants and 10 Rick+ nymphs reared on healthy plants using MICA. The results are plotted as the average intensity. Colors (A and B): red, Portiera; blue, Rickettsia.
FIG 5.
Change in the levels of Hamiltonella (A) and Portiera (B) in B. tabaci adults, as quantified by qPCR across different treatments. Adult whiteflies were reared on cotton, and the levels of Hamiltonella and Portiera were measured (cotton on the x axis). Part of the whiteflies were reared on cotton and were then transferred to tomato plants infected with TYLCV for 48 h (48 h TYLCV on the x axis). After 48 h on TYLCV-infected tomato plants, adult whiteflies were transferred to cotton for additional 48 h, and the levels of the two bacteria were then quantified (48 h TYLCV and 48 h cotton on the x axis). The levels of the two bacteria were also measured in whiteflies that were kept on TYLCV-infected tomato plants for 48 h and were then transferred to cotton for 5 days. Asterisks indicate statistically significant differences in the levels of the bacterium tested between the two strains.
The results presented in Fig. 3, 4, and 5 further demonstrate the influence of TYLCV or feeding on TYLCV-infected plants on the levels of Rickettsia in B. tabaci. An obvious role for other endosymbionts is not readily apparent, although the levels of Hamiltonella do increase in the Rick− strain after TYLCV acquisition. To test for further relationships between Rickettsia and TYLCV, virus levels were quantified in the same whitefly samples collected for Rickettsia quantification shown in Fig. 3. There was a significant negative correlation between Rickettsia and TYLCV (Fig. 6): virus levels are at their maximum when Rickettsia is at almost zero levels and vice versa. This result strongly suggests direct or indirect competition between the virus and the bacterium inside the whitefly.
FIG 6.
Relationships between levels of Rickettsia and TYLCV in Rick+ B. tabaci adults after 48-h AAP on TYLCV-infected tomato plants at the first time point, after 48 h on TYLCV-infected tomato plants and after an additional 48 h on cotton at the second time point, and after 48 h on TYLCV-infected tomato plants and an additional 5 days on cotton at the third time point. The levels of TYLCV and Rickettsia were measured using a qPCR approach as detailed in Materials and Methods.
Evidence for in vivo exclusion between Rickettsia and TYLCV.
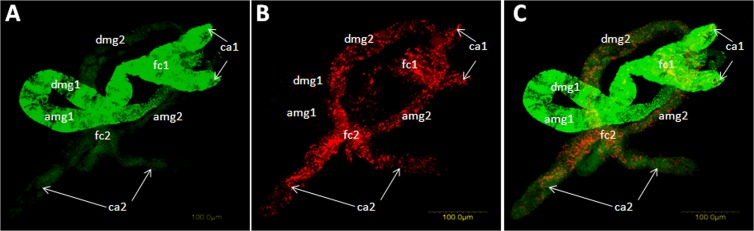
Collectively, the results support the hypothesis that significant direct or indirect, relationships between Rickettsia and TYLCV occur inside the whitefly. The fact that a negative correlation in Rickettsia and TYLCV amounts was observed (Fig. 6) supports the virus transmission experiments, which showed a higher ability of the Rick+ strain to transmit TYLCV (Fig. 1). These data suggest that infection with Rickettsia results in faster clearance of the virus, minimum contact with the insect tissue, and higher transmission ability. Rickettsia influences the biology of the whitefly, including altering stress- and immunity-related genes, such as the HSP70 gene (17, 25), whereas the virus was shown to negatively impact the whitefly biology (34, 35). To test further this hypothesis, double FISH analysis that targeted both Rickettsia and TYLCV was performed on dissected midguts from Rick+ whiteflies. Generally, midguts highly infected with Rickettsia contained lower levels of TYLCV, whereas midguts infected with lower levels of the bacterium contained much higher levels of the virus (Fig. 7). Interestingly, and upon close inspection, TYLCV in highly infected Rickettsia was located mainly in the filter chamber, the site in the midgut where significant amounts of TYLCV are absorbed into the hemolymph on the way to be transmitted (36, 37) (Fig. 7B). In comparison, midguts infected with low levels of Rickettsia showed an even distribution of the virus (Fig. 7B). As controls, midguts from the Rick− strain were processed using the same FISH analysis, and the results generally showed that TYLCV was evenly distributed, as seen in midgut 2 in Fig. 7. These results may explain the transmission results in which the Rick+ strain showed a greater ability to transmit TYLCV. The data suggest that infection with Rickettsia modifies the distribution of the virus, preferentially translocating virus to the filter chamber. Higher virus titers and preferred localization to the filter chamber may speed absorption into the hemolymph and along the transmission route, thus leading to greater transmission of the virus to tomato plants. Consequently, the virus levels in a midgut highly infected with Rickettsia are generally lower compared to a Rickettsia-free midgut (Fig. 7B).
