ABSTRACT
Sporadic activity by H5N2 influenza viruses has been observed in chickens in Taiwan from 2003 to 2012. The available information suggests that these viruses were generated by reassortment between a Mexican-like H5N2 virus and a local enzootic H6N1 virus. Yet the origin, prevalence, and pathogenicity of these H5N2 viruses have not been fully defined. Following the 2012 highly pathogenic avian influenza (HPAI) outbreaks, surveillance was conducted from December 2012 to July 2013 at a live-poultry wholesale market in Taipei. Our findings showed that H5N2 and H6N1 viruses cocirculated at low levels in chickens in Taiwan. Phylogenetic analyses revealed that all H5N2 viruses had hemagglutinin (HA) and neuraminidase (NA) genes derived from a 1994 Mexican-like virus, while their internal gene complexes were incorporated from the enzootic H6N1 virus lineage by multiple reassortment events. Pathogenicity studies demonstrated heterogeneous results even though all tested viruses had motifs (R-X-K/R-R) supportive of high pathogenicity. Serological surveys for common subtypes of avian viruses confirmed the prevalence of the H5N2 and H6N1 viruses in chickens and revealed an extraordinarily high seroconversion rate to an H9N2 virus, a subtype that is not found in Taiwan but is prevalent in mainland China. These findings suggest that reassortant H5N2 viruses, together with H6N1 viruses, have become established and enzootic in chickens throughout Taiwan and that a large-scale vaccination program might have been conducted locally that likely led to the introduction of the 1994 Mexican-like virus to Taiwan in 2003.
IMPORTANCE H5N2 avian influenza viruses first appeared in chickens in Taiwan in 2003 and caused a series of outbreaks afterwards. Phylogenetic analyses show that the chicken H5N2 viruses have H5 and N2 genes that are closely related to those of a vaccine strain originating from Mexico in 1994, while the contemporary duck H5N2 viruses in Taiwan belong to the Eurasian gene pool. The unusually high similarity of the chicken H5N2 viruses to the Mexican vaccine strain suggests that these viruses might have been introduced to Taiwan by using inadequately inactivated or attenuated vaccines. These chicken H5N2 viruses are developing varying levels of pathogenicity that could lead to significant consequences for the local poultry industry. These findings emphasize the need for strict quality control and competent oversight in the manufacture and usage of avian influenza virus vaccines and indicate that alternatives to widespread vaccination may be desirable.
INTRODUCTION
It is generally accepted that all H5 or H7 high-pathogenic avian influenza (HPAI) viruses were derived from low-pathogenic avian influenza (LPAI) viruses introduced to terrestrial birds from aquatic birds (1–4). After consecutive passages in the new hosts, LPAI H5 and H7 viruses may acquire additional basic amino acids at the connecting peptide of the hemagglutinin protein (HA) (5–7). The number of these basic amino acids is directly associated with the virulence of the viruses in chickens and other terrestrial poultry (8, 9). Generally, an H5 or H7 virus with a minimal motif of R-X-K/R-R at the cleavage site and an absence of carbohydrate modification in the vicinity of the connecting peptide will be considered potentially highly pathogenic (8, 10–13).
Since 1961, more than 28 outbreaks of HPAI H5 or H7 viruses have been documented worldwide (14, 15). Except for one outbreak in terns, these have occurred in terrestrial poultry, mainly chickens and turkeys (1, 16). In some of the outbreaks, both the highly pathogenic and progenitor viruses were identified in the field (2, 5, 17, 18). In Taiwan, despite its high-density practice of poultry farming, no activity by HPAI H5 or H7 viruses had been recorded before 2003.
In the last decade, H5N2 influenza virus activity has been observed occasionally in chickens in Taiwan. The initial outbreak of LPAI H5N2 viruses occurred in late 2003, and a second LPAI outbreak was reported in 2008 (19). Genetic analyses of publically available sequences of the Taiwanese H5N2 viruses suggested that their surface protein genes, HA and neuraminidase (NA), were not derived from Eurasian gene pool viruses but were closely related to an H5N2 virus isolated from chickens in Mexico during an outbreak in 1994 (19). Their internal genes originated from an H6N1 virus lineage that has been enzootic in chickens in Taiwan for the last 2 decades (19, 20).
