ABSTRACT
A natural subviral agent of human hepatitis B virus (HBV), hepatitis delta virus (HDV), requires only the envelope proteins from HBV in order to maintain persistent infection. HBV surface antigens (HBsAgs) can be produced either by HBV replication or from integrated HBV DNA regardless of replication. The functional properties of the integrant-generated HBsAgs were examined using two human hepatocellular carcinoma-derived cell lines, Hep3B and PLC/PRF/5, that contain HBV integrants but do not produce HBV virions and have no signs of HBV replication. Both cell lines were able to support HDV replication and assembly/egress of HDV virions. Neither of the cell lines was able to produce substantial amounts of the pre-S1-containing HDV particles. HDV virions assembled in PLC/PRF/5 cells were able to infect primary human hepatocytes, while Hep3B-derived HDV appeared to be noninfectious. These results correlate with the findings that the entire open reading frame (ORF) for the large (L) envelope protein that is essential for infectivity is present on HBV RNAs from PLC/PRF/5 cells, while an L protein ORF that was truncated and fused to inverted precore sequences was found using RNAs from Hep3B cells. This study demonstrates for the first time that at least some of the HBV DNA sequence naturally integrated during infection can produce functional small and large envelope proteins capable of the formation of infectious HDV virions. Our data indicate that in vivo chronic HDV infection can persist in the absence of HBV replication (or when HBV replication is profoundly suppressed) if functional envelope proteins are supplied from HBV integrants.
IMPORTANCE The study addresses the unique mechanism of HDV persistence in the absence of ongoing HBV replication, advances our understanding of HDV-HBV interactions, and supports the implementation of treatments directly targeting HDV for HDV/HBV-infected individuals.
INTRODUCTION
Hepatitis delta virus (HDV) is a significant human pathogen, with approximately 20 million people worldwide being chronic carriers. HDV is a natural subviral agent of human hepatitis B virus (HBV) that requires from its helper hepadnavirus only the envelope proteins in order to form virions and infect hepatocytes via the HBV receptor. In infected livers, HDV coexists with HBV. Chronic HBV infection remains a main risk factor for hepatocellular carcinoma (HCC) and is associated with more than half of all HCC cases (1–4). Concomitant HDV infection is able to inflict additional liver damage associated with accelerated liver disease, cirrhosis, liver failure, and HCC (5–11). Treatment with alpha interferon is beneficial for only a subset of HDV carriers. There are no treatments in clinical practice that directly target HDV, and practically none of the anti-HBV drugs blocks HDV infection (5, 12, 13). In livers chronically infected with HBV, as many as 90% of hepatocytes may appear to be free of HBV replication markers. HBV-infected individuals can support HDV infection regardless of the presence of HBV replication markers. Often, HDV/HBV carriers exhibit relatively high HDV levels, while HBV levels remain low or undetectable. In the HDV/HBV-infected human liver, a subpopulation of hepatocytes was positive only for HBV, a second subpopulation was positive only for HDV, and another subpopulation was positive for both viruses. Thus, a number of HDV-infected hepatocytes apparently may not display markers of HBV replication. In addition, HDV is able to suppress HBV replication (2, 14–24). The observations presented above indicate that HDV and HBV, while being present in the same liver, may not necessarily be present in the same cell. Furthermore, the data indicate that (i) HDV may not depend on ongoing HBV replication and (ii) chronic HDV infection may actually persist in the absence of HBV replication, if another alternative source of HBV envelope proteins is available.
The envelope proteins can be produced from integrated HBV DNA independently of hepadnavirus replication. During chronic HBV infection, a significant number of hepatocytes contain HBV DNA integrants. Normal hepatocytes that are apparently free of HBV replication markers but still express HBV envelope proteins can appear as a result of resolved HBV infection or via immune-mediated selection. In addition, HCC cells may apparently no longer support HBV replication but can still support the production of envelope proteins from HBV integrants (3, 14–16, 25–27). In this study, we examined the hypothesis that HDV can persist in the absence of HBV replication (or when HBV replication is profoundly suppressed) if functional envelope proteins are supplied from naturally integrated HBV DNA. Such a mechanism of HDV persistence has not been explored previously. A potential obstacle here is that deletions, insertions, and rearrangements are often observed in integrated HBV DNA (28–33). In addition, the functional properties of integrant-derived HBV surface antigens (HBsAgs) remain to be fully evaluated (16, 29–31, 34, 35).
We examined two human cell lines derived from HBV-induced HCCs, Hep3B and PLC/PRF/5 (Alexander cells), that bear HBV integrated DNA but do not produce HBV virions and display no markers of HBV replication (29–31, 36). Both cell lines were able to support HDV replication and assembly and secretion of HDV virions. The HDV virions produced by PLC/PRF/5 cells (HDV-PLC) were able to infect primary human hepatocytes (PHH). However, Hep3B-derived HDV (HDV-Hep3B) appeared to be noninfectious. Our results support the hypothesis that at least some of the natural HBV integrants produce functional large (L) and small (S) envelope proteins (the presence of which represents the minimal requirements for the formation of infectious virus particles) and thus can facilitate HDV persistence, regardless of ongoing HBV replication. Our findings also suggest that HDV is likely a more independent and more significant pathogen than is currently assumed and that HDV/HBV carriers will likely benefit from treatments directly targeting HDV.
MATERIALS AND METHODS
Plasmids, cell lines, and transfection.
The LMS vectors, pNF-HBV-LMS-B4, pNF-HBV-LMS-C5, pNF-HBV-LMS-D1, pNF-HBV-LMS-D3, and pNF-HBV-LMS-F1, were constructed to express the L, middle (M), and S envelope proteins that belong to natural variants of HBV genotypes B, C, D, and F (GenBank accession numbers D00330, AF411411 (M-minus variant), AB205127, V01460 and X69798, respectively). For each LMS vector, the designation at the end of the vector name refers to the HBV genotype and the number of a particular sequence among the variants of the given HBV genotype in the assembled collection of HBV sequences. HBV variants D1, B4, and C5 were acquired during the chronic stage of HBV infection. For variants D3 and F1, the information about their origin was not available (37–40; K. Abe, personal communication). The HBV sequences were inserted into the XhoI/XbaI sites of the pSVL vector (Pharmacia). Each HBV insert begins at the natural start codon of the L protein; contains the entire L protein-coding sequence along with the 3′-untranslated region (3′-UTR), which bears the posttranscriptional regulatory element (PRE); and ends about 100 nucleotides downstream of the PRE. For reference, in a typical HBV genotype A sequence, PRE occupies positions 1151 to 1684 (41–43). Hep3B (Hep3B2.1-7) and PLC/PRF/5 cells (29–31) were purchased from ATCC. To initiate HDV replication and assembly, Hep3B and Alexander cells were transfected with plasmid pSVLD3 (44). Each type of reference HDV virion was produced by cotransfection of Huh7 cells with a 1:1 mixture of pSVLD3 and one of the LMS vectors described above using the Fugene HD reagent (Roche) according to the instructions of the manufacturer.
Immunoprecipitation.
Immunoprecipitation (IP) experiments were conducted using the method that was described previously and employs Pansorbin cells (Calbiochem) (45). Rabbit polyclonal antimatrix antibodies were generated by GenScript. They were raised against the synthetic peptide that spans amino acid residues 91-IPPPASTNRQSGRQPTPISPPLRDSHPQAMQWNSTAFH-128 of the pre-S region of the L envelope protein (genotype A of HBV) and contains the conserved matrix domain (underlined) (45, 46). According to the software predicting the antigenic sites in proteins (mobyle.pasteur.fr/cgi-bin/portal.py?#forms::antigenic), the peptide described above harbors a single antigenic determinant, TPISPPLRDS, which is located within the pre-S1 domain. Thus, it was anticipated that IP with the antimatrix antibodies would specifically target only the pre-S1-containing HDV particles. The virions that contain the pre-S1 domain of the L envelope protein (which interacts with the HBV receptor [45]) on the outer surface are considered potentially infectious. In a separate study, we examined the specificity of the antimatrix antibodies. Two types of HDV virions were examined. The first type of HDV virion (HDV-D1 LMS) was assembled using the vector expressing the L, M, and S proteins of HBV variant D1 (i.e., pNF-HBV-LMS-D1). The second type (HDV-D1 L-minus) was prepared using the L-minus version of the above-mentioned vector, which was modified to express the M and S envelope proteins but not the L envelope protein. The antimatrix antibodies were compared to the S26 mouse monoclonal antibody that recognizes the QDPR sequence in the pre-S2 domain (47) and therefore is able to bind the L and M proteins. Using HDV-D1 L-minus virions, it was found that the antimatrix antibodies (compared to the S26 antibody) were unable to immunoprecipitate about 78% of the L− M+ S+ virions. This result suggests that the antimatrix antibodies very poorly recognize the M protein, which is exposed on the outer surfaces of the particles. Using HDV-D1 LMS virions, it was deduced that the fraction of the L protein-containing particles in the virions that were pulled down during the IP procedure by the antimatrix antibodies was at least 83%, while the rest of precipitated virions (about 17% or less) may consist of particles that (i) bear the pre-S2 sequences in the context of the M protein and (ii) do not contain the L protein (i.e., the virions were L− M+ S+) (data not shown). Because HDV virions containing the pre-S1 domain (pre-S1-HDVs) represented the predominant majority of the virions that were immunoprecipitated by the antimatrix antibodies, the contribution of virions that contained the M protein but not the L protein can be considered not significant. The virions that were immunoprecipitated by the antimatrix antibodies were designated pre-S1*-HDVs, because these antibodies demonstrated significant preferential but not exclusive specificity for the pre-S1-bearing virus particles. The use of the asterisk indicates that the virions which were immunoprecipitated by the antimatrix antibodies were predominantly pre-S1-HDVs with the addition of not significant amounts of particles that bore the M protein but not the L protein. The pre-S1*-HDVs therefore served as an acceptable approximation of the fraction of virions that are potentially infectious. The ratio of 3 μl of antimatrix antibodies per 10 μl of the concentrated HDV stock (prepared using polyethylene glycol [PEG] [48]) was found to be optimal in order to achieve the best performance of the IP procedure. In addition, the optimized IP procedure was tested using different HDV stocks that were prepared by employing the envelope proteins of 17 different HBV variants. Regardless of the presence of the variant-specific amino acid residues that were not identical to those in the region of the peptide sequence mentioned above (positions 91 to 128 in HBV genotype A), the IP procedure was able to pull down the pre-S1*-HDVs with an efficiency that was very similar for the different HDV stocks tested. It appears that approximately 90% (and, in some cases, >90%) of all the pre-S1*-HDVs were precipitated. In addition, no correlation was observed between the amounts of the immunoprecipitated pre-S1*-HDVs and the presence of particular nonidentical residues (or their combinations) in the region spanned by the peptide mentioned above (not shown). Therefore, it was concluded that antimatrix antibodies recognize the envelope proteins of different HBV variants with comparable efficiencies.