FIG 7.
Representative double FISH of TYLCV (red) and Rickettsia (green) in B. tabaci midguts dissected from Rick+ female whiteflies collected from TYLCV-infected tomatoes. (A) Two midguts, midgut 1 and midgut 2, are shown with only the green channel for Rickettsia. (B) Same two midguts as in panel A but shown with the red channed for TYLCV. (C) Overlay of panels A and B showing the two channels for Rickettsia and TYLCV. Abbreviations: amg, ascending midgut; dmg, descending midgut; fc, filter chamber; ca, ceca. At least 20 midguts were analyzed using this double FISH analysis method.
Rickettsia levels are dynamic in TYLCV-infected plants.
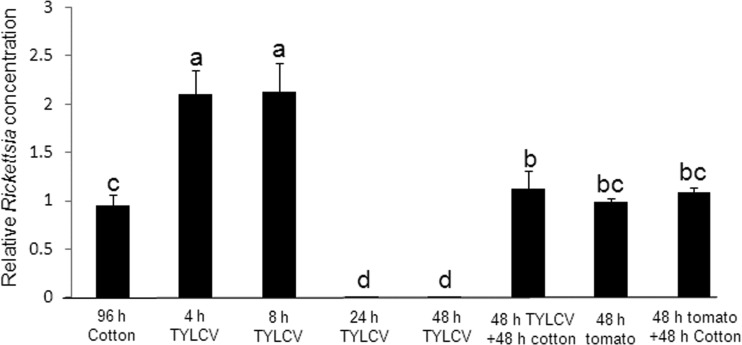
To further investigate the temporal relationship between Rickettsia and TYLCV, the levels of Rickettsia in response to the acquisition and retention of TYLCV in B. tabaci were investigated in a time course series. The levels of Rickettsia in 4-day-old adults were measured before being collected from cotton and then were measured in cohorts 4, 8, 24, and 48 h after they had been transferred and kept on TYLCV-infected tomato plants. The levels of the bacterium doubled after 4 and 8 h on infected plants and then decreased to zero levels after 24 and 48 h (Fig. 8). After 48 h on TYLCV, whiteflies were transferred back to cotton for 48 h, and the levels of the bacterium increased again and were comparable to those observed initially on cotton (Fig. 8). These levels were not significantly different from the levels of the bacterium in whiteflies that were kept for 48 h on healthy tomato plants or in whiteflies that were kept on healthy tomato plants and then were transferred to cotton for an additional 48 h (Fig. 8). These results confirm the qPCR and FISH results presented above and confirm the rapid response of Rickettsia to the presence of TYLCV.
FIG 8.
Change in the levels of Rickettsia in Rick+ B. tabaci adults that were reared for 96 h on cotton (most left column). Rickettsia levels were then measured in cohorts that were transferred to TYLCV-infected tomato plants for 4, 8, 24, and 48 h. The Rickettsia levels were also measured in additional cohorts that were reared for 48 h on healthy tomato plants and then transferred to cotton. Quantifications were performed using a qPCR approach as detailed in Materials and Methods.