Since January 2012, H5N2 virus activity has been observed almost every month and has covered many regions of Taiwan, resulting in significant mortality in chickens (15, 21). However, how this virus was generated and how it became prevalent in the field remain to be answered. To explore these questions, avian influenza surveillance was conducted from December 2012 to July 2013 at a wholesale live-poultry market in Taipei where birds were shipped from different areas of Taiwan. Our findings show that both H5N2 and H6N1 influenza viruses were cocirculating in chickens in Taiwan at that time. All Taiwan H5N2 viruses from chickens had multiple basic amino acids at the cleavage site of the HA connecting peptides, creating motifs that are associated with high pathogenicity in chickens. However, no additional basic amino acids were present, and the structurally adjacent glycosylation site that may influence pathogenicity was conserved in all viruses (10–13). Phylogenetic analyses revealed that these H5N2 viruses were introduced to this region from an external source but have undergone multiple reassortments with the enzootic H6N1 virus lineages.
MATERIALS AND METHODS
Surveillance and virus isolation.
Influenza surveillance of live poultry was conducted in Taipei at the wholesale market. Sampling was conducted weekly from apparently healthy chickens and ducks from December 2012 to July 2013 (Fig. 1). To avoid contamination and expand representation, no more than four fecal samples were collected from each cage and only fresh droppings were taken. Samples were kept in a cool box and shipped to the laboratory within 2 h. Virus isolation was conducted using 9- to 11-day-old embryonated chicken eggs. Hemagglutinin-positive isolates were subtyped by hemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays using a panel of reference sera, as described previously (22, 23).
FIG 1.
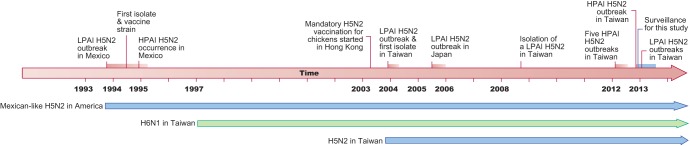
Timeline of the Mexican-like H5N2 lineage viruses and the H6N1 viruses from Taiwan. Events discussed in the text are marked on the timeline with the duration of H5N2 outbreaks in Mexico and Taiwan. Arrows below the timeline indicate the length of persistence of the main lineages in their regions.
Serological survey.
Thirty to 100 chicken blood samples were collected on each sampling occasion. Chicken sera were isolated and stored at −20°C. Serum samples collected from December 2012 to July 2013 were tested by HI for antibodies against Ck/TW/853/12 (TW853; H3N8), Dk/TW/1030/12 (TW1030; H6N8), Ck/TW/2593/12 (TW2593; H5N2), Ck/TW/2267/12 (TW2267; H6N1), and Ck/HK/NT155/08 (HK-NT155; H9N2). Prior to the tests, all chicken sera were treated by receptor-destroying enzyme (RDE; Denka Seiken Co., Ltd., Tokyo, Japan) and absorbed using fresh chicken red blood cells to remove nonspecific inhibitors and substances responsible for nonspecific agglutination. Sera were tested starting at a 1:10 dilution in phosphate-buffered saline (PBS).
Virus sequencing and phylogenetic analyses.
Full-genome sequences of all the H5 viruses from chickens (n = 16) and ducks (n = 12) isolated during the survey period and selected chicken H6 viruses (n = 7) from each sampling occasion were obtained as previously described (24). Sequencing was performed using a BigDye Terminator, version 3.1, cycle sequencing kit on an ABI 3730 genetic analyzer (Applied Biosystems) according to the manufacturer's instructions. DNA sequences were compiled and edited using Lasergene, version 8.0 (DNASTAR, Madison, WI). Multiple sequence alignments were compiled using MUSCLE, version 3.8 (25). The maximum-likelihood (ML) phylogeny of each gene segment was inferred using the GTR+I+Γ4 nucleotide substitution model (general time-reversible model with a proportion of invariant sites and gamma-distributed rate variation across sites) in PhyML (version 3.0) (26). Robustness of the ML topology was evaluated by 1,000 bootstrap replicates, and the Bayesian clade posterior probability was determined from the topologies sampled in the Bayesian molecular clock analysis.
Molecular clock analysis.