RNA isolation and real-time PCR (qPCR) assay.
Total RNA from cells or virions (including Pansorbin cell-bound particles) was isolated with the TRI Reagent (Molecular Research Center) and then treated with DNase (Life Technologies) prior to further analysis. The genomic (G) HDV RNA was measured using quantitative PCR (qPCR) as previously described (48). The copy numbers were quantified by using a 10-fold dilution series of an in vitro-transcribed, gel-purified, unit-length G HDV RNA standard in the range of 20 to 2 × 106 genome equivalents (GE) of HDV and by considering that 1 million HDV RNA molecules are equal to 1 pg of the RNA standard.
Infection of PHH in vitro.
The PHH were purchased in the format of 48-well plates from Life Technologies. HDV virions were concentrated approximately 100-fold before infection using PEG. The infection of PHH was performed in the presence of 5% PEG 8000 (Sigma), as described previously (48). Infected PHH were assayed at 9 days postinfection.
Immunofluorescence.
The immunofluorescence (IF) procedure is described in detail elsewhere (48, 49). PHH were fixed at room temperature with 4% paraformaldehyde for 30 min, washed two times with phosphate-buffered saline (PBS), and then permeabilized using 0.2% Triton X-100 solution in PBS. Mouse monoclonal antibody against human alpha-tubulin (Sigma) and polyclonal rabbit antibodies against the delta antigens (δAgs) were used to stain the PHH. Nuclear DNA was stained using 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) solution (Sigma). An inverted Nikon TE2000-U microscope with a ×20 objective was used for analysis of stained PHH.
Analysis of HBV integrant-derived RNAs using PCR, cloning, and sequencing.
To examine the presence of specific HBV sequences on the RNAs harvested from nontransfected Hep3B or Alexander cells, primer A (470-CGGGCAACATACCTTGATAATCCAGA-445), primer B (2813-ATTTTGTGGGTCACCATATTCTTGGGAACAA-2843), primer C (3186-TCTAAGAGACAGTCATCCTCAGGC-3209), primer D (138-GGGACCCTGCACCGAACATG-157), primer E1 (E1 was used only for the material from Alexander cells; 2878-CCTCTCGAGTGGGGGAAGAAT-2898), primer E2 (E2 was used only for the material from Hep3B cells; 2878-GCAAAGGCATGGGGACGAAT-2898), and primer F (1886-CAAGGCACAGCTTGGAGGCTT-1866) were used. Six PCR assays were conducted using the pairs of primers BA, CA, DA, E1A, E2A, and FA. Primer R (752-ACCACATCATCCATACTGAAAGCC-726) was used for reverse transcription. Primers B and A were also used for sequence analysis of the region coding for 211 N-terminal amino acids of the L protein. Primers G (801-ATTGTAACAGCGGTATAAAGGGA-740) and H (413-CTGCTGCTATGCCTCATCTTCTT-435) were utilized to analyze the sequences coding for amino acids 95 to 209 in the S domain. For the latter assay, primer K (873-GTTTAGGGAATAACCCCATCTCTTCGT-847) was used for cDNA synthesis. The nucleotide numbering is according to that for HBV genotype A (41–43). The PCR products obtained with primer pairs BA and GH were cloned into pCRII-TOPO vector (Life Technologies), and the resulting clones were sequenced using M13 universal primers.
To address further the functionality of the integrant-derived envelope proteins, total RNA from Alexander cells was used and the entire open reading frame (ORF) for the L envelope protein was amplified by reverse transcription-PCR using the following primers: (i) for cDNA synthesis, primer M (1190-TCAGCAAACACTTGGCACAG-1171); (ii) for the outer (first) PCR, primers B and N (1120-GAAAGGCCTTATAAGTTGGCGAGAA-1096); and (iii) for the inner (second) PCR, primers P (TCTAGTCGAC-2854-ATGGGGCTTTCTTGG-2868, which starts with an adaptor sequence containing the SalI site [underlined] and continues with the HBV-specific sequence) and Q (GTAGCTAGC-1084-GCTTAGCTTGTATACATGCATATA-1061, which starts with an adaptor sequence containing the NheI site [underlined] and continues with the HBV-specific sequence). The resulting product of the nested PCR spanning the entire ORF of the L protein was digested with the SalI and NheI restriction enzymes and cloned into the pSVL vector (Pharmacia) using the XhoI/XbaI sites. Next, to ensure the export of the mRNA into the cytoplasm and its proper translation, the fragment of the HBV genotype G genome (because the initial sequencing of the cloned ORFs for the L protein revealed that they belong to genotype G of HBV) that starts immediately downstream of the L protein ORF, includes the PRE, and finishes about 100 nucleotides downstream of the PRE (42, 43) was introduced immediately next to the 3′ end of the ORF of the L protein that was cloned into pSVL using the material from Alexander cells. To amplify the fragment described above, plasmid pNF-HBV-LMS-G1, which is a vector that expresses the L, M, and S envelope proteins of HBV genotype G, variant 1 (GenBank accession number AF405706), was used. The primers for this amplification were S (1032-CCAATGTGGTTACCCTGCCT-1047) and T (GCTTAGC-1788-GCCTACAGCCTCCTA-1773, which starts with an adaptor sequence containing the BlpI site [underlined] and continues with the HBV-specific sequence). Prior to the cloning into the construct bearing the sequences coding for the entire ORF of the L protein (from PLC/PRF/5 cells), the PCR product obtained with primer pair ST was digested with Eco91I. After completion of the cloning and sequencing, two sequences initially derived from RNA harvested from Alexander cells were selected and designated ORF52 and ORF62. Based on these sequences, two LMS vectors expressing ORF52 or ORF62 were constructed and were designated pNF-LMS-ORF52 and pNF-LMS-ORF62, respectively. These two LMS vectors were subsequently used to assemble two virus types, HDV-PLCORF52 and HDV-PLCORF62, respectively, as described above, using a strategy of cotransfection of Huh7 cells.
RESULTS
HDV assembly in Hep3B and Alexander cells.
Hep3B cells apparently contain at least two HBV integrants, while PLC/PRF/5 cells bear at least seven HBV inserts. Neither of the cell lines produces HBV nor exhibits markers of HBV replication (29–31). Since a main substrate for integration is a double-stranded linear (DSL) HBV genome, the production of the pregenomic and precore RNAs from such an integrant is not anticipated, while mRNAs encoding the L, M, and S envelope proteins can be transcribed as HBV-host hybrids harboring host sequences at the 3′-end immediately prior to the poly(A) tail. The functional properties of the polypeptides translated from these integrant-derived mRNAs have yet to be fully evaluated (16, 29–31, 34, 35). Cells of both of the cell lines, Hep3B and Alexander cells, were transfected with plasmid pSVLD3 (44) to initiate HDV replication. As a control, Huh7 cells were simultaneously transfected with pSVLD3 and the vector pNF-HBV-LMS-D3 that expresses functional L, M and S of HBV genotype D, variant 3.
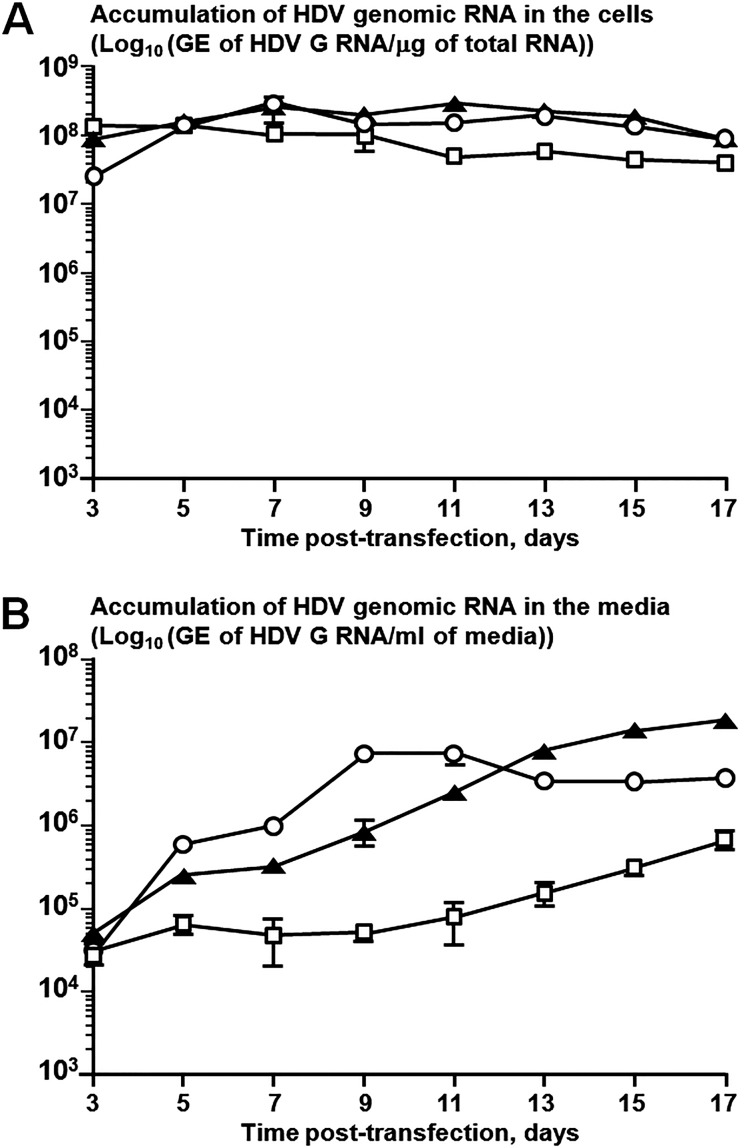
qPCR analysis of the intracellular accumulation of HDV G RNA between days 3 and 17 posttransfection revealed that both Hep3B and Alexander cells supported HDV replication at levels comparable to those supported by Huh7 cells (Fig. 1A). All three cell lines facilitated the assembly and secretion of HDV (Fig. 1B). At 3 days posttransfection, HDV secretion levels were comparable in all cell lines. Between days 5 and 11 posttransfection, HDV virions were produced the most efficiently by Huh7 cells. During this time period, Hep3B cells were producing about 2 to 12 times less HDV virions (HDV-Hep3B). Starting on day 13, Hep3B cells displayed levels of HDV secretion approximately 2 to 5 times higher than those for Huh7 cells. Between days 5 and 17, compared to Huh7 cells, Alexander cells secreted about 4 to 121 times less HDV (HDV-PLC), which were the lowest HDV yields observed among the three cell lines.
