ABSTRACT
Tomato spotted wilt virus (TSWV) is the type member of tospoviruses (genus Tospovirus), plant-infecting viruses that cause severe damage to ornamental and vegetable crops. Tospoviruses are transmitted by thrips in the circulative propagative mode. We generated a collection of NSs-defective TSWV isolates and showed that TSWV coding for truncated NSs protein could not be transmitted by Frankliniella occidentalis. Quantitative reverse transcription (RT)-PCR and immunostaining of individual insects detected the mutant virus in second-instar larvae and adult insects, demonstrating that insects could acquire and accumulate the NSs-defective virus. Nevertheless, adults carried a significantly lower viral load, resulting in the absence of transmission. Genome sequencing and analyses of reassortant isolates showed genetic evidence of the association between the loss of competence in transmission and the mutation in the NSs coding sequence. Our findings offer new insight into the TSWV-thrips interaction and Tospovirus pathogenesis and highlight, for the first time in the Bunyaviridae family, a major role for the S segment, and specifically for the NSs protein, in virulence and efficient infection in insect vector individuals.
IMPORTANCE Our work is the first to show a role for the NSs protein in virus accumulation in the insect vector in the Bunyaviridae family: demonstration was obtained for the system TSWV-F. occidentalis, arguably one of the most damaging combination for vegetable crops. Genetic evidence of the involvement of the NSs protein in vector transmission was provided with multiple approaches.
INTRODUCTION
Tomato spotted wilt virus (TSWV) is the type species of Tospovirus, the only plant-infecting genus in the family Bunyaviridae (1). Currently, TSWV ranks among the top 10 most economically important plant viruses worldwide (2). Distribution of TSWV extends globally, and susceptibility to TSWV infection is seen in as many as 1,000 plant species in different agricultural settings (3, 4).
Virus particles are quasispherical in shape, from 80 to 120 nm in diameter, and are surrounded by an envelope of host-derived lipids in which two glycoproteins, Gn and Gc, are embedded. They contain a tripartite negative/ambisense RNA genome composed of three single-stranded RNA molecules, designated long (L), medium (M), and small (S) RNAs, packaged by the nucleocapsid protein (N) and a low number of viral RNA-dependent RNA polymerases (RdRp) (3, 4). Two nonstructural proteins, NSm and NSs, are encoded in the viral sense by the M and S segments, respectively; NSm is the movement protein (5), and NSs was shown to suppress gene silencing in plant hosts (6, 7).
Tospoviruses are transmitted by insects belonging to the order Thysanoptera (8). The tospovirus-thrips relationship has a peculiar characteristic: tospoviruses are phylogenetically associated with insect-infecting viruses, and it has been hypothesized that during evolution, they adapted to the plant kingdom and became plant pathogens (1). This evolutionary pathway belongs to only two other genera in plant-infecting viruses (Rhabdovirus and Reovirus), while other insect-transmitted plant viruses do not replicate in the vector body and are not phylogenetically related to insect-infecting viruses (9). The western flower thrips, Frankliniella occidentalis (Pergande), is the most efficient vector of TSWV. Its extremely wide host range, broad geographical distribution, short reproductive cycle, and high fecundity have contributed to the success of this insect pest as an invasive species; since its introduction in Europe, TSWV has become one of the limiting factors in the production of several vegetable crops (4).
The virus is transmitted in a propagative and persistent manner by F. occidentalis (8). Acquisition by thrips that can result in final transmission is restricted to the first and early second larval stages, and acquisition efficiency decreases as development proceeds (8). Once virions are acquired from infected plants by larval thrips, the virus enters and replicates in the midgut epithelial cells, moves to the muscle cells of the gut, and eventually reaches and replicates in the salivary glands. Adults (and partly second-instar larvae) can transmit the virus by the injection of viruliferous saliva into plant tissues (8).
The M genomic segment has been previously associated with the efficiency of transmission by F. occidentalis. Several reports focused on the role of M-encoded glycoproteins embedded in the viral envelope as the first site of interaction between viral particles and thrips. Data were published supporting the model of receptor-mediated endocytosis for TSWV entry, showing interaction of Gn and Gc glycoproteins with 50-kDa (10, 11) and 96-kDa (12) thrips proteins, respectively, probably coordinating the entry process and circulation through the vector. Furthermore, binding of a Gn-soluble form to the thrips midgut inhibited transmission by F. occidentalis (13). The importance of the viral envelope and glycoproteins for interaction with the vector was also supported by further experimental evidence: (i) TSWV nucleocapsid preparations could not infect primary thrips cell cultures, whereas the same preparations could systemically infect mechanically inoculated plants, and (ii) thrips could not transmit an envelope-defective isolate (14). Previous authors (15) used reassortant analysis and sequencing to link and map for the first time specific mutations in the Gn/Gc open reading frame (ORF) to the loss of thrips transmissibility without inhibition of enveloped virion assembly. A TSWV-M nontransmissible isolate expressing a smaller amount of Gn protein was also reported, but sequence analysis was not conclusive for identification of the determinant of loss of transmission (16).
Besides acquisition, the rate of virus replication (17), migration from the midgut to visceral muscle cells and salivary glands (18, 19), and TSWV titer (20) have been reported as crucial factors for vector competence and transmission. In this respect, defective interfering L RNA has also been associated with a lack of transmission, due to the low number of infectious units available in the inoculum (14).
The scope of this work was the demonstration of the role of the NSs protein encoded by the S segment in thrips transmission. Given the absence of a reverse-genetic system for TSWV, we generated a collection of NSs-defective isolates from well-characterized and transmissible wild-type (WT) isolates. By using several approaches (i.e., leaf disc assays, quantification of virus titer, and confocal laser scanning microscopy), we showed that larvae were able to acquire and accumulate the NSs-defective isolate, but after pupation, adults carried a very limited amount of virus, resulting in the absence of transmission. Sequence analysis and generation of reassortant isolates allowed us to provide strong genetic evidence for the active role of NSs in maintaining efficient tissue colonization in the thrips vector, allowing transmission. Our results show for the first time that the S genomic segment carries a viral genetic determinant for transmission in the tospovirus-thrips interaction, and specifically, unprecedented in the Bunyaviridae family, that the NSs protein is necessary for efficient virus accumulation and transmission by insect vector individuals.
MATERIALS AND METHODS
Parental TSWV isolates.
Two well-characterized TSWV WT field isolates, p105 and p202/3WT, present in the Plant Virus Italy (PLAVIT) collection at our institute, were used as parents for generating NSs-truncated isolates. Preliminary to our work, we passaged such parental isolates three consecutive times through single-lesion mechanical inoculation on Nicotiana tabacum L. cv. White Burley. The isolates were then tested for positive thrips transmission and stored in liquid nitrogen until use. The ability to encode a WT NSs was determined by sequence analysis and Western blotting according to previous protocols (21).
Generation of NSs-defective isolates.
We forced the occurrence of NSs-defective mutant isolates from the two WT parental isolates by single-passage mechanical inoculation on Capsicum chinensis Jacq PI152225 carrying the Tsw gene, under greenhouse conditions, exploiting the ability of TSWV to overcome the Tsw resistance gene through mutations in the NSs-coding region under selective pressure (21). Among the resistance-breaking (RB) isolates obtained, we selected those with a recovery phenotype, as an indication of a loss of function of the silencing suppressor NSs (21). Such isolates were serially inoculated by three passages of single local lesions on N. tabacum cv. White Burley and then multiplied on Nicotiana benthamiana Domin and Datura stramonium L. in order to carry out quantification experiments and leaf disc assays with a population of F. occidentalis competent for transmission. In parallel, the parental WT isolates were also passaged mechanically three times to ensure the same number of mechanical inoculations for both groups of isolates. Molecular characterization of the NSs protein, Western blot analysis for its detection, and in vivo silencing suppression assays were performed exactly as previously detailed (21).
Virus quantification in plant hosts.
Double antibody sandwich-enzyme linked immunosorbent assay (DAS-ELISA) was performed as previously described for TSWV using the antinucleocapsid antibody A421VI (22), except that the samples were diluted 1/20 in order to be in a range where absorption values were linearly correlated to virus titer in the infected plants. At least five plants for each isolate were used in each experiment. In order to ensure equal amounts of inoculum for the WT and NSs-truncated isolates, we took into account the results of preliminary experiments to test viral titer in N. benthamiana for each strain and used that information to standardize the viral titer in the inocula in subsequent experiments. Two independent experiments were performed for each isolate pair. A five-point standard curve prepared from sap of a pool of p105-infected leaves (undiluted and 80%, 60%, 40%, and 20% dilutions in sap from healthy plants) was used for calculation of relative quantities from absorbance values. Significant differences between the relative quantities of the two isolates were determined by analysis of variance (ANOVA) and Tukey test (P ≤ 0.05) using SigmaStat analysis software v. 3.5.
