ABSTRACT
The increasing prevalence of ranavirus (RV; Iridoviridae) infections of wild and commercially maintained aquatic species is raising considerable concerns. While Xenopus laevis is the leading model for studies of immunity to RV, amphibian antiviral interferon (IFN) responses remain largely uncharacterized. Accordingly, an X. laevis type I interferon was identified, the expression of the gene for this IFN was examined in RV (frog virus 3 [FV3])-infected tadpoles and adult frogs by quantitative PCR, and a recombinant form of this molecule (recombinant X. laevis interferon [rXlIFN]) was produced for the purpose of functional studies. This rXlIFN protected the kidney-derived A6 cell line and tadpoles against FV3 infection, decreasing the infectious viral burdens in both cases. Adult frogs are naturally resistant to FV3 and clear the infection within a few weeks, whereas tadpoles typically succumb to this virus. Hence, as predicted, virus-infected adult X. laevis frogs exhibited significantly more robust FV3-elicited IFN gene expression than tadpoles; nevertheless, they also tolerated substantially greater viral burdens following infection. Although tadpole stimulation with rXlIFN prior to FV3 challenge markedly impaired viral replication and viral burdens, it only transiently extended tadpole survival and did not prevent the eventual mortality of these animals. Furthermore, histological analysis revealed that despite rXlIFN treatment, infected tadpoles had considerable organ damage, including disrupted tissue architecture and extensive necrosis and apoptosis. Conjointly, these findings indicate a critical protective role for the amphibian type I IFN response during ranaviral infections and suggest that these viruses are more pathogenic to tadpole hosts than was previously believed, causing extensive and fatal damage to multiple organs, even at very low titers.
IMPORTANCE Ranavirus infections are threatening wild and commercially maintained aquatic species. The amphibian Xenopus laevis is extensively utilized as an infection model for studying ranavirus-host immune interactions. However, little is known about amphibian antiviral immunity and, specifically, type I interferons (IFNs), which are central to the antiviral defenses of other vertebrates. Accordingly, we identified and characterized an X. laevis type I interferon in the context of infection with the ranavirus frog virus 3 (FV3). FV3-infected adult frogs displayed more robust IFN gene expression than tadpoles, possibly explaining why they typically clear FV3 infections, whereas tadpoles succumb to them. Pretreatment with a recombinant X. laevis IFN (rXlIFN) substantially reduced viral replication and infectious viral burdens in a frog kidney cell line and in tadpoles. Despite reducing FV3 loads and extending the mean survival time, rXlIFN treatments failed to prevent tadpole tissue damage and mortality. Thus, FV3 is more pathogenic than was previously believed, even at very low titers.
INTRODUCTION
Amphibian populations are facing a serious threat of extinction (1), with a decline of approximately one-third (32%) of species resulting from complex, poorly understood causes (2, 3). Due to the dramatic increases in the prevalence of ranavirus (RV; family Iridoviridae) infections and the resulting mortalities of aquatic species, these viruses are now believed to be a factor contributing to the amphibian decline and are recognized as ecologically and economically relevant agents (1–3). RVs are large, icosahedral, double-stranded DNA viruses that cause systemic diseases and mortalities resulting from hemorrhaging and necrotic death of multiple afflicted organs (1). These viruses infect amphibian species across the world, including Asia (4–6), Australia (7, 8), the United Kingdom (9, 10), and North America (11–14), and as such are considered emerging infectious diseases (3). Of the three RV species presently plaguing amphibians, as recognized by the International Committee on Taxonomy of Viruses, Bohle iridovirus (BIV) and Ambystoma tiginum virus (ATV) have thus far remained confined to infecting their respective natural host species (8, 14, 15). In contrast, frog virus 3 (FV3), initially isolated from the leopard frog, Rana (Lithobates) pipiens (4), is now recognized worldwide to be an amphibian pathogen with a threatening potential to cross multiple species barriers and establish infections to the detriment of new hosts (15–18).
The amphibian Xenopus laevis provides an ideal platform for studying RV-host immune interactions and elucidating the mechanisms governing amphibian tadpole susceptibility and adult resistance to RV infections (19). In fact, X. laevis has successfully been employed as an FV3 infection model to underline several key immune parameters that may contribute to these susceptibility differences (19–22). Unfortunately, studies of amphibian antiviral immunity have been limited by the largely uncharacterized frog antiviral defenses, especially those orchestrated by type I interferon (IFN) cytokines.
Notably, the cytokines belonging to the type I IFN family are central to vertebrate antiviral immunity. In mammals, these moieties are encoded by intronless genes that comprise the multigene alpha IFN (IFN-α) family (13 in humans) and the single IFN-β (reviewed in reference 23). Akin to mammals, reptiles and birds also express single exon-encoded type I IFNs (24, 25). In contrast, lower vertebrates, such as cartilaginous and bony fish as well as amphibians, are now known to possess a distinct type I IFN system. These lower vertebrate IFN homologs also comprise multiple members but are encoded on transcripts containing five exons and four introns (24–28). To date, only the teleost IFNs have been characterized in detail; they have now been subdivided into two groups (group I, 2 C residues; group II, 4 C residues) on the basis of cysteine patterns (28, 29) and further subdivided into four groups (IFNa to IFNd) according to phylogeny (26, 29). Interestingly, whereas the mammalian multigene IFNs all utilize the same receptor complex for cell signaling (30, 31), the fish group I and II IFNs are thought to act through distinct receptor complexes (32).
Functional studies have predominantly been performed on group I, type I fish IFNs (28, 32–36), where these cytokines have a range of antiviral activity, such as through induction of hallmark antiviral genes (the genes for MX1, viperin, and PKR) and antiviral protection (32, 35, 37, 38). Interestingly, it has recently been shown that salmonid IFNs a to d are under distinct transcriptional regulation and possess different antiviral capacities (39). In fact, some of these IFN cytokines display potent antiviral effects, while at present, others are believed not to confer any antiviral properties (39).
Amphibians such as Xenopus represent a key stage in the evolution of vertebrate antiviral defenses. However, to date this arm of the amphibian immune response has remained largely uncharacterized. Accordingly, we identified, cloned, and produced a recombinant form of an X. laevis type I interferon (rXlIFN), examined its gene expression in ranavirus (FV3)-infected tadpoles and adult frogs, and assessed its capacity to protect Xenopus cells in vitro and tadpoles in vivo against FV3 infection.
MATERIALS AND METHODS
Animals.
Outbred premetamorphic (stages 54 to 56) tadpoles and metamorphic (stage 64) and adult (2 years old) frogs were obtained from our X. laevis research resource for immunology at the University of Rochester (http://www.urmc.rochester.edu/smd/mbi/xenopus/index.htm). Experiments involving frogs and tadpoles were carried out according to the Animal Welfare Act from the United States Department of Agriculture (USDA), Public Health Service policy (A-3292-01), and the Public Health Act of New York State. Animal care and all the protocols were reviewed and approved by the University of Rochester Committee on Animal Resources (approval number 100577/2003-151).
Identification of X. laevis type I IFN.
Partial X. laevis IFN cDNA was identified using primers against X. tropicalis IFN5. Rapid amplification of cDNA ends (RACE)-PCR was performed in accordance with the manufacturer's directions (Clontech) to identify the 5′ and 3′ regions of the X. laevis cDNA transcript.
In silico analyses.
Protein sequence alignments were performed using Clustal W software (http://www.ebi.ac.uk/clustalw/). Signal peptide regions were identified using the SignalP (version 3.0) server (http://www.cbs.dtu.dk/services/SignalP/). Phylogenetic analysis was performed with Clustal X software using the neighbor-joining method and bootstrapping 10,000 times, with values expressed as percentages.
FV3 stocks and animal infections.
Fathead minnow (FHM) cells (ATCC CCL-42; American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 30°C with 5% CO2. FV3 was grown by a single passage in FMH cells and purified via ultracentrifugation on a 30% sucrose cushion. Tadpole kidneys and A6 cells to be assessed for infectious FV3 burdens were subjected to 3 rounds of sequential freeze-thaw lysis and intermittent repeated passages through a 24-gauge needle; the resulting homogenates were examined by plaque assays. All plaque assays were performed on BHK cell monolayers under an overlay of 1% methylcellulose, as previously described (22).
All tadpole infections were performed by intraperitoneal (i.p.) injection of 1 × 104 PFU of FV3 in 10-μl volumes. Adult frogs were infected i.p. with 5 × 106 PFU of FV3 in 100-μl volumes. At the desired times postinfection, frogs were euthanized by immersion in 0.5% tricaine methane sulfonate (MS-222), and tissues were removed and processed for RNA and DNA isolation.
Semiquantitative (RT) and quantitative PCR gene expression analysis.
Total RNA and DNA were extracted from frog tissues using the TRIzol reagent following the manufacturer's directions (Invitrogen). All cDNA syntheses were performed using an iScript cDNA synthesis kit according to the manufacturer's directions (Bio-Rad, Hercules, CA) and 500 ng of total DNase (Ambion)-treated RNA. Reverse transcription (RT)-PCR and quantitative RT-PCR (qRT-PCR) analyses were performed using 2.5 μl of cDNA templates and 50 ng of DNA templates. RT-PCR products were resolved on 1.5% agarose gels, visualized with ethidium bromide, and compared against a 1-kb-plus DNA marker (Invitrogen).