For the HA and NA genes, the genetic distance from the common ancestral node of the lineage to each viral isolate was measured from the ML tree and plotted against the sample collection dates. Linear regression was used to indicate the rate of accumulation of mutations over time. A more detailed evolutionary time scale for each virus gene phylogeny, with confidence limits, was obtained using relaxed molecular clocks under uncorrelated lognormal (UCLD) and exponential (UCED) rate distributions, implemented in a Bayesian Markov chain Monte Carlo (BMCMC) statistical framework (27), using BEAST, version 1.8 (28). The SRD06 nucleotide substitution model (29) and Bayesian Skyride demographic model (30) were used. Multiple runs were performed for each data set, giving a total of 6 × 107 states (with 1 × 107 states discarded as burn-in) that were summarized to compute statistical estimates of the parameters. Convergence of the BMCMC analysis was assessed in Tracer, version 1.6 (A. Rambaut M. Suchard, and A. J. Drummond, 2013 [http://tree.bio.ed.ac.uk/software/tracer/]).
Pathogenicity study.
Three H5N2 isolates (from groups A and B of the H5N2 viruses), with two different motifs (R-E-K-R or R-K-K-R) at the cleavage site of the HA connecting peptide, were selected to determine the intracerebral pathogenicity index (ICPI) (31). Freshly harvested allantoic fluid from embryonated chicken eggs containing viruses of a hemagglutination titer of 1,024 was diluted to 1:10 with sterile PBS, and 50 μl was intracerebrally inoculated into each of 10 1-day-old chicks (24 to 36 h after hatching). Mock-infected birds were inoculated with 50 μl of PBS. All birds were examined every 24 h for 8 days and scored as 0 if normal, 1 if sick, and 2 if dead. The mean scores per bird per observation over the 8-day period were calculated. A virus with a value of ≥0.7 is considered pathogenic, and a virus with a value of ≥1.5 is considered highly pathogenic.
Animal experimental protocols were approved by the Institutional Ethical Review Board of Shantou University Medical College (reference no. SUMC2012-134).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study are available from GenBank under accession numbers KJ162585 to KJ162864.
RESULTS
Virus isolation and prevalence.
From 8,659 chicken dropping swabs, 16 H5N2 (∼0.2%), 46 H6N1 (∼0.5%), and a total of 5 H3 viruses were isolated. Avian paramyxovirus type 1 (APMV-1) was also detected in chickens and ducks with isolation rates of 0.3% and 3.7%, respectively. Most of these isolates were detected in the cooler months from December to March (Table 1). Samples from domestic ducks (n = 2,580) showed a slightly higher prevalence of influenza viruses, with 12 H5 (∼0.5%), 35 H6 (∼1.4%), and 85 (∼3.3%) of other subtypes isolated. These findings suggest that the viruses in chickens are less diverse than those in ducks and that H5 viruses are prevalent at a low level in both ducks and chickens.
TABLE 1.
Isolation of influenza viruses and APMV-1 in domestic ducks and chickens in Taiwana
| Host and sampling time | No. of samples | No. of influenza virus isolatesb |
No. of APMV-1 isolates | |||
|---|---|---|---|---|---|---|
| Total | H5 type | H6 type | Other subtype | |||
| Chicken | ||||||
| December 2012 | 2,607 | 22 | 4 | 14 | 4 | 2 |
| 2013 | ||||||
| January | 1,771 | 18 | 5 | 12 | 1 | 7 |
| February | 517 | 4 | 0 | 4 | 0 | 1 |
| March | 1,339 | 14 | 6 | 8 | 0 | 9 |
| April | 535 | 0 | 0 | 0 | 0 | 7 |
| May | 814 | 7 | 1 | 6 | 0 | 1 |
| June | 477 | 2 | 0 | 2 | 0 | 2 |
| July | 599 | 0 | 0 | 0 | 0 | 0 |
| Total | 8,659 | 67 | 16 | 46 | 5 | 29 |
| Duck | ||||||
| December 2012 | 286 | 18 | 0 | 3 | 15 | 1 |
| 2013 | ||||||
| January | 106 | 17 | 10 | 2 | 5 | 17 |
| February | 137 | 0 | 0 | 0 | 0 | 11 |
| March | 557 | 49 | 0 | 3 | 46 | 52 |
| April | 193 | 0 | 0 | 0 | 0 | 0 |
| May | 539 | 9 | 0 | 1 | 8 | 4 |
| June | 352 | 3 | 0 | 0 | 3 | 4 |
| July | 410 | 36 | 2 | 26 | 8 | 7 |
| Total | 2,580 | 132 | 12 | 35 | 85 | 96 |
APMV-1, type 1 avian paramyxovirus.
Three subtypes of influenza viruses were detected in chickens. These were H3, H5N2, and H6N1. All of the H5 viruses isolated from ducks belonged to the Eurasian gene pool.
Serological survey of influenza viruses in chickens.