Transmission experiments with leaf disc assays.
Northwestern Italian populations of F. occidentalis were reared on pollen and green bean pods, as food source and oviposition sites, in gauze-covered glass jars, with corrugated cardboard on the bottom to provide pupation sites. Mass rearing was conducted in growth chambers at 25 ± 1°C and 65% ± 5% rH, with a 16:8 light-dark photoperiod (17, 23). Systemically infected N. benthamiana leaves were carefully washed to minimize the glandular hair effect on thrips and were used for acquisition. Cohorts of F. occidentalis larvae not more than 2 to 3 h old were placed in a Plexiglas Tashiro cage and left to feed on the infected leaves for 72 h. The larvae were then transferred to other cages on green bean pods to complete their development. Adults were individually tested for virus transmission in plastic tubes (1.5 ml) using a 12-mm-diameter disc of D. stramonium leaves with two inoculation access periods (IAPs) of 48 h each. After 72 h of floating on water in 24-well plates, the leaf discs were analyzed by DAS-ELISA as previously described in detail (22).
Quantification of virus titer in second-instar larvae and adult thrips.
Systemically infected leaves from N. benthamiana plants mock inoculated or inoculated with WT and NSs-defective isolates were used for acquisition (72 h). Larvae were then maintained on healthy plants for 24 h; cohorts of 8 to 10 individuals were collected and processed for quantitative reverse transcription (qRT)-PCR virus RNA quantification, or alternatively, left to complete development until the adult stage. Adults alive after the two IAPs were also processed in cohorts of 8 to 10 individuals for qRT-PCR. Total RNA was prepared using TRIzol reagent (Invitrogen). Nine hundred microliters of TRIzol reagent was added in plastic tubes (1.5 ml) containing the insects and 1/3 volume of 0.5-mm-diameter silica beads (BioSpec Products) and frozen in liquid nitrogen. The content was ground using a bead beater homogenizer (FastPrep-24 instrument; MP Biomedicals), taking care not to overheat the sample, and incubated at 25°C for 5 min. After addition of 100 μl of chloroform, tubes were vortexed for 5 s, incubated at 25°C for 3 min, and centrifuged at 12,000 × g for 15 min in a microcentrifuge (Eppendorf). The supernatant (400 μl) was transferred into a new tube and extracted again using 400 μl of chloroform as described above. The supernatant was transferred into a new tube along with 500 μl of 99% isopropanol, 1 μl of 1-mg ml−1 glycogen, and 5 μl of 3 M sodium acetate, pH 5.2, and incubated on ice for 10 min. After centrifugation at 12,000 × g for 15 min, the pellet was washed with 900 μl of 70% ethanol and centrifuged at 12,000 × g for 1 min; after removal of the ethanol, the pellet was centrifuged at 12,000 × g for 1 min, dried in a vacuum chamber, resuspended in 20 μl of water, and centrifuged at 12,000 × g for 5 s. First-strand cDNA was synthesized with 10 μl of total RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems); qRT-PCR was performed using TaqMan gene expression master mix (Applied Biosystems), following the protocol and amplification conditions suggested by the manufacturer with the StepOne Plus real-time PCR system (Applied Biosystems), using published primers (24). The thrips actin gene (24) was used for normalization and ΔΔCT calculation as defined in the Step One software v. 2.0 manual (Applied Biosystems). Significant differences between relative quantities of the two isolates in insects were determined by ANOVA (P ≤ 0.05) using SigmaStat analysis software v. 3.5.
Confocal microscope observation of immunofluorescent insect sections.
Thrips from cohorts from the same experiments as used for virus titer quantification and transmission were processed for immunofluorescent analyses. Larvae and adults were fixed overnight at 4°C in 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4). After washing with PBS, they were embedded in 8% (wt/vol) low-melting-point agarose. Longitudinal sections, 100 μm thick, were obtained with a vibrating-blade microtome (Leica VT1000S). Sections were blocked at room temperature for 30 min with 1% bovine serum albumin (BSA) in PBS and incubated overnight at 4°C with the primary antibody. The primary antibody was a polyclonal antinucleocapsid antibody (A421VI) from the Istituto di Virologia Vegetale (IVV) collection. The sections were then washed three times with PBS, blocked at room temperature for 30 min with 1% BSA in PBS, and then incubated for 2 h with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG antibody (Sigma). Sections not treated with primary antibody or secondary antibody served as negative controls (data not shown). Sections were examined with a Leica TCS SP2 confocal microscope. Laser intensity and detector gain were first set on uninfected samples, and then the same settings were kept for all subsequent observations.
Reassortment experiments and sequence analysis of genome segments.
Reassortment experiments were carried out as previously described (21), using isolate p202/3RB together with a phylogenetically distant but thrips-transmissible Brazilian isolate available in our collection (BR20). During the selection for putative reassortant isolates, we used the recovery phenotype on C. chinensis Jacq PI152225 as a marker for the presence of an S segment originating from p202/3RB, whereas molecular characterization of the M and L segments was carried out as previously described (21), except that the PCR fragments were directly sequenced instead of using restriction fragment length polymorphism (RFLP) markers. The S segment was sequenced using previously described primers and RT-PCR conditions (21). The oligonucleotides used for RT-PCR amplification and sequencing of the M segment are available upon request.
Electron microscopy.
Systemically infected leaves of N. benthamiana were crushed and homogenized in 0.1 M phosphate buffer, pH 7.0, containing 2% polyvinylpyrrolidone (PVP). A drop of the crude extract was allowed to adsorb for about 3 min on carbon- and Formvar-coated grids and then rinsed several times with water. Grids were negatively stained with 0.5% uranyl acetate, and excess fluid was removed with filter paper. Observations and photographs were made using a CM 10 electron microscope (Philips).
Detection of glycoproteins.
A triple antibody sandwich (TAS) ELISA was performed as previously described (25) for the detection of glycoproteins and nucleocapsids as an internal reference. Plates were coated with a mixture of polyclonal antisera against TSWV virions from the IVV collection (A292, A293, and A421), and after overnight incubation with samples, monoclonal antibody specific for N protein (4F2; DSMZ AS0106) or Gn protein (APO-2B3/2D11; IVV collection) was used. The tertiary antibody was an alkaline phosphatase-conjugated rabbit anti-mouse (Sigma). Systemically infected leaves of N. benthamiana were diluted 1:100 (wt/vol) in ELISA sample buffer. The experiment was repeated twice.
Nucleotide sequence accession numbers.
Sequences determined in this study were deposited in the GenBank database and are described in Table SA1 in the supplemental material. Accession numbers are as follows: HQ830185 to HQ830188, HQ839729 to HQ839731, and HQ914636 to HQ914647.
RESULTS
Generation of a library of putative NSs-defective TSWV isolates.
The lack of a reverse-genetic system dependent on a cDNA infectious clone for tospoviruses has limited the functional study of virus-encoded proteins to mutants naturally occurring in nature or artificially generated in a controlled environment through successive mechanical inoculations. No mutant defective in functional NSs accumulation was described in more than 30 years of tospovirus molecular studies. We and, more recently, other authors have demonstrated that mutations in a functional NSs protein are responsible for overcoming the resistance provided by the Tsw gene in pepper (21, 26). Among the RB isolates we described, a subset was characterized by deletions in the NSs coding region that caused a frameshift and expression of truncated proteins. Such isolates were able to systemically infect resistant C. chinensis PI152225 and D. stramonium immediately after inoculation but could not maintain infection in newly emerging leaves, resulting in recovery (21).
Based on these results, in this new work we forced the occurrence of NSs-defective isolates from well-characterized WT and thrips-transmissible isolates through single-passage mechanical inoculation on resistant pepper and by screening for the systemic infection turning into recovery phenotype. This approach allowed direct comparison of WT and derived NSs-defective isolates by sequence analysis and thrips transmission assays. We used as parental strains two different well-characterized WT isolates, p105 and p202/3WT, whose competence for efficient thrips transmission was determined (Table 1). We derived the RB recovery isolates p105RBMar, p105RBMaxI, and p105-803RB from isolate p105 and isolate p202/3RB from isolate p202/3WT (Table 1). The p105RBMar, p105RBMaxI, p105-803RB, and p202/3RB isolates could be mechanically transmitted to a set of common TSWV host plant species and caused systemic infection. However, symptoms on C. chinensis PI152225 and D. stramonium became milder after an initial severe systemic infection, and in newly emerging leaves symptoms recovered (Fig. 1A to E) and the virus was absent, as shown by Western blotting of systemically infected and recovered tissue (Fig. 1F).
TABLE 1.
