ABSTRACT
Human and mouse SAMHD1 proteins block human immunodeficiency virus type 1 (HIV-1) infection in noncycling human monocytic cells by reducing the intracellular deoxynucleoside triphosphate (dNTP) concentrations. Phosphorylation of human SAMHD1 at threonine 592 (T592) by cyclin-dependent kinase 1 (CDK1) and cyclin A2 impairs its HIV-1 restriction activity, but not the dNTP hydrolase activity, suggesting that dNTP depletion is not the sole mechanism of SAMHD1-mediated HIV-1 restriction. Using coimmunoprecipitation and mass spectrometry, we identified and validated two additional host proteins interacting with human SAMHD1, namely, cyclin-dependent kinase 2 (CDK2) and S-phase kinase-associated protein 2 (SKP2). We observed that mouse SAMHD1 specifically interacted with cyclin A2, cyclin B1, CDK1, and CDK2. Given the role of these SAMHD1-interacting proteins in cell cycle progression, we investigated the regulation of these host proteins by monocyte differentiation and activation of CD4+ T cells and examined their effect on the phosphorylation of human SAMHD1 at T592. Our results indicate that primary monocyte differentiation and CD4+ T-cell activation regulate the expression of these SAMHD1-interacting proteins. Furthermore, our results suggest that, in addition to CDK1 and cyclin A2, CDK2 phosphorylates T592 of human SAMHD1 and thereby regulates its HIV-1 restriction function.
IMPORTANCE SAMHD1 is the first dNTP triphosphohydrolase found in mammalian cells. Human and mouse SAMHD1 proteins block HIV-1 infection in noncycling cells. Previous studies suggested that phosphorylation of human SAMHD1 at threonine 592 by CDK1 and cyclin A2 negatively regulates its HIV-1 restriction activity. However, it is unclear whether human SAMHD1 interacts with other host proteins in the cyclin A2 and CDK1 complex and whether mouse SAMHD1 shares similar cellular interacting partners. Here, we identify five cell cycle-related host proteins that interact with human and mouse SAMHD1, including three previously unknown cellular proteins (CDK2, cyclin B1, and SKP2). Our results demonstrate that several SAMHD1-interacting cellular proteins regulate phosphorylation of SAMHD1 and play an important role in HIV-1 restriction function. Our findings help define the role of these cellular interacting partners of SAMHD1 that regulate its HIV-1 restriction function.
INTRODUCTION
SAM domain- and HD domain-containing protein 1 (SAMHD1) inhibits replication of human immunodeficiency virus type 1 (HIV-1) in noncycling myeloid cells and resting CD4+ T cells by blocking the process of reverse transcription (reviewed in references 1, 2, 3, and 4). Degradation of SAMHD1 by Vpx proteins from HIV-2 and some simian immunodeficiency viruses (SIVs) increases HIV-1 infection in myeloid cells (5–7). SAMHD1 functions as a deoxynucleoside triphosphate triphosphohydrolase (dNTPase), which hydrolyzes dNTPs in vitro (8, 9) and reduces the intracellular dNTP pool in noncycling cells (10–14). The HD domain of SAMHD1 encompasses the dNTPase activity and is sufficient to mediate HIV-1 restriction in noncycling cells (15). Overexpression of full-length human SAMHD1 in dividing cells reduces the dNTP pool but does not block HIV-1 infection (14), suggesting that SAMHD1 activity may be regulated in cycling cells. SAMHD1 also has nucleic acid binding and exonuclease activities in vitro (16–18). Thus, it is possible that mechanisms beyond its dNTPase function can regulate SAMHD1-mediated HIV-1 restriction.
Human and mouse SAMHD1 proteins (hSAMHD1 and mSAMHD1, respectively) block HIV-1 infection in noncycling human monocytic cells (13, 19). Recent studies using samhd1-null mice confirmed that mSAMHD1 acts as a dNTPase to reduce the intracellular dNTP concentrations and thereby contributes to its HIV-1 restriction in vivo (20, 21). SAMHD1 is a phosphoprotein, and its HIV-1 restriction function is inversely regulated by phosphorylation (22–24). It has been shown that phosphorylation of hSAMHD1 at T592 by CDK1 and cyclin A2 negatively regulates its HIV-1 restriction activity (22, 23). CDK1 is known to complex with cyclins A2 and B1 (25, 26), while cyclin A2 also interacts with CDK2 (27). However, it is unknown whether hSAMHD1 interacts with other cellular proteins in the cyclin A2 and CDK1 complex and whether mSAMHD1 shares similar cellular interacting partners.
In the present study, we sought to identify and characterize cellular proteins interacting with hSAMHD1 and mSAMHD1. By overexpressing hSAMHD1 or mSAMHD1 in a human cell line and using coimmunoprecipitation (co-IP) and mass spectrometry, we identified and validated two additional host proteins interacting with hSAMHD1, CDK2 and SKP2. We found that mSAMHD1 specifically interacts with cyclin A2, cyclin B1, CDK1, and CDK2. We investigated the changes in expression of these SAMHD1-interacting proteins by differentiation of monocytic cells as well as the activation of CD4+ T cells and peripheral blood mononuclear cells (PBMCs). Moreover, we examined the effects of these SAMHD1-interacting proteins on the phosphorylation of hSAMHD1 at T592 and their ability to block HIV-1 infection. Our results suggest that several SAMHD1-interacting cellular proteins collectively regulate phosphorylation of hSAMHD1 at T592 and its HIV-1 restriction function.
MATERIALS AND METHODS
Plasmids.
HIV-1 proviral vector pNL-Luc-E−R+ containing a firefly luciferase reporter gene (28) and the pLenti vectors expressing hemagglutinin (HA)-tagged hSAMHD1 or mSAMHD1 (isoform 1) and the empty vector control (13) were kind gifts from Nathaniel Landau (New York University). The pLenti vector expressing HA-tagged mutant SAMHD1 (T592A) was generated using a QuikChange mutagenesis kit (Agilent Technologies) based on the pLenti vector expressing wild-type (WT) hSAMHD1. The green fluorescent protein (GFP)-expressing control vector was previously described (29). The pCMV-HA construct expressing HA-tagged mouse CD9 was generated by standard cloning of mouse cDNA of CD9.
Cell culture.
Human healthy donors' PBMCs were isolated from the buffy coat of healthy blood donors as previously described (30). Monocyte-derived dendritic cells (DCs) were generated using monocytes isolated from PBMCs by treatment with granulocyte-macrophage colony-stimulating factor and interleukin-4 (IL-4) (50 ng/ml) as described previously (14). Primary CD4+ T-lymphocytes were isolated from PBMCs as described previously (30, 31). DCs and CD4+ T-lymphocytes generated using these methods were more than 98% pure by flow cytometry analysis of surface markers as described previously (31–33). Primary resting CD4+ T cells were cultured in the presence of 20 IU/ml of recombinant IL-2 (obtained from the NIH AIDS Research and Reference Reagent Program) and activated by 5 μg/ml phytohemagglutinin (PHA; Sigma-Aldrich) for 72 h as described previously (14). CD4+ T cells were also activated by T-cell receptor (TCR) cross-linking using anti-CD3 and anti-CD28 (1 μg/ml; BD Pharmingen) for 72 h in the presence of IL-2 (20 U/ml). T-cell activation was confirmed by immunostaining for CD25 as described previously (10). Human embryonic kidney cell line HEK293T cells and the HIV-1 indicator cell line GHOST/X4/R5 have been previously described (33, 34). Monocytic U937 and THP-1 cell lines were cultured as described previously (30, 35). These cells were treated with 30 ng/ml phorbol 12-myristate 13-acetate (PMA) for 36 h to differentiate the cells into noncycling, macrophage-like cells as described previously (5). U937 cell lines stably expressing wild-type and the T592A mutant hSAMHD1 were generated by spinoculation of parental U937 cells with concentrated lentiviral vectors and subjecting to puromycin selection as described previously (19).