Evidence of the prevalence of influenza viruses in chickens was also assessed by serological measurements against recent Taiwanese duck or chicken viruses (Table 2). Of 894 samples, none were seropositive to the chicken H3N8 virus (TW853) even though H3 viruses were isolated on three sampling occasions from December 2012 to January 2013. The seropositive rate (with HI titers of >1:20) to a recent chicken H5N2 virus (TW2593) was 8.3%, which was much higher than the virus isolation rate in this survey period. About 3.5% of serum samples were seropositive (with HI titers of >1:20) to a 2012 H6N1 chicken virus (TW2267) from the enzootic lineage in Taiwan. Only a small number (1.4%) reacted at marginal levels (HI titer of 1:10 to 1:40) with an H6N8 virus (TW1030) from the gene pool, which was very likely a cross-reaction with the chicken H6N1 virus as each of these samples had a high titer (HI of >1:40) against TW2267 (data not shown). In contrast to these results, 82.1% of the tested sera reacted with an HI titer higher than 1:20 against chicken H9N2 virus (HK-NT155) from the Ck/Bei/1/94 lineage that has been enzootic in chickens in mainland China for 2 decades even though H9N2 viruses have not previously been reported (nor were they detected in our current surveillance) in chickens in Taiwan.
TABLE 2.
Serological survey of chicken in Taiwan as determined by HI testsa
| HI titer | TW853 (H3N8) | TW1030 (H6N8) | TW2267 (H6N1) | TW2593 (H5N2) | HK-NT155 (H9N2) |
|---|---|---|---|---|---|
| 1,280 | 0 | 0 | 5 | 0 | 7 |
| 640 | 0 | 0 | 5 | 0 | 5 |
| 320 | 0 | 0 | 9 | 0 | 21 |
| 160 | 0 | 0 | 4 | 1 | 142 |
| 80 | 0 | 0 | 6 | 53 | 321 |
| 40 | 0 | 1 | 2 | 20 | 238 |
| 20 | 0 | 2 | 7 | 12 | 133 |
| 10 | 0 | 6 | 7 | 7 | 22 |
| <10 | 894 | 885 | 849 | 801 | 5 |
A hemagglutination inhibition (HI) titer of ≥1:40 can be considered positive. A total of 894 sera were tested. TW853, Ck/TW/853/12; TW1030, Dk/TW/1030/12; TW2593, Ck/TW/2593/12; TW2267, Ck/TW/2267/12, HK-NT155, Ck/HK/NT155/08.
Phylogenetic analyses.
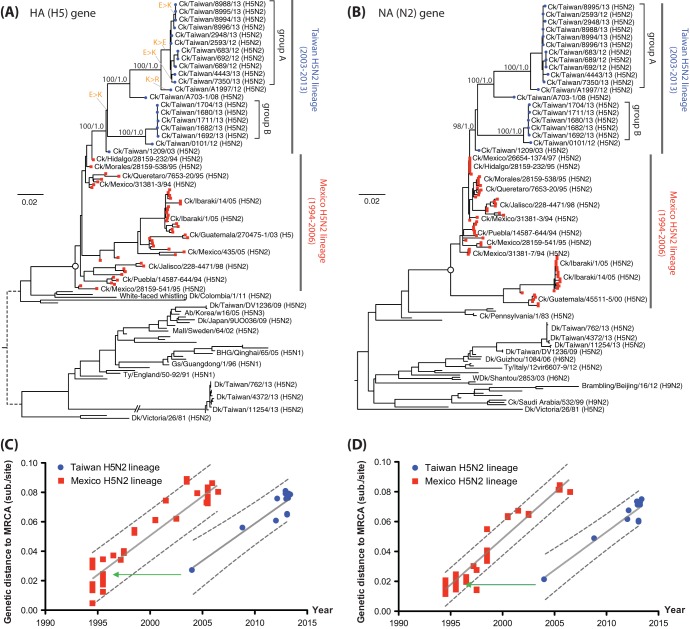
The HA gene of the duck H5 viruses from Taiwan clustered with Eurasian gene pool H5 viruses (Fig. 2). However, both the HA and NA of the chicken H5N2 viruses, isolated in our surveillance and earlier in Taiwan, formed a well-supported monophyletic lineage (ML bootstrap support of 98 to 100% and Bayesian clade posterior probability of 1.0) that descended from the HA and NA genes of Ck/Hidalgo/28159-232/94 (Fig. 2A and B). This H5N2 virus from the 1994 outbreak in Mexico was used as a commercial vaccine strain (32, 33). These findings suggest that the H5N2 viruses in chickens were introduced to Taiwan from Mexico either by bird movement or by inadequately inactivated or attenuated vaccines.