Characteristics of the TSWV isolates derived for this study and used in the transmission experiments by leaf disc assay with Frankliniella occidentalis
| Expta | Isolate | Genotype features | Description of specific mutation | Total no. of tested thrips | No. of transmitting thrips | % transmission |
|---|---|---|---|---|---|---|
| I | p105 | Wild type, NSs of 467 aa | 44 | 11 | 25.0 | |
| p202 | RB, truncated NSs of 399 aa (7) | Double base deletion (position 1171) causing frameshift | 24 | 0 | 0 | |
| II | p202/3WT | Wild type, NSs of 467 aa | 220 | 108 | 49.0 | |
| p202/3RB | RB, NSs truncated of 443 aa | Single base deletion (position 1371) causing frameshift | 257 | 0 | 0 | |
| III | p105 | Wild type, NSs of 467 aa | 173 | 72 | 41.6 | |
| p105RBMar | RB, truncated NSs of 443 aa | Single base deletion (position 1363) causing frameshift | 118 | 0 | 0 | |
| p105RBMaxI | RB, truncated NSs of 323 aa | For mutation (G1058T) introducing stop codon | 119 | 0 | 0 | |
| p105WTMaxII | Wild type, full length NSs of 467 aa | Mutation (T1058C) stop codon mutated to Q | 112 | 48 | 43.0 | |
| IV | p105 | Wild type, NSs of 467 aa | 14 | 4 | 28.6 | |
| p105RBMar | RB, truncated NSs of 443 aa | Single base deletion (position 1363) causing frameshift | 27 | 0 | 0 | |
| Mock inoculated | 19 | 0 | 0 | |||
| V | p105 | Wild type, NSs of 467 aa | 72 | 52 | 72.2 | |
| p105RBMar | RB, truncated NSs of 443 aa | Single base deletion (position 1363) causing frameshift | 69 | 0 | 0 | |
| p105-803RB | RB, truncated NSs of 382 aa | Two single base deletions (positions 1232 and 1382) causing frameshift | 40 | 0 | 0 | |
| Mock inoculated | 73 | 0 | 0 | |||
| VI | BR20 | Wild-type Brazilian parental isolate, NSs of 467 aa | 100 | 26 | 26.0 | |
| p202/3RB | RB, truncated NSs of 443 aa | Single base deletion (position 1371) causing frameshift | 100 | 0 | 0 | |
| Reassortant A | S p202/3RB M p202/3RB L BR20 | 107 | 0 | 0 | ||
| Reassortant B | S p202/3RB M p202/3RB L BR20 | 138 | 0 | 0 |
The acquisition access period for experiment I was 48 h; for experiments II to VI, it was 72 h.
FIG 1.
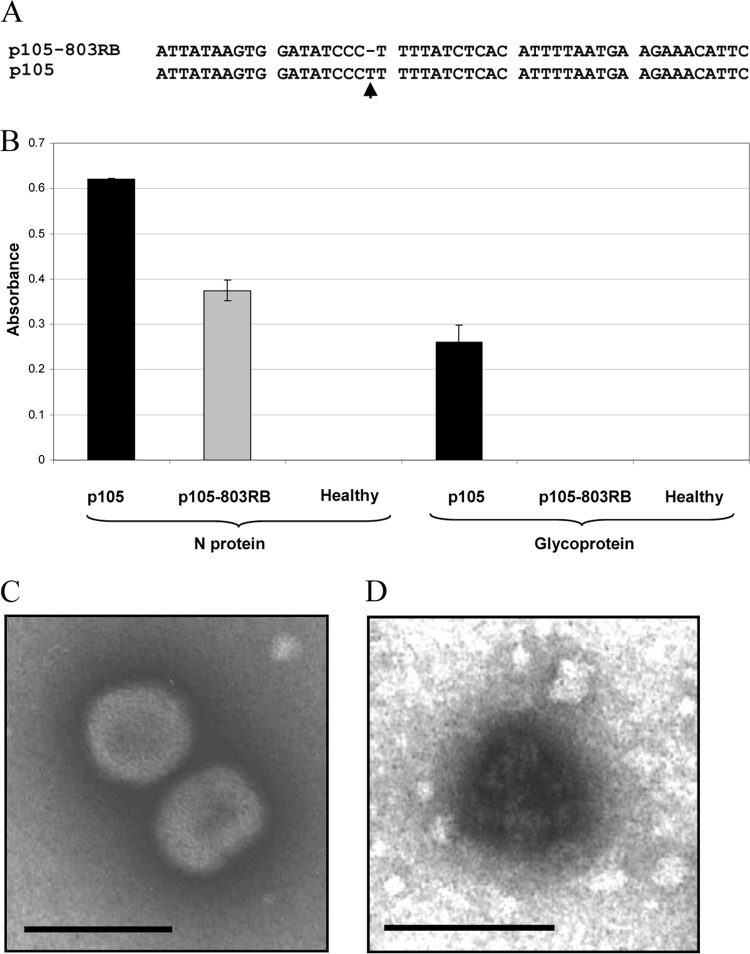
Symptoms and nucleocapsid accumulation on Capsicum chinensis PI152225 and Datura stramonium infected with different TSWV isolates. (A to E) C. chinensis carrying the Tsw resistance gene (top pictures) and D. stramonium (bottom pictures) photographed 14 days postinoculation; the inset in the upper right corner of each picture shows newly emerging leaves. The wild-type parental isolates p202/3WT and p105 induced only local necrotic lesions on C. chinensis (A and C); the same isolates infected systemically D. stramonium, displaying severe symptoms (A and C). Isolates p202/3RB, p105-803RB, and p105RBMar (B, D, and E) were able to infect systemically C. chinensis PI152225 and D. stramonium but could not maintain infection in newly emerging leaves. (F) Western blot analysis of total leaf extracts from systemically infected (Syst) and recovered (Rec) tissues. Detection was performed using a polyclonal rabbit immune serum against the nucleocapsid protein of TSWV. Below each Western blot panel, a Coomassie-stained gel is shown to display total protein loadings.
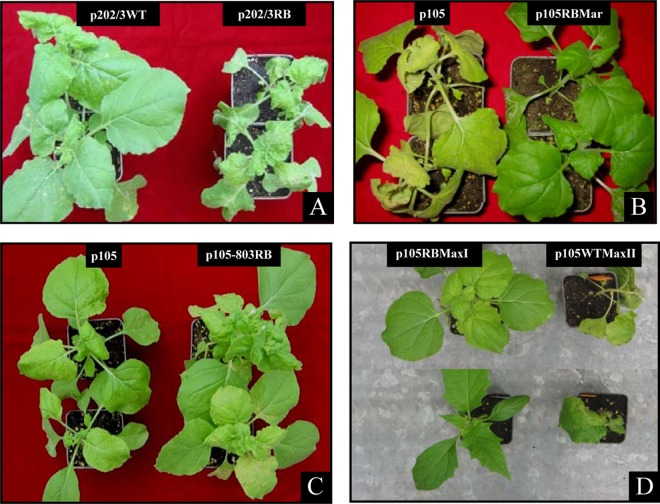
In N. benthamiana, symptoms of the various NSs-defective isolates varied in intensity: isolate p202/3RB displayed more severe symptoms than did p202/3WT, resulting in lethal necrosis 10 days after inoculation (Fig. 2A); isolates p105RBMar and p105RBMaxI had milder symptoms than did p105 but maintained constant symptom expression throughout infection, without recovery (Fig. 2B and D); and isolate p105-803RB had lower infection ability (judged as success of mechanical inoculation) and showed a very delayed time course of infection, with final recovery (Fig. 2C). During the mechanical inoculation passages of p105RBMaxI (see Materials and Methods), we observed a sudden increase in symptom severity in a single N. benthamiana plant and loss of the recovery phenotype after a subsequent inoculation on D. stramonium (Fig. 2D); this phenotypic revertant WT isolate was designated p105WTMaxII.
FIG 2.
Tomato spotted wilt virus isolates carrying different truncations in the NSs protein display diverse symptom severity in Nicotiana benthamiana. (A to C) Symptoms following mechanical inoculation of isolates p202/3RB (A), p105RBMar (B), and p105-803RB (C) in comparison with their parental isolates p202/3WT and p105. (D) Sudden increased severity of symptoms in N. benthamiana (top) and loss of the recovery phenotype on Datura stramonium (bottom) observed for the phenotypic revertant isolate p105WTMaxII, derived through serial mechanical inoculation passages of p105RBMaxI.
The resistance-breaking/recovery TSWV isolates code for defective NSs protein.