Relative qRT-PCR gene expression analyses of IFN and MX1 were performed via the ΔΔCT threshold cycle (CT) method, with the level of expression being examined relative to that of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) endogenous control and normalized against the lowest observed level of expression. To measure FV3 loads and viral DNA (vDNA) polymerase II (Pol II) gene expression, absolute qRT-PCR was performed on DNA and cDNA using a serially diluted standard curve. Briefly, an FV3 DNA Pol II PCR fragment was cloned into the pGEM-T Easy vector (Promega), and this construct was amplified in bacteria, quantified, and serially diluted to yield 1010 to 101 plasmid copies of the vDNA Pol II. These dilutions were employed to create a standard curve in subsequent absolute qRT-PCR experiments to assess viral genome and vDNA Pol II transcript copy numbers relative to those on this standard curve. All experiments were performed using an ABI 7300 real-time PCR system, PerfeCTa SYBR green FastMix, and carboxy-X-rhodamine (Quanta). Expression analysis was performed using ABI sequence detection system (SDS) software. All primers were validated prior to use. Primer sequences are available upon request.
Production of rXlIFN.
The portion of the X. laevis type I IFN sequence corresponding to the signal peptide-cleaved fragment was ligated into the pMIB/V5 His A insect expression vector (Invitrogen) and introduced into Sf9 insect cells (Cellfectin II reagent; Invitrogen). The rXlIFN-positive transfectants were selected using 10 μg/ml blasticidin, scaled up into 500-ml liquid cultures, and grown for 5 days. The rXlIFN-containing supernatants were concentrated against polyethylene glycol flakes (8 kDa), dialyzed against 150 mM sodium phosphate, and passed through Ni-nitrilotriacetic acid agarose columns (Qiagen) to bind the recombinant cytokine. The column was washed twice with 10 volumes of high-stringency wash buffer (0.5% Tween 20, 50 mM sodium phosphate, 500 mM sodium chloride, 100 mM imidazole) and five times with 10 volumes of low-stringency wash buffer (like the high-stringency wash buffer described above but with 40 mM imidazole). rXlIFN was eluted in fractions using 500 mM imidazole, and the purity of the recombinant protein was assessed by SDS-PAGE and Western blotting against the V5 epitope on rXlIFN. The rXlIFN-containing fractions were pooled, and a sample was taken to determine the protein concentration by the Bradford protein assay (Bio-Rad). After addition of a protease inhibitor cocktail (Roche), the rXlIFN preparation was aliquoted and stored at −20°C until use.
The vector control was derived by transfecting Sf9 cells with an empty expression vector and following the methodology described for the generation and isolation of rXlIFN.
The antiviral activity of rXlIFN was assessed by pretreatment of A6 cell cultures at various doses (0.05, 5, 50, or 500 ng/ml rXlIFN) for 8 h, followed by FV3 infection at multiplicities of infection (MOIs) of 0.04, 0.2, and 1 for 24 h. A6 cells were harvested, lysed, and assessed by plaque assays (as described above). For all MOIs, the virus loads were drastically (but not completely) reduced at both the 50- and 500-ng/ml doses and still significantly decreased at 5 ng/ml (data not shown). Thus, 50 ng/ml was used for subsequent in vitro experiments, whereas 500 ng of total rXlIFN protein was used in tadpole protection studies.
Cell culture medium.
The ASF culture medium used in these studies has been previously described (19). All cell cultures were established using ASF supplemented with 10% fetal bovine serum, 20 μg/ml kanamycin, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco). Amphibian phosphate-buffered saline (APBS) has been previously described (19).
A6 cell maintenance, rXlIFN stimulation, and FV3 infection.
A6 cell cultures were maintained in the medium described above with weekly passages.
For A6 cell-MX1 gene expression studies, 1 × 106 A6 cells were seeded into individual wells of 24-well plates and incubated for 1, 3, 6, or 12 h with 50 ng/ml rXlIFN or an equal volume of the vector control, RNA was isolated, and cDNA was synthesized using the methods described above.
On the day prior to FV3 infection, A6 cells were seeded onto sterile microscope slides in 6-well plates at approximately 80% confluence. On the following day, cells were incubated for 8 h with medium alone (mock infection), 50 ng/ml rXlIFN, or an equal volume of the vector control and subsequently infected with FV3 at an MOI of 0.3. At 1 and 3 days postinfection (dpi), cells were fixed and stained using rabbit anti-FV3 53R primary antibody (Ab; generously provided by Greg V. Chinchar) and fluorescein isothiocyanate (FITC)-labeled goat antirabbit secondary Ab. Cellular nuclei were visualized using a Hoechst DNA stain. Slides were mounted onto microscope slides and examined using an Axiovert 200 inverted microscope and Infinity 2 digital camera (objective, ×40/0.6; Zeiss). Digital images were analyzed using Image-Pro Plus and ImageJ software.
Tadpole rXlIFN stimulation and FV3 infection.
For tadpole MX1 expression studies, tadpoles were injected i.p. with 500 ng of rXlIFN protein or an equal volume of the vector control. On the following day, tadpoles were euthanized in 0.5% tricaine methane sulfonate (MS-222), and tissues were removed and processed for RNA.
For short-term rXlIFN protection studies, stage 54 tadpoles (n = 6 tadpoles per treatment group) were injected i.p. with 500 ng of rXlIFN or an equal volume of the vector control and 8 h later were injected with either 104 PFU FV3 in APBS or APBS alone. At 6 days following treatment and FV3 infection, peritoneal leukocytes (PLs) were isolated by i.p. flushing with APBS using fine-tipped pulled-glass needles. Tissues were removed, and the RNA, DNA, and cDNA were derived as described above. PLs from 6 tadpoles were pooled to obtain sufficient RNA and DNA for analysis.
For tadpole survival studies, stage 50 tadpoles (n = 11 tadpole per treatment group) were injected as described above and monitored over the course of 50 days. Tadpoles were checked twice daily, and dead animals were immediately frozen and stored at −20°C for DNA isolation.
Histology.
Tadpoles were preinjected with the vector control or rXlIFN (500 ng/tadpole), infected with FV3 or mock infected by injection of APBS as described above, and reared until they displayed characteristic signs of terminal infection, including irregular swimming and trouble maintaining balance. At this point, the animals were euthanized in 0.5% tricaine methane sulfonate and fixed in buffered 10% formalin. Fixed tadpoles were rinsed with phosphate-buffered saline (PBS), immersed in successively higher ethanol solutions (30, 50, 70%), embedded in paraffin, sectioned (5-μm sections), and hematoxylin-eosin stained. The resulting histology slides were examined using a Niko Eclipse E200 phase-contrast microscope, and images were taken using a Nikon SPOT Idea digital camera and analyzed using SPOT Imaging software.
Statistical analysis.
Statistical analysis was performed using a one-way analysis of variance (ANOVA) and Tukey's post hoc test. A two-sample F test was performed on the A6 cell MX1 gene expression data. A probability level of P equal to <0.05 was considered significant. The Vassar Stat program was used for statistical computation (http://faculty.vassar.edu/lowry//anova1u.html).
Nucleotide sequence accession number.
The X. laevis cDNA sequence determined in this study was deposited in GenBank under accession number KF597522.
RESULTS
Identification and in silico and phylogenetic analysis of an X. laevis type I IFN.
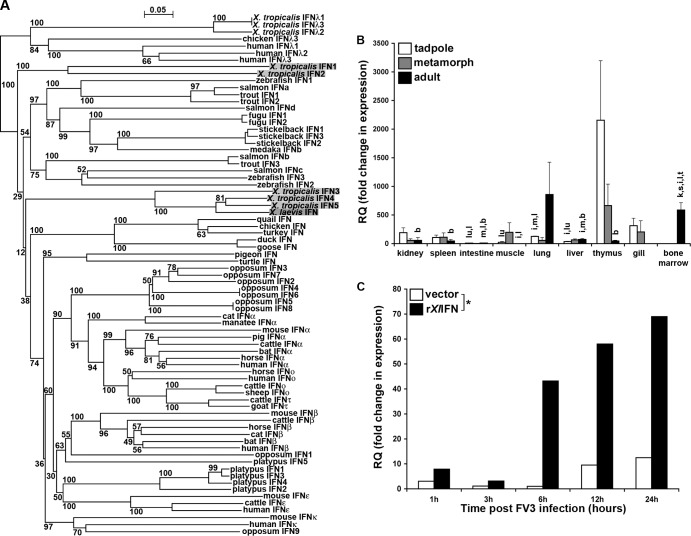
To investigate amphibian antiviral immunity against FV3, we sought to identify an X. laevis type I IFN. Accordingly, we designed primers against the X. tropicalis type I IFN sequences (GenBank accession no. BN001171), and via conventional and RACE-PCR, we successfully identified the full-length cDNA transcript of an X. laevis IFN (data not shown). This X. laevis transcript encodes a 189-residue protein with a signal peptide, four structurally conserved cysteines, six mRNA instability motifs (ATTTA) (40, 41), and a conventional polyadenylation signal (AATAAA; data not shown).