FIG 1.
Replication of the HDV genome and assembly of HDV virions in cells transfected with plasmid pSVLD3. Hep3B (Hep3B2.1-7) and PLC/PRF/5 (Alexander) cells were transfected with plasmid pSVLD3 to initiate HDV replication (44). As a control, Huh7 cells were simultaneously transfected with equal mass amounts of the vectors pSVLD3 and pNF-LMS-D3 that express the L, M, and S proteins of HBV genotype D, variant 3 (GenBank accession number V01460). Each transfection at each time point was performed in duplicate. (A) Replication of HDV genomes. Total RNA was isolated from transfected cells at days 3, 5, 7, 9, 11, 13, 15, and 17 posttransfection and analyzed by an HDV-specific qPCR (48). The vertical axis represents the levels of HDV G RNA accumulation in Huh7 cells (open circles), Hep3B cells (black triangles), or Alexander cells (open squares). (B) Assembly of HDV virions. Total RNA was isolated from the media of the transfected cells at days 3, 5, 7, 9, 11, 13, 15, and 17 posttransfection and analyzed by qPCR for the G RNA of HDV (48). The vertical axis presents the levels of HDV G RNA accumulation in the media of Huh7 cells (open circles), Hep3B cells (black triangles), or Alexander cells (open squares). In the case of Hep3B and Alexander cells, each number represents the average value obtained by analysis of two independent transfections. In each qPCR assay, each RNA sample was analyzed in triplicate. The standard errors of the means are indicated in both panels.
Assembled HDV-Hep3B and HDV-PLC virions were then compared to the reference HDV types, each of which was produced in Huh7 cells by cotransfection with pSVLD3 and the LMS vector expressing the envelope proteins of a particular natural HBV variant. Those were HDVs coated with the envelope proteins of either HBV genotype (i) B (HDV-B4), (ii) C (HDV-C5), (iii) D (HDV-D1), or (iv) F (HDV-F1). Thus, all of the HDV types, including HDV-Hep3B and HDV-PLC, contained the same HDV ribonucleoprotein inside the virions and differed only by the origin of the HBV envelope proteins employed for assembly. Prior to further analysis, all the HDV stocks described above were concentrated about 100-fold using polyethylene glycol (48). The results of comparison of the concentrated virus stocks are summarized in Table 1. HDV-F1 displayed the highest total concentration of HDV virions. The total concentration of HDV-D1 virions was approximately 5-fold lower. For HDV-Hep3B and HDV-C5, the total HDV levels were 11.7% and 5.7%, respectively, compared to that of HDV-F1. The concentrations of HDV-B4 and HDV-PLC were 1.1% and 2.1%, respectively. Therefore, the total HDV yields facilitated by HBsAgs expressed in Hep3B and Alexander cells were within the range observed for the envelope proteins of natural HBV variants. The fraction of pre-S1-containing and therefore potentially infectious virions (because the N terminus of the pre-S1 domain harbors the receptor-binding region [50]) was analyzed using immunoprecipitation (IP) with antimatrix antibodies against the peptide harboring the conserved HBV matrix domain (3, 46, 50). As described in Materials and Methods, the antimatrix antibodies displayed a strong preference but not an exclusive specificity for the pre-S1 domain. In a separate study, it was determined that the virions immunoprecipitated using the antimatrix antibodies were predominantly (greater than 80%) pre-S1-containing HDVs along with a not significant fraction (less than 20%) of L− M+ S+ virions (not shown). The HDV virions pulled down with the antimatrix antibodies were designated pre-S1*-HDVs, indicating that the predominant majority of the precipitated virions were pre-S1-HDVs. Pre-S1*-HDV quantification therefore served as an acceptable approximation of the content of potentially infectious HDV virions. The results of the IP-based quantification of pre-S1*-HDVs are presented in Table 1. The largest fractions of pre-S1*-HDVs were found in HDV-D1 and HDV-F1, 32.1% and 27.3%, respectively. For HDV-B4 and HDV-C5, the percentages of pre-S1*-HDVs were 16.1% and 12.4%, respectively. The lowest yields of pre-S1*-HDVs were measured in HDV-Hep3B (0.5%) and HDV-PLC (1.0%). The percentages of the pre-S1*-HDVs measured for HBV-Hep3B and HDV-PLC were not significantly higher than the cutoff value of the sensitivity of the IP procedure, which was determined using HDV coated with the S envelope protein only (i.e., 0.2%).
TABLE 1.
Parameters of concentrated virus stocks of different HDV typesf
| HDV typee | Total yield |
Pre-S1* HDV yield |
||
|---|---|---|---|---|
| GE/μla | %b | GE/μlc | %d | |
| HDV-PLCg | (1.80 ± 0.41) × 105 | 2.1 | (1.72 ± 0.43) × 103 | 1.0 |
| HDV-Hep3Bh | (1.02 ± 0.06) × 106 | 11.7 | (4.64 ± 0.29) × 103 | 0.5 |
| HDV-D1i | (1.59 ± 0.46) × 106 | 18.3 | (5.10 ± 1.57) × 105 | 32.1 |
| HDV-F1j | (8.71 ± 0.85) × 106 | 100.0 | (2.38 ± 0.32) × 106 | 27.3 |
| HDV-B4i | (9.28 ± 2.18) × 104 | 1.1 | (1.49 ± 0.89) × 104 | 16.1 |
| HDV-C5i | (4.98 ± 1.93) × 105 | 5.7 | (6.19 ± 2.11) × 104 | 12.4 |
Total yield values represent the numbers of HDV genome-containing particles (i.e., the numbers of HDV GE) per microliter of stock for any given type of HDV virion. All virions contained the S protein, while only a fraction of potentially infectious virions contained the molecules of the L protein with the pre-S1 domain exposed on the outer surfaces.
The highest total HDV yield observed for HDV-F1 type was used as the value of 100% for comparison of the efficiency of virion production for the other HDV types.
The pre-S1*-HDV yield values represent the amounts of virions that were immunoprecipitated using the antimatrix antibodies (HDV GE/μl of stock). The antimatrix antibodies displayed strong preferential specificity in binding to the pre-S1 sequences, and the values of pre-S1*-HDV yield serve as a good approximation for the quantification of potentially infectious (L protein-containing) particles. The asterisk indicates that the predominant majority of precipitated particles were pre-S1-HDVs (greater than 80%), and the rest (less than 20%) were apparently L− M+ S+ virions (see Materials and Methods).
The percentage of pre-S1*-HDVs shows the fraction of pre-S1*-HDVs relative to the total number of secreted virions for any given HDV type.
HDV-Hep3B, HDV virions assembled in Hep3B cells; HDV-PLC, HDV virions assembled in PLC/PRF/5 cells. The other HDV virion types were assembled in Huh7 cells using the cotransfection approach described in Materials and Methods. HDV-B4, virions coated with the envelope proteins of HBV genotype B, variant 4; HDV-C5, virions coated with the envelope proteins of HBV genotype C, variant 5; HDV-F1, virions assembled with the envelope proteins of HBV genotype F, variant 1; HDV-D1, virions assembled with the envelope proteins of HBV genotype D, variant 1.
The HDV stocks were concentrated using PEG, as described in Materials and Methods. The data presented here reflect the averages obtained by analysis of several independent preparations of the virus stocks, as indicated in footnotes g to j. To calculate the copy numbers of genomic (G) HDV RNA, the in vitro-transcribed, gel-purified, unit-length G HDV RNA standard was used in the G HDV RNA strand-specific qPCR assay (48). In each qPCR assay, each RNA sample was analyzed in triplicate. The standard errors of the means for both kinds of measurements (total HDV yield and yield of pre-S1*-HDVs) are indicated.
Seven independent preparations were used.
Three independent preparations were used.
Four independent preparations were used.
Two independent preparations were used.
Infectivity of HDV virions coated with the envelope proteins produced from integrated HBV DNA.
