Abstract
Varicella-zoster virus (VZV) infection causes varicella, after which the virus becomes latent in ganglionic neurons. In tissue culture, VZV-infected human neurons remain viable at 2 weeks, whereas fibroblasts develop cytopathology. Next-generation RNA sequencing was used to compare VZV transcriptomes in neurons and fibroblasts and identified only 12 differentially transcribed genes of the 70 annotated VZV open reading frames (ORFs), suggesting that defective virus transcription does not account for the lack of cell death in VZV-infected neurons in vitro.
TEXT
Varicella-zoster virus (VZV) is an exclusively human neurotropic alphaherpesvirus that usually produces varicella (chickenpox) upon primary infection, after which the virus becomes latent in ganglionic neurons along the entire neuraxis. In tissue culture, VZV-infected neurons remain viable 2 weeks after infection, while all nonneuronal cells develop a cytopathic effect (CPE). Our earlier studies of VZV-infected neurons derived from induced pluripotent stem (iPS) cells revealed that neurons appeared healthy 2 weeks later, with no detectable infectious virus in the tissue culture medium; analysis of the neurons revealed VZV DNA, transcripts, and proteins corresponding to the VZV immediate early, early, and late kinetic phases of replication (1). Furthermore, ultrastructural examination revealed few complete virions and numerous aberrant viral particles (2). Together, these findings indicate that VZV is not latent in these neurons, despite the lack of CPE, and suggest a deficiency in replication and viral assembly in neurons during the productive infection. Thus, next-generation RNA sequencing (NextGen RNA-seq) was used to examine all annotated VZV transcripts in both cell types.
Terminally differentiated human neurons derived from human-induced pluripotent stem cells (iCell; Cellular Dynamics International, Madison, WI) and human fetal lung (HFL) fibroblasts were infected at a multiplicity of infection (MOI) of 1 × 10−3 cell-free vOka strain VZ virions (Zostavax; Merck, Whitehouse Station, NJ) as described previously (2). Total RNA was extracted from VZV-infected fibroblasts when ∼80% CPE was reached and from VZV-infected neurons 14 days postinfection (dpi), at which time 5 to 10% of the neurons were infected. cDNA libraries were constructed for each sample (3 neuron, 3 fibroblast samples) using the Illumina TruSeq stranded-mRNA sample preparation kit (Illumina, San Diego, CA). The six independently and uniquely indexed libraries were pooled and loaded onto a single lane of a HiSeq2000 flow cell for paired-end, 100-bp DNA sequencing using an Illumina HiSeq2000.
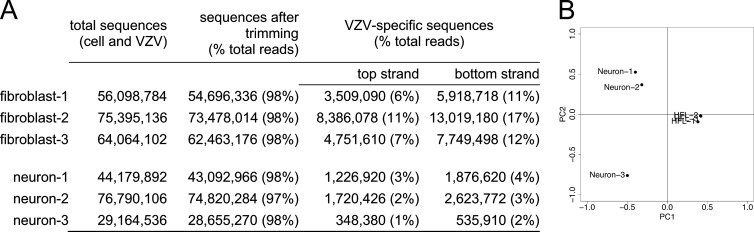
NextGen RNA-seq of six cDNA libraries yielded 29.2 to 76.8 million total reads per sample (Fig. 1A). Removal of low-quality bases (Phred score < 15 [3, 4]) using a custom Python script reduced total sequence data by ≤4%. The remaining sequences were mapped to the annotated VZV genome (Dumas; NCBI accession number NC_001348) using GSNAP (Genomic Short-Read Nucleotide Alignment Program) (5). Viral sequences comprised 3 to 7% of all reads from VZV-infected neurons and 17 to 29% of reads from VZV-infected fibroblasts. The bidirectional, strand-specific cDNA library construction protocol permitted alignment of sequences to either the annotated Dumas strain (top strand) or the complementary (bottom) strand (Fig. 1A) of the VZV genome using Cufflinks (6). After strand alignment of VZV sequences, the number of fragments per kilobase of exon per million mapped reads (FPKM; a relative expression level normalized to the sum of all viral sequences divided by specific open reading frame [ORF] length) was determined. FPKMs from all six libraries were analyzed using the statistical transformation technique of principal-component analysis (PCA) to visualize the differences between samples (Fig. 1B). Samples were first separated by the largest component of variance (principal component 1 [PC1]) and then by the next-largest and independent component of variance (PC2). Based on PCA results and the low number of reads from neuron 3, it was considered to be an outlier and removed from further analysis.