To examine the evolutionary relationships among vertebrate type I IFN proteins, we performed phylogenetic analyses (Fig. 1A). Interestingly, X. tropicalis IFN1 and IFN2 branched as a separate clade from X. tropicalis IFNs 3, 4, and 5 and the X. laevis IFN, albeit with low bootstrap values (Fig. 1A). Furthermore, X. tropicalis IFNs 1 and 2 branched ancestral to fish group I and II IFNs, X. tropicalis IFNs 3, 4, and 5, and the X. laevis IFN (Fig. 1A), suggesting that the former may have been retained from an ancestral species, while the latter diverged with evolutionary time. The fish and amphibian IFNs branched ancestral from the reptile, avian, monotreme, marsupial, and mammalian type I IFNs (Fig. 1A).
FIG 1.
Analysis of X. laevis IFN phylogeny (A), quantitative tissue gene expression (B), and capacity to elicit A6 kidney cell MX1 gene expression (C). (A) The phylogenetic tree was constructed from a multiple-sequence alignment using the neighbor-joining method and bootstrapped 10,000 times; the bootstrap values are expressed as percentages. (B) Tissues from outbred premetamorphic (stages 54 to 56) tadpoles and metamorphic (stage 64) and adult (2 years old) frogs were assessed. Tissues from 3 individuals of each stage were examined. The individual letters above the bars correspond to tissues exhibiting significantly different (P < 0.05) IFN gene expression: k, kidney; s, spleen; i, intestine; m, muscle; lu, lung; l, liver; t, thymus; g, gill; b, bone marrow. (C) A6 cells (1 × 106) were incubated for the indicated times with 50 ng/ml rXlIFN or an equal volume of the vector control, harvested, and assessed by qRT-PCR for MX1 gene expression. *, statistically significant difference (P < 0.05) in variance between the vector- and rXlIFN-treated A6 cell cultures over time. The level of expression of both genes relative to that of the GAPDH endogenous control was examined (B and C). RQ, relative quantification values.
Quantitative analysis of IFN gene expression in tissues of tadpoles and metamorphic and adult X. laevis frogs.
We performed qRT-PCR gene expression analysis of X. laevis IFN in the tissues of tadpoles (stage 54) and metamorphic (stage 64) and adult frogs (Fig. 1B). Most tissues examined exhibited modest levels of IFN transcripts, irrespective of developmental stage (Fig. 1B). However, IFN gene expression was strikingly high in tadpole thymi and declined through metamorphosis to much lower levels in adult frogs (Fig. 1B). Notably, considerably greater IFN transcript levels were seen in the lungs of adults than in the lungs of tadpoles and metamorphs (Fig. 1B). Finally, relatively robust IFN gene expression was detected in the adult bone marrow (Fig. 1B).
rXlIFN enhances MX1 gene expression in A6 cells.
To assess whether the identified X. laevis IFN possesses antiviral activity, we produced a recombinant form of the signal sequence-cleaved X. laevis IFN protein (rXlIFN) using an insect protein expression system. We incubated X. laevis kidney-derived A6 cell line cultures with either this purified rXlIFN (50 ng/ml) or a vector control (supernatants from Sf9 insect cells transfected with an empty vector and processed as described above for the rXlIFN isolation procedure). Cells were harvested at 1, 3, 6, 12, and 24 h following the treatment, and the expression of the antiviral MX1 gene was examined (Fig. 1C). We observed time-dependent rXlIFN-induced MX1 gene expression in A6 cells, beginning at 6 h poststimulation (Fig. 1C), indicating that rXlIFN possessed antiviral properties in vitro.
rXlIFN confers antiviral protection against FV3 infection of A6 cells.
To examine the antiviral protective capacity of rXlIFN in vitro, we preincubated A6 cell cultures for 8 h with the recombinant cytokine (50 ng/ml) or with the vector control and then infected the cells with FV3 at an MOI of 0.3 and examined the efficacy of infection using anti-FV3 53R Ab (42) (Fig. 2). After 1 day of FV3 infection, a substantial proportion of the vector-treated A6 cells showed cytopathicity (Fig. 2A, first column, phase-contrast images) and anti-FV3 53R Ab staining (Fig. 2A, middle column), indicative of infection. Conversely, rXlIFN-pretreated cultures infected in parallel exhibited very low numbers of FV3-infected cells and almost no cytopathicity (Fig. 2A, bottom). By 3 dpi, the majority of cells in vector-treated A6 cell cultures were dead and lysed, with the remainder exhibiting extreme cytopathicity and irregular, clumped growth with active FV3 infection being seen in over 70% of the remaining cells (Fig. 2B, top, and C). In stark contrast, at 3 dpi, rXlIFN-treated A6 cell cultures showed no cytopathicity and no detectable viral antigen (Fig. 2B, bottom, and C).
FIG 2.
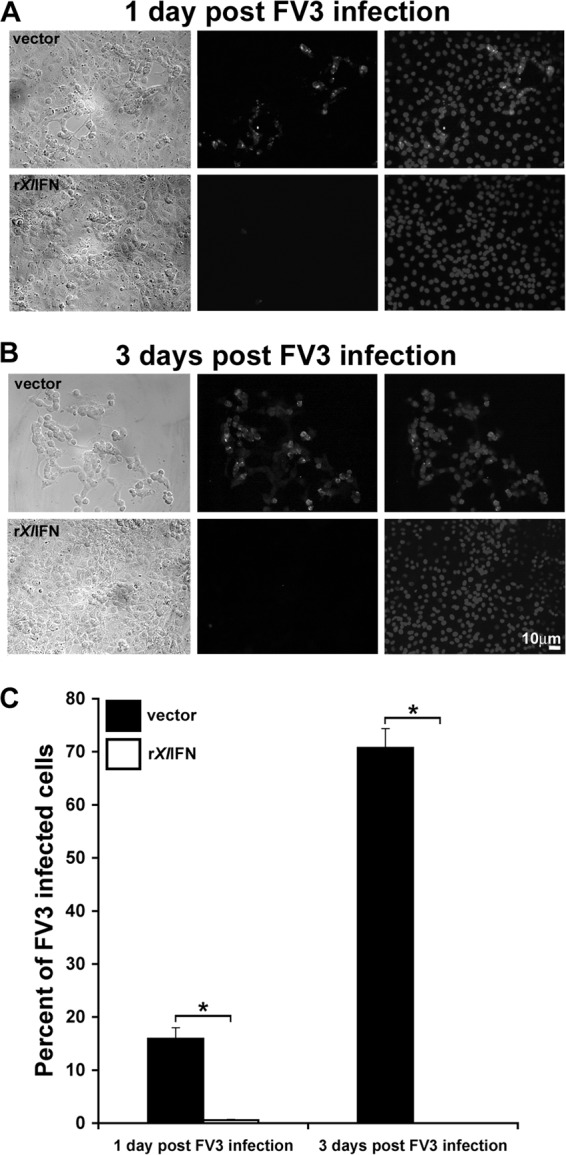
Treatment of A6 cells with rXlIFN is protective against FV3 infection. A6 cells were cultured for 8 h with medium alone (mock infection; not shown), 50 ng/ml rXlIFN, or an equal volume of the vector control, subsequently infected with FV3 at an MOI of 0.3, and processed at 1 dpi (A) and 3 dpi (B). Cells were stained using rabbit anti-FV3 53R primary Ab and FITC-labeled goat antirabbit secondary Ab. Cellular nuclei were visualized using the Hoechst DNA stain. First column, phase-contrast images of A6 cells pretreated with the vector control or rXlIFN prior to infection for 1 or 3 days with FV3; second column, anti-FV3 53R Ab immunofluorescence images corresponding to those presented in the first column; third column, merged images of the respective Hoechst- and anti-FV3 53R Ab-stained cultures presented in the first column. (C) Percentage of FV3-infected A6 cells. Digital images from the experiments represented in panels A and B were analyzed using Image-Pro Plus and ImageJ software. Results are presented as mean percentages ± SEMs of infected cells (of total) per 10 fields. *, significant difference between vector and rXlIFN treatment groups (P < 0.05).
rXlIFN induces tadpole MX1 gene expression and ablates FV3 replication and transcriptional activity.
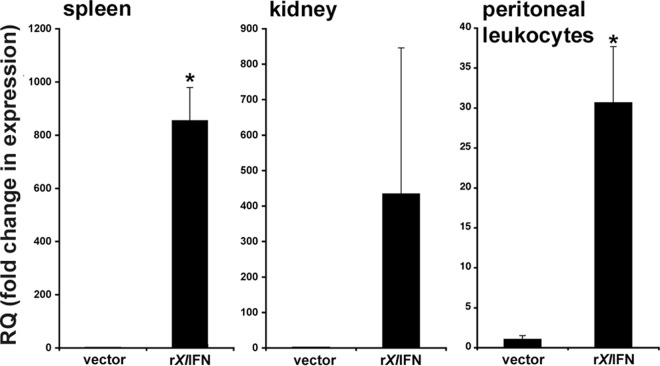
To examine whether rXlIFN could confer antiviral effects in vivo, we injected X. laevis tadpoles i.p. with the recombinant cytokine (500 ng/tadpole) and examined MX1 gene expression in the spleen (a central immune organ), kidney (the primary site of FV3 replication), and PLs (the first cells to encounter the virus) 1 day after rXlIFN administration. Interestingly, MX1 transcript levels were significantly elevated in the spleens and PLs of rXlIFN-injected animals, whereas the kidney expression of MX1 was variable (Fig. 3).