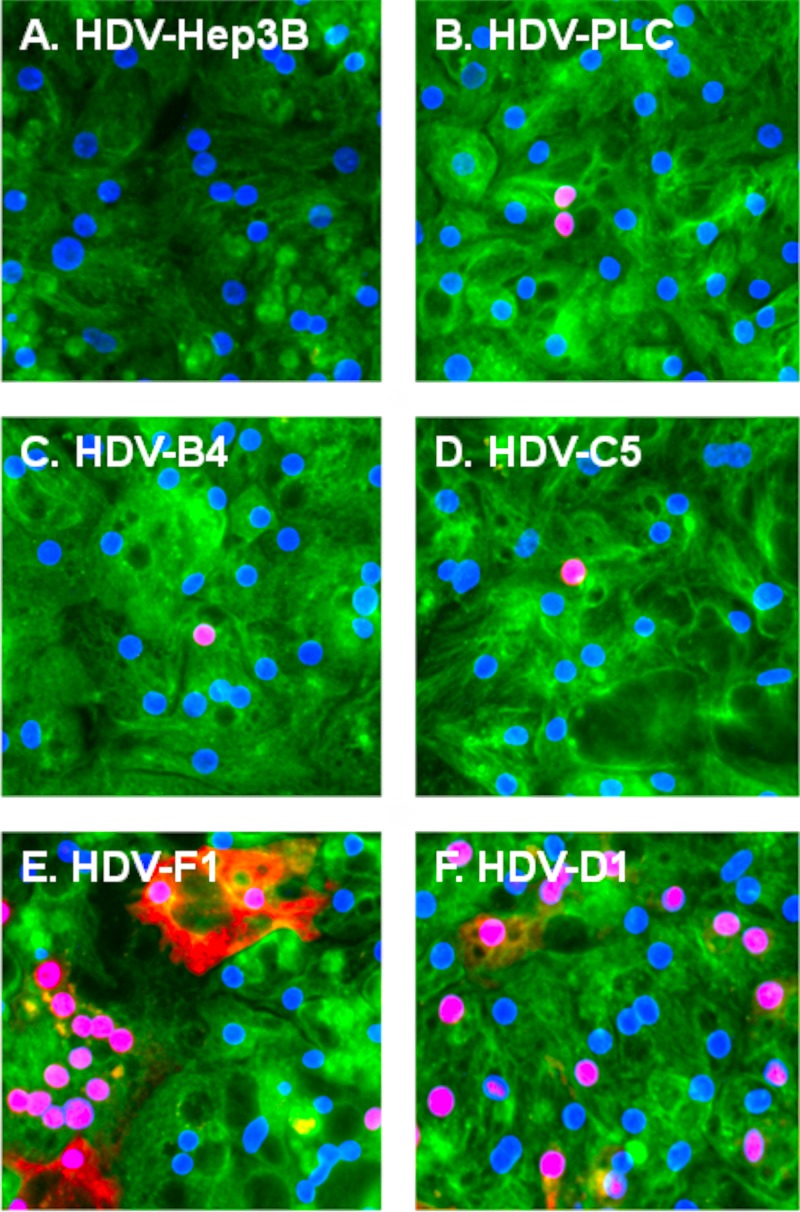
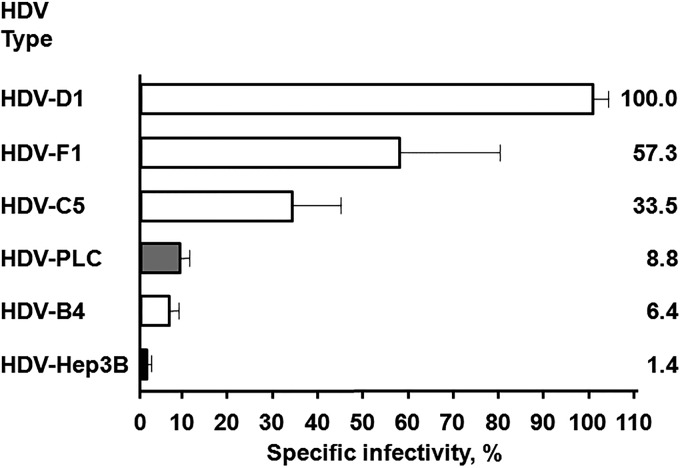
The infectivity of HDV-Hep3B and HDV-PLC was compared to that of HDV-B4, HDV-C5, HDV-D1, and HDV-F1 using in vitro infection of PHH. Four independent infections were conducted using four different PHH lots. For the experiments detecting HDV-positive hepatocytes using immunofluorescence (IF), the multiplicity of infection (MOI) was in the range of 25 to 100 total HDV virions per hepatocyte. HDV-infected cells were detected using rabbit polyclonal antibodies against recombinant small delta antigen (δAg) (48). Representative IF images are shown in Fig. 2. No HDV-positive cells were observed during any of the infections among PHH inoculated with HDV-Hep3B virions (Fig. 2A). Therefore, HDV-Hep3B was considered noninfectious. Unlike HDV-Hep3B, HDV-PLC (Fig. 2B) was infectious on all occasions. HDV-PLC was able to infect between 0.43% and 1.96% of PHH (MOI, 25 to 50). Similarly, HDV-positive PHH were observed during all infections for HDV-B4, HDV-C5, HDV-F1, and HDV-D1 (Fig. 2C to F, respectively). For HDV-D1, the percentage of infected PHH was in the range of 16.60% to 28.25% (MOI, 100). For HDV-F1, the fraction of infected hepatocytes was 3.71 to 23.41% (MOI, 100). For HDV-B4 (MOI, 30) and HDV-C5 (MOI, 50 to 100), the observed fractions of infected cells were 0.52 to 6.18% and 1.94 to 18.99%, respectively. In agreement with previous observations (48), two typical patterns of δAg subcellular localization in infected PHH were observed (Fig. 2). The major pattern was nucleoplasmic staining, and the minor one included additional staining in the cytoplasm. Next, the extent of HDV replication in infected PHH was measured by a qPCR that specifically assayed the number of HDV genomic RNA copies in isolated total cellular RNA (48). Based on the qPCR data, the specific infectivity (SI) values were calculated. The SI reflects the number of HDV genomes produced as a result of infection per average hepatocyte, which is normalized by the pre-S1*-HDV MOI (the number of pre-S1*-HDVs per hepatocyte used in the inoculum). The average SI determined for HDV-D1 was the highest and was used as the 100% value. All other SI values were expressed relative to the SI of HDV-D1 (Fig. 3). HDV-F1 and HDV-C5 had intermediate SI levels of 57.3% and 33.5%, respectively, while HDV-B4 displayed a relatively low SI level of 6.4%. For HDV-PLC, the average SI value observed was 8.8%. As expected, HDV-Hep3B had the lowest SI value of 1.4%. Overall, the generated data demonstrate that HDV-PLC was infectious, while Hep3B appeared to be noninfectious.
FIG 2.
Detection of PHH infected with different types of HDV virions using immunofluorescence. At day 9 postinfection, PHH were fixed, permeabilized, and analyzed for HDV-infected cells by immunofluorescence using rabbit polyclonal antibodies raised against recombinant small delta antigen (δAg) (48). Red staining is for the δAg accumulated in infected PHH. For detection of the cellular α-tubulin (green staining), mouse monoclonal antibody (Fisher Scientific) was used. Blue staining (DAPI) represents nuclear DNA. PHH were infected with HDV assembled in Hep3B cells (HDV-Hep3B) (A), HDV assembled in PLC/PRF/5 cells (HDV-PLC) (B), HDV coated with the envelope proteins of HBV genotype B, variant 4 (HDV-B4) (C), HDV coated with the envelope proteins of HBV genotype C, variant 5 (HDV-C5) (D) HDV coated with the envelope proteins of HBV genotype F, variant 1 (HDV-F1) (E), and HDV coated with the envelope proteins of HBV genotype D, variant 1 (HDV-D1) (F).
FIG 3.
Specific infectivity (SI) of different HDV types. The specific infectivity reflects the number of new HDV genomes per hepatocyte produced as a result of infection normalized by the pre-S1*-HDV MOI. The pre-S1*-HDV MOI is the number of the pre-S1*-HDVs per hepatocyte used in the inoculum. The numbers of HDV genomes in the inoculum and in total RNA harvested from infected primary human hepatocytes were determined using an HDV-specific qPCR assay (48). Averaged data from four independent infection experiments are shown (HDV-B4 was assayed in three out of four infections). The average SI for HDV-D1 was the highest and was used as the 100% value. All other SI values are presented as percentages relative to the SI of HDV-D1. The SI percentage values are shown as horizontal bars. The SI percentage scale is shown at the bottom. The HDV type is indicated on the left. The SI percentage value for each HDV type is indicated on the right. The standard errors of the means are provided.
To examine further the infectivity of HDV-PLC, the entire ORF for the L envelope protein was PCR amplified using total RNA from nontransfected Alexander cells and then cloned into the vector that enables the expression of the L, M, and S proteins after transfection. Two different ORF sequences were used to produce HDV-PLCORF52 and HDV-PLCORF62 virus stocks using the above-described strategy of cotransfection of Huh7 cells. The total numbers of HDV virions/μl in the concentrated stocks of HDV-PLCORF52 and HDV-PLCORF62 were 3.51 × 105 and 7.97 × 105, respectively. These values correspond to 4.0% and 9.2%, respectively, compared to the yield of HDV-F1 virions (see Table 1 for comparison), and are similar to the total yield of HDV facilitated by a natural variant of HBV genotype G (GenBank accession number AF405706), which was 7.53 × 105 GE/μl (in a concentrated stock) (not shown). The measured percentages of pre-S1*-HDVs were 9.3% for HDV-PLCORF52 and 33.0% for HDV-PLCORF62. The increased fractions of pre-S1*-HDVs (compared to HDV-PLC [Table 1]) are likely a result of efficient L protein expression from a strong simian virus 40 late promoter located upstream of the cloned ORFs in the expression vectors. Given that the expression of the S protein in both vectors occurred from the authentic HBV promoter, the results presented above suggest that in Alexander cells, the S proteins per se are likely not deficient in the formation and egress of the L-S complexes, and the low percentage of pre-S1-HDVs in HDV-PLC could be explained at least in part by the low rates of L production/accumulation. At an MOI of 100, HDV-PLCORF52 and HDV-PLCORF62 infected 4.17% and 10.42% of PHH, respectively, which is consistent with the higher percentage of pre-S1-HDVs in the inoculum. The SI values for HDV-PLCORF52 and HDV-PLCORF62 were 24.84% and 10.42%, respectively, compared to the SI value for HDV-D1 (see Fig. 3 for comparison). These results suggest that the L proteins encoded by ORF52 and ORF62 likely have no obvious defects in supporting HDV infectivity. Since these SI values were similar to or less than 3-fold higher than the SI value for HDV-PLC (Fig. 3), it is possible that HDV-PLC consists of virions, the individual infectivities of which do not differ significantly.