Transfections and immunoblotting.
HEK293T cells were transfected using a calcium phosphate method to overexpress human and mouse SAMHD1 or vector controls, and cells were processed for downstream applications at 24 h posttransfection. Cells were harvested as indicated and lysed in cell lysis buffer (Cell Signaling) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Cell lysates were subjected to immunoblotting as described previously (36). Restore Western blot stripping buffer (Pierce) was used to strip antibodies from probed membranes. SuperSignal chemiluminescence substrate (Pierce) was used to detect horseradish peroxidase-conjugated secondary antibodies. Polyclonal mouse antibody reactive to SAMHD1 (ab67820) was purchased from Abcam and used at 1 μg/ml in 5% milk in Tris-buffered saline-Tween. Rabbit polyclonal antibody specific to phosphorylated SAMHD1 at residue T592 has been previously described (23). Antibodies specific to the HA epitope (Ha.11 clone 16B12) and FLAG epitope (M2) were purchased from Covance and Sigma-Aldrich, respectively. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (AbD serotec) antibody probing served as a loading control. Immunoblotting images were captured and analyzed using the Luminescent Image analyzer (LAS 4000) and Multi Gauge V3.0 software (Fuji Film) as described previously (14).
Coimmunoprecipitation and mass spectrometry.
HEK293T cells were transfected with an empty vector or constructs expressing HA-tagged mouse CD9 (a negative control) and hSAMHD1 as described previously (13, 14). At 24 h posttransfection, cells were harvested, washed in phosphate-buffered saline (PBS), and lysed in 1% digitonin lysis buffer on ice. Lysates were centrifuged, and the supernatants were collected and incubated with anti-HA agarose conjugates (Sigma-Aldrich) for 2 h at 4°C. Proteins bound to the agarose conjugate were pelleted and washed 3 times in 0.1% digitonin. Bound proteins were eluted using an HA peptide (Sigma-Aldrich) or by boiling in SDS buffer, prior to mass spectrometry or immunoblotting. Eluted lysates were used to validate mass spectrometry results by performing co-IP using an anti-HA antibody and immunoblotting for the identified cellular proteins with antibodies specific to human cyclin A2, CDK2 (Santa Cruz Biotechnology), SKP2, cyclin B1, and CDK1 (Cell Signaling).
For protein identification, co-IP products were separated by SDS-PAGE and gel slices were cut and digested with trypsin overnight at 37°C. Extracted peptides were analyzed using an LTQ-Orbitrap mass spectrometer at the Mass Spectrometry Core Facility at the Ohio State University. Protein assignments were made using Mascot software, and data analysis was performed using Scaffold software. Proteins not identified in the CD9 control lane but having spectral counts greater than 10 in the hSAMHD1 or mSAMHD1 lanes were considered for follow-up studies. Additionally, proteins with spectral counts more than 10-fold higher in the hSAMHD1 or mSAMHD1 lanes than in the CD9 control lane were also considered candidate interacting proteins for follow-up analysis.
HIV-1 stocks and viral infection assays.
Single-cycle, luciferase reporter HIV-1 pseudotyped with vesicular stomatitis virus G protein (HIV-Luc/VSV-G) was generated by calcium phosphate cotransfection of HEK293T cells with the pNL-Luc-E−R+ and pVSV-G as described previously (37). All virus stocks were harvested 2 days posttransfection and filtered through a 0.45-μm filter. The infectious units of virus stocks were evaluated by limiting dilution on GHOST/X4/R5 cells as described previously (33). HIV-1 infection assays using luciferase reporter viruses were performed using a multiplicity of infection (MOI) range of 0.5 to 2 as described previously (33, 38). Cell lysates were obtained 1 day postinfection and analyzed for luciferase activity using a commercially available kit (Promega) according to the manufacturer's instructions. Total cell protein was quantified using a bicinchoninic acid assay (BCA; Pierce), and all luciferase results were normalized to total protein content. HIV-1 infection using U937 stable cell lines was performed after PMA differentiation (100 ng/ml) for 24 h. Cells were infected at an MOI of 2 for 2 h. HIV-1 infection was determined by luciferase assays performed 24 h postinfection. THP-1 cells pretreated with dimethyl sulfoxide (DMSO) controls or CDK1 and CDK2 inhibitors for 24 h were infected with HIV-1 at an MOI of 0.5 for 2 h, after which virus was washed and cells were recultured in fresh media containing inhibitors. HIV-1 infection was determined by luciferase assay performed 24 h postinfection.
Treatment of CDK1, CDK2, and SKP2 inhibitors.
HEK293T and THP-1 cells were treated with specific inhibitors to CDK1 (22) (CGP74514A from EMD Millipore), CDK2 (39) (inhibitor II from Santa Cruz Biotech), and SKP2 E3 Ligase (40) (inhibitor I C1 from EMD Millipore) at the concentrations indicated in the figures or treated with DMSO controls. HEK293T cells (6 × 105) were pretreated for 6 h with DMSO or inhibitors and transfected with the pLenti-hSAMHD1 plasmid to overexpress hSAMHD1. The inhibitors were present during transfection, and at 18 h posttransfection fresh media containing inhibitors were added. Cells were incubated for a further 6 h and lysed for immunoblotting. THP-1 cells (1 × 106) were treated with DMSO controls or inhibitors to CDK1, CDK2, and SKP2 at the same concentrations as indicated in the figures for a total of 30 h prior to harvesting lysates for immunoblotting (23). The 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (CellTiter cell proliferation assay; Promega) was used to measure cell viability after the inhibitor treatment.
Expression of WT and dominant negative (DN) mutants of CDK1, CDK2, and SKP2 in HEK293T cells.
The plasmids encoding HA-tagged WT and the DN mutants (D146N) of CDK1 and CDK2 (41) were obtained from Addgene. The plasmid encoding FLAG-tagged WT and the DN mutant of SKP2 [F-box-deleted SKP2, or (ΔF) SKP2] (42) were kind gifts from Michele Pagano (New York University). Plasmid DNA encoding the above specific proteins or empty vector controls were cotransfected with pLenti-SAMHD1 into HEK293T cells as described previously (14). In brief, pLenti-hSAMHD1 (0.25 μg) was cotransfected into HEK293T cells (each well of 12-well plates) with either 0.25 μg or 0.75 μg of CDK1, CDK2, or SKP2 expression plasmids. All transfection samples were made to a final amount of 1 μg DNA using the empty vector. Expression of these proteins was detected by immunoblotting using anti-HA or anti-FLAG as described previously (41, 42).
siRNA-mediated knockdown of CDK1, CDK2, and cyclin A2.
HEK293T cells were transfected with SMARTpool On-Target plus small interfering RNA (siRNA) specific for CDK1 (L003224), CDK2 (L003236), cyclin A2 (L003205), or a scramble control (Dharmacon) using Trans-IT TKO (Mirus) according to the manufacturer's instructions. Cells were transfected with 100 nM siRNA and incubated for 24 h, after which a second-round cotransfection was performed with 100 nM siRNA and 0.25 μg pLenti-hSAMHD1 plasmid DNA and left for a further 24 h prior to harvesting of lysates for immunoblotting.
Statistical analysis.
Data were analyzed using the Student t test, and statistical significance was defined at P values of <0.05.
RESULTS
Identification of cellular proteins interacting with human and mouse SAMHD1.