FIG 3.
Administration of rXlIFN induces MX1 gene expression in tadpole spleens, kidneys, and peritoneal leukocytes. Tadpoles (stages 54 to 56) were injected i.p. with 500 ng of rXlIFN or an equal volume of the vector control. Twenty-four hours later, tissues and cells were isolated for qRT-PCR analysis. Kidney and spleen tissues from 4 individuals were examined. Peritoneal leukocytes were isolated from 3 individual tadpoles and pooled; 3 distinct pools were examined. Results are means ± SEMs, and the levels of MX1 gene expression relative to the level of expression of the GAPDH endogenous control were examined. *, significant difference from vector-treated controls (P < 0.05).
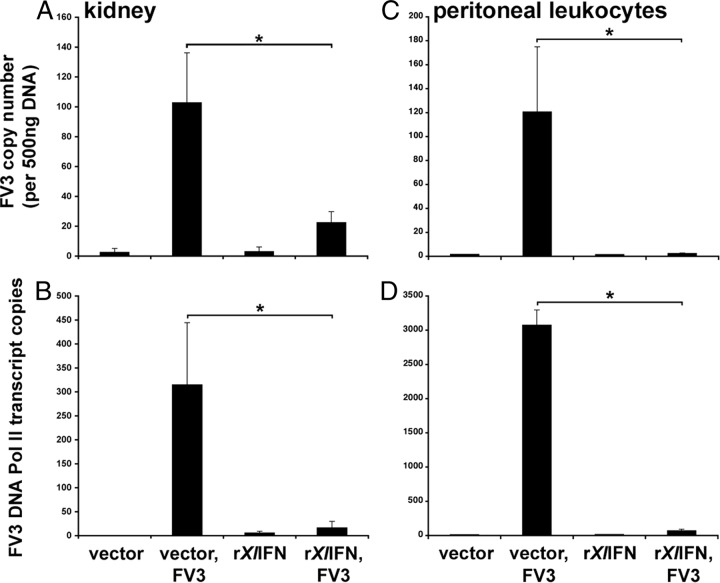
Since rXlIFN conferred potent antiviral effects on A6 cells in vitro (Fig. 1C and 2) and elicited significant increases in MX1 gene expression in tadpoles in vivo (Fig. 3), we next examined if this cytokine could also protect the highly susceptible X. laevis tadpoles against FV3 infection. Accordingly, we pretreated tadpoles by i.p. injection of rXlIFN (500 ng/tadpole) 8 h prior to infection with FV3 (104 PFU) and at 6 dpi assessed the viral loads and viral transcription by qRT-PCR (Fig. 4). Notably, both the kidneys and PLs from tadpoles that had been preadministered the rXlIFN showed significantly reduced FV3 loads compared to the vector-injected controls (Fig. 4A and C). Furthermore, rXlIFN treatment also hampered FV3 gene expression, as shown by the significantly diminished levels of the essential FV3 transcript vDNA Pol II (Fig. 4B and D).
FIG 4.
Administration of rXlIFN to tadpoles prior to FV3 infection reduces viral burdens and viral DNA Pol II expression. Stage 54 tadpoles (n = 6 per treatment group) were injected i.p. with 500 ng of rXlIFN or an equal volume of the vector control. After 8 h, tadpoles were injected i.p. with 104 PFU FV3 in APBS or APBS alone. At 6 dpi, peritoneal leukocytes and kidneys were removed for RNA and DNA isolation. Peritoneal leukocytes were isolated from 3 individual tadpoles and pooled to obtain sufficient RNA and DNA for analysis, and 3 distinct pools were examined. The vDNA Pol II copy number in the derived cDNA and DNA samples was measured by absolute qRT-PCR (using a vDNA Pol II standard curve) to determine FV3 gene expression and viral loads in kidneys (A and B) and PLs (C and D). Results are means ± SEMs. *, significant difference from vector-treated controls (P < 0.05).
The observations presented above indicate that rXlIFN efficiently triggers X. laevis tadpole antiviral immunity, effectively abrogating FV3 replication and transcriptional activity.
Pretreatment of tadpoles and A6 cells with rXlIFN reduces their infectious FV3 burdens.
In order to examine the effects of rXlIFN on the infectivity of FV3, we pretreated A6 cells with the recombinant protein (or vector control) as described above, harvested and homogenized cells at 1 and 3 dpi, and examined their intracellular infectious viral burdens by plaque assays (Fig. 5A). In corroboration of our prior findings, pretreatment of A6 cell cultures with rXlIFN significantly reduced the intracellular FV3 burdens (in numbers of PFU/ml) at 1 and 3 dpi compared to those for vector-pretreated FV3-infected parallel A6 cell cultures (Fig. 5A).
FIG 5.
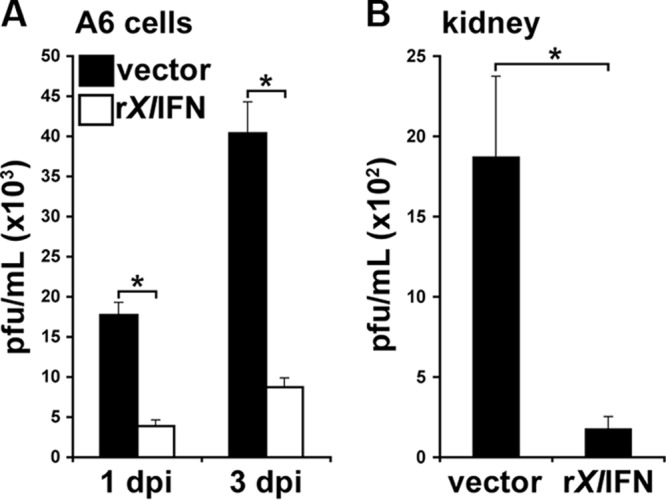
Pretreatment of A6 cell cultures and tadpoles with rXlIFN reduces infectious viral yields. (A) A6 cells were treated for 8 h with 50 ng/ml rXlIFN or an equal volume of the vector control, subsequently infected with FV3 at an MOI of 0.3, and processed at 1 and 3 dpi. The results are the means ± SEMs of the PFU/ml (103) from triplicate experiments. (B) Stage 54 tadpoles (n = 3/treatment group) were injected i.p. with 500 ng of rXlIFN or an equal volume of the vector control. After 8 h, tadpoles were injected i.p. with 104 PFU of FV3 in APBS or APBS alone. At 6 dpi, kidneys were removed and plaque assays were performed on their homogenates. Results are means ± SEMs of the PFU/ml (102). *, significant difference from vector-treated controls (P < 0.05).
In order to extend these in vitro findings to in vivo conditions, tadpoles were pretreated with rXlIFN and infected with FV3 as described above and sacrificed after 6 dpi, and their kidneys were isolated, homogenized, and also assessed for intracellular viral burdens by plaque assays (Fig. 5B). Consistent with the protective properties conferred by rXlIFN in other assays, kidneys from animals pretreated with the antiviral cytokine contained significantly fewer PFU/ml of FV3 than vector control-pretreated infected animals (Fig. 5B).
X. laevis tadpoles and adults exhibit distinct IFN gene expression patterns and viral burdens during FV3 infection.
Since rXlIFN pretreatment significantly abolished viral replication and gene expression in tadpoles upon FV3 infection, we wanted to determine whether larval susceptibility to FV3 (compared to adult frog resistance) correlated with differences in IFN gene expression. Accordingly, we infected tadpoles and adults with FV3 and examined IFN gene expression in their kidney, spleen, and liver tissues at 0, 1, 3, and 6 dpi (Fig. 6). In adult spleens and kidneys, IFN gene expression was barely detectable at 1 dpi, was dramatically elevated at 3 dpi, and diminished closer to the baseline level by 6 dpi (Fig. 6A and B). The IFN gene expression kinetics were relatively delayed in adult X. laevis frog livers, with IFN mRNA levels being significantly increased at 3 dpi and further increased by 6 dpi (Fig. 6C). In contrast, kidney IFN gene expression in infected tadpoles was substantially delayed (the peak was at 6 dpi rather than 3 dpi in the adults) and very modest (6 times lower than that in the adults, on average) compared to that in the adults (Fig. 6A). The changes to the tadpole spleen IFN gene expression kinetics were even more modest following viral infection (Fig. 6B). This is particularly telling of the tadpole/adult FV3 susceptibility paradigm, since the amphibian kidney is the primary site of ranaviral replication, whereas the spleen represents the central immune organ of these animals. Hence, the degree of expression of an antiviral cytokine at these sites is critical for the efficacy and outcomes of an antiranaviral response. Interestingly, the baseline level of IFN expression in the liver was significantly greater in tadpoles than adults, while the FV3-elicited upregulation of this gene, albeit very modest, occurred earlier in tadpoles (1 dpi) than in adult frogs (3 dpi) (Fig. 6C). Despite this, at 3 days after FV3 infection, the IFN transcript levels seen in adult liver tissues were significantly greater than those seen in tadpole liver tissues (Fig. 6C). The adult liver IFN mRNA levels seen at 6 dpi were even greater than those seen at 3 dpi, while the tadpole IFN transcript levels were comparable to those detected at 3 dpi (Fig. 6C).