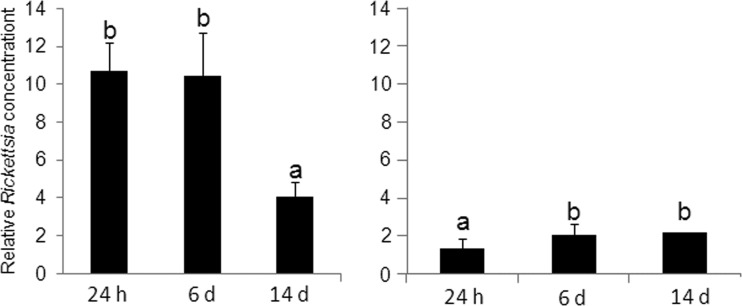
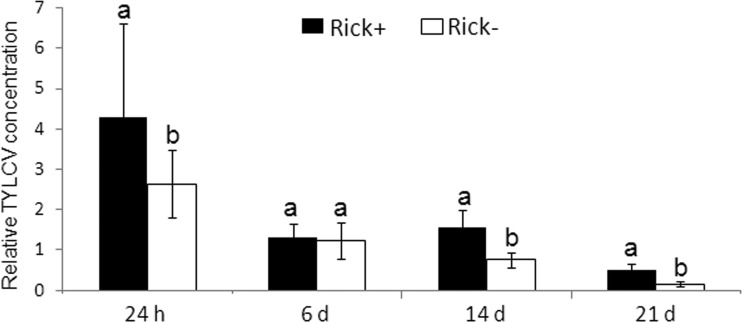
The results suggest that the interaction between Rickettsia and TYLCV might be mediated, in part, by the infected plant. To further test whether these interactions are dynamic and whether the levels of Rickettsia respond to the change in the virus titers in the infected plant, the levels of the bacterium were measured in adult whiteflies reared on newly TYLCV-infected tomato plants, in which the virus titer gradually increased postinfection. The progress of infection was accompanied by a decrease in Rickettsia levels (Fig. 9, left panel). In comparison, the levels of Rickettsia increased in adults that were reared continuously on TYLCV-infected plants and were then transferred to cotton upon emergence as adults for up to 14 days (Fig. 9, right panel). In these adults, the levels of the virus decreased, although to a slightly lower extent in Rick+ whiteflies compared to whiteflies from the Rick− strain, as seen from the increased retention of virus at the 14- and 21-day time points (Fig. 10).
FIG 9.
Rickettsia levels in Rick+ adult whiteflies after different periods on tomato plants newly infected with TYLCV (left panel) during a 14-day time period. The right panel shows the increase in Rickettsia levels in viruliferous adult whiteflies reared on cotton for 14 days.
FIG 10.
TYLCV concentration in TYLCV-infected Rick+ and Rick− adult whiteflies after different periods on cotton. TYLCV-infected whiteflies (from populations grown on TYLCV tomatoes) were collected and placed on cotton. The virus concentrations were determined at different time points using qPCR. A total of 135 adult whiteflies from each population were used for each time point.