FIG 1.
Analysis of NextGen RNA-seq data quality. (A) Quantification of viral transcripts identified by RNA-seq from VZV-infected fibroblasts and human neurons. (B) Variance among all six samples was analyzed. FPKMs from all libraries were separated using principal-component analysis (PCA). Principal component 1 (PC1) separated samples by their largest variance and resulted in separation between cell types. PC2 separated samples by the next-largest variance, independent of the first, and resulted in separation of samples within cell types. PCA analysis indicated a lack of variation between all three fibroblast samples, whereas neuron 3 was different from neurons 1 and 2. PCA analysis showed that neuron 3 was an outlier.
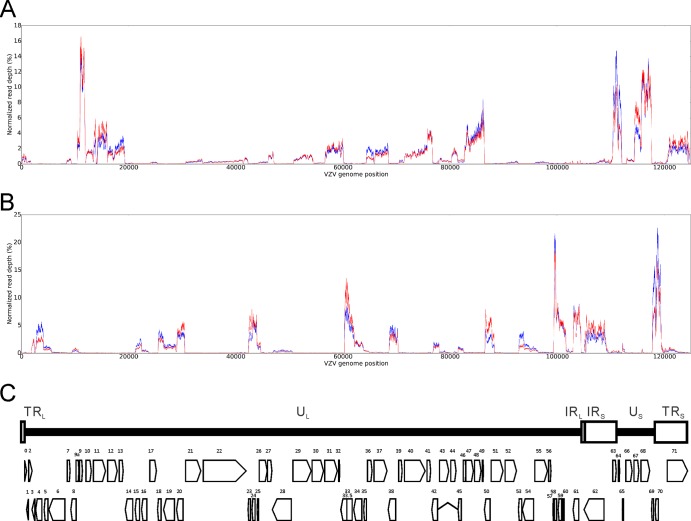
After mapping of the five remaining libraries (two neuron, three fibroblast libraries), the average read depth per nucleotide was normalized to the total number of reads across the entire virus genome for each sample; read depths were averaged for all neuron (Fig. 2, blue) or all fibroblast (Fig. 2, red) samples and plotted by top (Fig. 2A) and bottom (Fig. 2B) strands. Figure 2C shows the relative locations of all annotated VZV ORFs and the directions of transcription.
FIG 2.
Normalized average read depths per base. The total number of times that each nucleotide was sequenced (raw DNA read depth) was normalized to the sum of numbers of all VZV transcripts in neurons (blue) and fibroblasts (red). Sequence reads were plotted for the top (A) or bottom (B) strands of the VZV genome. (C) Positions of both viral ORFs and physical features of VZV DNA. UL and US, unique long and short sequences, respectively; TRL and TRS, terminal repeats of UL and US, respectively; IRL and IRS, internal repeats of UL and US, respectively.
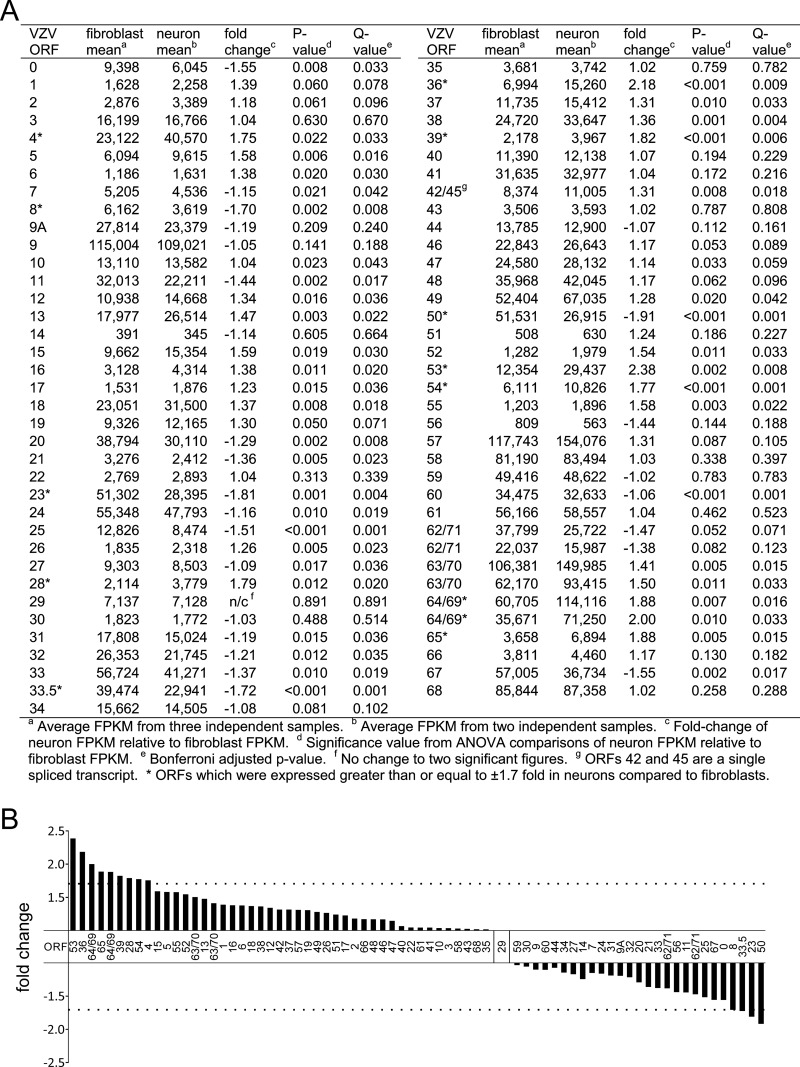