FIG 6.
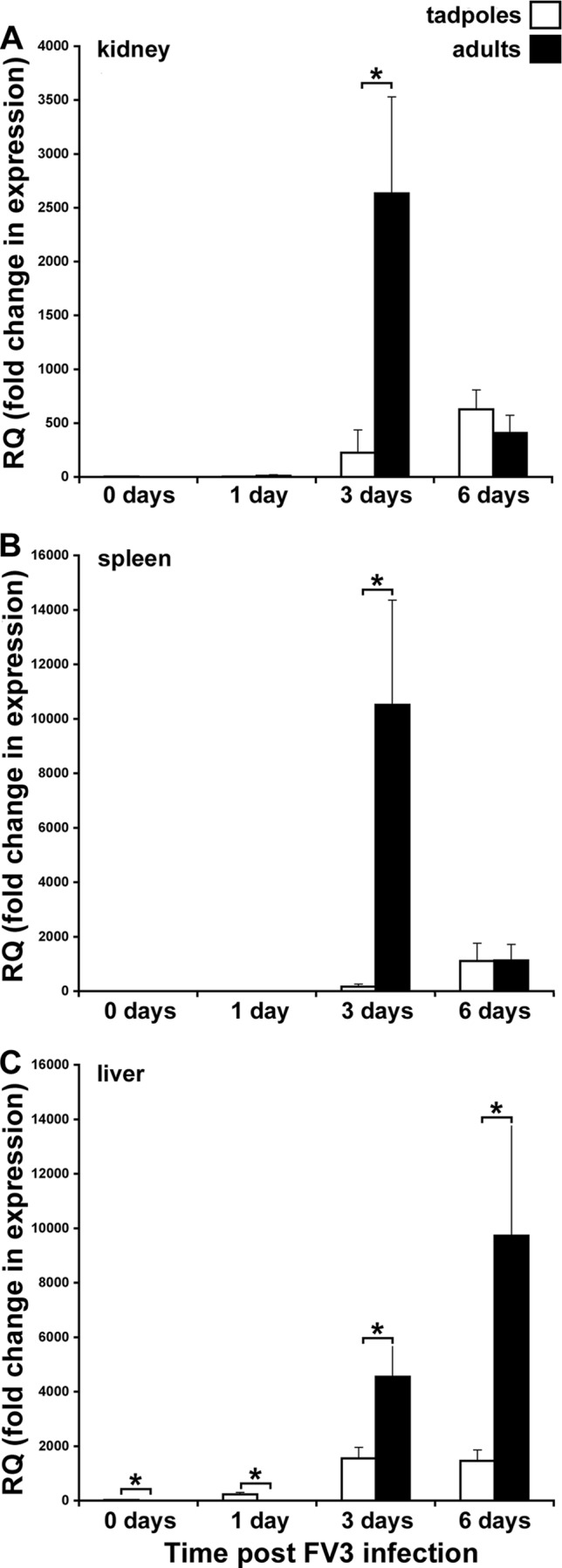
FV3-infected X. laevis adults express higher levels of the type I IFN gene in their tissue than virally infected tadpoles. Tadpoles were infected i.p. with 1 × 104 PFU of FV3 (21). Adult frogs were infected i.p. with 5 × 106 PFU of FV3 (22). At the indicated times, animals were euthanized and tissues were collected. The levels of IFN gene expression relative to the expression levels of the GAPDH endogenous control in kidney (A), spleen (B), and liver (C) tissues from three individuals per treatment group were determined. Results are means ± SEMs. *, significant difference between the tadpole and adult stages (P < 0.05).
To elucidate the possible consequences of the differences in IFN gene expression upon FV3 replication in tadpoles and adult frogs, we assessed the FV3 loads in the kidney and liver tissues of these animals by qRT-PCR (Fig. 7). Surprisingly and contrary to the notion that susceptible animals are expected to possess higher viral burdens, we observed that at all infection times examined, tadpoles had substantially lower (as much as 10 times lower) FV3 loads in kidney and liver tissues than adult frogs (Fig. 7A and B).
FIG 7.
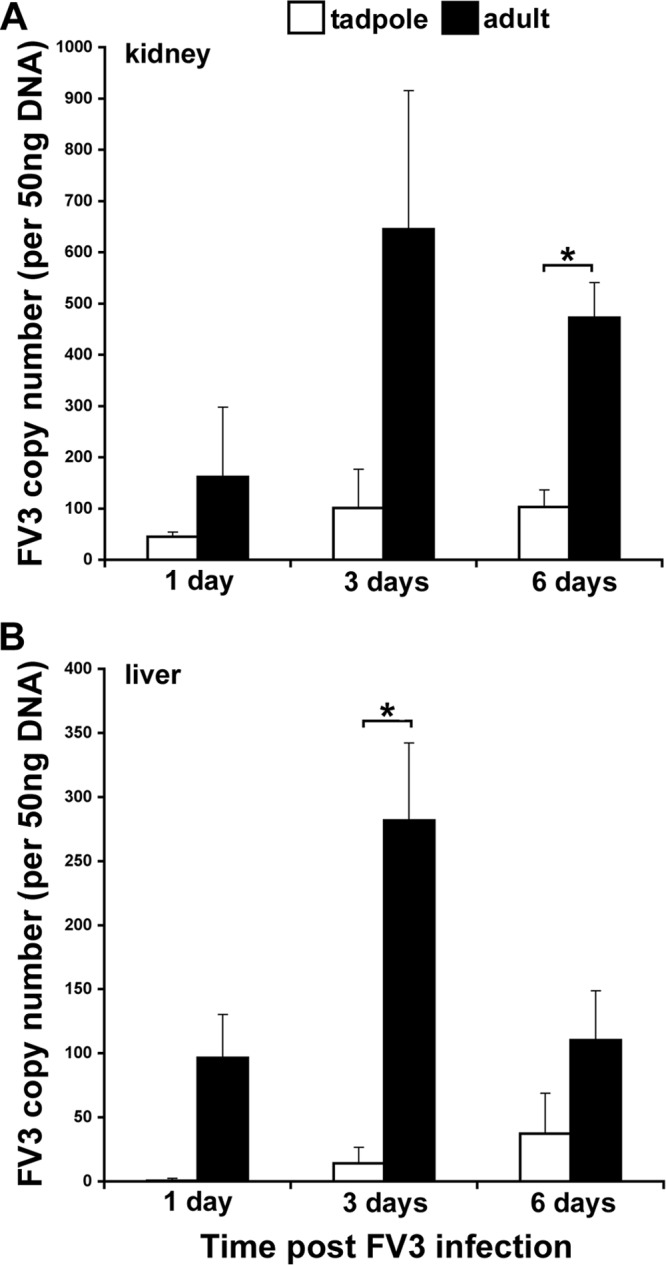
FV3-infected X. laevis adults exhibit greater kidney and liver viral burdens than virus-infected tadpoles. Tadpoles were infected i.p. with 1 × 104 PFU of FV3; adult frogs were infected i.p. with 5 × 106 PFU of FV3. At the indicated times, animals were euthanized, tissues were collected, and DNA was isolated. Viral loads were measured by absolute qRT-PCR (vDNA Pol II) of 50 ng total DNA derived from kidney (A) and liver (B) tissues. Tissues from three individuals per treatment group were examined. Results are means ± SEMs. *, significant difference between the tadpole and adult stages (P < 0.05).
X. laevis tadpoles administered rXlIFN exhibit prolonged mean survival times and lower viral burdens during FV3 infection.
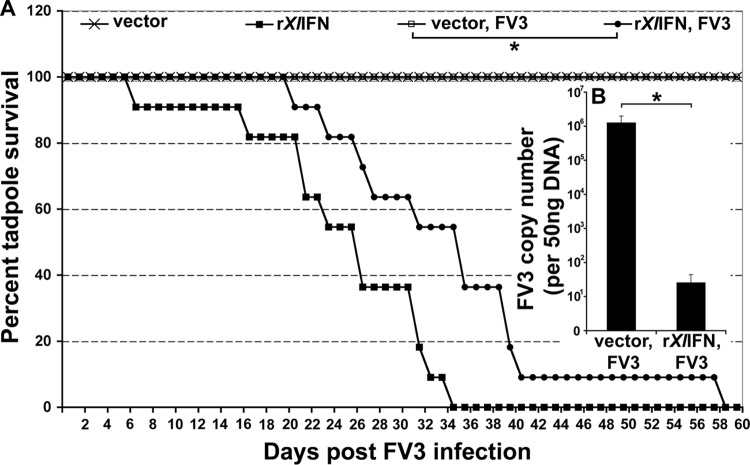
Our initial rationale that tadpole viral susceptibility stems from insufficient IFN responses during FV3 infection (compared to that in adults) was supported by the significant decreases in FV3 burdens observed following administration of rXlIFN to tadpoles prior to FV3 infection (Fig. 4). We therefore anticipated that tadpoles prestimulated with rXlIFN should not succumb to FV3. To test this hypothesis, we injected tadpoles with rXlIFN (500 ng/tadpole) or the vector control 8 h prior to FV3 infection (104 PFU) and monitored tadpole survival daily for 60 days (Fig. 8A). Contrary to our prediction, although tadpoles prestimulated with rXlIFN had significantly extended mean survival times, the majority of these animals still died within 2 months of the time of infection (Fig. 8A).