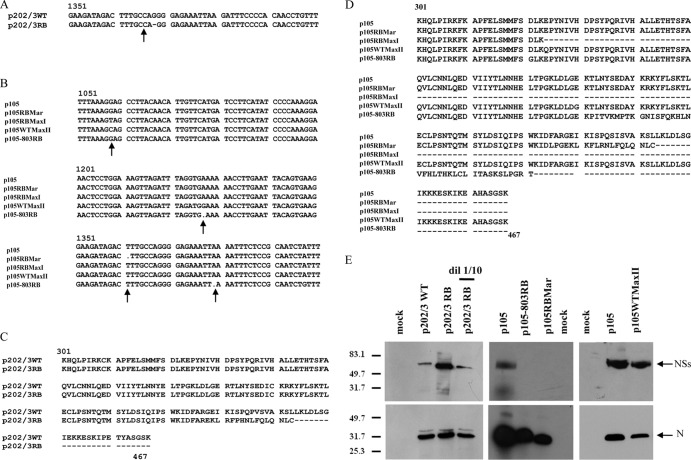
Sequencing of NSs coding sequences of the five RB TSWV isolates revealed, in two cases, p202/3RB and p105RBMar, a single nucleotide deletion: a G at position 1371 for isolate p202/3RB and a T at position 1363 for isolate p105RBMar (Fig. 3A and B), which resulted in a frameshift coding for a truncated protein of 443 amino acids (aa) (Fig. 3C and D). Isolate p105-803RB showed two single nucleotide deletions (Fig. 3B) at positions 1232 and 1382 that resulted in frameshifts and a truncated protein of 382 aa (Fig. 3D). In the case of p105RBMaxI, a G-to-T mutation at position 1058 (Fig. 3B) resulted in an early stop codon in the reading frame and hence a truncated protein of 323 aa (Fig. 3D). The TSWV isolate p105WTMaxII showed a further mutation at the same site from a T to a C (Fig. 3B), which changed the stop codon into a glutamine residue, giving rise to a new full-length NSs allele (Fig. 3D) different from the original p105 NSs allele. We performed Western blot analysis on plant extracts infected with three defective isolates selected for further in-depth analysis; a truncated NSs protein was clearly detected for isolate p202/3RB (Fig. 3E), while no NSs-specific band could be detected for either p105RBMar or p105-803RB, confirming that the NSs protein was altered or not present in these isolates. We also tested silencing suppression ability with a standard in vivo assay; the new defective NSs alleles derived in this study failed to function as silencing suppressors in planta (Fig. 4 and data not shown).
FIG 3.
Molecular characterization of Tomato spotted wilt virus resistance-breaking isolates showing a recovery phenotype obtained in controlled conditions from defined wild-type field isolates. (A and B) Nucleotide alignments of portions of the S genomic segment; mutations occurring in each of the derived isolates are indicated by arrows. (C) Amino acid sequence alignment of the nonstructural protein of isolates p202/3WT and p202/3RB. A single deletion in the nucleotide sequence of p202/3RB resulted in a frameshift and a truncated protein of 443 aa. (D) Amino acid sequence alignment of all the different NSs alleles present in isolates obtained from the WT isolate p105. In isolate p105RBMar, a nucleotide deletion resulted in a frameshift and a truncated protein of 443 aa. A nucleotide mutation in isolate p105RBMaxI resulted in a stop codon, whereas a further mutation resulted in a full-length NSs in isolate p105WTMaxII with an E-to-Q substitution at position 324. Two single nucleotide deletions in p105-803RB caused a frameshift and a truncated protein of 382 aa. (E) Western blot analysis of total protein leaf extracts from Nicotiana benthamiana infected by different isolates, using antinucleocapsid and anti-NSs antibodies.
FIG 4.
Test for silencing suppression activity for three allelic version of NSs protein (isolates p105WTMaxII, p105RBMaxI, and p105RBMar) derived from the original allele present in isolate p105. Full-length NSs allelic variants were cloned in pBin61 vector and transiently expressed through agroinfiltration together with pBin-GFP in16C transgenic Nicotiana benthamiana.
Virus titer of NSs-defective isolates in N. benthamiana and D. stramonium plants.
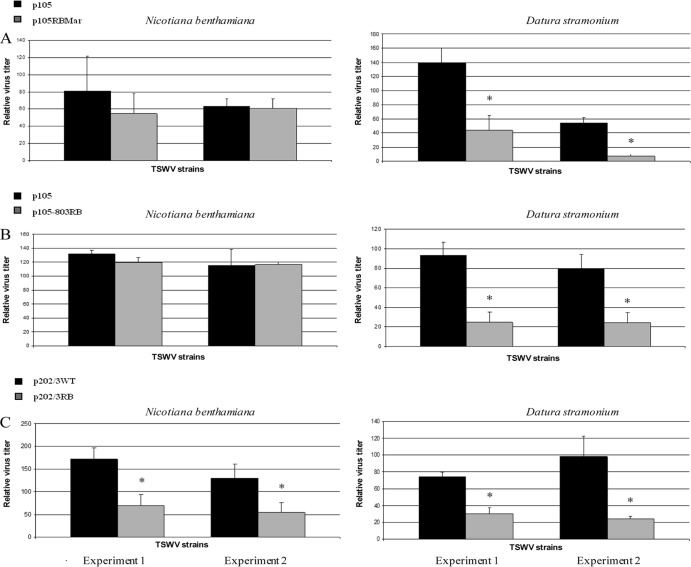
Virus titer in the plant could potentially strongly influence efficiency of acquisition from the insect vector (27, 28). Given the wide variety of symptom severities displayed by our library of NSs-defective mutants, often associated with differential quantitative viral accumulation, we quantified the virus titer in N. benthamiana and D. stramonium, in order to select the best plant host as a source of inoculum. In fact, the lack of an active silencing suppressor can result in lower viral accumulation (as suggested by the recovery phenotype observed in D. stramonium plants), possibly to the point of limiting acquisition by the vector. Therefore, for all the NSs-defective isolates used in successive quantification experiments in thrips, we proceeded to quantify the virus in leaves randomly sampled from hosts before being used for acquisition experiments. In the case of p105RBMar, D. stramonium leaves contained an amount of virus that was statistically significantly lower than that of the WT, whereas no significant differences were observed in N. benthamiana (P = 0.17 and P = 0.73 for the two experimental replicates) (Fig. 5A). A similar profile was observed for p105-803RB, which displayed significant changes in abundance in D. stramonium but not in N. benthamiana (P = 0.18 and P = 0.90 for the two replicates) (Fig. 5B). In contrast, a significant difference in titer in both plant hosts was observed for the p202/3WT and p202/3RB isolates, with higher abundance of the former (Fig. 5C); this was in contrast with symptom severity on N. benthamiana, which was stronger for the latter isolate (Fig. 2A). Considering these results, N. benthamiana was used as the acquisition host in all further experiments with thrips.
FIG 5.
Quantification of virus titer in Nicotiana benthamiana and Datura stramonium suggests the former as a better acquisition host in transmission experiments with thrips. Plants were mechanically inoculated with isolates p105 and p105RBMar (A), p105 and p105-803RB (B), and p202/3WT and p202/3RB (C). Systemically infected leaves displaying severe symptoms were sampled 7 days postinoculation and used for acquisition (72 h). Double antibody sandwich-enzyme linked immunosorbent assay was performed using the antinucleocapsid antibody A421VI. A five-point standard curve prepared from sap of a pool of p105-infected leaves from the same plants (undiluted and 80%, 60%, 40%, and 20% dilutions with sap from healthy plants) was used for relative virus titer determination based on absorbance values. Statistical analysis was performed by ANOVA using SigmaStat analysis software v. 3.5. An asterisk indicates that differences are statistically significant (P < 0.05).
NSs-defective TSWV isolates are not transmitted by F. occidentalis.
A preliminary leaf disc experiment examining transmission by thrips was carried out using the NSs-defective isolate p202 that we characterized in previous work (21). This isolate was not transmitted by a population of F. occidentalis, which in contrast efficiently transmitted p105, a closely related WT isolate from Italy (Table 1, experiment I). For p202, we had no record of the WT isolate from which it was derived directly, and therefore, further comparative analysis was impaired. Instead, in this work, we could perform thrips transmission experiments for each of the newly obtained NSs-defective isolates in direct comparison with their WT parental isolates. We observed that F. occidentalis individuals completely failed to transmit p202/3RB, p105RBMar, p105RBMaxI, and p105-803RB, while they were able to transmit the two WT parental isolates, with efficiencies of 49% for p202/3WT and 46% for p105 (Table 1, experiments I to V). In the case of the revertant isolate p105WTMaxII, derived from p105RBMaxI through the serendipitous selection of an isolate from a single severely infected plant and coding for a full-length NSs, the transmission efficiency was restored to 43% (Table 1, experiment III). Overall, these data showed that none of the NSs-defective isolates could be transmitted by a competent population of F. occidentalis.