Analysis of RNA sequences transcribed from integrated HBV DNAs.
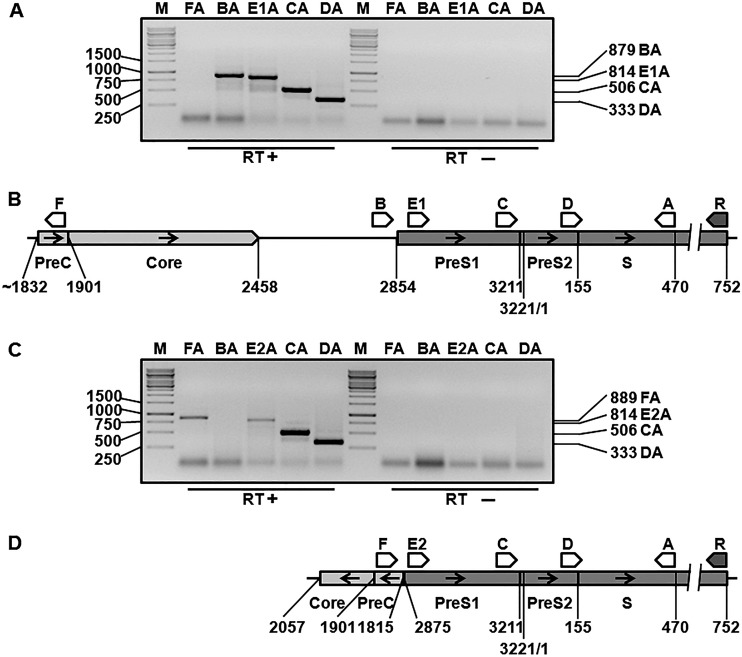
Next, we conducted further examination of the HBV RNAs transcribed from the integrated DNAs using reverse transcription-PCR, followed by cloning and sequencing, in order to better understand the results of the infectivity assays. Several different PCR assays were developed to examine the presence of specific HBV sequences on mRNAs accumulated in untransfected Hep3B and Alexander cells (Fig. 4). Primers A, B, and R spanned sequences that are conserved among different HBV genotypes, A to I. The design of the other primers, C, D, F, E1, and E2, incorporated the information about HBV integrants found in Alexander cells and Hep3B cells (30, 34, 35, 51) and our data acquired by partial cloning/sequencing of RNA sequences accumulated in the cell lines mentioned above (not shown). The PCR products BA, E1A, CA, and DA were observed for RNAs harvested from Alexander cells, which was consistent with the presence of the 5′-UTR and sequences coding for the entire pre-S1 and pre-S2 regions and for a considerable N-terminal portion of the S domain (Fig. 4A and B). In contrast, using total RNA from Hep3B cells, we observed the absence of the PCR product BA and the presence of the E2A, CA, DA, and FA products (Fig. 4C and D), suggesting that the 5′-UTR and sequences coding for the N terminus of the pre-S1 region are not expressed in Hep3B cells. The results presented in Fig. 4 and subsequent examination of the sequences which were amplified by a PCR assay with primer pair FA using RNA harvested from Hep3B cells (Fig. 5) were consistent with the description of an integrant in Hep3B cells that has (i) a large deletion that includes the pre-S1 promoter, the 5′-UTR, and the N terminus of the pre-S1 region and (ii) a rearrangement which resulted in the joining of the remainder of the truncated pre-S1 domain to the inverted precore/core sequences (Fig. 4D) (30). Interestingly, the junction site mapped on RNAs accumulated in Hep3B cells, precore position 1819 (1818)–pre-S1 position 2877 (2878) (the positions in parentheses are shown because the second T of the TT dinucleotide at the junction site could belong to either the precore or the pre-S1 region) was not the exact match for the junction that was previously determined on the DNA integrant, which was precore position 1815–pre-S1 position 2875 (Fig. 5) (30). All these data (Fig. 4 and 5) argue against expression of the L protein in Hep3B cells and are consistent with HDV-Hep3B being noninfectious. The existence of the PCR product FA for RNA from Hep3B cells (Fig. 4C) suggests that transcription of the RNA bearing the inverted precore/core sequences joined to the truncated pre-S1 domain is driven by an unknown cellular promoter.
FIG 4.
Analysis of the presence of specific HBV regions on integrant-derived RNAs accumulated in Hep3B and PLC/PRF/5 cells. Total RNA from nontransfected Alexander cells or Hep3B cells was extracted with the TRI Reagent, reverse transcribed using primer R, and then assayed for the presence of the specific HBV sequences related to the production of the envelope proteins using six different PCR assays, which employed the pairs of primers FA, BA, E1A, E2A, CA, and DA. The numbering for the primers' locations and for other indicated HBV-specific boundaries is based on the genomic organization of a typical genotype A of HBV (41). Primers A, B, and R were based on the sequences conserved among different HBV genotypes, A to I. Primers C, D, E1 (E1 was used only for the material from Alexander cells), E2 (E2 was used only for the material from Hep3B cells), and F were designed on the basis of information (i) that we acquired by partial sequencing of the RNAs from Hep3B and Alexander cells and (ii) that was previously published for HBV integrants found in the above-mentioned cell lines (30, 34, 35, 51). Primer B is placed in the conserved 5′-UTR of L protein mRNA. The occurrence of the PCR product BA should reveal the presence of the pre-S1 domain and most of the open reading frame for the L protein on RNA regardless of the origin of the HBV sequence, because the PCR assay BA is based on sequences conserved among different HBV genotypes, A to I. (A and C) PCR analysis of total RNA harvested from Alexander cells (A) and Hep3B cells (C). Lanes M, double-stranded DNA markers. Selected molecular sizes (in base pairs) are indicated to the left of the gel images. The presence or absence (negative controls) of reverse transcriptase (RT) during cDNA synthesis is indicated at the bottom of the gel images. The types of the PCR assays that were conducted are indicated at the top of the images above the corresponding lanes. For example, BA indicates that the PCR using the B and A primers was conducted. Note that in panel A, one of the PCR assays was E1A (specific for RNAs from Alexander cells), while in panel C, it was replaced by the PCR assay E2A (specific for RNAs from Hep3B cells). The sizes (in base pairs) of the observed PCR products BA, E1A (or E2A), CA, DA, and FA are indicated on the right. Note in panel A that the PCR product FA was not observed, as expected (see the diagram in panel B). The results shown in panel A indicate (i) the presence of the 5′-UTR and sequences coding for the pre-S1 and pre-S2 regions and for a considerable part of the N terminus of the S domain on RNAs accumulated in Alexander cells, as well as (ii) the absence of rearrangements in the assayed area on the corresponding integrated HBV DNA. The PCR products observed in panel A are consistent with the diagram presented in panel B. In panel C, the PCR product BA was not observed. The results shown are consistent with (i) the absence of the 5′-UTR and the sequences coding for the N terminus of the pre-S1 region on RNAs expressed in Hep3B cells and (ii) a deletion/rearrangement in the corresponding integrated DNA that resulted in the loss of (a) the pre-S1 promoter, (b) the 5′-UTR, (c) the N terminus of the pre-S1 domain, and (d) a considerable region upstream of the pre-S1 promoter, which led to the fusion of the remainder of the pre-S1 domain to the inverted precore (PreC)/core sequences. The occurrence of the FA PCR product strongly suggests that the corresponding RNA sequences were transcribed using an unknown cellular (non-HBV) promoter. The PCR products observed in panel C are consistent with the diagram presented in panel D. (B) Locations and orientations of the primers relative to nonrearranged HBV integrant, which is the likely template for the synthesis of HBV RNAs producing the L, M, and S envelope proteins in Alexander cells. The diagram is consistent with the description of one of the integrants found in Alexander cells, which has no alterations or rearrangements in the assayed area, bears the entire L protein open reading frame (51), and therefore likely facilitates the production of at least functional L and S proteins. The left-hand side position of the integrant is shown to be at approximately nucleotide 1832, which reflects the fact that the main substrate for integration is the DSL HBV DNA genome (16). (D) Locations and orientations of the primers relative to the integrated HBV DNA, which is the likely template for the synthesis of HBV RNAs encoding the envelope proteins in Hep3B cells. The diagram is consistent with the description of the integrant found in Hep3B cells, which has a deletion/rearrangement that resulted in the loss of the pre-S1 promoter, the N terminus of the pre-S1 domain, and fusion of the remainder of the pre-S1 sequences to the inverted precore/core sequences. The positions of the precore and pre-S1 sequences at the junction site are shown in the integrant, as previously described (30).
FIG 5.
Sequences at and around the inverted precore/truncated pre-S1 junction site found on RNAs from Hep3B cells. The product obtained by reverse transcription-PCR with primer pair FA (see the legend to Fig. 4) using total RNA from Hep3B cells was cloned into pCRII-TOPO vector, and the resulting plasmids were sequenced to reveal the sequences at and around the inverted precore/truncated pre-S1 junction site. The alignment, obtained using Clustalw2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/), reflects the comparison of seven different sequences generated by cloning to the reference HBV nucleotide sequence (GenBank accession number X02763) of Valenzuela et al. (57), which is shown at the top. The numbering is that for a typical HBV genotype A genome (41). The nucleotide positions are indicated on the top row. The nucleotides identical to those in the reference sequence are shown as dashes, and nonidentical nucleotides are shown as single letters. At the bottom, identical nucleotides are marked with asterisks, purine-to-purine substitutions are marked by dots, and the A-to-T change is indicated by a colon. The number of identical sequences obtained by cloning is indicated on the left. The obtained sequences demonstrate the deletion and rearrangement which resulted in the junction of the inverted precore/core sequences to the truncated pre-S1 region. The figure demonstrates the fragment of the inverted precore nucleotide sequences (positions 1865 to 1819 [1818]) joined to the fragment of the truncated open reading frame for the L envelope protein (nucleotide positions 2877 [2878] to 3221 and 1 to 208 are shown). Note that because the second T of the TT dinucleotide at the junction site could belong to either the precore or the pre-S1 sequence, the adjoining positions are shown as 1819 (1818) for the precore sequence and 2877 (2878) for the pre-S1 sequence. The precore position 1819 (1818)–pre-S1 position 2877 (2878) junction site that was mapped on the expressed RNA is not an exact match to the junction that was previously mapped on the DNA integrant, which was precore position 1815–pre-S1 position 2875 (30).