FIG 8.
Pretreatment of tadpoles with rXlIFN prior to FV3 infection increases animal mean survival time and lowers postmortem viral burdens. Stage 50 tadpoles (n = 11 per treatment group) were injected with rXlIFN (500 ng/tadpole) or an equal volume of the vector control and 8 h later were infected i.p. with 1 × 104 PFU of FV3 (in APBS) or an equal volume of APBS. (A) Animal survival was monitored over the course of 60 days. Tadpoles were checked twice daily, and dead animals were immediately frozen and stored at −20°C for DNA isolation. DNA was isolated from whole tadpoles postmortem, and FV3 loads were determined by absolute qRT-PCR. (B) FV3 copy numbers from 11 vector-injected and FV3-infected tadpoles and 10 rXlIFN-injected and FV3-infected tadpoles are represented as mean log numbers ± SEMs. *, significant difference between the vector and rXlIFN treatment groups (P < 0.05).
When we examined the postmortem FV3 loads in the carcasses of the tadpoles described above, we discovered that the tadpoles that had been preinjected with rXlIFN prior to FV3 infection had FV3 loads several log units lower than those of the vector control tadpoles (Fig. 8B). This finding suggests that tadpole susceptibility and mortalities are likely the result of variables unrelated to high viral burdens.
rXlIFN does not protect X. laevis tadpoles against FV3-induced kidney and liver pathology.
Since tadpoles that had been preinjected with rXlIFN prior to FV3 inoculation still succumbed to infection, even though IFN significantly lowered the viral burdens (Fig. 8), we examined whether the rXlIFN-protected tadpoles were dying from possible FV3-induced tissue damage. Accordingly, tadpoles were rXlIFN pretreated and FV3 infected as described above and then harvested once they displayed terminal signs of infection (irregular swimming, balance problems) and processed for histology analysis (Fig. 9). Compared to mock-infected tadpoles that had been pretreated with the vector control, tadpoles infected with FV3 after vector control treatment exhibited significant damage to their livers (Fig. 9A and E, respectively) and kidneys (Fig. 9B and F, respectively). These injuries included the loss of tissue architecture, immune cell infiltration, and the formation of intracytoplasmic inclusion bodies (Fig. 9G and H), characteristic of RV pathology (43). Intriguingly, in corroboration of the rXlIFN-diminished FV3 loads (Fig. 8B), pretreatment of animals with rXlIFN reduced the severity of the tissue damage incurred from subsequent FV3 infections (Fig. 9). However, tadpoles injected with this potent antiviral cytokine prior to viral infection nonetheless incurred liver and kidney tissue damage, including substantial cytopathology and a loss of tissue architecture (Fig. 9C and D, respectively), which were not found in vector-pretreated uninfected controls (Fig. 9A and B, respectively) or rXlIFN-pretreated uninfected animals (data not shown). These observations suggest that the extensive FV3-induced damage to multiple organs is a major contributor to the mortality of infected animals, including those pretreated with rXlIFN.
FIG 9.
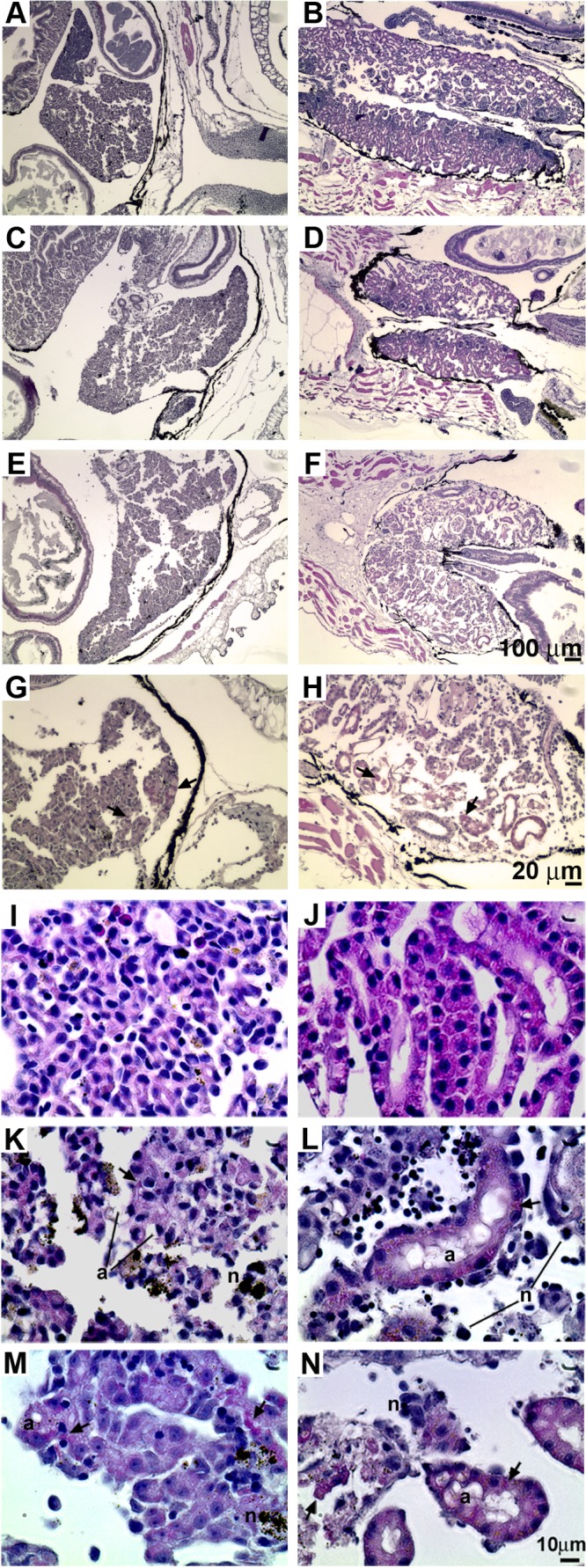
Pretreatment of tadpoles with rXlIFN does not prevent FV3 infection-induced liver and kidney damage. Tadpoles were preinjected with 500 ng of total rXlIFN or an equal volume of the vector control and 8 h later were infected with FV3 or mock infected by APBS injection. Animals were reared until they displayed characteristic signs of terminal infection, sacrificed, and prepared for histology analysis. The cellular compositions were as follows: vector, APBS, liver (A); vector, APBS, kidney (B); rXlIFN, FV3, liver (C); rXlIFN, FV3, kidney (D); vector, FV3, liver (E); vector, FV3, kidney (F); higher magnification, vector, FV3, liver (G); and higher magnification, vector, FV3, kidney (H). Arrows, intracytoplasmic inclusions characteristic of ranavirus-induced pathology. (I to N) Pretreatment of tadpoles with rXlIFN does not prevent FV3 infection-induced cellular damage, necrosis, and apoptosis. Tadpoles were preinjected with rXlIFN (500 ng) or the vector control 8 h before infection with FV3. Animals were reared until they displayed characteristic signs of terminal infection, sacrificed, and prepared for histology analysis. The cellular compositions were as follows: vector, APBS, liver (I); vector, APBS, kidney (J); rXlIFN, FV3, liver (K); rXlIFN, FV3, kidney (L); vector, FV3, liver (M); and vector, FV3, kidney (N). Arrows, FV3-induced intracytoplasmic inclusions; n, necrotic cells; a, apoptotic cells. Bars, 100 μm (A to F), 20 μm (G, H), and 10 μm (I to N).
Histological examination at higher magnification revealed that rXlIFN treatment prior to FV3 infection resulted in fewer intracytoplasmic inclusion bodies (typical of FV3 infection) in liver and kidney tissues (Fig. 9K and L, respectively) than the number in vector-treated FV3-infected animals (Fig. 9M and N, respectively). This corroborates our earlier findings that rXlIFN stimulation of tadpoles significantly diminishes their viral loads. However, despite the diminished viral burdens and irrespective of the treatment (rXlIFN or vector), infection of tadpoles with FV3 resulted in severe disruption of the cellular organization in both liver and kidney tissues as well as the presence of major necrotic cell death (dark or black-stained cells and debris; indicated by the letter n) and apoptotic cell death (vacuolated and blebbed cells; indicated by the letter a) in liver tissue (Fig. 9K and M, respectively) and kidney tissue (Fig. 9L and N, respectively). None of these pathologies were observed in the liver and kidney tissues of the tadpoles treated with the saline vector or tadpoles pretreated with rXlIFN and injected with the vehicle (Fig. 9I and J, respectively, and data not shown). Finally, the degree of immune infiltration seen within the tissues of afflicted animals (irrespective of treatment) was modest, considering the severity of the FV3-induced destruction (Fig. 9).