The NSs-defective isolates (p105RBMar and p202/3RB) can be acquired and accumulate in second-instar larvae, but their titer diminishes in adult thrips.
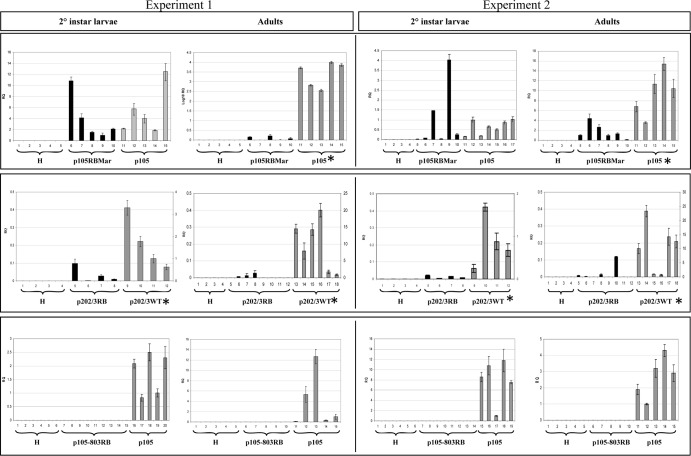
Leaf disc assays clearly showed that NSs-defective isolates could not be transmitted by the thrips vector. We then investigated if the loss of transmission corresponded to the loss of acquisition by larvae, as previously shown for glycoprotein and envelope-deficient mutants (14, 15). For this purpose, we set up experiments for TSWV quantification by real-time PCR in second-instar larvae using isolates p105RBMar, p202/3RB, and p105-803RB in comparison with the related WT isolate. For each isolate, we performed at least two distinct experiments with replicates within each experiment. Thrips feeding on leaves from noninoculated plants were always included in each experiment as a negative control. In the case of the comparison between isolates p105RBMar and p105, both virus isolates could be detected in second-instar larvae cohorts, and the relative accumulations between the NSs-defective and the WT isolates were not significantly different (P = 0.64 and P = 0.57, for the two experimental replicates) (Fig. 6). Regarding the comparison between p202/3RB and p202/3WT, we could detect both isolates in all second-instar larvae cohorts, with a significant difference in titer between the two variants (Fig. 6), likely mimicking the significant differences observed in the acquisition host (Fig. 5C). In both experiments, no virus could be detected in thrips feeding on healthy plants.
FIG 6.
NSs-defective isolates can be acquired and accumulate in second-instar larvae, but virus titer is lower in adult thrips. We compared virus relative abundances in cohorts of 8 to 10 individual larvae or adults of p105 and p105RBMar, p202/3WT and p202/3RB, and p105 and p105-803RB. H corresponds to thrips feeding on healthy leaves. For each comparison, we show the results of two independent experiments. Relative quantification of TSWV isolates in second-instar larvae and adults of Frankliniella occidentalis was performed by real-time TaqMan qPCR, using the thrips actin gene for normalization and ΔΔCT calculation using Step One software v. 2.0 (Applied Biosystems). RQ, relative quantity. A random sample was used as a reference (RQ = 1) in each experiment. Significant differences between the relative quantities of the two isolates were determined by ANOVA (P ≤ 0.05) using SigmaStat analysis software v. 3.5. An asterisk indicates that differences are statistically significant. In case of p202/3RB and WT experiments, the RQ scales are different for each of the two isolates: the left scale refers to p202/3RB, whereas the right scale refers to p202/3WT.
Given that transmission rate was shown to depend on virus titer in adult thrips (20), we proceeded to evaluate virus accumulation in cohorts of adults from the same experiments as used for titer evaluation in larvae. In the case of p105RBMar, the accumulation shown in larvae (statistically not different from the corresponding WT isolate p105) was not confirmed in adult thrips, in which the NSs-defective isolate accumulated only minimally, with a significant difference (Fig. 6). In adult thrips fed on p202/3RB-infected plants, virus accumulation was detected only occasionally, in 3 out of 8 cohorts in the first experiment and 4 out of 8 cohorts in the second experiment (Fig. 6). Moreover, as shown by the different scales in the y axes among larvae and adults for p202/3WT, the relative accumulations were an order of magnitude higher for the adults than for second-instar larvae (Fig. 6), indicating an erratic and much lower accumulation of the RB isolate in the adults. These results showed that the defective viruses could be acquired by the vector larvae and accumulated in them with a titer mimicking the titer in the acquisition host, but virus accumulation in adult thrips was minimal.
In the case of isolate p105-803RB, we could not detect virus accumulation either in second-instar larvae or in adults (Fig. 6). This unexpected result suggested the inability of the virus to be acquired. We therefore checked for possible mutations in the glycoprotein coding region that could directly affect acquisition. In fact, sequencing of the glycoprotein precursor encoded by the M segment revealed a single nucleotide mutation (T deletion) (Fig. 7A), which resulted in a frameshift and a predicted truncated protein of 152 aa. As confirmation, TAS-ELISA of leaf extracts from N. benthamiana symptomatic plants was negative for Gn glycoprotein for p105-803RB, while high absorbance was present for the control isolate p105; using the same extracts, a TAS-ELISA for detection of the nucleocapsid protein displayed a strong reaction for both isolates (Fig. 7B). Furthermore, morphological analysis by transmission electron microscopy on crude sap obtained from systemically infected N. benthamiana leaves showed typical enveloped particles for the p105 isolate (Fig. 7C), while only nucleocapsids could be observed for the p105-803RB isolate (Fig. 7D).
FIG 7.
Tomato spotted wilt virus isolate p105-803RB does not code for glycoproteins and is not enveloped. (A) Nucleotide alignment of a portion of the M segment showing the single nucleotide deletion in the glycoprotein precursor coding sequence for the p105-803RB isolate. (B) Absorbance values of triple antibody sandwich-enzyme linked immunosorbent assay of extracts from Nicotiana benthamiana infected by p105 and p105-803RB isolates, using antinucleocapsid and anti-Gn monoclonal antibodies. (C) Transmission electron microscope image of a leaf dip extract of the p105 isolate showing accumulation of enveloped viral particles in systemically infected leaves of N. benthamiana. (D) Transmission electron microscope image of a leaf dip extract of isolate p105-803RB showing only accumulation of nucleocapsids in systemically infected leaves of N. benthamiana. Bars represent 100 nm.
Immunostaining of thrips sections confirmed that the nucleocapsid of NSs-defective isolates accumulates in tissues and organs of second-instar larvae but not in adults.
In parallel with virus titer quantification experiments, a number of second-instar larvae and adult thrips from the same acquisition experiments were used for immunostaining of the nucleocapsid protein in thrips sections observed with a confocal laser scanning microscope. Isolates p105 and p202/3WT were detected along the alimentary canal of larvae and along the alimentary canal and visceral muscles of adults. Isolates p105RBMar and p202/3RB were clearly detected along the alimentary canal only in larvae; in contrast, fluorescence on adults was rare and very faint (Fig. 8). These results confirmed that NSs-truncated isolates could be acquired and accumulate in second-instar larvae but not in adults. In the case of p105-803RB, we failed to detect specific fluorescence in both larvae and adults (not shown). Percentages of virus-positive thrips are given in Table 2.
FIG 8.
TSWV infection of thrips vectors demonstrated by immunofluorescence on sections of larvae and adults. Primary antibodies were against the TSWV nucleocapsid protein. Each image is presented as the overlay of fluorescence and bright-field images. Images shown are taken from different individuals and are representative of all the observations performed (see Table 2). White bars represent 50 μm; black bars represent 100 μm. Abbreviations: ab, abdomen; d, dorsum; he, head; mg, midgut; pr, pronotum; th, thorax; v, ventre; vm, visceral muscles.
TABLE 2.
Tomato spotted wilt virus positivity in thrips as determined by confocal microscope analysis using a polyclonal antinucleocapsid antibody (A421VI from IVV collection)
| TSWV strain | thrips life stage | Total no. of tested thrips | Total no. of positive thrips | % positive |
|---|---|---|---|---|
| p105WT | Larva | 67 | 16 | 24 |
| Adult | 84 | 24 | 29 | |
| p105RBMar | Larva | 25 | 4 | 16 |
| Adult | 12 | 1 | 8 | |
| p105-803RB | Larva | 30 | 0 | 0 |
| Adult | 34 | 0 | 0 | |
| p202/3WT | Larva | 41 | 16 | 39 |
| Adult | 16 | 8 | 50 | |
| p202/3RB | Larva | 32 | 4 | 13 |
| Adult | 38 | 1 | 3 |
Formal genetic demonstration that the mutation in the NSs coding region is responsible for the lack of transmission in NSs-defective isolates.