Human and mouse SAMHD1-mediated restriction of HIV-1 is limited to noncycling cells (13, 19). We hypothesized that SAMHD1-mediated HIV-1 restriction is negatively regulated in proliferating cells that are permissive to HIV-1 infection, possibly through its interactions with certain cellular protein(s). Thus, we sought to identify such host protein(s) that interact with SAMHD1 and to examine their expression in HIV-1-permissive and -nonpermissive cell types.
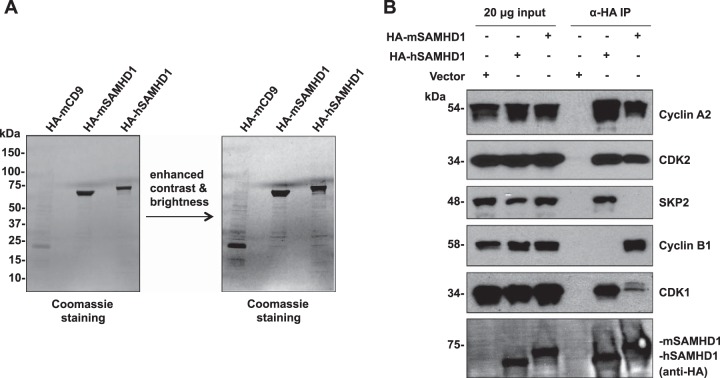
To identify cellular proteins interacting with SAMHD1, we overexpressed HA-tagged hSAMHD1 and mSAMHD1 in HEK293T cells, performed co-IP, eluted target proteins using an HA peptide, and identified SAMHD1-interacting proteins by mass spectrometry. Coomassie blue staining of the co-IP products confirmed successful IP of the HA-tagged SAMHD1 and host proteins that potentially interact with SAMHD1 (Fig. 1A). We identified four proteins interacting with hSAMHD1 and mSAMHD1, all of which are associated with cell cycle regulation (Table 1 and Fig. 1B).
FIG 1.
Identification of cellular proteins interacting with human and mouse SAMHD1. (A) HEK293T cells were transfected with constructs expressing HA-tagged human or mouse SAMHD1 (HA-hSAMHD1 and HA-mSAMHD1, respectively) or HA-tagged mouse CD9 (HA-mCD9) as a negative control. Cell lysates were harvested the following day and coimmunoprecipitated using anti-HA-coated agarose. Interacting proteins were eluted using an HA peptide, and samples were analyzed by SDS-PAGE and Coomassie blue staining. Gel slices were excised and processed for mass spectrometry analysis. (B) HEK293T cells were transfected as described for panel A, and the empty vector was used as a negative control. Eluted lysates were used to validate mass spectrometry results by performing coimmunoprecipitation using anti-HA and immunoblotting for the identified cellular proteins using antibodies specific to human cyclin A2, CDK2, SKP2, cyclin B1, and CDK1 (the order is based on the peptide abundance identified by mass spectrometry [Table 1]).
TABLE 1.
Human or mouse SAMHD1-interacting cellular proteins identified in HEK293T cells
| Cellular protein identified | Size (kDa) | No. of identifications by MSa |
||
|---|---|---|---|---|
| Mouse CD9 | Human SAMHD1 | Mouse SAMHD1 | ||
| Cyclin A2 (CCNA2) | 54 | NDb | 16 | 1 |
| Cyclin-dependent kinase 2 (CDK2) | 34 | ND | 11 | ND |
| S-phase kinase-associated protein 2 (SKP2) | 48 | ND | 10 | ND |
| G2/mitotic-specific cyclin B1 (CCNB1) | 58 | ND | ND | 24 |
| Cyclin-dependent kinase 1 (CDK1) | 34 | 1 | 8 | 22 |
HA-tagged mouse CD9 (a negative control), human SAMHD1, and mouse SAMHD1 were overexpressed in HEK293T cells. The values represent the number of times a peptide was identified and assigned to that protein in mass spectrometric analysis (MS).
ND, the protein was not detected.
To validate the mass spectrometry data, we repeated the co-IPs and confirmed the interacting proteins by immunoblotting (14). The results showed that cyclin A2, CDK2, SKP2, and CDK1 interact specifically with hSAMHD1 (Fig. 1B). Compared to hSAMHD1, mSAMHD1 was found to interact less strongly with cyclin A2, CDK2, and CDK1. However, cyclin B1 interacted specifically with mSAMHD1 only, and SKP2 did not interact with mSAMHD1 (Fig. 1B), suggesting that the different species of SAMHD1 could have their function regulated through specific interactions with different host proteins. As expected, the vector controls did not interact with any of these identified host proteins (Fig. 1B), and similar results were obtained using HA-tagged CD9 as a control (data not shown).
Regulation of SAMHD1-interacting proteins by monocyte differentiation and T-cell activation.
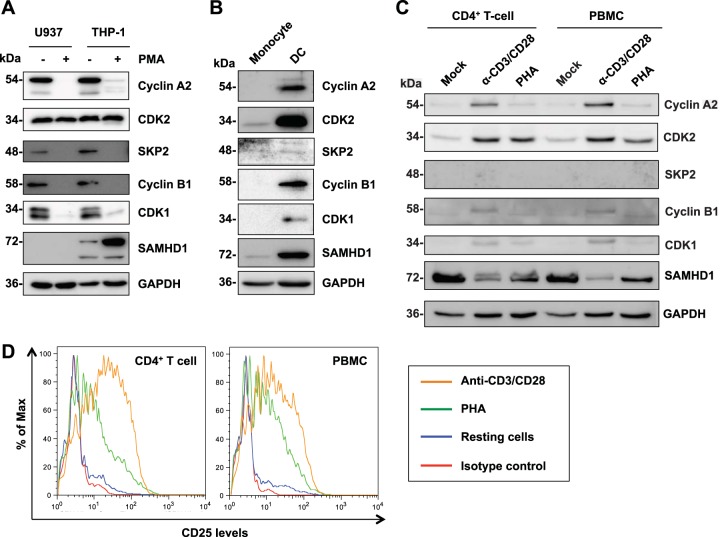
Previous studies have indicated that cyclin A2 and CDK1 expression is downregulated upon differentiation of noncycling monocytic cells (22). We thus examined expression levels of the identified interacting proteins in cell types that are known to be permissive or restrictive to HIV-1 infection. We found that all of the interacting proteins identified were expressed in cycling monocytic U937 and THP-1 cells (Fig. 2A), which are permissive to HIV-1 infection (5). PMA treatment of these monocytic cell lines that express high levels of hSAMHD1 differentiates the cells into a noncycling status and potentiates SAMHD1-mediated HIV-1 restriction (5). Upon PMA treatment, the expression level of cyclin A2/B1, SKP2, and CDK1 was decreased or below the detection limit, while CDK2 expression remained constant (Fig. 2A). As expected, U937 cells did not express endogenous SAMHD1, and we observed an increase in SAMHD1 levels in PMA-treated THP-1 cells (Fig. 2A).
FIG 2.
Regulation of SAMHD1-interacting proteins by monocyte differentiation and T-cell activation. (A) Regulation of the expression of SAMHD1 and its interacting proteins in monocytic cell lines by PMA treatment for 36 h. Cells were cultured and mock or PMA treated. Cell lysates (20 μg) were used for protein detection by immunoblotting, and GAPDH was used as a loading control. (B) Regulation of the expression of SAMHD1 and its interacting proteins in primary monocytes and monocyte-derived dendritic cells (DCs). DCs were differentiated from autologous monocytes, and cell lysates were analyzed by immunoblotting. Data shown represent one of two donors analyzed. (C) Regulation of the expression of SAMHD1 and its interacting proteins by activation of CD4+ T cells and PBMCs by either anti-CD3/CD28 or PHA treatment for 72 h. Cell lysates (10 μg) were analyzed by immunoblotting. Data shown represent one of two donors analyzed. (D) Activation of primary resting CD4+ T cells and PBMCs. Primary CD4+ T cells and PBMCs were isolated from healthy donors and activated by treatment with PHA or anti-CD3/CD28 in the presence of IL-2. Immunostaining of the T-cell activation markers CD25 was analyzed by flow cytometry.