DISCUSSION
This article presents the first reported study of amphibian type I IFN immunity. This is particularly relevant considering the key place in vertebrate evolution represented by amphibian species such as X. laevis. Interestingly, a hallmark characteristic of fish and amphibian IFNs is the five-exon/four-intron genomic organization, which is very distinct from the intronless type I IFN transcripts encoded by reptiles, birds, and mammals (24, 29, 36). In fact, there has been considerable debate whether the fish type I IFNs are ancestral homologues of the higher vertebrate type I or type III IFNs. Notably, the fish counterparts exhibit a five-exon/four-intron gene organization akin to that of the mammalian type III IFNs but possess prototypic sequence patterns, such as cysteine positioning and the C-terminal CAWE motifs conserved in higher vertebrate type I IFNs (24, 27, 28, 44). In fact, since fish are currently believed not to possess type III IFNs, it is intriguing that amphibians possess both type I IFNs akin to those of fish and bona fide type III IFNs (27). This suggests that the divergence of type I and III IFNs occurred prior to the appearance of tetrapods (27) and brings to question the relative biological roles of the amphibian type I IFNs compared to those of fish, which are currently thought to lack the type III antiviral cytokines.
The efficacy of function of the four fish IFN subgroups range from highly antiviral to nonfunctional (39) and are believed to signal through at least two distinct receptor complexes (32). It is noteworthy that the recombinant form of the X. laevis IFN identified here displayed very potent antiviral properties. In addition, our phylogenetic analysis suggests that X. tropicalis IFNs 1 and 2 may have been conserved from an ancestral IFN, whereas X. tropicalis IFNs 3, 4, and 5, the X. laevis IFN, and the fish IFNs diverged more recently. Possibly, as in fish, the amphibian type I IFNs may possess distinct functional properties and interact with different cognate receptors. With regard to the position of amphibians in the context of the evolution of type I IFN defenses, it will be intriguing to learn if, indeed, the individual Xenopus type I IFNs display a disparity of antiviral functions akin to those seen in teleosts or act more like the higher vertebrate IFNs and signal through a single receptor complex.
During our analysis of X. laevis type I IFN gene expression in the tissues of virus-infected tadpoles and adult frogs, we observed that despite similar FV3 expansion kinetics across tissues, the liver IFN gene expression was delayed in comparison to that seen in the kidneys of infected animals, particularly in adult frogs. It is difficult to speculate on the precise explanation for this discrepancy. Presumably, these observations reflect the distinct immune cell compositions of these tissues, where the liver contains large quantities of immunomodulatory Kupffer cells (reviewed in reference 45), which would likely skew the immune outcomes within this site of infection.
This work is the first to demonstrate the roles of the amphibian type I IFNs in immunity against the etiologically relevant ranavirus FV3. Our findings underline both the efficacy with which the X. laevis IFN is able to deter infections with a potent virus like FV3 and the resilience of this pathogen, where, despite significant rXlIFN-induced perturbation of viral replication and expansion, FV3 was still ultimately lethal to the tadpole hosts. Indeed, we observed that tadpoles mounted relatively delayed and meager FV3-elicited IFN responses compared to those induced by the virus in adult frogs. This difference is a likely factor contributing to the FV3 susceptibility of tadpoles and the adult resistance to this virus. Furthermore, as nothing is known regarding the receptor system employed by the amphibian type I IFN, it is possible that the tadpole susceptibility is further compounded at the receptor level.
Considering the notion that X. laevis adults mount robust and presumably effective IFN responses to FV3 that contribute to the effective clearance of this pathogen within 2 weeks, it is intriguing that (at least during the acute infection) adults exhibit significantly greater FV3 loads than tadpoles, which are more susceptible to and typically succumb to infection. This suggests that the inability to contain and/or minimize the expansion of FV3 is not the mechanism resulting in tadpole mortality. Conversely, our observations underline the inherent capacity of adult frogs to tolerate much greater FV3 burdens. This idea is substantiated by the findings that while the rXlIFN-pretreated tadpoles possessed viral loads several log units lower than those of the adults, the rXlIFN-pretreated tadpoles still eventually succumbed to FV3 infection. This mortality, despite the extended mean survival time and considerably diminished viral burdens in tadpoles, suggests that even at significantly reduced titers, FV3 confers irreversible damage to multiple tadpole organs (as seen in our histology studies), including an extensive loss of tissue architecture and cellular organization through necrotic and apoptotic cell death. Furthermore, the meager immune infiltration of these severely compromised tissues suggests that this damage is likely incurred relatively early in the infection and/or results primarily from virus-mediated cytopathy rather than inflammation.
It is noteworthy that FV3 infection of rats was employed as a model for hepatitis over 30 years ago (46–48). In these experiments, rat inoculation with FV3 resulted in necrotic Kupffer cell death, cessation of hepatic clearance, and subsequent toxicity, culminating in severe hepatitis, inflammation, and animal deaths (46). Although the temperature of 37°C used for these mammalian studies was not permissive to ranaviral replication (49), FV3 particles were detected in phagocytic vacuoles and endocytic compartments of mammalian Kupffer cells, where approximately a quarter of these virions exhibited viral core membrane-host membrane fusion and viral core material release into cell cytoplasms (50). This nonspecific capacity to infiltrate phagocytic cells of otherwise resistant hosts may explain the recent increase in the prevalence and expansion of the host tropisms of ranaviruses. Furthermore, the inability of FV3 to replicate at nonpermissive mammalian body temperatures implies that the pathogenic effects described above were not mediated through cell lytic mechanisms (at least in the animal models described above) but instead were mediated through prepackaged factors already present in the FV3 virion. In support of this, early research on FV3 in mammalian models also revealed that this virus induces rapid cellular RNA, DNA, and protein synthesis arrest (51) resulting from structural proteins that can be solubilized from the viral particles (52) and that are sufficient for inhibiting host cell nucleic acid synthesis (49). Our findings indicate that the amphibian tadpole susceptibility to FV3 is most likely not the result of extensive viral burdens. Rather, we propose that prepackaged FV3 components and/or FV3 products generated during early phases of ranaviral infection culminate in compounding long-term tissue damage and organ failure, prominently contributing to tadpole mortality.
ACKNOWLEDGMENTS
We thank Daria Krenitsky for her assistance with histology analysis. We thank Greg V. Chinchar for generously providing the anti-FV3 53R Ab. We thank Tina Martin and David Albright for animal husbandry. We thank Eva-Stina Eldhom for her critical review of the manuscript.
This work was supported by grants R24-AI-059830 and IOB-074271 from NIH and NSF, respectively. L.G. was supported by an NSERC postdoctoral fellowship program and an LSRF postdoctoral fellowship program from the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Chinchar VG. 2002. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 147:447–470. 10.1007/s007050200000 [DOI] [PubMed] [Google Scholar]
- 2.Williams T, Barbosa-Solomieu V, Chinchar VG. 2005. A decade of advances in iridovirus research. Adv. Virus Res. 65:173–248. 10.1016/S0065-3527(05)65006-3 [DOI] [PubMed] [Google Scholar]
- 3.Chinchar VG, Hyatt A, Miyazaki T, Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr. Top. Microbiol. Immunol. 328:123–170 [DOI] [PubMed] [Google Scholar]
- 4.Granoff A, Came PE, Breeze DC. 1966. Viruses and renal carcinoma of Rana pipiens. I. The isolation and properties of virus from normal and tumor tissue. Virology 29:133–148 [DOI] [PubMed] [Google Scholar]
- 5.Granoff A, Came PE, Rafferty KA., Jr 1965. The isolation and properties of viruses from Rana pipiens: their possible relationship to the renal adenocarcinoma of the leopard frog. Ann. N. Y. Acad. Sci. 126:237–255. 10.1111/j.1749-6632.1965.tb14278.x [DOI] [PubMed] [Google Scholar]
- 6.Zhan QY, Xiao F, Li ZQ, Gui JF, Mao J, Chinchar VG. 2001. Characterization of an iridovirus from the cultured pig frog Rana grylio with lethal syndrome. Dis. Aquat. Organ. 48:27–36. 10.3354/dao048027 [DOI] [PubMed] [Google Scholar]
- 7.Speare R, Freeland WJ, Bolton SJ. 1991. A possible iridovirus in erythrocytes of Bufo marinus in Costa Rica. J. Wildl. Dis. 27:457–462. 10.7589/0090-3558-27.3.457 [DOI] [PubMed] [Google Scholar]
- 8.Speare R, Smith JR. 1992. An iridovirus-like agent isolated from the ornate burrowing frog Limnodynastes ornatus in northern Australia. Dis. Aquat. Organ. 14:51–57. 10.3354/dao014051 [DOI] [Google Scholar]