FIG 2.
Phylogenies of the H5 and N2 genes. (A and B) Maximum-likelihood phylogenies of H5 and N2 genes showing the chicken H5N2 viruses from Taiwan. Bootstrap support and Bayesian posterior clade probabilities of selected lineages are shown. Taxa from the H5N2 outbreak in Mexico and the related viruses found in Japan are indicated. The most recent common ancestor (MRCA) of the Mexican-origin H5N2 lineage is indicated by the open circles in the trees. Changes of the amino acid at position -3 of HA1 (Table 3) are indicated in orange and a connecting thin bar. In the HA tree, the long branch leading to the Taiwan H5N2 duck viruses reported in this study has a length of 0.249 substitutions/site. (C and D) Linear regression plots of genetic distances from the MRCA of the Mexican-origin H5N2 lineage against isolation time of the taxa. The estimate and 95% confidence intervals for the evolutionary rate are shown by solid and dashed lines, respectively. Green arrows indicate the isolation dates of Mexican viruses that have similar genetic distances to the MRCA as the 2003 isolate from Taiwan.
Within the Taiwan chicken H5N2 virus lineage, both the HA and NA genes of the viruses diverged into at least two subclades. One includes viruses isolated in 2008 and from 2012 to 2013 (group A), while the other contains viruses isolated from 2012 to 2013 (group B). After the initial outbreak in 2003, the Mexico-like H5N2 viruses might have been reintroduced to chickens on at least two independent occasions or diverged quickly after their introduction into separate subclades that have persisted independently.
Relative to the inferred most recent common ancestor (MRCA) of the Taiwanese and Mexican H5N2 viruses (circles in Fig. 2A and B), both lineages demonstrated similar accumulations of genetic substitutions over time, indicating similar evolutionary rates (Fig. 2C and D). However, the Taiwanese lineage showed an apparent shift of 10 years in its accumulation of differences from the common ancestor, consistent with the phylogenetic trees of the HA and NA genes, where the 2003 H5N2 virus from Taiwan appeared more similar to the 1994 than to the contemporary H5N2 Mexican viruses. Molecular clock models showed lower evolutionary rates in the branches of the HA and NA trees at or near the transition of viruses from Mexico to Taiwan (data not shown). These data are supportive of the unnatural introduction of a Mexican virus, originating around 1994, to Taiwan in 2003.