Comparison between donor-matched primary monocytes and DCs revealed that monocytes, which are highly refractory to HIV-1 infection (7, 29), had very low levels of CDK2 and SKP2 expression (Fig. 2B). None of the other identified proteins were detectable (Fig. 2B). In contrast, DCs, which support HIV-1 infection to a very low extent (5, 14), expressed detectable levels of all the proteins (Fig. 2B). Of note, DCs expressed higher levels of SAMHD1 than did undifferentiated monocytes; however, SAMHD1 has been previously reported to restrict HIV-1 in undifferentiated primary monocytes (7). This could suggest that SAMHD1-mediated restriction is tightly regulated and highly active in monocytes or that there are additional, unknown mechanisms of HIV-1 restriction in monocytes compared to DCs.
SAMHD1 also acts as a restriction factor for HIV-1 in resting CD4+ T cells (10, 12). We thus compared the expression of SAMHD1-interacting proteins in resting CD4+ T cells and PBMCs to either PHA- or anti-CD3/CD28-activated cells. We found that in resting cells, SAMHD1-interacting proteins were not detectable (SKP2, cyclin B1, and CDK1) or were expressed at very low levels (cyclin A2 and CDK2) (Fig. 2C). Interestingly, upon activation, expression of each protein was increased, which was more pronounced in anti-CD3/CD28-activated cells than in PHA-activated cells and correlated with the activation status of the cells (Fig. 2C and D). SKP2 remained undetectable despite activation of the cells. We also noted a decrease in SAMHD1 levels in activated CD4+ T cells and PBMCs, particularly in anti-CD3/CD28-activated cells (Fig. 2C), which is consistent with our previous results (14) and a recent report (43) and suggests a mechanism of SAMHD1 downregulation by T-cell activation. Together, these results demonstrate that monocyte differentiation and T-cell activation can significantly regulate the expression of several SAMHD1-interacting proteins.
Inhibitors of CDK1 and CDK2 block T592 phosphorylation of SAMHD1 and reduce HIV-1 infection in THP-1 cells.
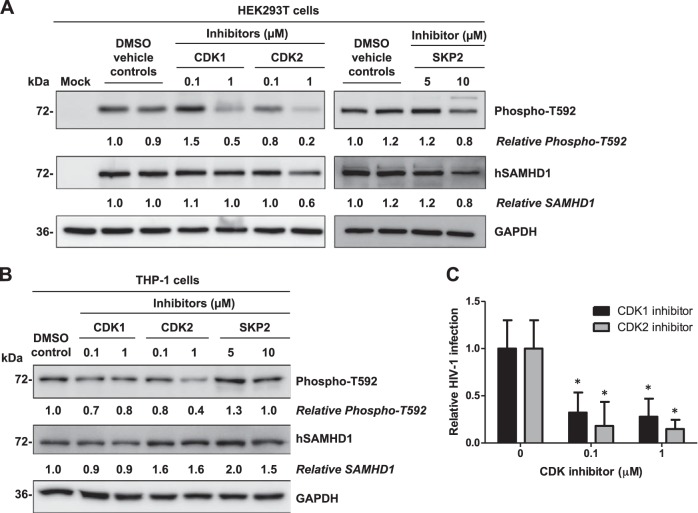
A previous study demonstrated that inhibition of CDK1 impairs phosphorylation of SAMHD1 at T592 in U937 cells stably expressing hSAMHD1 (22). To examine whether inhibition of CDK1, CDK2, and SKP2 activity affected the phosphorylation of SAMHD1 at T592, we pretreated HEK293T cells with specific inhibitors to CDK1, CDK2, and SKP2 prior to overexpressing hSAMHD1. These inhibitors were used below concentrations reported to have off-target effects (manufacturers' data) and were used for a duration of time deemed to not have cytotoxic effects, as determined by cell viability assays (data not shown). Both CDK1 and CDK2 inhibitors (at 1 μM) reduced phosphorylation of SAMHD1 at T592 by 50% and 80%, respectively (Fig. 3A). Inhibition of SKP2 activity with the specific inhibitor (at 5 or 10 μM) did not have a significant effect on the level of T592-phosphorylated SAMHD1 protein (Fig. 3A).
FIG 3.
Inhibitors of CDK1 and CDK2 block T592 phosphorylation of SAMHD1 and reduce HIV-1 infection in THP-1 cells. (A) HEK293T cells were pretreated with either DMSO (vehicle control) or inhibitors to CDK1, CDK2, and SKP2 at the assigned concentrations and transiently transfected to overexpress SAMHD1. The effect of each individual inhibitor on the phosphorylation of SAMHD1 at T592 was assessed at 30 h post-inhibitor treatment by immunoblotting. (B) THP-1 cells were treated with either DMSO or inhibitors to CDK1, CDK2, and SKP2 at the concentrations indicated for 30 h. Endogenous total or T592-phosphorylated SAMHD1 protein levels were determined by immunoblotting using specific antibodies. GAPDH was used as a loading control. Relative phospho-T592 levels represent T592-phosphorylated hSAMHD1 normalized to GAPDH. Relative hSAMHD1 levels represent hSAMHD1 normalized to GAPDH. Protein bands were quantified and normalized to the respective DMSO-treated controls, which were set to 1. The data presented are representative of 3 independent experiments. (C) Inhibitors of CDK1 and CDK2 reduce HIV-1 infection in THP-1 cells. Cells were pretreated with either DMSO control or inhibitors of CDK1 or CDK2 and then cultured in the presence of the inhibitors for 24 h at the concentrations indicated. At 24 h post-inhibitor treatment, cells were challenged with single-cycle HIV-Luc/VSV-G at an MOI of 0.5 for 2 h; cells were then washed and cultured in fresh media containing inhibitor for 24 h, and lysates were harvested for luciferase assay. All infections were calculated relative to the DMSO controls, which were set as 1. Data presented are representative of 2 independent experiments, and error bars show the standard deviations of triplicate samples. *, P < 0.05 (compared to the DMSO controls without inhibitors).
We also determined the effects of these inhibitors on T592 phosphorylation of endogenous hSAMHD1 protein in undifferentiated THP-1 cells. In keeping with the results obtained using hSAMHD1-overexpressing HEK293T cells, we found that treatment of THP-1 cells with CDK1- and CDK2-specific inhibitors reduced T592 phosphorylation of hSAMHD1 by 20 to 30% and 20 to 60%, respectively (Fig. 3B), suggesting that both CDK1 and CDK2 mediate T592 phosphorylation of endogenous hSAMHD1 in the cells. The treatment of THP-1 cells with a SKP2 inhibitor had no significant effect on T592 phosphorylation of hSAMHD1 (Fig. 3B), which was consistent with our findings in HEK293T cells (Fig. 3A). Given that CDK1 and CDK2 are important for SAMHD1 phosphorylation in THP-1 cells, we questioned whether blocking CDK1 or CDK2 kinase activity and thereby SAMHD1 phosphorylation at T592 might render THP-1 cells less permissive to HIV-1 infection. To this end, we treated THP-1 cells with CDK1 and CDK2 inhibitors and challenged the treated cells with a single-cycle luciferase reporter HIV-Luc/VSV-G. Inhibition of both CDK1 and CDK2 at 0.1 and 1.0 μM concentrations significantly reduced HIV-1 infection 3- to 4-fold and 5.5- to 6.8-fold compared to DMSO controls (P < 0.05), respectively (Fig. 3C). Cell viability assays confirmed that the inhibitor concentrations used for the duration of the infection experiment (2 days) were not significantly cytotoxic in THP-1 cells (>90% viable cells), which ruled out a possible reduction in HIV-1 infection resulting from inhibitor-impaired cell viability. Thus, our data demonstrate a correlation between decreased T592 phosphorylation and reduced HIV-1 infection, suggesting an important role for CDK1- and CDK2-mediated SAMHD1 phosphorylation at T592 in negatively regulating HIV-1 infection.