The results so far discussed were derived through an experimental approach that could not exclude the possibility of further mutations randomly occurring elsewhere in the genome causing the lack of transmission, as was the case with isolate p105-803RB. Nevertheless, two different experimental approaches allowed us to demonstrate that the mutation in the NSs region caused the transmissibility defect. In one case, a serendipitously derived revertant (p105WTMaxII) restored transmission (Table 1, experiment III). A second approach took advantage of derived reassortant isolates, in combination with sequence analyses of the whole M segment. In order to exclude a role for the L genomic segment as the determinant of transmission in our case, we generated reassortant isolates using as parental isolates p202/3RB together with a phylogenetically distant but thrips-transmissible Brazilian isolate available in our collection (BR20). We were able to identify and characterize two different reassortant isolates, named A and B, carrying the L segment of BR20 and the S and M segments of p202/3RB. These reassortants were not competent for transmission (Table 1, experiment VI), showing that the p202/3RB L segment did not carry the genetic determinant for the loss of transmission competence.
We then fully sequenced the M and S genomic segments of the original p202/3WT isolate and p202/3RB. Sequence comparison showed 100% identity for the M segment (GenBank accession numbers HQ830188 and HQ830185), thus excluding mutations in the movement protein, glycoprotein precursor, and noncoding regions. The S segment differed only in the G nucleotide deletion in the NSs coding sequence, described above (GenBank accession numbers HQ830187 and HQ830186). Overall, the combination of reassortant analysis and the full-length sequences of the S and M segments allowed us to specifically associate the loss of transmission competence in leaf disc assays with the mutations impeding accumulation of full-length NSs in infected thrips.
DISCUSSION
In this work, we have identified genetically a new molecular determinant for transmission of TSWV by thrips: the NSs protein encoded by the S genomic segment is dispensable for acquisition by larvae but is necessary for abundant accumulation in adults, resulting in transmission. This is a new paradigm in the tospovirus-thrips interaction; in fact, all the previous work indicated virus-encoded glycoproteins as necessary for virus acquisition, whereas in this study, we identified a virus determinant necessary for abundant accumulation in the thrips once the virus is acquired. The NSs gene of tospovirus-related animal viruses, including phleboviruses and orthobunyaviruses, was previously reported to be critical for virulence, but all studies were carried out on model cell lines (29, 30, 31, 32, 33, 34). No investigation for the S segment has been carried out on individual insect vectors. In previous literature, the M segment of La Crosse and snowshoe hare bunyavirus was shown to cosegregate with the transmission phenotype to laboratory mice by natural mosquito vectors (35, 36). In this context, our work shows, for the first time in the Bunyaviridae family, an essential role for the NSs protein in virulence (inferred in this work from virus titer) in insects in vivo.
Given the absence of a reverse-genetic system for TSWV (as well as for the entire Tospovirus genus), mutant analysis, reassortment experiments, and comparison of genomic sequences are the only available tools to associate mutations with specific phenotypes. With these approaches, a groundbreaking work associated specific mutations in the Gn-Gc ORF with the loss of acquisition and therefore transmissibility by thrips (15). Mimicking this experimental approach, we generated a library of NSs-defective isolates derived from well-characterized WT isolates under greenhouse conditions, and with a combination of sequencing and reassortment analysis, we were able to genetically link defects in NSs production to the loss of transmission. Pivotal to this study was previous work in which we observed the phenotype of a subset of RB isolates from a collection generated through mechanical inoculation: isolates displaying a recovery phenotype after the initial systemic infection on resistant pepper carried deletions in the NSs coding region that made it nonfunctional (21). In this new study, we derived five RB isolates showing recovery on C. chinensis PI152225 and D. stramonium from two well-characterized transmissible isolates; these mutants allowed direct comparison by sequence analysis and for competence of thrips transmissibility, giving us the unique opportunity to monitor the role of the NSs protein in thrips infection and transmission. All NSs nucleotide changes observed in our RB isolates were different from those observed in the NSs-defective isolates previously reported (21). These results further confirm that several different mutations evenly distributed in the NSs coding region are related to the RB phenotype (21, 26).
Effects of deletion in the NSs coding region on symptoms and virus titers in different plant hosts.
Our collection of isolates allowed us to monitor the effect of NSs deletions on symptom development in different hosts. For pepper and datura, we had already described a recovery phenotype for NSs-defective isolates (21), but in this study we characterized, for the first time, such recovery as virus clearance, since in symptomless newly emerging tissue, the virus could not be detected (Fig. 1). In the literature, recovery from symptoms can be associated with different viral expression levels in different virus-host interactions (37, 38, 39). It is interesting that virus clearance occurred only in datura and pepper and not in N. benthamiana, in which symptoms could be stronger or milder than with the original WT, according to the specific isolate. In case of the p202/3RB isolate, we observed much stronger symptoms on N. benthamiana (Fig. 2) and higher accumulation of the NSs protein than for the WT isolate (Fig. 3). In spite of the strain's high virulence, the titer of the p202/3RB isolate (judged as nucleocapsid accumulation) was three times lower in N. benthamiana, showing that a lower virus concentration does not necessarily correlate with lower symptom severity, which could be instead a specific effect of abundant accumulation of the truncated version of the NSs protein.
Previous results showed a correlation between the amount of NSs protein and the severity of disease symptoms, with severe TSWV isolates expressing a larger amount of the NSs protein (40), suggesting a role for the NSs protein in pathogenesis. The function of the NSs protein in TSWV pathogenesis needs further understanding. It is known that NSs can aggregate as fibrous structures in the plant cell cytoplasm (40), and it functions as a suppressor of posttranscriptional gene silencing (PTGS) (6, 7), likely by sequestering long double-stranded RNA (dsRNA) and small interfering RNA (siRNA) molecules before they are uploaded into the RNA-induced silencing complex (41). The analysis of several NSs constructs showed the importance of a few amino acids in the N-terminal domain for both of these activities (26). In all the isolates we monitored, a functional NSs was important for abundant virus titer in D. stramonium. Such a strict requirement could come from its role as a silencing suppressor, which is important in hosts in which such defense is very active (datura and pepper in our experimental system) but is less relevant in N. benthamiana, in which silencing defense is somewhat impaired (42, 43). Such a difference was recently pointed out in comparison of siRNA profiles of TSWV-infected N. benthamiana and tomato plants: small RNAs (21 to 22 nucleotides) were much less abundant in the former host (44). This characteristic is also likely correlated to the finding of the revertant strain p105WTMaxII derived from p105RBMaxI in N. benthamiana and not in the other plant hosts (D. stramonium or C. chinensis) in which replication and accumulation are strongly impaired by the effective silencing defense, making the occurrence of a revertant less likely. Similarly, in the insects, replication and accumulation were generally low for NSs-defective isolates and limited in time, likely impairing the chances of occurrence of revertants in the insects.
In this work, we have also for the first time serendipitously generated a virus isolate that is deficient in two virus-encoded proteins. Isolate p105-803RB was originally selected because of its recovery phenotype and confirmed to carry a deletion in the NSs protein, but the difficulties in maintaining a high number of infected plants and its total inability to be acquired by thrips suggested the possible occurrence of further mutations in the genome, specifically in the region coding for determinants of acquisition, such as glycoproteins. In fact, with different approaches we showed that not only was the NSs coding region affected but also the glycoprotein precursor was not detected, and the isolate was without an envelope (Fig. 7D). To our knowledge, this is the first time that a member of the Bunyaviridae family without both NSs and glycoproteins has been shown to be competent for infection in its primary host.
Effects of deletion in the NSs coding region on virus titer in thrips.
A possible role for the NSs protein in thrips infection was hypothesized in previous works; in fact, NSs is known to be expressed in infected insects (45) and to accumulate abundantly in salivary gland tissues (17). We set up leaf disc assays of transmission by F. occidentalis for all the isolates in the study and found an absence of transmission for all the NSs-defective isolates. Interestingly, the p105WTMaxII isolate, a revertant of isolate p105RBMaxI encoding a full-length NSs, was transmissible and displayed a functional and silencing active NSs, providing strong evidence of genetic control of transmissibility by NSs.