VZV ORFs that were transcribed differently in neurons than in fibroblasts were identified based on their calculated fold changes in FPKMs and then by analysis of variance (ANOVA) using a two-neuron- by three-fibroblast-library design (Fig. 3A). Alignment of the fold changes in transcript levels from highest (greater transcription in neurons) to lowest (greater transcription in fibroblasts) revealed a break at a ±1.70-fold change (Fig. 3B, dotted horizontal lines). Based on a P value of ≤0.05 and a ±1.70-fold change, transcription of eight VZV ORFs was increased in neurons compared to in fibroblasts, and four VZV ORFs were transcribed less in neurons than in fibroblasts (Fig. 3A, asterisks).
FIG 3.
Fold changes in numbers of VZV transcripts in human neurons from their numbers in fibroblasts. (A) ANOVA of VZV transcript numbers from VZV-infected fibroblasts and human neurons. (B) Fold changes in the numbers of VZV transcripts for all ORFs in infected human neurons from those in fibroblasts, arranged from highest (greater in neurons) to lowest (greater in fibroblasts). Dotted horizontal lines denote a ±1.70-fold change in transcript levels between cell types. ORFs 53, 36, 64/69, 65, 39, 28, 54, and 4 were transcribed more in neurons, and ORFs 50, 23, 33.5, and 8 were transcribed more in fibroblasts.
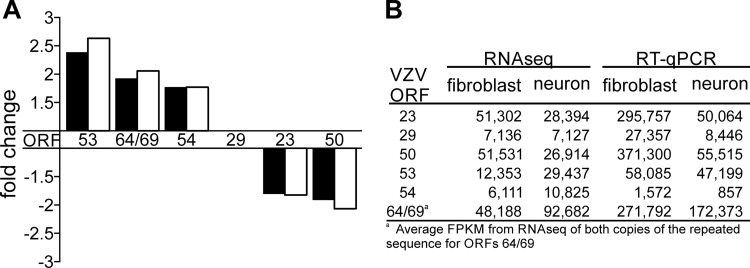
To validate NextGen RNA-seq results, reverse transcription quantitative PCR (RT-qPCR) was performed on five differentially transcribed VZV ORFs as described previously (7) using the same RNAs used for RNA-seq. VZV ORF 53, 54, and 64/69 transcripts were more abundant in neurons than in fibroblasts, while VZV ORF 23 and 50 transcripts were more abundant in fibroblasts (Fig. 3B). Since RNA-seq data were normalized to total VZV transcript levels (FPKM), each RT-qPCR also required normalization. For this, VZV ORF 29 was used since the ratio of the level of this transcript in virus-infected neurons to its level in fibroblasts was 1 (i.e., the transcript abundances did not differ between cell types) (Fig. 3). After normalization, the fold change of the level of transcription of each VZV ORF in infected neurons from the level of transcription in infected fibroblasts was determined (Fig. 4A, white bars). RT-qPCR results were the same as those found using NextGen RNA-seq technology (Fig. 4A, black bars).
FIG 4.