In summary, the results presented here support the hypothesis that infection with Rickettsia spp., a facultative endosymbiont of B. tabaci, is instrumental in altering the insect interaction with TYLCV, an ssDNA virus transmitted by B. tabaci. The results specifically show that Rickettsia-infected whiteflies acquire more TYLCV, retain the virus longer, and thus exhibit nearly twice the transmission efficiency of a Rickettsia-uninfected strain, which originated from the same genetic background. The results further show that some level of antagonistic relationships exists within the whitefly between Rickettsia and TYLCV. Competition between Rickettsia and the virus is indicated by data showing that they become spatially segregated in midguts when both are present and that Rickettsia levels plummet during virus acquisition, suggesting that both the virus and the Rickettsia are competing for host molecular machinery. These results offer some insights into the involvement of an additional facultative bacterial endosymbiont in the ability of B. tabaci to serve as a vector for circulative plant viruses. The results presented here enable us to draw further parallels between vectors of plant and animal viruses. The best example is the endosymbiont Wolbachia, which is common among insects and, like Rickettsia in whiteflies, manipulates reproductive fitness and insect host physiology. Recent studies have demonstrated that through maternal inheritance and reproductive incompatibility, a Wolbachia strain introduced from Drosophila melanogaster (wMel) invaded natural populations of the mosquito Aedes aegypti in Australia and reached near fixation within a few months (38). This is important because Wolbachia has been shown to prime the mosquito innate immune system, thus limiting the ability of pathogens such as dengue virus, Chikungunya virus, and Plasmodium to replicate in mosquito cells (39, 40). Limiting the replication of dengue virus in the mosquito vector was shown to be possible using host microRNAs that manipulate host gene expression (41). Additional studies have shown that upregulation of the innate immunity system in the mosquito by Wolbachia inhibits the development of filarial nematodes (42). A more significant effect of Wolbachia on its mosquito insect host was shown to be the significant shortening of its life span by half, possibly as a consequence of the continued activation of the immune system (43). In this context, our results obtained with Rickettsia, which exhibits significant effects on B. tabaci and may activate responses similar to those activated in mosquitoes by Wolbachia, suggest the value of reexamining the unprecedented role of this endosymbiont and possibly other endosymbionts in shaping the biology of whiteflies in general and their role in maintaining viable interactions with circulative plant viruses. Future studies of the interplay between Rickettsia and its whitefly host may lead to the identification of genes that maintain these interactions. Targeting such genes could influence these symbiotic interactions, which may lead to significant effects on the whitefly host.
ACKNOWLEDGMENTS
We gratefully acknowledge members of the Cilia lab for helpful discussions and Eduard Belausov for technical assistance with the confocal microscope.
Footnotes
Published ahead of print 5 March 2014
REFERENCES
- 1.Czosnek H, Ghanim M. 2012. Back to basics: are begomoviruses whitefly pathogens? J. Integrative Agric. 11:225–234. 10.1016/S2095-3119(12)60007-0 [DOI] [Google Scholar]
- 2.Ghanim M, Morin S, Zeidan M, Czosnek H. 1998. Evidence for transovarial transmission of tomato yellow leaf curl virus by its vector, the whitefly Bemisia tabaci. Virology 240:295–303. 10.1006/viro.1997.8937 [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Preisser EL, Chu D, Pan H, Xie W, Wang S, Wu Q, Zhou X, Zhang Y. 2013. Multiple forms of vector manipulation by a plant-infecting virus: Bemisia tabaci and Tomato yellow leaf curl virus. J. Virol. 87:4929–4937. 10.1128/JVI.03571-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghanim M, Morin S, Czosnek H. 2001. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188–196. 10.1094/PHYTO.2001.91.2.188 [DOI] [PubMed] [Google Scholar]