A closer look at the infection cycle in thrips by leaf disc assay showed that the three RB isolates had different behaviors: while two isolates could be acquired and accumulated in the young larvae, p105-803RB could not be acquired by thrips and accumulation was not observed in larvae or in adults. Such behavior also worked as a proof of concept for showing that mere feeding on virus-infected leaves by larvae followed by maintenance on healthy beans for 24 h, as in our experimental system, does not result in detection inside the thrips by qRT-PCR or immunostaining. Taking this into consideration, the detection of isolates p105RBMar and p202/3RB in thrips could only be attributed to an effective acquisition step by the thrips, implying that the lack of transmission observed for these isolates was not due to the lack of acquisition but to further impairment after this step.
qRT-PCR coupled with confocal microscope observations of thrips sections gave us further elements to hypothesize a possible mechanism for the lack of transmission of these isolates. Titer quantification and immunofluorescence of NSs-defective isolates p105RBMar and p202/3RB showed, in both cases, the accumulation of viruses in second-instar larvae, while very low accumulation was observed in adults, compared with their parental isolates. Specifically, in the case of p105RBMar the difference in accumulation titer was statistically significant only in adults and not in second-instar larvae. For isolate p202/3RB we observed a statistically significant difference from the WT isolate p202/3WT also in the larval stage. Nevertheless, as for p105RBMar, the accumulation in adults was much lower, and specifically a number of adult cohorts were even virus negative, showing also for this isolate pair that the requirement for NSs was more stringent in adults. A possible explanation for the behavior of p202/3RB could take into account a lower acquisition efficiency due to the lower titer in the plant host used for acquisition. Therefore, our data showed that the NSs protein was not important for a relatively abundant accumulation in thrips larvae but became essential for maintaining efficient accumulation of virus in adult thrips. Interestingly, such behavior mimics what happens in silencing-competent plant hosts such as C. chinensis PI152225 and D. stramonium, which can be systemically infected immediately after inoculation with NSs-defective isolates but show recovery and no virus accumulation in newly emerging tissues.
Our data do not provide a demonstration of the molecular role of NSs in thrips. Nevertheless, we can envision, among others, different scenarios: the putative mechanism governing infection efficiency could be related to the possible role of NSs as a silencing suppressor in thrips.
Until now, no direct proof of thrips mounting an RNA interference (RNAi) response against invading virus has been provided. However, a recent high-throughput analysis showed expression of components of the RNAi silencing machinery in F. occidentalis infected by TSWV (46), and recent findings seem to support the function of NSs as a silencing suppressor in the insect vector. A recombinant Semliki Forest virus expressing the NSs of TSWV was able to suppress silencing in tick cells (47). The NSs of TSWV also enhanced baculovirus replication in different lepidopteran cell lines (48) as well as in insects in vivo (R. Oliveira Resende, Universidade de Brasilia, personal communication). A role for the TSWV NSs protein in suppressing silencing in both plant and insect hosts is also supported by the ability of the homologous NS3 protein of Rice hoja blanca tenuivirus (a phylogenetically related virus) to bind to siRNA and microRNA (miRNA) in both hosts (49, 50). Similarly, the NSs of La Crosse virus (LACV), a mosquito-transmitted member of the Bunyaviridae family, the Orthobunyavirus genus, has been demonstrated to markedly inhibit RNAi in mammalian cell cultures (51).
In our work, we found that clones expressing the truncated forms of NSs failed to show suppression of gene silencing in planta. The C terminus of the protein seems to be important for this function, as even a deletion of 27 aa (p105MarRB) abolished the ability. Accordingly, a recent work based on the generation of different NSs constructs reported that deletion of the C-terminal domain rendered NSs completely dysfunctional as a silencing suppressor (26). The lack of silencing suppression activity can be speculated also for the thrips vector, although a different NSs molecular mechanism cannot be ruled out. This topic presents an interesting research subject to understand the molecular mechanisms involved in this virus-vector interaction, given that TSWV activates the thrips vector immune system (52), and specific transcripts and proteins associated with antiviral defense in insects have been reported to be expressed in infected thrips (46, 53). In this context, we cannot exclude for NSs a role different from silencing suppressor in evading a not-yet-defined antiviral defense system, such as Jak-STAT, Toll, immunodeficiency, and apoptosis signaling pathways (46), or alternatively, a truly active role in pathogenesis that implies not antiviral defense but other vector-specific steps, such as determining tissue tropism in the insect.
In conclusion, the combination of several approaches has allowed us to gain new valuable insights into TSWV pathogenesis. Understanding the molecular basis of tospovirus-thrips interaction will lead to the elucidation of the roles of specific viral proteins in the pathogenic process and will be crucial to improving our ability to control TSWV infections in the future. This work also highlights that tospoviruses and their thrips vectors represent an ideal system for studying processes of virus infection and host interaction that could be extended to other viruses of the Bunyaviridae family of importance to human and animal health.
Supplementary Material
Footnotes
Published ahead of print 12 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00079-14.
REFERENCES
- 1.Goldbach R, Peters D. 1996. Molecular and biological aspects of tospoviruses, p 129–157 In Elliot RM. (ed), The Bunyaviridae. Plenum Press, New York, NY [Google Scholar]
- 2.Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD. 2011. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12:938–954. 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappu HR, Jones RAC, Jain RK. 2009. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141:219–236. 10.1016/j.virusres.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 4.Turina M, Tavella L, Ciuffo M. 2012. Tospoviruses in the Mediterranean area. Adv. Virus Res. 84:403–437. 10.1016/B978-0-12-394314-9.00012-9 [DOI] [PubMed] [Google Scholar]
- 5.Kormelink R, Storms M, Vanlent J, Peters D, Goldbach R. 1994. Expression and subcellular location of the NSm protein of Tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200:56–65. 10.1006/viro.1994.1162 [DOI] [PubMed] [Google Scholar]
- 6.Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T. 2002. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 532:75–79. 10.1016/S0014-5793(02)03632-3 [DOI] [PubMed] [Google Scholar]
- 7.Bucher E, Sijen T, De Haan P, Goldbach R, Prins M. 2003. Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 77:1329–1336. 10.1128/JVI.77.2.1329-1336.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield AE, Ullman DE, German TL. 2005. Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43:459–489. 10.1146/annurev.phyto.43.040204.140017 [DOI] [PubMed] [Google Scholar]
- 9.Hull R. 2002. Matthews' plant virology, p 485–531 Academic Press, San Diego, CA [Google Scholar]
- 10.Bandla MD, Campbell LR, Ullman DE, Sherwood JL. 1998. Interaction of Tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathology 88:98–104. 10.1094/PHYTO.1998.88.2.98 [DOI] [PubMed] [Google Scholar]
- 11.Medeiros RB, Ullman DE, Sherwood JL, German TL. 2000. Immunoprecipitation of a 50-kDa protein: a candidate receptor component for tomato spotted wilt tospovirus (Bunyaviridae) in its main vector, Frankliniella occidentalis. Virus Res. 67:109–118. 10.1016/S0168-1702(00)00123-4 [DOI] [PubMed] [Google Scholar]
- 12.Kikkert M, Meurs C, van de Wetering F, Dorfmuller S, Peters D, Kormelink R, Goldbach R. 1998. Binding of tomato spotted wilt virus to a 94-kDa thrips protein. Phytopathology 88:63–69. 10.1094/PHYTO.1998.88.1.63 [DOI] [PubMed] [Google Scholar]
- 13.Whitfield AE, Kumar NKK, Rotenberg D, Ullman DE, Wyman EA, Zietlow C, Willis DK, German TL. 2008. A soluble form of the Tomato spotted wilt virus (TSWV) glycoprotein G(N)(G(N)-S) inhibits transmission of TSWV by Frankliniella occidentalis. Phytopathology 98:45–50. 10.1094/PHYTO-98-1-0045 [DOI] [PubMed] [Google Scholar]
- 14.Nagata T, Inoue-Nagata AK, Prins M, Goldbach R, Peters D. 2000. Impeded thrips transmission of defective Tomato spotted wilt virus isolates. Phytopathology 90:454–459. 10.1094/PHYTO.2000.90.5.454 [DOI] [PubMed] [Google Scholar]