Validation of RNA-seq by RT-qPCR. RNA used in RNA-seq analysis was reverse transcribed with oligo(dT), and primers and cDNA were analyzed by RT-qPCR. Primer/probe sets were designed for five VZV ORFS that exceeded the ±1.70-fold cutoff; three genes (ORFs 53, 64/69, and 54) were transcribed more in neurons, and two genes (ORFs 23 and 50) were transcribed less in neurons. Each RT-qPCR mixture contained a primer/probe set for ORF 29 for normalization. (A) Fold changes (from neurons to fibroblasts) in the levels of transcription of the six ORFs from RNA-seq analysis (black bars) or by RT-qPCR after normalization to ORF 29 (white bars). (B) Raw data from RNA-seq (FPKMs) and RT-qPCR (copy numbers) used to construct the graph in panel A.
Herein, next-generation RNA sequencing was used to better understand the absence of CPE during productive VZV infection of human neurons compared to the effects of infection of fibroblasts by determining the complete virus transcriptome in each cell type. Surprisingly, only 12 of the 70 VZV ORFs showed differences in transcript abundance between the two cell types.
We found that VZV-infected human neurons in cell culture transcribed every annotated ORF, unlike the limited viral transcription present in human and monkey ganglia latently infected with varicella-zoster virus (8–10). This confirmed a significant difference in virus gene transcription in vitro from that in human neurons latently infected with VZV. A comparison of the 12 differentially transcribed VZV ORFs to orthologous herpes simplex virus 1 (HSV-1) genes revealed that they did not belong to a unique class of virus genes: one was immediate early (ORF 4), three were early (ORFs 8, 28, and 36), and eight were late (ORFs 23, 33.5, 39, 50, 53, 54, 64/69, and 65); six were essential (ORFs 4, 28, 33.5, 39, 53, and 54) and six were nonessential (ORFs 8, 23, 36, 50, 64/69, and 65); and nine mapped to the bottom strand (ORFs 4, 8, 23, 28, 33.5, 50, 53, 54 and 65), two to the top strand (ORFs 36 and 39), and one to both strands (ORF 64/69). Overall, viral transcription in neurons that survive 2 weeks after VZV infection does not appear to be defective compared to that in fibroblasts. Further studies are needed to compare the rates of VZV DNA replication in neurons and fibroblasts.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AG006127 (D.G.), AG032958 (D.G. and R.J.C.), and NS082228 (R.J.C.) from the National Institutes of Health. N. L. Baird was supported by training grant NS007321 to D. Gilden from the National Institutes of Health. K. L. Jones and RNA sequencing were supported in part by the Genomics and Biostatistics/Bioinformatics Shared Resources of Colorado's NIH/NCI Cancer Center (support grant P30CA046934).
We thank Marina Hoffman for editorial review and Lori DePriest for manuscript preparation.
Footnotes
Published ahead of print 5 March 2014
REFERENCES
- 1.Yu X, Seitz S, Pointon T, Bowlin JL, Cohrs RJ, Jonjic S, Haas J, Wellish M, Gilden D. 2013. Varicella zoster virus infection of highly pure terminally differentiated human neurons. J. Neurovirol. 19:75–81. 10.1007/s13365-012-0142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grose C, Yu X, Cohrs RJ, Carpenter JE, Bowlin JL, Gilden D. 2013. Aberrant virion assembly and limited glycoprotein C production in varicella-zoster virus-infected neurons. J. Virol. 87:9643–9648. 10.1128/JVI.01506-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 4.Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 5.Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohrs RJ, Gilden DH. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 81:2950–2956. 10.1128/JVI.02745-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer C, Kerns A, Barron A, Kreklywich C, Streblow DN, Messaoudi I. 2011. Simian varicella virus gene expression during acute and latent infection of rhesus macaques. J. Neurovirol. 17:600–612. 10.1007/s13365-011-0057-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagel MA, Choe A, Traktinskiy I, Cordery-Cotter R, Gilden D, Cohrs RJ. 2011. Varicella-zoster virus transcriptome in latently infected human ganglia. J. Virol. 85:2276–2287. 10.1128/JVI.01862-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouwendijk WJ, Choe A, Nagel MA, Gilden D, Osterhaus AD, Cohrs RJ, Verjans GM. 2012. Restricted varicella-zoster virus transcription in human trigeminal ganglia obtained soon after death. J. Virol. 86:10203–10206. 10.1128/JVI.01331-12 [DOI] [PMC free article] [PubMed] [Google Scholar]