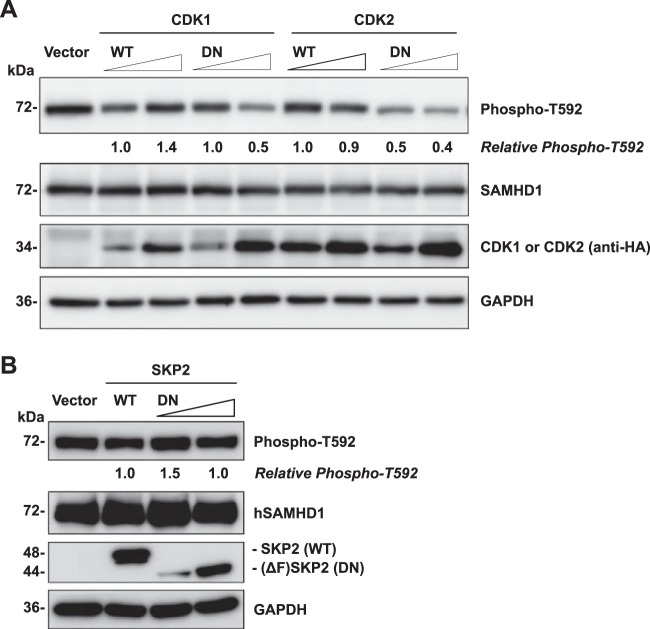
Expression of DN mutants of CDK1 and CDK2 reduces T592 phosphorylation of hSAMHD1.
To confirm the effects of inhibitors to CDK1, CDK2, and SKP2 on SAMHD1 phosphorylation and eliminate any potential off-target effects of the inhibitors, we coexpressed hSAMHD1 with DN mutants of CDK1, CDK2, and SKP2 in HEK293T cells and examined T592-phosphorylated SAMHD1. The DN mutants (D146N) of CDK1 and CDK2 are CDK-defective variants of their active sites, which specifically inhibit the function of endogenous CDK1 and CDK2, respectively, when overexpressed in cells (41). The SKP2 DN mutant is an F-box-deleted variant that is unable to bind components of the E3 ligase complex and therefore can inhibit endogenous SKP2 activity (42). The expression of WT and DN mutants of CDK1, CDK2, and SKP2 in transfected HEK293T cells was confirmed by immunoblotting (Fig. 4A and B). As expected, the DN mutant of SKP2 with F-box deletion (44 kDa) was smaller than the WT protein (48 kDa) (Fig. 4B). The levels of phosphorylated SAMHD1 at position T592 were decreased by 50% only when the CDK1 DN mutant was overexpressed at a higher level, compared to the empty vector or WT CDK1 controls, without affecting total SAMHD1 protein levels (Fig. 4A). A stronger effect was observed for the CDK2 DN mutant, which reduced SAMHD1 phosphorylation at T592 by 50 and 60% under low and high CDK2 DN overexpression conditions, respectively. However, this enhanced effect could be attributed to the expression level of the DN mutant of CDK2 being higher than that of CDK1 (Fig. 4A). These data corroborate that CDK1 is involved in T592 phosphorylation of hSAMHD1 (22, 23) and further implicate CDK2 as a host protein that mediates SAMHD1 phosphorylation at T592. Overexpression of a DN mutant of SKP2 did not have a significant effect on T592 phosphorylation of hSAMHD1 (Fig. 4B), in agreement with the results obtained using SKP2 inhibitor (Fig. 3). These data suggest that SKP2 is not directly involved in T592 phosphorylation of hSAMHD1, though the interaction may serve a separate role.
FIG 4.
Expression of dominant negative (DN) mutants of CDK1 and CDK2 reduces T592 phosphorylation of hSAMHD1. (A and B) Overexpression of hSAMHD1 and wild-type (WT) or DN mutants of HA-tagged CDK1 or CDK2 (A) or FLAG-tagged SKP2 (B) in HEK293T cells. Empty vector was used as a negative control. A 2-fold increase of plasmid DNA-expressing DN mutants was applied in the cotransfection as indicated. At 24 h posttransfection, cell lysates were harvested for immunoblotting to determine overexpression of transfected DNA using either HA- or FLAG-specific antibodies. The effect on phosphorylated SAMHD1 levels was determined using a phospho-T592-specific antibody. GAPDH was used as a loading control. Relative phospho-T592 levels represent T592-phosphorylated hSAMHD1 normalized to GAPDH. Protein bands were quantified and normalized to the respective WT controls, which were set to 1. The data presented are representative of 3 independent experiments.
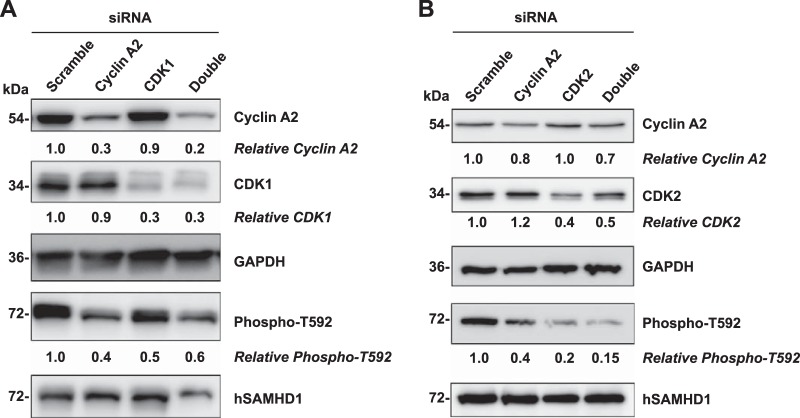
Knockdown of CDK1, CDK2, and cyclin A2 in HEK293T cells reduces T592 phosphorylation of hSAMHD1.
Previous studies suggested that cyclin A2 and CDK1 are responsible for hSAMHD1 phosphorylation at T592 (22, 23). To examine whether knockdown of CDK1 and cyclin A2 in dividing cells affects phosphorylation of SAMHD1 at T592, siRNA-mediated knockdown of CDK1 and cyclin A2 was performed in HEK293T cells. The results showed that both cyclin A2 and CDK1 protein levels were efficiently reduced, by 70 to 80%, using specific siRNAs, compared to the scramble control (Fig. 5A). Immunoblotting revealed that in the cyclin A2- and CDK1-specific knockdown cells, phosphorylation of overexpressed hSAMHD1 at T592 was significantly reduced by approximately 40 to 60% compared to total SAMHD1 protein (Fig. 5A). To further establish a role for CDK2 in SAMHD1 phosphorylation, we also performed siRNA-mediated knockdown of CDK2, either alone or in combination with cyclin A2. The data show that siRNA knockdown of CDK2 reduced protein levels by 60% compared to the scrambled siRNA control (Fig. 5B). A similar reduction in CDK2 protein levels was also observed when CDK2 was knocked down in combination with cyclin A2. CDK2 knockdown resulted in an 80 to 85% reduction in SAMHD1 phosphorylation at T592 (Fig. 5B). Notably, siRNA knockdown of CDK2 had a more pronounced effect on the reduction of SAMHD1 phosphorylation at T592 than CDK1, which is consistent with our findings using CDK1/2 inhibitors (Fig. 3) and DN mutants (Fig. 4). Taken together, our results provide direct evidence that CDK2 can also partner with cyclin A2 to phosphorylate hSAMHD1 at T592.