We further analyzed (i) the sequence of the N-terminal part of the L envelope protein that contains the HBV receptor-binding site and also (ii) the sequence of the S domain at and around the HDV-binding site (HDV-BS) that is essential for HDV assembly (45, 52). As described above, the intact sequences coding for the pre-S1 domain were found only on RNAs from Alexander cells and not on RNAs from Hep3B cells (Fig. 4). Figure 6A shows the N-terminal region of the L protein (amino acids 1 to 211) determined using HBV RNAs from Alexander cells. No deletions or other rearrangements were observed. The closest match to the most abundant sequence was a variant of genotype G (GenBank accession number BAG50284.1; 88% sequence identity). The most abundant sequence found in Alexander cells (7/14 sequences) was identical to the ORF52 sequence, and one of the least abundant sequences was a match for the ORF62 sequence (1/14 sequences), which means that ORF52 and ORF62 together represent the majority (8/14) of RNA sequences recovered from Alexander cells. Therefore, our data regarding the infectivity of HDV-PLCORF52 and HDV-PLCORF62 are likely representative for the majority of HDV-PLC virions. In addition, taking into consideration (i) the above-mentioned total yields of HDV virions facilitated by ORF52 and ORF62 and (ii) the fact that the sequences of ORF52 and ORF62 correspond to the majority of sequences recovered from Alexander cells, it became apparent that the majority of the L protein mRNAs in Alexander cells encode envelope proteins that are likely not defective in supporting assembly of infectious HDV. Within the N terminus of the pre-S1 domain (up to position 86 for HBV genotypes bearing the pre-S1 domain of 119 amino acids [45]), which is critical for the infectivity of HBV and HDV, all the recovered sequences were identical, which suggests that if the variants of the L envelope protein presented in Fig. 6A would have facilitated different levels of HDV infectivity, this should have been mediated by residues outside the N terminus of the pre-S1 domain. Curiously, the entire pre-S1 domain differed by a single amino acid at position 101 (the position that is dispensable for HDV infectivity [45, 53]) between ORF52 and ORF62, in which the former had K and the latter had G. Given the variety of recovered RNA sequences, it is conceivable that in Alexander cells, more than one HBV integrant facilitates production of the L envelope protein.
FIG 6.
Examination of amino acid sequences encoded by HBV mRNAs transcribed from the integrated viral DNAs. Total RNA was extracted from Hep3B or Alexander cells and analyzed by reverse transcription-PCR, followed by cloning and sequencing in order to examine two regions. The first region spans the pre-S1, pre-S2, and N terminus of the S domain. The second region covers areas at and around the HDV-binding site (HDV-BS), including the major antigenic loop (MAL). The details are described in Materials and Methods. The results of sequencing are expressed as translated amino acid sequences. The alignments were obtained using Muscle alignment software (http://www.ebi.ac.uk/Tools/msa/muscle/). The residues identical to those in the reference sequences (top lines) are shown as dashes, and nonidentical residues are shown as a single-letter code. The numbers of recovered identical sequences are shown on the left. At the bottom of each alignment, identical amino acids are marked as asterisks, conserved amino acids are marked as colons, and semiconserved amino acids are marked as periods. (A) For the first region, in agreement with the data presented in Fig. 4 and 5, the complete sequences that begin at the N terminus of the pre-S1 domain were obtained only for RNAs from Alexander cells. The displayed region covers positions 1 to 211, which includes the entire pre-S1 and pre-S2 domains and the first 38 N-terminal residues of the S domain. The first N-terminal residue of the pre-S1 domain is position 1 (the numbering is for HBV genotype G [41]). The reference sequence (GenBank accession number BAG50284.1; HBV genotype G) is shown on the top. This sequence is the closest match (88% identity) to the most abundant sequence recovered from Alexander cells (the second line from the top, in which the sequence is presented as a single-letter amino acid code). The most abundant sequence (marked with ρ) is identical to the ORF52 sequence. The other less abundant sequences are placed in the lines below. One of the least abundant sequences (bottom line) is marked with ρ and is identical to the ORF62. (B and C) Sequences spanning the area at and around the HDV-binding site for RNA from PLC/PRF/5 cells (B) and RNA from Hep3B cells (C). The displayed region spans from amino acids 95 to 209 of the small envelope protein (the numbering is for HBV genotype A [41] and is given by counting the first N-terminal residue of the S domain as position 1), which includes the C terminus of the second transmembrane domain (C-TMII); the major antigenic loop (MAL); the third transmembrane domain (TMIII); the small cytosolic loop, which is the HDV-binding site (HDV-BS); and the N terminus of the fourth transmembrane domain (N-TMIV) (58). For panel B, the reference sequence (shown on the top) is the closest match (99%) to the majority of the sequences recovered from Alexander cells (GenBank accession number AAA45516.1; HBV genotype A, subtype A1). A single sequence (marked with β) had a downstream deletion which should result in a frameshift and translation of the hybrid sequence that is different from the S sequence of HBV downstream of amino acid 209. For panel C, the reference sequence (on the top) is a 100% match to the majority of the sequences recovered from Hep3B cells (GenBank accession number CAC51288.1; HBV genotype A, subtype A2). The least abundant sequence (marked with β) had a T(113)TT insertion and a unique deletion, which should result in translation of the non-HBV sequences starting at position 161 and the possible production of a hybrid protein. Four premature stop codons (marked as X's) were found in the non-HBV part of this sequence.
The alignment presented in Fig. 6A allows us to conduct an additional analysis of the sequences that were found in Alexander cells in terms of their ability to react with the antimatrix antibodies. Compared to the sequence of the peptide that was originally used to produce the antimatrix antibodies, there were a few residues present in the alignment that were not identical to the above-mentioned peptide sequence. Clearly, V(90) and T(100) (the numbering is for a typical genotype G sequence; Fig. 6A) do not influence the interactions with the antimatrix antibodies. They are both present on the ORF62 sequence, for which we already determined a relatively high percentage (i.e., 33.0%) of pre-S1*-HDVs in the HDV-PLCORF62 virus stock. Only 3/14 sequences bore the unique L(120) residue, while a very abundant nonidentical residue, K(101), was present on 10/14 sequences shown in the alignment. As stated above, this is the only position in which nonidentical residues were found within the entire pre-S1 domain in the sequences of ORF52 [which contains K(101)] and ORF62 [which bears G(101)]. Therefore, the change G(101)K was introduced into the ORF62 sequence to create the ORF62G(101)K mutant. When the two newly made virus stocks, HDV-PLCORF62 and HDV-PLCORF62G(101)K, were assayed by IP with the antimatrix antibodies, practically no differences were observed. The fraction of pre-S1*-HDVs in HDV-PLCORF62 in the new experiment was measured to be 37.5%, while for HDV-PLCORF62G(101)K, the percentage of pre-S1*-HDVs was 34.8%. The result suggests that there was no defect in the recognition of pre-S1*-HDVs by the antimatrix antibodies in the HDV-PLCORF62G(101)K stock. It also became apparent that the observed differences in the percentages of the pre-S1*-HDVs between HDV-PLCORF52 and HDV-PLCORF62 are not related to the above-mentioned K residue at position 101. In addition, using the data presented in Fig. 5, the sequences of the C terminus of the pre-S1 domain were deduced from the corresponding nucleotide sequences found in Hep3B cells. Within the region that is recognized by the antimatrix antibodies, no residues that could potentially alter the interactions with the antimatrix antibodies were found (not shown). Therefore, it was interpreted that if the L protein would have been expressed in Hep3B cells, it would be easily identified by IP using the antimatrix antibodies. Overall, considering these new findings and the above-mentioned data from an analysis of different HBV variants that was conducted in a separate study, it was concluded that in Hep3B and Alexander cells there was no considerable number of sequences found that would not be efficiently recognized by the “anti-matrix” antibodies.
Next, we examined the sequences coding for amino acids 95 to 209 of the S domain spanning the region of the major antigenic loop (MAL) that is important for infectivity and HDV-BS (45, 52). In Alexander cells (Fig. 6B), 12 sequences were 99% identical to another HBV sequence (GenBank accession number FJ692561.1; HBV genotype A, subtype A1). All the sequences had a single substitution [A(166)G] in the MAL. One sequence had a substitution [Q(101)R] in the C terminus of the second transmembrane domain, one had a change [W(191)R] in the C terminus of a third transmembrane domain, and another one had a substitution [Y(200)C] in HDV-BS (the numbering considered the N-terminal position of the S protein [S domain] to be position 1). The functional consequences of these changes are yet to be determined. Another sequence from Alexander cells was identical to the above-mentioned 12/16 sequences in the region of positions 95 to 209, but it had a single nucleotide deletion immediately downstream of residue 209, which would result in the appearance of a non-HBV amino acid sequence downstream of this position (Fig. 6B). In Hep3B cells (Fig. 6C), 11 sequences were identical to a previously reported HBV sequence (GenBank accession number GQ477489.1; HBV genotype A, subtype A2) and displayed no mutations in the MAL and HDV-BS. The 12th sequence had an insertion [T(113)TT] and a unique deletion, which may lead to the translation of non-HBV sequences starting at position 161 and the possible production of a hybrid protein. The deletion mentioned above also resulted in four premature stop codons downstream of position 160, which could explain the low abundance of this sequence. In addition, the latter sequence may indicate mRNA transcription from a different HBV integrant. Overall, the majority of sequences recovered from both cell lines had an unchanged HDV-BS sequence, including three tryptophan residues that are critical for HDV assembly (1, 52). These findings are in agreement with the observation that both cell lines support the assembly and secretion of HDV. The data also suggest that the low efficiency of HDV secretion by Alexander cells (Fig. 1 and Table 1) is not a result of a compromised interaction of HDV-BS with the HDV ribonucleoprotein. It also appears that the differences in infectivity between HDV-Hep3B and HDV-PLC were not mediated by the sequences of the MAL, which has the ability to influence the overall infectivity of the virions (52), and likely reflect the absence of the L protein in HDV-Hep3B virions (Fig. 4 and 5) (30, 54).