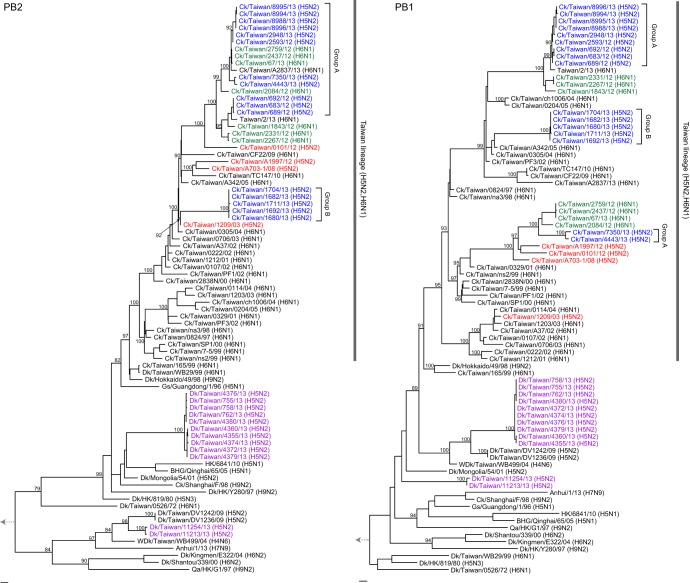
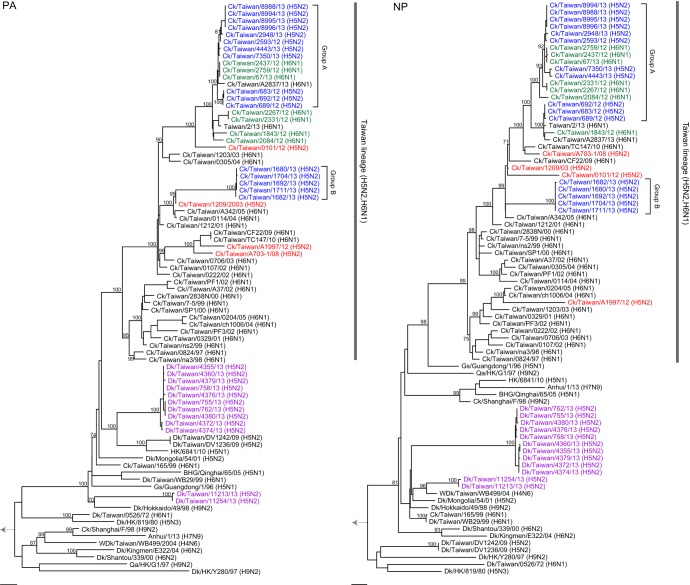
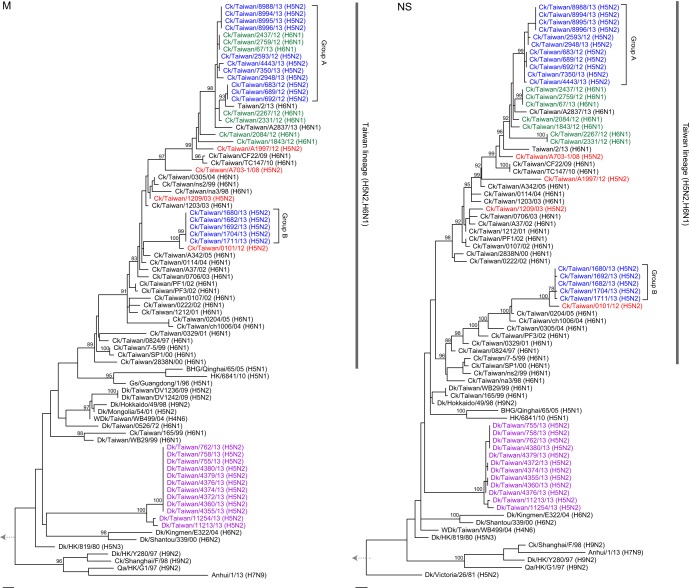
Phylogenetic analyses of each of the six internal genes (Fig. 3, 4, and 5) showed that all Taiwanese chicken H5N2 viruses grouped with the H6N1 viruses that have been enzootic in chickens in Taiwan since 1997 (20). Two major groups, consistent with the groups in the HA and NA trees (Fig. 2A and B), were seen in the phylogenies of the internal gene trees of the H5N2 viruses (Fig. 3, 4, and 5). The larger group (group A) clustered with contemporary H6N1 viruses that were also isolated in our surveillance and with the recent human H6N1 virus (34), while the second group (group B) clustered with H6N1 viruses isolated from 2003 to 2005. A third subgroup of two group A H5N2 isolates with recent H6N1 viruses was found in the PB1 phylogeny but not in the phylogenies of the other genes. The internal genes did not show the evolutionary rate discrepancy observed in the HA and NA genes (Fig. 2 to 5), suggesting that the H5N2 viruses might have reassorted near the time of their introduction with contemporary H6N1 chicken viruses. Overall, these findings suggest that these H5N2 viruses were generated through several independent reassortment events with enzootic H6N1 viruses.
FIG 3.
Maximum-likelihood phylogenies of the PB2 and PB1 genes. Taxon names of sequences reported in this study are shown in blue (chicken H5N2 lineage), green (enzootic chicken H6N1 lineage), and purple (duck H5N2). The chicken H5N2 viruses initially isolated from Taiwan are in red. Virus subtypes are indicated in parentheses. Bootstrap support values (percents) from 1,000 pseudoreplicates are shown for selected lineages. The scale bar to the left of each tree represents 0.01 substitutions per site. Abbreviations for host species: Ck, chicken; Dk, duck; WDk, wild duck; Qa, quail; Gs, Goose; BHG, bar-headed goose. Human isolates do not use any host specification. HK, Hong Kong.
FIG 4.
Maximum-likelihood phylogenies of the PA and NP gene segments. Other details are as described in the legend of Fig. 3.
FIG 5.
Maximum-likelihood phylogenies of the M and NS gene segments. Other details are as described in the legend of Fig. 3.
Molecular characterization.