FIG 5.
Knockdown of CDK1, CDK2, and cyclin A2 in HEK293T cells reduces T592 phosphorylation of SAMHD1. siRNA-mediated knockdown of cyclin A2, CDK1, or both (double) (A) and cyclin A2, CDK2, or both (double) (B) in HEK293T cells was performed using a two-round transfection protocol. At 24 h after the second round of siRNA transfection, cell lysates were harvested for immunoblotting to determine efficient knockdown of target proteins. GAPDH was used as a loading control. Relative cyclin A2 and CDK1 levels shown represent GAPDH-normalized densitometry compared to scramble siRNA control. Cells that had undergone a single round of siRNA transfection were then dually transfected with a second round of siRNA as well as a plasmid to overexpress wild-type hSAMHD1. Cell lysates were harvested at 24 h after the second round of siRNA transfection, and levels of phosphorylated (Phospho-T592) and total hSAMHD1 proteins were determined by immunoblotting. Relative phosphorylated T592 levels represent phosphorylated hSAMHD1 normalized to total hSAMHD1, compared to scramble control, which was set as 1. Membranes to be immunoblotted were cut and probed for each antibody accordingly as shown (cyclin A2, CDK1, CDK2, GAPDH, T592 phosphorylated hSAMHD1, and total hSAMHD1). The data presented are representative of 3 independent experiments.
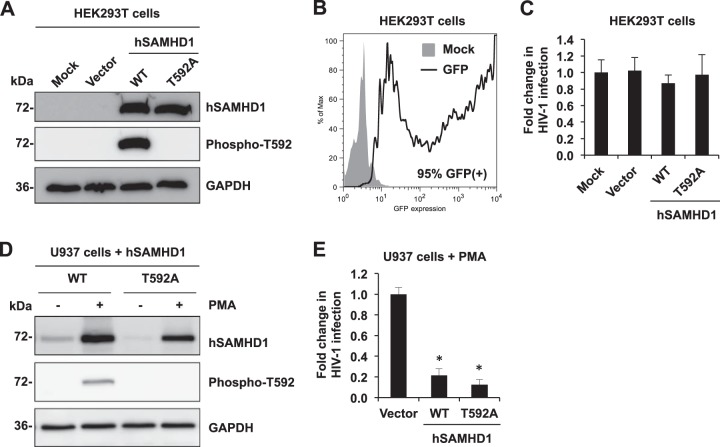
The unphosphorylated form of hSAMHD1 at T592 restricts HIV-1 infection in differentiated U937 cells but not in cycling HEK293T cells.
To directly test whether hSAMHD1 phosphorylation at T592 regulates its HIV-1 restriction function, we compared the HIV-1-restrictive function of WT hSAMHD1 and the phospho-ablative T592A mutant in cycling HEK293T cells and in a differentiated U937 cell line expressing hSAMHD1. HEK293T cells were transfected with either vector control, WT, or T592A to overexpress hSAMHD1. Overexpressed WT hSAMHD1 in cycling HEK293T cells was confirmed to be phosphorylated at T592. As expected, the T592A mutant SAMHD1 could not be detected using a phosphospecific antibody (Fig. 6A). The transfection efficiency of HEK293T cells was 95% as indicated by the expression of GFP from a control vector (Fig. 6B). To determine the effect of the T592A mutant on HIV-1 infection, transfected HEK293T cells were then infected with HIV-Luc/VSV-G. As expected, vector control and WT SAMHD1 overexpression did not affect HIV-1 infection in HEK293T cells (Fig. 6C), which is likely due to insignificant depletion of the dNTP pool in the cells (14). Interestingly, overexpression of T592A hSAMHD1 in HEK293T cells did not result in HIV-1 restriction (Fig. 6C), which is consistent with the results of the T592V mutant of hSAMHD1 in HeLa cells (23). Together, these data suggest that abolishing the phosphorylation of SAMHD1 at T592 is not sufficient to induce a SAMHD1-specific restriction phenotype in cycling cells.
FIG 6.
Unphosphorylated form of hSAMHD1 at T592 restricts HIV-1 infection in differentiated U937 cells, but not in cycling HEK293T cells. (A and B) HEK293T cells were transfected with vector control or plasmids expressing wild-type (WT) hSAMHD1 or a T592A mutant. (A) At 24 h posttransfection, lysates were harvested for immunoblotting to confirm overexpression of total hSAMHD1 and T592 phosphorylated hSAMHD1 (Phospho-T592). (B) High efficiency of transfection in HEK293T cells. Cells transfected with a GFP-expressing vector were harvested and analyzed by flow cytometry at 24 h posttransfection to determine GFP expression (95% cells were positive for GFP). (C) Transfected HEK293T cells were infected with single-cycle HIV-Luc/VSV-G at an MOI of 0.5. At 24 h postinfection, cell lysates were harvested for a luciferase assay to determine HIV-1 infection. The fold change of HIV-1 infection is shown. The data presented are representative of 2 independent experiments. (D) U937 cells stably expressing hSAMHD1 (WT) and hSAMHD1 (T592A) mutant were treated with PMA for 24 h and subjected to immunoblotting to assess the expression level of WT and T592A mutant proteins. (E) A parallel set of samples that were PMA treated in a similar manner were infected with HIV-Luc/VSV-G at an MOI of 2, and infection levels were assessed at 24 h by measuring the luciferase activity. The data presented are representative of 3 independent experiments, and error bars show the standard deviations of triplicate samples. *, P < 0.001 (compared to the vector control).
Given that SAMHD1 restricts HIV-1 in PMA-treated U937-SAMHD1 stable cells (5, 23), we questioned whether the phosphorylation-defective T592A mutant of SAMHD1 would block HIV-1 more effectively than wild-type SAMHD1 using this cell model. Expression of the WT and the T592A mutant in the PMA-treated U937 stable cell lines was confirmed by immunoblotting using SAMHD1- and phospho-specific antibodies (Fig. 6D). Consistent with previous published results (13), we observed significant induction of WT and T592A expression upon PMA treatment, which has been attributed to PMA-mediated transcriptional upregulation of the cytomegalovirus (CMV) promoter that drives expression of WT and T592A from the integrated pLenti-based proviral vector (13). Single-cycle HIV-1 infection of the PMA-treated U937-SAMHD1 (WT) stable cell line revealed a 5-fold reduction in infection compared to the vector control cell line (P = 0.00012), and PMA-treated U937-SAMHD1 (T592A) cells restricted HIV-1 efficiently as evidenced by a 6.7-fold decrease (P = 0.00005) in infection (Fig. 6E). There is no statistic difference (P = 0.136) between WT and T592A in restricting HIV-1 infection in PMA-treated U937 cells (Fig. 6E). However, this restriction phenotype was not able to be efficiently further enhanced, most likely due to overexpression of WT SAMHD1 alone rendering the cells highly restrictive to HIV-1 infection. These results suggest the involvement of cell cycle-related CDK/cyclin complexes in posttranslationally regulating hSAMHD1 through phosphorylation at T592 in noncycling cells, which could be important for the anti-HIV-1 activity of hSAMHD1.
DISCUSSION
Our study confirmed recent findings that hSAMHD1 interacts with CDK1 and cyclin A2 (22, 23) and identified additional cell cycle regulatory proteins (CDK2 and SKP2) that interact with hSAMHD1. We also identified that mSAMHD1 specifically interacts with cell cycle-related proteins, including cyclin A2, cyclin B1, CDK1, and CDK2. The identification of these SAMHD1-interacting proteins suggests that the dNTPase activity of SAMHD1 or its retroviral restriction activity may be tightly regulated in cycling and noncycling cells.