- 9.Cunningham AA, Langton TE, Bennett PM, Lewin JF, Drury SE, Gough RE, Macgregor SK. 1996. Pathological and microbiological findings from incidents of unusual mortality of the common frog (Rana temporaria). Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:1539–1557. 10.1098/rstb.1996.0140 [DOI] [PubMed] [Google Scholar]
- 10.Drury SE, Gough RE, Cunningham AA. 1995. Isolation of an iridovirus-like agent from common frogs (Rana temporaria). Vet. Rec. 137:72–73. 10.1136/vr.137.3.72 [DOI] [PubMed] [Google Scholar]
- 11.Bollinger TK, Mao J, Schock D, Brigham RM, Chinchar VG. 1999. Pathology, isolation, and preliminary molecular characterization of a novel iridovirus from tiger salamanders in Saskatchewan. J. Wildl. Dis. 35:413–429. 10.7589/0090-3558-35.3.413 [DOI] [PubMed] [Google Scholar]
- 12.Donnelly TM, Davidson EW, Jancovich JK, Borland S, Newberry M, Gresens J. 2003. What's your diagnosis? Ranavirus infection. Lab. Anim. (NY) 32:23–25 [DOI] [PubMed] [Google Scholar]
- 13.Greer AL, Berrill M, Wilson PJ. 2005. Five amphibian mortality events associated with ranavirus infection in south central Ontario, Canada. Dis. Aquat. Organ. 67:9–14. 10.3354/dao067009 [DOI] [PubMed] [Google Scholar]
- 14.Jancovich JK, Davids EW, Seiler A, Jacobs BL, Collins JP. 2001. Transmission of the Ambystoma tigrinum virus to alternative hosts. Dis. Aquat. Organ. 46:159–163. 10.3354/dao046159 [DOI] [PubMed] [Google Scholar]
- 15.Jancovich JK, Bremont M, Touchman JW, Jacobs BL. 2010. Evidence for multiple recent host species shifts among the ranaviruses (family Iridoviridae). J. Virol. 84:2636–2647. 10.1128/JVI.01991-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker JA, Tweedie A, Gilligan D, Asmus M, Whittington RJ. 2003. Experimental infection of Australian freshwater fish with epizootic haematopoietic necrosis virus (EHNV). J. Aquat. Anim. Health 25:66–76. 10.1080/08997659.2012.747451 [DOI] [PubMed] [Google Scholar]
- 17.Haislip NA, Gray MJ, Hoverman JT, Miller DL. 2011. Development and disease: how susceptibility to an emerging pathogen changes through anuran development. PLoS One 6:e22307. 10.1371/journal.pone.0022307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt AD, Gould AR, Zupanovic Z, Cunningham AA, Hengstberger S, Whittington RJ, Kattenbelt J, Coupar BE. 2000. Comparative studies of piscine and amphibian iridoviruses. Arch. Virol. 145:301–331. 10.1007/s007050050025 [DOI] [PubMed] [Google Scholar]
- 19.Robert J, Abramowitz L, Gantress J, Morales HD. 2007. Xenopus laevis: a possible vector of Ranavirus infection? J. Wildl. Dis. 43:645–652. 10.7589/0090-3558-43.4.645 [DOI] [PubMed] [Google Scholar]
- 20.Chen G, Robert J. 2011. Antiviral immunity in amphibians. Viruses 3:2065–2086. 10.3390/v3112065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Jesus Andino F, Chen G, Li Z, Grayfer L, Robert J. 2012. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus frog-virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology 432:435–443. 10.1016/j.virol.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morales HD, Abramowitz L, Gertz J, Sowa J, Vogel A, Robert J. 2010. Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J. Virol. 84:4912–4922. 10.1128/JVI.02486-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. 2011. Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17:2619–2627. 10.1158/1078-0432.CCR-10-1114 [DOI] [PubMed] [Google Scholar]
- 24.Robertsen B. 2006. The interferon system of teleost fish. Fish Shellfish Immunol. 20:172–191. 10.1016/j.fsi.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 25.Zou J, Secombes CJ. 2011. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 35:1376–1387. 10.1016/j.dci.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 26.Chang M, Nie P, Collet B, Secombes CJ, Zou J. 2009. Identification of an additional two-cysteine containing type I interferon in rainbow trout Oncorhynchus mykiss provides evidence of a major gene duplication event within this gene family in teleosts. Immunogenetics 61:315–325. 10.1007/s00251-009-0366-y [DOI] [PubMed] [Google Scholar]
- 27.Qi Z, Nie P, Secombes CJ, Zou J. 2010. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 184:5038–5046. 10.4049/jimmunol.0903374 [DOI] [PubMed] [Google Scholar]
- 28.Zou J, Tafalla C, Truckle J, Secombes CJ. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 179:3859–3871 [DOI] [PubMed] [Google Scholar]
- 29.Sun B, Robertsen B, Wang Z, Liu B. 2009. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev. Comp. Immunol. 33:547–558. 10.1016/j.dci.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. 2008. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 377:715–724. 10.1016/j.jmb.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809. 10.1128/CMR.14.4.778-809.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggad D, Mazel M, Boudinot P, Mogensen KE, Hamming OJ, Hartmann R, Kotenko S, Herbomel P, Lutfalla G, Levraud JP. 2009. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 183:3924–3931. 10.4049/jimmunol.0901495 [DOI] [PubMed] [Google Scholar]
- 33.Altmann SM, Mellon MT, Distel DL, Kim CH. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 77:1992–2002. 10.1128/JVI.77.3.1992-2002.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long S, Wilson M, Bengten E, Bryan L, Clem LW, Miller NW, Chinchar VG. 2004. Identification of a cDNA encoding channel catfish interferon. Dev. Comp. Immunol. 28:97–111. 10.1016/S0145-305X(03)00122-8 [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Munoz A, Roca FJ, Meseguer J, Mulero V. 2009. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J. Immunol. 182:3440–3449. 10.4049/jimmunol.0802528 [DOI] [PubMed] [Google Scholar]
- 36.Robertsen B, Bergan V, Rokenes T, Larsen R, Albuquerque A. 2003. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 23:601–612. 10.1089/107999003322485107 [DOI] [PubMed] [Google Scholar]
- 37.Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P, Lutfalla G. 2007. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 178:4385–4394 [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Xu X, Huang L, Wu J, Lu Q, Xiang Z, Liao J, Weng S, Yu X, He J. 2010. Administration of recombinant IFN1 protects zebrafish (Danio rerio) from ISKNV infection. Fish Shellfish Immunol. 29:399–406. 10.1016/j.fsi.2010.04.020 [DOI] [PubMed] [Google Scholar]
- 39.Svingerud T, Solstad T, Sun B, Nyrud ML, Kileng O, Greiner-Tollersrud L, Robertsen B. 2012. Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J. Immunol. 189:5912–5923. 10.4049/jimmunol.1201188 [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki ES, Ladner MB, Wang AM, Van Arsdell J, Warren MK, Coyne MY, Schweickart VL, Lee MT, Wilson KJ, Boosman A. 1985. Molecular cloning of a complementary DNA encoding human macrophage-specific colony-stimulating factor (CSF-1). Science 230:291–296. 10.1126/science.2996129 [DOI] [PubMed] [Google Scholar]
- 41.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246–254. 10.1093/nar/29.1.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitley DS, Yu K, Sample RC, Sinning A, Henegar J, Norcross E, Chinchar VG. 2010. Frog virus 3 ORF 53R, a putative myristoylated membrane protein, is essential for virus replication in vitro. Virology 405:448–456. 10.1016/j.virol.2010.06.034 [DOI] [PubMed] [Google Scholar]
- 43.Kik M, Martel A, Sluijs AS, Pasmans F, Wohlsein P, Grone A, Rijks JM. 2011. Ranavirus-associated mass mortality in wild amphibians, the Netherlands, 2010: a first report. Vet. J. 190:284–286. 10.1016/j.tvjl.2011.08.031 [DOI] [PubMed] [Google Scholar]
- 44.Lutfalla G, Roest Crollius H, Stange-Thomann N, Jaillon O, Mogensen K, Monneron D. 2003. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4:29. 10.1186/1471-2164-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon LJ, Barnes M, Tang H, Pritchard MT, Nagy LE. 2013. Kupffer cells in the liver. Compr. Physiol. 3:785–797. 10.1002/cphy.c120026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gut JP, Anton M, Bingen A, Vetter JM, Kirn A. 1981. Frog virus 3 induces a fatal hepatitis in rats. Lab. Invest. 45:218–228 [PubMed] [Google Scholar]
- 47.Kirn A, Bingen A, Steffan AM, Wild MT, Keller F, Cinqualbre J. 1982. Endocytic capacities of Kupffer cells isolated from the human adult liver. Hepatology 2:216–222 [DOI] [PubMed] [Google Scholar]
- 48.Kirn A, Steffan AM, Bingen A. 1980. Inhibition of erythrophagocytosis by cultured rat Kupffer cells infected with frog virus 3. J. Reticuloendothel. Soc. 28:381–388 [PubMed] [Google Scholar]
- 49.Aubertin AM, Hirth C, Travo C, Nonnenmacher H, Kirn A. 1973. Preparation and properties of an inhibitory extract from frog virus 3 particles. J. Virol. 11:694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gendrault JL, Steffan AM, Bingen A, Kirn A. 1981. Penetration and uncoating of frog virus 3 (FV3) in cultured rat Kupffer cells. Virology 112:375–384. 10.1016/0042-6822(81)90284-1 [DOI] [PubMed] [Google Scholar]
- 51.Elharrar M, Hirth C, Blanc J, Kirn A. 1973. Pathogenesis of the toxic hepatitis of mice provoked by FV 3 (frog virus 3): inhibition of liver macromolecular synthesis. Biochim. Biophys. Acta 319:91–102. 10.1016/0005-2787(73)90044-0 (In French.) [DOI] [PubMed] [Google Scholar]
- 52.Bingen-Brendel A, Batzenschlager A, Gut JP, Hirth C, Vetter JM, Kirn A. 1972. Histological and virological study of acute degenerative hepatitis produced by the FV3 (frog virus 3) in mice. Ann. Inst. Pasteur (Paris) 122:125–142 (In French.) [PubMed] [Google Scholar]