DISCUSSION
HDV persistence depends on the availability of functional L and S envelope proteins of HBV in order to maintain the assembly and infectivity of the virions (50, 52). Since HDV requires only the envelope proteins from HBV, ongoing HBV replication may not be a prerequisite for maintenance of persistent HDV infection. The following observations support this notion: HDV-infected individuals may have high levels of HDV replication and significantly decreased or undetectable levels of HBV replication, most anti-HBV drugs that successfully suppress HBV replication do not block HDV infection, HDV can suppress HBV replication, and HDV-infected hepatocytes may appear to be free of the signs of HBV replication (2, 5, 17, 19–23). A production of HBsAgs from integrated HBV DNA can be an alternative to the replication source of the envelope proteins. HBV DNA integration occurs throughout the course of infection. Since the DSL HBV genome is a main substrate for integration, the precore and pregenomic HBV RNAs are not expected to be transcribed from the integrants, while the synthesis of mRNAs for the envelope proteins can occur regardless of HBV replication (3, 16). Therefore, hepatocytes in the liver, where HBV infection is resolved or HBV replication is significantly suppressed (by drug treatment, for example), may still be able to support persistent HDV infection by providing HBV envelope proteins from naturally integrated DNA. However, the functional properties of HBV integrant-derived envelope proteins were not fully evaluated previously, and no data exist regarding their ability to support the production of infectious virions (16, 29–31, 34, 35). In the current study, using two HCC-derived cell lines that bear integrated HBV DNA and have no signs of HBV replication (29–31), we demonstrated for the first time that at least some of the HBV DNA integrant(s) expresses functional envelope proteins that facilitate the assembly of infectious HDV.
As expected, after transfection with an HDV-expressing construct, Hep3B and Alexander cells supported the replication, assembly, and release of HDV (Fig. 1 and Table 1). The immunoprecipitation results suggested a very low yield of pre-S1-containing HDV virions for both cell lines (Table 1). Regardless of the considerable levels of HDV replication, Alexander cells facilitated the lowest yield of HDV virions compared to those from transfected Huh7 and Hep3B cells (Fig. 1). The sequencing data revealed that the majority of the integrant-derived HBV mRNAs in both cell lines code for the functional HDV-binding sequence (Fig. 6). Alexander cells likely have a problem with the efficiency of L protein production and not with the egress of the S-L protein complexes, since when we expressed the sequences of ORF52 and ORF62 from the plasmids with a strong promoter for the L mRNA, we observed the yields of preS1*-HDVs that were 9.3 times higher and 33 times higher, respectively, than that for HDV-PLC. This issue, however, is outside the scope of the current study.
Virions secreted from Alexander cells were able to infect PHH, while Hep3B-derived HDV appeared to be noninfectious (Fig. 2 and 3). Our analysis of HBV RNAs expressed in Hep3B and PLC/PRF/5 cells suggests the likely reasons for that. Using reverse transcription-PCR, we found on RNAs accumulated in Alexander cells the presence of sequences coding for the entire pre-S1 and pre-S2 regions along with a considerable N-terminal part of the S domain. Taken together with the fact that Alexander cells support the assembly and egress of HDV virions and considering the results of the infectivity assays, we conclude that Alexander cells express at least functional L and S proteins in quantities that are sufficient to facilitate the production of infectious HDV. Unlike for Alexander cells, our results suggest that the L protein is likely not produced in Hep3B cells, since our assay did not find full-size intact sequences coding for the pre-S1 region on the accumulated HBV RNAs, which was consistent with HDV-Hep3B being noninfectious. Our data are consistent with previous reports stating that (i) Alexander cells contain at least one integrant bearing the entire ORF for the L protein, which could facilitate the production of L, (ii) while in Hep3B cells, five different HBV-specific transcripts were detected, three of which (one predominant and two minor) were likely initiated at the pre-S2/S promoter and the other two minor transcripts might cover most of the pre-S region (at the same time, no evidence that the last two RNAs might bear the intact and complete pre-S1 domain was obtained). In addition, in the integrant that was characterized earlier in Hep3B cells, the sequences coding for the N terminus of the pre-S1 domain were deleted and the reminder of the pre-S1 domain was fused to the inverted precore/core sequences (30, 34, 35, 51, 55).
The D1, B4, and C5 variants used in this study were collected from chronic carriers of HBV, and the envelope proteins of at least two of them, D1 and C5, supported considerable levels of HDV infectivity (Fig. 3). This observation may indicate that if there could be a tendency of diminishing the overall infectivity of HDV particles over the course of HDV infection, such possible decrease of HDV infectivity may not be mediated by the functional deficiency of available HBV envelope proteins. Further studies are needed to examine whether during the chronic stage of HDV infection, HBV envelope proteins are able to support considerable levels of HDV infectivity.
Overall, our study demonstrated for the first time that in cells of HBV-induced HCCs, at least some HBV integrants, which were naturally incorporated into the host DNA during the infection, can produce functional L and S envelope proteins competent for facilitating the assembly of infectious HDV virions in the absence of ongoing HBV replication. The results obtained therefore provide novel information regarding the functionality of the integrant-derived HBV envelope proteins and the mechanisms of HBV-HDV interactions. As expected and in agreement with previous reports demonstrating the alterations in integrated HBV DNAs (29, 30, 35, 51), not all integrants were able to support the production of functional HBV envelope proteins. Thus, none of the integrants in Hep3B cells was able to support the production of infectious HDV. In Alexander cells, only one out of seven HBV integrants described so far (51) may apparently facilitate the production of the functional L protein that is essential for the virions' infectivity. The generated data further support our hypothesis that HDV persistent infection in vivo may not require ongoing HBV replication, as long as functional HBsAgs are supplied from the integrated HBV DNA. Furthermore, our results (i) suggest that HDV is likely a more independent and more significant pathogen than is currently appreciated and (ii) support the implementation of medical interventions that directly target HDV and should be used along with anti-HBV drugs for HDV/HBV carriers.
In addition, taking together our recently published data indicating that HDV in vivo infects the cells of hepadnavirus-induced HCCs (56) and the results of the current study, we conclude that HBV-induced HCCs are likely able to support the entire HDV life cycle, including attachment, entry, trafficking to replication sites, and the assembly and egress of new infectious HDV virions. Therefore, our study further advances our understanding of the mechanism of the relationship between HDV infection and HCC.
ACKNOWLEDGMENTS
S.O.G. and S.M. were supported by NIH grant NCI R01CA166213. S.O.G. was also supported by NIH grants NIAID R21AI099696, NIAID R21AI097647, and NCRR P20RR016443, by a University of Kansas Research Institute bridging grant, and by the University of Kansas Endowment Association.
We thank Stefan Wieland, Frank Chisari, Kenji Abe, Yu-mei Wen, and Stephan Schaefer for providing us with the constructs harboring sequences of different HBV variants. We acknowledge the help of Megan Dudek.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Taylor JM. 2006. Hepatitis delta virus. Virology 344:71–76. 10.1016/j.virol.2005.09.033 [DOI] [PubMed] [Google Scholar]
- 2.Smedile A, Rizzetto M. 2011. HDV: thirty years later. Dig. Liver Dis. 43(Suppl 1):S15–S18. 10.1016/S1590-8658(10)60687-1 [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51–68. 10.1128/MMBR.64.1.51-68.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupberger J, Hildt E. 2007. Hepatitis B virus-induced oncogenesis. World J. Gastroenterol. 13:74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koytak ES, Yurdaydin C, Glenn JS. 2007. Hepatitis D. Curr. Treat. Options Gastroenterol. 10:456–463. 10.1007/s11938-007-0045-8 [DOI] [PubMed] [Google Scholar]
- 6.Verme G, Brunetto MR, Oliveri F, Baldi M, Forzani B, Piantino P, Ponzetto A, Bonio F. 1991. Role of hepatitis delta virus infection in hepatocellular carcinoma. Dig. Dis. Sci. 36:1134–1136. 10.1007/BF01297460 [DOI] [PubMed] [Google Scholar]
- 7.Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realdi G, Schalm SW. 2000. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut 46:420–426. 10.1136/gut.46.3.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michielsen PP, Francque SM, van Dongen JL. 2005. Viral hepatitis and hepatocellular carcinoma. World J. Surg. Oncol. 3:27. 10.1186/1477-7819-3-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romeo R, Del Ninno E, Rumi M, Russo A, Sangiovanni A, de Franchis R, Ronchi G, Colombo M. 2009. A 28-year study of the course of hepatitis delta infection: a risk factor for cirrhosis and hepatocellular carcinoma. Gastroenterology 136:1629–1638. 10.1053/j.gastro.2009.01.052 [DOI] [PubMed] [Google Scholar]
- 10.Fattovich G, Stroffolini T, Zagni I, Donato F. 2004. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 127:S35–S50. 10.1053/j.gastro.2004.09.014 [DOI] [PubMed] [Google Scholar]
- 11.Vassilopulos D, Hadziyannis SJ. 2006. Clinical features of hepatitis delta virus, p 76–80 In Handa H, Yamaguchi Y. (ed), Hepatitis delta virus. Springer Science+Business Media, New York, NY [Google Scholar]
- 12.Niro GA, Ciancio A, Gaeta GB, Smedile A, Marrone A, Olivero A, Stanzione M, David E, Brancaccio G, Fontana R, Perri F, Andriulli A, Rizetto M. 2006. Pegylated interferon alpha-2b as monotherapy or in combination with ribavirin in chronic hepatitis delta. Hepatology 44:713–720. 10.1002/hep.21296 [DOI] [PubMed] [Google Scholar]