Interestingly, we observed some differences in interacting proteins between human and mouse SAMHD1. Of note, previous studies indicated that mSAMHD1 isoform 1 (used in this study), but not isoform 2, has a phosphorylation site at threonine 603 (44, 45). It would be interesting to investigate whether phosphorylation of mSAMHD1 isoform 1 at this site is regulated by CDK1/2, cyclin A2, or cyclin B1 and whether this phosphorylation is critical for mSAMHD1-mediated retroviral restriction. However, due to the lack of an antibody to specifically recognize phosphorylation of mSAMHD1, we did not examine the effect of the mSAMHD1-interacting proteins on phosphorylation and restriction function of mSAMHD1.
Our observation that the SAMHD1-interacting proteins are downregulated in HIV-1 restrictive, noncycling cells, such as monocytes or resting CD4+ T cells, suggests that in cycling, HIV-1 permissive cells, these proteins could negatively regulate SAMHD1 function through its phosphorylation. Interestingly, in DCs, expression of all interacting proteins was detectable, suggesting a potential equilibrium between the unphosphorylated and phosphorylated forms of SAMHD1, which is consistent with previous findings (22) and helps explain the very low levels of HIV-1 infection observed in DCs (5, 14). CDK2 was the only interacting protein whose expression level was consistent in cycling or noncycling U937 and THP-1 cells, although it was increased slightly in activated CD4+ T cells and PBMCs.
Although CDK1 has previously been identified to phosphorylate SAMHD1 at T592 (22, 23), we hypothesized that CDK2 also partners with one of the cyclins to phosphorylate hSAMHD1 at T592. Using an inhibitor to CDK2 in two different cell types (Fig. 3), we found that CDK2 is able to mediate phosphorylation of SAMHD1 at T592, indicating that CDK1 is not the only protein interacting with hSAMHD1 that can mediate its phosphorylation status at T592. Using DN mutants of CDK1 and CDK2 in HEK293T cells (Fig. 4A), we further confirmed that both CDK1 and CDK2 are also able to mediate T592 phosphorylation of hSAMHD1. Our new findings suggest that SAMHD1 phosphorylation in vivo could be mediated by multiple host kinases. To evaluate the importance of CDK1- and CDK2-mediated phosphorylation of SAMHD1 for HIV-1 infection, we pretreated THP-1 cells with inhibitors to CDK1 and CDK2 prior to challenge with HIV-1 (Fig. 3C). Our results suggest that dephosphorylation of SAMHD1 is correlated with SAMHD1-mediated HIV-1 restriction.
SKP2 is a component of the SKP1-Cullin-1-F-box ubiquitin E3 ligase complex and has been shown to interact with cyclin A2 and CDK2 (46). Treatment of SAMHD1-expressing THP-1 cells and HEK293T cells with a SKP2 inhibitor did not reduce levels of T592-phosphorylated hSAMHD1 (Fig. 3), suggesting that SKP2 is unlikely to be involved in T592 phosphorylation of SAMHD1. Our results of DN mutants of SKP2 further confirmed this observation (Fig. 4B). However, given that SKP2 has a role in regulating cell survival and apoptosis (47), its interaction with hSAMHD1 may serve another potential cellular role.
Although we did not observe cytotoxic effects, overexpression of CDK1/CDK2 and DN interference may have potential global effects on the cell cycle. We thus confirmed our results using an siRNA knockdown approach. We observed that siRNA knockdown of CDK1, CDK2, and cyclin A2 in HEK293T cells expressing WT hSAMHD1 reduced SAMHD1 T592 phosphorylation, which further confirms the results using CDK1/2 inhibitors and DN mutants. However, siRNA knockdown of these proteins in HEK293T cells did not affect HIV-1 infection (data not shown), suggesting that the phosphorylation status of overexpressed hSAMHD1 alone is not sufficient to mediate HIV-1 restriction in cycling cells. These results were further supported by the lack of HIV-1 restriction observed with the phospho-ablative T592A mutant in HEK293T cells (Fig. 6A and C). Our previous work indicated that overexpression of hSAMHD1 in HEK293T cells is not capable of reducing the dNTP pool below the Km of HIV-1 reverse transcriptase (14). Thus, we speculate that a sufficiently decreased dNTP pool and dephosphorylation of SAMHD1 at T592 are required in conjunction to mediate HIV-1 restriction in noncycling cells. Evidence for this was provided when we compared the HIV restriction capacity of the T592A mutant with WT SAMHD1 in PMA-differentiated monocytic cells. Upon PMA treatment, we observed that although SAMHD1 WT protein is not completely unphosphorylated (Fig. 6D), both WT hSAMHD1 and T592A mutant efficiently restricted HIV-1 infection (Fig. 6E). These results suggest that an equilibrium between T592 unphosphorylated and phosphorylated forms of hSAMHD1 exists in PMA-treated U937-SAMHD1 (WT) cells, but the levels of phosphorylated SAMHD1 may not be high enough to significantly affect HIV-1 restriction function of SAMHD1. This could explain the slightly lower level of restriction of HIV-1 by WT hSAMHD1 than by the T592A mutant. In addition, as PMA treatment of SAMHD1-expressing U937 cells renders the cells highly restrictive to HIV-1 infection, it is plausible that the restriction effect cannot be further enhanced in this cellular model.
Our identification of SAMHD1-interacting cellular proteins reveals CDK2 as an additional cofactor contributing to T592 phosphorylation of hSAMHD1 and thereby regulating SAMHD1-mediated HIV-1 restriction function in nondividing cells. A recent study indicated that endogenous SAMHD1 expression in human fibroblasts is regulated by the cell cycle, with maximal expression during quiescence and minimal expression during S phase (48). Thus, SAMHD1 interactions with the cellular cofactors may have a significant role in regulating its biological function, particularly in relation to the cell cycle. Although previous studies have indicated that the dNTPase activity of hSAMHD1 is independent of hSAMHD1 phosphorylation (23, 24), our results suggest that both sufficient dNTPase activity and unphosphorylated hSAMHD1 at T592 are likely to be required for hSAMHD1 retroviral restriction activity.
ACKNOWLEDGMENTS
We thank Nathaniel Landau (New York University) for SAMHD1-expressing constructs, Vineet KewalRamani (National Cancer Institute) for U937 and THP-1 cell lines, and Michele Pagano for the SKP2-expressing constructs. We thank Gustavo Leone and James Dowdle (Ohio State University) for sharing expertise in the blotting of cell cycle proteins. We thank Heather Hoy for technical assistance and the members of the Wu laboratory for helpful discussions. Recombinant IL-2 was obtained from the AIDS Research and Reference Reagent Program of the National Institutes of Health (NIH).