As observed in the Mexican-like viruses, the Taiwan H5N2 viruses also had a 20-amino-acid (aa) deletion at the stalk region of the NA protein (deletion of residues 63 to 82), indicating that these viruses were adapted to chickens (35). An S31N substitution was found in the M2 protein of the Taiwan H5N2 and most H6N1 viruses, suggesting resistance to adamantanes (36). Truncation at position 218 of the C terminus of the NS1 protein was also present in the group B chicken Taiwan H5N2 viruses, as seen in the recent H7N9 viruses and many H9N2 and H6N1 viruses from terrestrial poultry (37, 38). The host specificity markers of the PB2 protein, positions 627 and 701, remained as E and D residues, consistent with what was commonly seen in avian viruses (39).
Although a full-length PB1-F2 protein is normally found in avian influenza virus isolates, the H5N2 and related H6N1 viruses from Taiwan had variable lengths for this protein. The group A viruses had a PB1-F2 protein of 57 amino acids (aa), the group B had a full-length protein of 90 aa, and the third group in the PB1 tree (Fig. 2) had a protein of 87 aa, while the 2003 isolate had a PB1-F2 of 34 aa. Truncation of PB1-F2 may moderate disease progression (40).
Three different motifs, R-E-K-R, R-K-K-R, and R-R-K-R, which are consistent with the patterns observed in highly pathogenic viruses (7, 12, 13), occurred at the HA connecting peptide of the chicken H5N2 viruses from Taiwan isolated since 2003 (Table 3). In our survey period, only the R-E-K-R and R-K-K-R motifs at the HA0 cleavage site were found, with both being isolated in the same sample set on only one occasion (March 2013) (Table 3). All of the H5N2 chicken viruses from Taiwan have the potential glycosylation site at residues 11 to 13 of HA1 (H3 mature peptide numbering) that has been associated with preventing a virus with a 4-amino-acid connecting peptide from being highly pathogenic (10, 11, 41).
TABLE 3.
Amino acid residues at the cleavage site of the HA protein of avian H5N2 virusesa
| Isolation date (mo/yr) | Strainb | Amino acid residue at: |
||||||
|---|---|---|---|---|---|---|---|---|
| HA1 position |
HA2 position +1 | |||||||
| −6 | −5 | −4 | −3 | −2 | −1 | |||
| 1994 | Ck/Hidalgo/28159-232/94 | P | Q | R | E | T | R | G |
| 2005 | Ck/Ibaraki/1/05 | P | Q | R | E | T | R | G |
| 12/2003 | Ck/TW/1209/03 | P | Q | R | E | K | R | G |
| 10/2008 | Ck/TW/A703-1/08 | P | Q | R | K | K | R | G |
| 1/2012 | Ck/TW/0101/12 | P | Q | R | K | K | R | G |
| 2/2012 | Ck/TW/A1997/12 | P | Q | R | R | K | R | G |
| 12/2012 | Ck/TW/683/12-like (n = 3) | P | Q | R | E | K | R | G |
| Ck/TW/2593/12 | P | Q | R | E | K | R | G | |
| 1/2013 | Ck/TW/1680/13-like (n = 5) | P | Q | R | K | K | R | G |
| 3/2013 | Ck/TW/2948/13 | P | Q | R | E | K | R | G |
| Ck/TW/7350/13 | P | Q | R | K | K | R | G | |
| Ck/TW/8994/13-like (n = 3) | P | Q | R | K | K | R | G | |
| Ck/TW/8996/13 | P | Q | R | E | K | R | G | |
| 5/2013 | Ck/TW/4443/13 | P | Q | R | E | K | R | G |
The glycosylation site at +11 (H3 numbering) is conserved in all viruses in this table.
Three viruses (Ck/TW/1680/13, Ck/TW/8994/13, and Ck/TW/8996/13) were selected for determination of pathogenicity in day-old chickens using the intracerebral pathogenicity index (ICPI).
The residue at position −3 of HA1 had mutated from E in the Mexican and the 2003 Taiwan H5N2 viruses to K (or R) on several occasions from 2008 to 2013 (Fig. 2A and Table 3). Overall, the sequences from our survey in Taiwan show seven cases of E and nine of K at position -3. The HA phylogenetic tree indicates repeated mutation from E to K (or R) at position -3 in the Taiwan H5N2 lineage, highlighting the possibility of multiple introductions of the virus into chickens in Taiwan.
Pathogenicity study.