- 5.Cicero JM, Brown JK. 2011. Functional anatomy of whitefly organs associated with Squash leaf curl virus (Geminiviridae: Begomovirus) transmission by the B biotype of Bemisia tabaci (Aleyrodidae: Hemiptera). Ann. Entomol. Soc. Am. 104:261–279. 10.1603/AN10075 [DOI] [Google Scholar]
- 6.Cicero JM, Brown JK. 2011. The anatomy of the accessory salivary glands of the whitefly Bemisia tabaci (Aleyrodidae: Hemiptera), and correlations to begomovirus transmission. Ann. Entomol. Soc. Am. 104:280–286. 10.1603/AN10171 [DOI] [Google Scholar]
- 7.Brown JK, Czosnek H. 2002. Whitefly- transmitted viruses, p 65–100 In Advances in botanical research. Academic Press, Inc, New York, NY [Google Scholar]
- 8.Czosnek H, Ghanim M. 2002. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci: insights from studies with Tomato yellow leaf curl virus. Ann. Appl. Biol. 140:215–231. 10.1111/j.1744-7348.2002.tb00175.x [DOI] [Google Scholar]
- 9.Hunter WB, Hiebert E, Webb SE, Tsai JK, JEP 1998. Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae). Plant Dis. 82:1147–1151. 10.1094/PDIS.1998.82.10.1147 [DOI] [PubMed] [Google Scholar]
- 10.Cicero JM, Hiebert E, Webb SE. 1995. The alimentary canal of Bemisia tabaci and Trialeurodes abutilonea (Homoptera, Sternorrhyncha): histology, ultrastructure, and correlations to function. Zoomorphology 115:31–39. 10.1007/BF00397932 [DOI] [Google Scholar]
- 11.Burrows ME, Caillaud MC, Smith DM, Benson EC, Gildow FE, Gray SM. 2006. Genetic regulation of polerovirus and luteovirus transmission in the aphid Schizaphis graminum. Phytopathology 96:828–837. 10.1094/PHYTO-96-0828 [DOI] [PubMed] [Google Scholar]
- 12.Ghanim M, Rosell RC, Campbell LR, Czosnek H, Brown JK, Ullman DE. 2001. Digestive, salivary, and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) B type. J. Morphol. 248:22–40. 10.1002/jmor.1018 [DOI] [PubMed] [Google Scholar]
- 13.Gildow FE, Rochow WF. 1980. Role of accessory salivary glands in aphid transmission of Barley yellow dwarf virus. Virology 104:97–108. 10.1016/0042-6822(80)90368-2 [DOI] [PubMed] [Google Scholar]
- 14.Caciagli P, Medina Piles V, Marian D, Vecchiati M, Masenga V, Mason G, Falcioni T, Noris E. 2009. Virion stability is important for the circulative transmission of Tomato yellow leaf curl Sardinia virus by Bemisia tabaci, but virion access to salivary glands does not guarantee transmissibility. J. Virol. 83:5784–5795. 10.1128/JVI.02267-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavez JD, Cilia M, Weisbrod CR, Ju HJ, Eng JK, Gray SM, Bruce JE. 2012. Cross-linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus-plant interactions. J. Proteome Res. 11:2968–2981. 10.1021/pr300041t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cilia M, Tamborindeguy C, Fish T, Howe K, Thannhauser TW, Gray S. 2011. Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J. Virol. 85:2148–2166. 10.1128/JVI.01504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götz M, Popovski S, Kollenberg M, Gorovitz R, Brown JK, Cicero J, Czosnek H, Winter S, Ghanim M. 2012. Implication of Bemisia tabaci heat shock protein 70 in begomovirus-whitefly interactions. J. Virol. 86:13241–13252. 10.1128/JVI.00880-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seddas P, Boissinot S, Strub JM, Van Dorsselaer A, Van Regenmortel MH, Pattus F. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325:399–412. 10.1016/j.virol.2004.05.014 [DOI] [PubMed] [Google Scholar]
- 19.Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189. 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, Tzuri G, Horowitz AR, Belausov E, Mozes-Daube N, Kontsedalov S, Gershon M, Gal S, Katzir N, Zchori-Fein E. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646–3652. 10.1128/AEM.72.5.3646-3652.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Kontsedalov S, Skaljac M, Brumin M, Sobol I, Czosnek H, Vavre F, Fleury F, Ghanim M. 2010. The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 84:9310–9317. 10.1128/JVI.00423-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brumin M, Levy M, Ghanim M. 2012. Transovarial transmission of Rickettsia spp. and organ-specific infection of the whitefly Bemisia tabaci. Appl. Environ. Microbiol. 78:5565–5574. 10.1128/AEM.01184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E. 2008. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 22:2591–2599. 10.1096/fj.07-101162 [DOI] [PubMed] [Google Scholar]