This work was supported by NIH grants AI098524, AI102822, and AI104483 to L.W. and AI095348 to J.S.Y. T.E.W. and F.D.-G. were supported by NIH grant AI087390 to F. D.-G. L.W. and J.S.Y. are supported in part by the Public Health Preparedness for Infectious Diseases Program of The Ohio State University.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.St Gelais C, Wu L. 2011. SAMHD1: a new insight into HIV-1 restriction in myeloid cells. Retrovirology 8:55. 10.1186/1742-4690-8-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L. 2012. SAMHD1: a new contributor to HIV-1 restriction in resting CD4+ T-cells. Retrovirology 9:88. 10.1186/1742-4690-9-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L. 2013. Cellular and biochemical mechanisms of the retroviral restriction factor SAMHD1. ISRN Biochem. 2013:11. 10.1155/2013/728392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu L. 2013. Samhd1 knockout mice: modeling retrovirus restriction in vivo. Retrovirology 10:142. 10.1186/1742-4690-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7:e1002425. 10.1371/journal.ppat.1002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- 9.Powell RD, Holland PJ, Hollis T, Perrino FW. 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286:43596–43600. 10.1074/jbc.C111.317628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18:1682–1687. 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9:87. 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228. 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9:105. 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. 2013. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436:81–90. 10.1016/j.virol.2012.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288:8101–8110. 10.1074/jbc.M112.431148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T. 2012. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to Aicardi-Goutieres syndrome-associated mutations. Hum. Mutat. 33:1116–1122. 10.1002/humu.22087 [DOI] [PubMed] [Google Scholar]
- 18.Tungler V, Staroske W, Kind B, Dobrick M, Kretschmer S, Schmidt F, Krug C, Lorenz M, Chara O, Schwille P, Lee-Kirsch MA. 2013. Single-stranded nucleic acids promote SAMHD1 complex formation. J. Mol. Med. (Berl.) 91:759–770. 10.1007/s00109-013-0995-3 [DOI] [PubMed] [Google Scholar]
- 19.Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR. 2012. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86:12552–12560. 10.1128/JVI.01657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, Bieniasz PD, Towers GJ, Moita LF, Crow YJ, Bonthron DT, Reis ESC. 2013. SAMHD1-dependent retroviral control and escape in mice. EMBO J. 32:2454–2462. 10.1038/emboj.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrendt R, Schumann T, Gerbaulet A, Nguyen LA, Schubert N, Alexopoulou D, Berka U, Lienenklaus S, Peschke K, Gibbert K, Wittmann S, Lindemann D, Weiss S, Dahl A, Naumann R, Dittmer U, Kim B, Mueller W, Gramberg T, Roers A. 2013. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 4:689–696. 10.1016/j.celrep.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3:1036–1043. 10.1016/j.celrep.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 23.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. 10.1016/j.chom.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welbourn S, Dutta SM, Semmes OJ, Strebel K. 2013. Restriction of virus infection but not catalytic dNTPase activity are regulated by phosphorylation of SAMHD1. J. Virol. 87:11516–11524. 10.1128/JVI.01642-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Draetta G, Luca F, Westendorf J, Brizuela L, Ruderman J, Beach D. 1989. Cdc2 protein kinase is complexed with both cyclin A and B: evidence for proteolytic inactivation of MPF. Cell 56:829–838. 10.1016/0092-8674(89)90687-9 [DOI] [PubMed] [Google Scholar]
- 26.Pines J, Hunter T. 1989. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58:833–846. 10.1016/0092-8674(89)90936-7 [DOI] [PubMed] [Google Scholar]
- 27.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376:313–320. 10.1038/376313a0 [DOI] [PubMed] [Google Scholar]
- 28.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. 10.1006/viro.1995.1016 [DOI] [PubMed] [Google Scholar]
- 29.Dong C, Kwas C, Wu L. 2009. Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J. Virol. 83:3518–3527. 10.1128/JVI.02665-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JH, Janas AM, Olson WJ, KewalRamani VN, Wu L. 2007. CD4 coexpression regulates DC-SIGN-mediated transmission of human immunodeficiency virus type 1. J. Virol. 81:2497–2507. 10.1128/JVI.01970-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JH, Janas AM, Olson WJ, Wu L. 2007. Functionally distinct transmission of human immunodeficiency virus type 1 mediated by immature and mature dendritic cells. J. Virol. 81:8933–8943. 10.1128/JVI.00878-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong C, Janas AM, Wang JH, Olson WJ, Wu L. 2007. Characterization of human immunodeficiency virus type 1 replication in immature and mature dendritic cells reveals dissociable cis- and trans-infection. J. Virol. 81:11352–11362. 10.1128/JVI.01081-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Martin TD, Vazeux R, Unutmaz D, KewalRamani VN. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76:5905–5914. 10.1128/JVI.76.12.5905-5914.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Bashirova AA, Martin TD, Villamide L, Mehlhop E, Chertov AO, Unutmaz D, Pope M, Carrington M, KewalRamani VN. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. U. S. A. 99:1568–1573. 10.1073/pnas.032654399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Silva S, Hoy H, Hake TS, Wong HK, Porcu P, Wu L. 2013. Promoter methylation regulates SAMHD1 gene expression in human CD4+ T cells. J. Biol. Chem. 288:9284–9292. 10.1074/jbc.M112.447201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janas AM, Wu L. 2009. HIV-1 interactions with cells: from viral binding to cell-cell transmission. Curr. Protoc. Cell Biol. Chapter 26:Unit 26.5. 10.1002/0471143030.cb2605s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Silva S, Planelles V, Wu L. 2012. Differential effects of Vpr on single-cycle and spreading HIV-1 infections in CD4+ T-cells and dendritic cells. PLoS One 7:e35385. 10.1371/journal.pone.0035385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JH, Kwas C, Wu L. 2009. Intercellular adhesion molecule 1 (ICAM-1), but not ICAM-2 and -3, is important for dendritic cell-mediated human immunodeficiency virus type 1 transmission. J. Virol. 83:4195–4204. 10.1128/JVI.00006-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bramson HN, Corona J, Davis ST, Dickerson SH, Edelstein M, Frye SV, Gampe RT, Jr, Harris PA, Hassell A, Holmes WD, Hunter RN, Lackey KE, Lovejoy B, Luzzio MJ, Montana V, Rocque WJ, Rusnak D, Shewchuk L, Veal JM, Walker DH, Kuyper LF. 2001. Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis. J. Med. Chem. 44:4339–4358. 10.1021/jm010117d [DOI] [PubMed] [Google Scholar]
- 40.Rico-Bautista E, Wolf DA. 2012. Skipping cancer: small molecule inhibitors of SKP2-mediated p27 degradation. Chem. Biol. 19:1497–1498. 10.1016/j.chembiol.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 41.van den Heuvel S, Harlow E. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050–2054. 10.1126/science.8266103 [DOI] [PubMed] [Google Scholar]
- 42.Carrano AC, Eytan E, Hershko A, Pagano M. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193–199. 10.1038/12013 [DOI] [PubMed] [Google Scholar]
- 43.Bloch N, O'Brien M, Norton TD, Polsky SB, Bhardwaj N, Landau NR. 2014. HIV type 1 infection of plasmacytoid and myeloid dendritic cells is restricted by high levels of SAMHD1 and cannot be counteracted by Vpx. AIDS Res. Hum. Retroviruses 30:195–203. 10.1089/aid.2013.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweet SM, Bailey CM, Cunningham DL, Heath JK, Cooper HJ. 2009. Large scale localization of protein phosphorylation by use of electron capture dissociation mass spectrometry. Mol. Cell. Proteomics 8:904–912. 10.1074/mcp.M800451-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanivan S, Gnad F, Wickstrom SA, Geiger T, Macek B, Cox J, Fassler R, Mann M. 2008. Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J. Proteome Res. 7:5314–5326. 10.1021/pr800599n [DOI] [PubMed] [Google Scholar]
- 46.Yam CH, Ng RW, Siu WY, Lau AW, Poon RY. 1999. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19:635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan CH, Morrow JK, Zhang S, Lin HK. 2014. Skp2: a dream target in the coming age of cancer therapy. Cell Cycle 154:556–568. 10.1016/j.cell.2013.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franzolin E, Pontarin G, Rampazzo C, Miazzi C, Ferraro P, Palumbo E, Reichard P, Bianchi V. 2013. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 110:14272–14277. 10.1073/pnas.1312033110 [DOI] [PMC free article] [PubMed] [Google Scholar]