- 15.Sin SH, McNulty BC, Kennedy GG, Moyer JW. 2005. Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. Proc. Natl. Acad. Sci. U. S. A. 102:5168–5173. 10.1073/pnas.0407354102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naidu RA, Sherwood JL, Deom CM. 2008. Characterization of a vector-non-transmissible isolate of Tomato spotted wilt virus. Plant Pathol. 57:190–200. 10.1111/j.1365-3059.2007.01707.x [DOI] [Google Scholar]
- 17.Wijkamp I, Vanlent J, Kormenlink R, Goldbach R, Peters D. 1993. Multiplication of Tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J. Gen. Virol. 74:341–349. 10.1099/0022-1317-74-3-341 [DOI] [PubMed] [Google Scholar]
- 18.Nagata T, Inoue-Nagata AK, van Lent J, Goldbach R, Peters D. 2002. Factors determining vector competence and specificity for transmission of Tomato spotted wilt virus. J. Gen. Virol. 83:663–671 http://vir.sgmjournals.org/content/83/3/663.full.pdf [DOI] [PubMed] [Google Scholar]
- 19.Nagata T, Inoue-Nagata AK, Smid HM, Goldbach R, Peters D. 1999. Tissue tropism related to vector competence of Frankliniella occidentalis for Tomato spotted wilt tospovirus. J. Gen. Virol. 80:507–515 [DOI] [PubMed] [Google Scholar]
- 20.Rotenberg D, Krishna Kumar NK, Ullman DE, Montero-Astua M, Willis DK, German TL, Whitfield AE. 2009. Variation in Tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathology 99:404–410. 10.1094/PHYTO-99-4-0404 [DOI] [PubMed] [Google Scholar]
- 21.Margaria P, Ciuffo M, Pacifico D, Turina M. 2007. Evidence that the non-structural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol. Plant Microbe Interact. 20:547–558. 10.1094/MPMI-20-5-0547 [DOI] [PubMed] [Google Scholar]
- 22.Mautino GC, Sacco D, Ciuffo M, Turina M, Tavella L. 2012. Preliminary evidence of recovery from Tomato spotted wilt virus infection in Frankliniella occidentalis individuals. Ann. Appl. Biol. 161:266–276. 10.1111/j.1744-7348.2012.00571.x [DOI] [Google Scholar]
- 23.Tedeschi R, Ciuffo M, Mason G, Roggero P, Tavella L. 2001. Transmissibility of four tospoviruses by a thelytokous population of Thrips tabaci from Liguria, northwestern Italy. Phytoparasitica 29:37–45. 10.1007/BF02981812 [DOI] [Google Scholar]
- 24.Boonham N, Smith P, Walsh K, Tame J, Morris J, Spence N, Bennison J, Barker I. 2002. The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan). J. Virol. Methods 101:37–48. 10.1016/S0166-0934(01)00418-9 [DOI] [PubMed] [Google Scholar]
- 25.Roggero P, Dellavalle G, Ciuffo M, Pennazio S. 1999. Effects of temperature on infection in capsicum sp. and Nicotiana benthamiana by Impatiens necrotic spot tospovirus. Eur. J. Plant Pathol. 105:509–512. 10.1023/A:1008742516820 [DOI] [Google Scholar]
- 26.de Ronde D, Butterbach P, Lohuis D, Hedil M, Van Lent JWM, Kormelink R. 2013. Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 14:405–415. 10.1111/mpp.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferriol I, Rubio L, Perez-Panades J, Carbonell EA, Davino S, Belliure B. 2013. Transmissibility of Broad bean wilt virus 1 by aphids: influence of virus accumulation in plants, virus genotype and aphid species. Ann. Appl. Biol. 162:71–79. 10.1111/j.1744-7348.2012.00579.x [DOI] [Google Scholar]
- 28.Okazaki S, Okuda M, Komi K, Yamasaki S. 2011. The effect of virus titer on acquisition efficiency of Tomato spotted wilt virus by Frankliniella occidentalis and the effect of temperature on detectable period of the virus. Australas. Plant Pathol. 40:120–125. 10.1007/s13313-010-0020-z [DOI] [Google Scholar]
- 29.Vialat P, Billecocq A, Kohl A, Bouloy M. 2000. The S segment of Rift Valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 74:1538–1543. 10.1128/JVI.74.3.1538-1543.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806. 10.1128/JVI.78.18.9798-9806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 81:4991–4999. 10.1128/JVI.01933-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669. 10.1073/pnas.98.2.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikegami T, Peters CJ, Makino S. 2005. Rift Valley fever virus non-structural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606–5615. 10.1128/JVI.79.9.5606-5615.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le May N, Dubaele S, De Santis LP, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. 10.1016/S0092-8674(04)00132-1 [DOI] [PubMed] [Google Scholar]
- 35.Beaty BG, Holterman M, Tabachnick W, Shope RE, Rozhon EJ, Bishop DH. 1981. Molecular basis of bunyavirus transmission by mosquitoes: role of the middle-sized RNA segment. Science 211:1433–1435. 10.1126/science.6781068 [DOI] [PubMed] [Google Scholar]
- 36.Beaty BG, Miller BR, Shope RE, Rozhon EJ, Bishop DH. 1982. Molecular basis of bunyavirus per os infection of mosquitoes: role of the middle-sized RNA segment. Proc. Natl. Acad. Sci. U. S. A. 79:1295–1297. 10.1073/pnas.79.4.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratcliff FG, MacFarlane SA, Baulcombe DC. 1999. Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell 11:1207–1216. 10.1105/tpc.11.7.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin HW, Ding SW. 2003. Identification and molecular characterization of a naturally occurring RNA virus mutant defective in the initiation of host recovery. Virology 317:253–262. 10.1016/j.virol.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 39.Jovel J, Walker M, Sanfaçon H. 2007. Recovery of Nicotiana benthamiana plants from a necrotic response induced by a nepovirus is associated with RNA silencing but not with reduced virus titer. J. Virol. 81:12285–12297. 10.1128/JVI.01192-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kormelink R, Kitajima EW, Dehaan P, Zuidema D, Peters D, Goldbach R. 1991. The nonstructural protein (NSs) encoded by the ambisense-s RNA segment of Tomato spotted wilt virus is associated with fibrous structures in infected-plant cells. Virology 181:459–468. 10.1016/0042-6822(91)90878-F [DOI] [PubMed] [Google Scholar]
- 41.Schnettler E, Hemmes H, Huismann R, Goldbach R, Prins M, Kormelink R. 2010. Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J. Virol. 84:11542–11554. 10.1128/JVI.00595-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, Carter SA, Cole AB, Cheng N, Nelson R. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. U. S. A. 101:6297–6302. 10.1073/pnas.0304346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodin M, Zaitlin D, Naidu RA, Lommel SA. 2008. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21:1015–1026. 10.1094/MPMI-21-8-1015 [DOI] [PubMed] [Google Scholar]
- 44.Mitter N, Koundal V, Williams S, Pappu H. 2013. Differential expression of Tomato Spotted Wilt Virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS One 8(10):e76276. 10.1371/journal.pone.0076276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullman DE, German TL, Sherwood JL, Westcot DM, Cantone FA. 1993. Tospovirus replication in insect vector cells—immunocytochemical evidence that the nonstructural protein encoded by the S-RNA of Tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456–463. 10.1094/Phyto-83-456 [DOI] [Google Scholar]
- 46.Rotenberg D, Whitfield AE. 2010. Analysis of expressed sequence tags for Frankliniella occidentalis, the western flower thrips. Insect Mol. Biol. 19:537–551. 10.1111/j.1365-2583.2010.01012.x [DOI] [PubMed] [Google Scholar]
- 47.Garcia S, Billecocq A, Crance JM, Prins M, Garin D, Bouloy M. 2006. Viral suppressors of RNA interference impair RNA silencing induced by a Semliki Forest virus replicon in tick cells. J. Gen. Virol. 87:1985–1989. 10.1099/vir.0.81827-0 [DOI] [PubMed] [Google Scholar]
- 48.Oliveira VC, Bartasson L, de Castro MEB, Correa JR, Ribeiro BM, Resende RO. 2011. A silencing suppressor protein (NSs) of a tospovirus enhances baculovirus replication in permissive and semipermissive insect cell lines. Virus Res. 155:259–267. 10.1016/j.virusres.2010.10.019 [DOI] [PubMed] [Google Scholar]
- 49.Hemmes H, Kaaij L, Lohuis D, Prins M, Goldbach R, Schnettler E. 2009. Binding of small interfering RNA molecules is crucial for RNA interference suppressor activity of Rice hoja blanca virus NS3 in plants. J. Gen. Virol. 90:1762–1766. 10.1099/vir.0.010488-0 [DOI] [PubMed] [Google Scholar]
- 50.Hemmes H, Lakatos L, Goldbach R, Burgyán J, Prins M. 2007. The NS3 protein of Rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both siRNAs and miRNAs. RNA 13:1079–1089. 10.1261/rna.444007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soldan SS, Plassmeyer ML, Matukonis NK, Gonzalez-Scarano F. 2005. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 79:234–244. 10.1128/JVI.79.1.234-244.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medeiros RB, Resende RD, De Avila AC. 2004. The plant virus Tomato spotted wilt tospovirus activates the immune system of its main insect vector, Frankliniella occidentalis. J. Virol. 78:4976–4982. 10.1128/JVI.78.10.4976-4982.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Badillo-Vargas IE, Rotenberg D, Schneweis DJ, Hiromasa Y, Tomich JM, Whitfield AE. 2012. Proteomic analysis of Frankliniella occidentalis and differentially expressed proteins in response to Tomato spotted wilt virus infection. J. Virol. 86:8792–8809. 10.1128/JVI.00285-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.