- 13.Casey J, Cote PJ, Toshkov IA, Chu CK, Gerin JL, Hornbuckle WE, Tennant BC, Korba BE. 2005. Clevudine inhibits hepatitis delta virus viremia: a pilot study of chronically infected woodchucks. Antimicrob. Agents Chemother. 49:4396–4399. 10.1128/AAC.49.10.4396-4399.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason WS, Litwin S, Jilbert AR. 2008. Immune selection during chronic hepadnavirus infection. Hepatol. Int. 2:3–16. 10.1007/s12072-007-9024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Yamamoto T, Zhou T, Aldrich CE, Frank K, Cullen JM, Jilbert AR, Mason WS. 2007. The liver of woodchucks chronically infected with the woodchuck hepatitis virus contains foci of virus core antigen-negative hepatocytes with both altered and normal morphology. Virology 359:283–294. 10.1016/j.virol.2006.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. 2010. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J. Virol. 84:8308–8315. 10.1128/JVI.00833-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowans EJ, Baroudy BM, Negro F, Ponzetto A, Purcell RH, Gerin JL. 1988. Evidence for replication of hepatitis delta virus RNA in hepatocyte nuclei after in vivo infection. Virology 167:274–278. 10.1016/0042-6822(88)90078-5 [DOI] [PubMed] [Google Scholar]
- 18.Wu JC. 2006. Functional and clinical significance of hepatitis D virus genotype II infection. Curr. Top. Microbiol. Immunol. 307:176–186. 10.1007/3-540-29802-9_9 [DOI] [PubMed] [Google Scholar]
- 19.Govindarajan S, Fields HA, Humphrey CD, Margolis HS. 1986. Pathologic and ultrastructural changes of acute and chronic delta hepatitis in an experimentally infected chimpanzee. Am. J. Pathol. 122:315–322 [PMC free article] [PubMed] [Google Scholar]
- 20.Tavanez JP, Cunha C, Silva CM, David E, Monjardino J, Carmo-Fonseca M. 2002. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA 8:637–646. 10.1017/S1355838202026432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollicino T, Raffa G, Santantonio T, Gaeta GB, Iannello G, Alibrandi A, Squadrito G, Calvi C, Colucci G, Levrero M, Raimondo G. 2011. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J. Virol. 85:432–439. 10.1128/JVI.01609-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JC, Chen PJ, Kuo MY, Lee SD, Chen DS, Ting LP. 1991. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 65:1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sureau C, Taylor J, Chao M, Eichberg JW, Lanford RE. 1989. Cloned hepatitis delta virus cDNA is infectious in the chimpanzee. J. Virol. 63:4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardi R, Rodriquez F, Buti M, Costa X, Cotrina M, Galimany R, Esteban R, Guardia J. 2001. Role of hepatitis B, C and D viruses in dual and triple infection: influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology 34:404–410. 10.1053/jhep.2001.26511 [DOI] [PubMed] [Google Scholar]
- 25.Krogsgaard K, Wantzin P, Aldershvile J, Kryger P, Andersson P, Nielsen JO. 1986. Hepatitis B virus DNA in hepatitis B surface antigen-positive blood donors: relation to the hepatitis B e system and outcome in recipients. J. Infect. Dis. 153:298–303. 10.1093/infdis/153.2.298 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Wu MC, Sham JS, Tai LS, Fang Y, Wu WQ, Xie D, Guan XY. 2002. Different expression of hepatitis B surface antigen between hepatocellular carcinoma and its surrounding liver tissue, studied using a tissue microarray. J. Pathol. 197:610–616. 10.1002/path.1150 [DOI] [PubMed] [Google Scholar]
- 27.Mason WS, Litwin S, Xu C, Jilbert AR. 2007. Hepatocyte turnover in transient and chronic hepadnavirus infections. J. Viral Hepat. 14(Suppl 1):22–28. 10.1111/j.1365-2893.2007.00911.x [DOI] [PubMed] [Google Scholar]
- 28.Von Loringhoven AF, Koch S, Hofschneider PH, Koshy R. 1985. Co-transcribed 3′ host sequences augment expression of integrated hepatitis B virus DNA. EMBO J. 4:249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaul Y, Ziemer M, Garcia PD, Crawford R, Hsu H, Valenzuela P, Rutter WJ. 1984. Cloning and analysis of integrated hepatitis virus sequences from a human hepatoma cell line. J. Virol. 51:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su TS, Hwang WL, Yauk YK. 1998. Characterization of hepatitis B virus integrant that results in chromosomal rearrangement. DNA Cell Biol. 17:415–425. 10.1089/dna.1998.17.415 [DOI] [PubMed] [Google Scholar]
- 31.Twist EM, Clark HF, Aden DP, Knowles BB, Plotkin SA. 1981. Integration pattern of hepatitis B virus DNA sequences in human hepatoma cell lines. J. Virol. 37:239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takada S, Gotoh Y, Hayashi S, Yoshida M, Koike K. 1990. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J. Virol. 64:822–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokino T, Fukushige S, Nakamura T, Nagaya T, Murotsu T, Shiga K, Aoki N, Matsubara K. 1987. Chromosomal translocation and inverted duplication associated with integrated hepatitis B virus in hepatocellular carcinomas. J. Virol. 61:3848–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou J, Rutter WJ. 1985. Hybrid hepatitis B virus-host transcripts in a human hepatoma cell. Proc. Natl. Acad. Sci. U. S. A. 82:83–87. 10.1073/pnas.82.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziemer M, Garcia P, Shaul Y, Rutter WJ. 1985. Sequence of hepatitis B virus DNA incorporated into genome of a human hepatoma cell line. J. Virol. 53:885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barin F, Goudeau A, Brechot C, Romet-Lemonne J, Sureau C, Lesage G. 1983. Further studies on production and characterization of HBsAg derived from a human hepatoma cell line (PLC/PRF/5). Dev. Biol. Stand. 54:81–92 [PubMed] [Google Scholar]
- 37.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. 1979. Nucleotide sequence of hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646–650. 10.1038/281646a0 [DOI] [PubMed] [Google Scholar]
- 38.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69:2575–2583. 10.1099/0022-1317-69-10-2575 [DOI] [PubMed] [Google Scholar]
- 39.Lin X, Yuan ZH, Wu L, Ding JP, Wen YM. 2001. A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency. J. Virol. 75:11827–11833. 10.1128/JVI.75.23.11827-11833.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi K, Brotman B, Usuda S, Mishiro S, Prince AM. 2000. Full-genome sequence analyses of hepatitis B virus (HBV) strains recovered from chimpanzees infected in the wild; implications for an origin of HBV. Virology 267:58–64. 10.1006/viro.1999.0102 [DOI] [PubMed] [Google Scholar]
- 41.Kramvis A, Kew M, Francois G. 2005. Hepatitis B genotypes. Vaccine 23:2409–2423. 10.1016/j.vaccine.2004.10.045 [DOI] [PubMed] [Google Scholar]
- 42.Panjaworayan N, Roessner SK, Firth AE, Brown CM. 2007. HBVRegDB: annotation, comparison, detection and visualization of regulatory elements in hepatitis B virus sequences. Virol. J. 4:136. 10.1186/1743-422X-4-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panjaworayan N, Payungporn S, Poovorawan Y, Brown CM. 2010. Identification of an effective siRNA target site and functional regulatory elements, within the hepatitis B virus posttranscriptional regulatory element. Virol. J. 7:216. 10.1186/1743-422X-7-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo M, Chao M, Taylor J. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchet M, Sureau C. 2007. Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81:5841–5849. 10.1128/JVI.00096-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruss V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sominskaya I, Bichko V, Pushko P, Dreimane A, Snikere D, Pumpens P. 1992. Tetrapeptide QDPR is a minimal immunodominant epitope within the preS2 domain of hepatitis B virus. Immunol. Lett. 33:169–172. 10.1016/0165-2478(92)90043-N [DOI] [PubMed] [Google Scholar]
- 48.Gudima SO, He Y, Meier A, Chang J, Chen R, Jarnik M, Nicolas E, Bruss V, Taylor J. 2007. Assembly of hepatitis delta virus: particle characterization including the ability to infect primary human hepatocytes. J. Virol. 81:3608–3617. 10.1128/JVI.02277-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gudima SO, He Y, Chai N, Bruss V, Urban S, Mason W, Taylor J. 2008. Primary human hepatocytes are susceptible to infection by hepatitis delta virus assembled with envelope proteins of woodchuck hepatitis virus. J. Virol. 82:7276–7283. 10.1128/JVI.00576-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glebe D, Urban S. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shamay M, Agami R, Shaul Y. 2001. HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene 20:6811–6819. 10.1038/sj.onc.1204879 [DOI] [PubMed] [Google Scholar]
- 52.Sureau C. 2006. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 307:113–131. 10.1007/3-540-29802-9_6 [DOI] [PubMed] [Google Scholar]
- 53.Gudima SO, Meyer A, Dunbrack R, Taylor J, Bruss V. 2007. Two potentially important elements of the hepatitis B virus large envelope protein are dispensable for infectivity of hepatitis delta virus. J. Virol. 81:4343–4347. 10.1128/JVI.02478-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou CK, Su TS, Chang C, Hu CP, Huang MY, Suen CS, Chou NW, Ting LP. 1989. Insulin suppresses hepatitis B surface antigen expression in human hepatoma cells. J. Biol. Chem. 264:15304–15308 [PubMed] [Google Scholar]
- 55.Su TS, Lin LH, Chou CK, Chang C, Ting LP, Hu C, Han SH. 1986. Hepatitis B virus transcripts in a human hepatoma cell line, Hep 3B. Biochem. Biophys. Res. Commun. 138:131–138. 10.1016/0006-291X(86)90256-1 [DOI] [PubMed] [Google Scholar]
- 56.Freitas N, Salisse J, Cunha C, Toshkov I, Menne S, Gudima SO. 2012. Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology 56:76–85. 10.1002/hep.25663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valenzuela P, Quiroga M, Zalvidar J, Gray P, Rutter WJ. 1980. The nucleotide sequence of hepatitis B viral genome and identification of the major viral genes, p 57–70 In Fields BN, Jaenisch R, Fox CF. (ed). Animal virus genetics. Academic Press, New York, NY [Google Scholar]
- 58.Blanchet M, Sureau C. 2006. Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity. J. Virol. 80:11935–11945. 10.1128/JVI.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]