- 24.Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, Tabashnik BE, Chiel E, Duckworth VE, Dennehy TJ, Zchori-Fein E, Hunter MS. 2011. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332:254–256. 10.1126/science.1199410 [DOI] [PubMed] [Google Scholar]
- 25.Brumin M, Kontsedalov S, Ghanim M. 2011. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 18:57–66. 10.1111/j.1744-7917.2010.01396.x [DOI] [Google Scholar]
- 26.Sánchez-Campos S, Navas-Castillo J, Camero R, Soria C, Diaz J, Moriones E. 1999. Displacement of tomato yellow leaf curl virus (TYLCV)-Sr by TYLCV-Is in tomato epidemics in Spain. Phytopathology 89:1038–1043. 10.1094/PHYTO.1999.89.11.1038 [DOI] [PubMed] [Google Scholar]
- 27.Gray SM, Power AG, Smith DM, Seaman AJ, Altman NS. 1991. Aphid transmission of barley yellow dwarf virus: acquisition access periods and virus concentration requirements. Phytopathology 81:539–545. 10.1094/Phyto-81-539 [DOI] [Google Scholar]
- 28.Chay CA, Gunasinge UB, Dinesh-Kumar SP, Miller WA, Gray SM. 1996. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219:57–65. 10.1006/viro.1996.0222 [DOI] [PubMed] [Google Scholar]
- 29.Lucio-Zavaleta E, Smith DM, Gray SM. 2001. Variation in transmission efficiency among barley yellow dwarf virus-RMV isolates and clones of the normally inefficient aphid vector, Rhopalosiphum padi. Phytopathology 91:792–796. 10.1094/PHYTO.2001.91.8.792 [DOI] [PubMed] [Google Scholar]
- 30.Lee L, Palukaitis P, Gray SM. 2002. Host-dependent requirement for the Potato leafroll virus 17-kDa protein in virus movement. Mol. Plant-Microbe Interact. 15:1086–1094. 10.1094/MPMI.2002.15.10.1086 [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Walker G. 2003. Electrical penetration graphs of the nymphal stage of Bemisia argentifolii. Entomol. Exp. Appl. 109:101–111. 10.1046/j.1570-7458.2003.00093.x [DOI] [Google Scholar]
- 32.Kontsedalov S, Zchori-Fein E, Chiel E, Gottlieb Y, Inbar M, Ghanim M. 2008. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest. Manag. Sci. 64:789–792. 10.1002/ps.1595 [DOI] [PubMed] [Google Scholar]
- 33.Skaljac M, Zanic K, Ban SG, Kontsedalov S, Ghanim M. 2010. Coinfection and localization of secondary symbionts in two whitefly species. BMC Microbiol. 10:142. 10.1186/1471-2180-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinstein G, Czosnek H. 1997. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity, and fecundity. J. Gen. Virol. 78(Pt 10):2683–2689 [DOI] [PubMed] [Google Scholar]
- 35.Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, Zhang CX, Liu SS, Wang XW. 2011. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 85:3330–3340. 10.1128/JVI.02507-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghanim M, Brumin M, Popovski S. 2009. A simple, rapid and inexpensive method for localization of Tomato yellow leaf curl virus and Potato leafroll virus in plant and insect vectors. J. Virol. Methods 159:311–314. 10.1016/j.jviromet.2009.04.017 [DOI] [PubMed] [Google Scholar]
- 37.Skaljac M, Ghanim M. 2010. Coinfection and localization of secondary symbionts in two whitefly species. Isr. J. Plant Sci. 58:103–111. 10.1560/IJPS.58.2.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, Greenfield M, Durkan M, Leong YS, Dong Y, Cook H, Axford J, Callahan AG, Kenny N, Omodei C, McGraw EA, Ryan PA, Ritchie SA, Turelli M, O'Neill SL. 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476:454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- 39.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139:1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- 40.Rances E, Ye YH, Woolfit M, McGraw EA, O'Neill SL. 2012. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 8:e1002548. 10.1371/journal.ppat.1002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang G, Hussain M, O'Neill SL, Asgari S. 2013. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to gangue virus inhibition in Aedes aegypti Proc. Natl. Acad. Sci. U. S. A. 110:10276–10281. 10.1073/pnas.1303603110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kambris Z, Cook PE, Phuc HK, Sinkins SP. 2009. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326:134–136. 10.1126/science.1177531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh E, Mercer DR, Fu Y, Dobson SL. 2009. Pathogenicity of life-shortening Wolbachia in Aedes albopictus after transfer from Drosophila melanogaster. Appl. Environ. Microbiol. 75:7783–7788. 10.1128/AEM.01331-09 [DOI] [PMC free article] [PubMed] [Google Scholar]