To evaluate the pathogenicity conferred by the different HA cleavage motifs, three H5N2 isolates (Ck/TW/1680/13, Ck/TW/8994/13, and Ck/TW/8996/13), which fell into groups A and B (Fig. 2A) and had either an R-K-K-R or R-E-K-R motif (Table 3), were inoculated into 1-day-old chickens to determine the ICPI of the viruses. Ck/TW/8996/13 (R-E-K-R, group A) was not pathogenic or lethal in chicks (ICPI of 0), while the highly related Ck/TW/8994/13 (R-K-K-R, group A), which was isolated from the same sampling occasion, was fatal in 3 of the 10 chicks, giving an ICPI of 0.3375 (Table 4). The group B virus, Ck/TW/1680/13 (R-K-K-R), had an ICPI of 1.675 and killed all chickens by day 5 postinoculation (Table 4). Therefore, neither the R-E-K-R nor the R-K-K-R motif, in the context of a nearby glycosylation site, is sufficient to cause high pathogenicity, as measured by the ICPI. While a basic amino acid at position -3 of HA1 might be sufficient to cause some lethality to chicks, other molecular characteristics may be involved in the higher pathogenicity of Ck/TW/1680/13.
TABLE 4.
Determination of the ICPI of the chicken H5N2 viruses prevalent in Taiwan
| Virus | Clinical sign | No. of chickens with specific sign by day postinoculation |
ICPIa | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| Ck/TW/1680/13 | Normal | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.675 |
| Sick | 4 | 7 | 4 | 1 | 0 | 0 | 0 | 0 | ||
| Dead | 1 | 3 | 6 | 9 | 10 | |||||
| Ck/TW/8994/13 | Normal | 10 | 10 | 9 | 8 | 7 | 7 | 7 | 7 | 0.3375 |
| Sick | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | ||
| Dead | 0 | 0 | 0 | 1 | 2 | 3 | 3 | 3 | ||
| Ck/TW/8996/13 | Normal | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 0 |
| Sick | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Dead | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
ICPI, intracerebral pathogenicity index.
DISCUSSION
Our surveillance study has revealed that H5N2 viruses are prevalent at low levels in the chicken population of Taiwan. Phylogenetic analyses of these viruses show that they were generated by reassortment between an H5N2 virus that originated from Mexico in 1994 and a locally enzootic H6N1 virus lineage. Apparently, this kind of reassortment has occurred on multiple occasions. How this Mexican-like H5N2 virus entered Taiwan is still not clear.
Introduction of influenza viruses from one region to another is usually associated with poultry movement, bird migration, or escape from an insufficiently inactivated or attenuated vaccine strain (22, 42, 43). Even though Taiwan is located on the East Asian migratory bird flyway and although intercontinental movement of influenza viruses related to flyways has occurred previously (44–46), influenza viruses from the North American lineage had not been documented in Taiwan before this H5N2 virus was detected in 2003. The Taiwanese Mexican-like H5N2 viruses that emerged around 2003 did not have genes of contemporary Mexican-like viruses but had surface genes more similar to a 1994 virus, Ck/Hidalgo/28159-232/94, that was used as a commercial vaccine strain (19) (Fig. 2A and B). Their internal genes were obtained by reassortment with the contemporary enzootic H6N1 viruses of Taiwan (20) (Fig. 3, 4, and 5). This time discrepancy between the emergence dates of the Mexican and Taiwanese viruses is reflected in the discordance of the dates on the phylogenetic tree and accumulation of substitutions from the lineage common ancestor (Fig. 2). A similar pattern occurred in the reemergence of a 1950s-like seasonal human H1N1 virus in 1977 (47).
LPAI Mexican-like H5N2 viruses caused an outbreak in chickens in Japan from 2005 to 2006 (48). These LPAI viruses clustered, over all their segments, with contemporary Mexican H5N2 viruses but have not been detected in aquatic birds in Japan. These observations are highly suggestive that the H5N2 virus was artificially introduced to chickens rather than being introduced by migratory bird flyways, most probably as an escape from a vaccination program. Mexican-like H5N2 vaccines have been used previously in this region, for example, in Hong Kong from 2003 to 2013 (49), although their use in Japan or Taiwan has not been officially reported.
Our phylogenetic analyses revealed that current Taiwan H5N2 viruses diverged into groups A and B and were distinct from the earlier isolates from 2003 and 2008, raising the possibility of multiple introductions of the viruses into Taiwan or a single introduction of a virus that persisted undetected and continuously evolved in the field. In the connecting peptide of these viruses, the residue at position −3 of HA1 varies between E and K (with one instance of R) (Table 3). Changes to basic amino acids are commonly observed at the connecting peptide when a virus changes from low to high pathogenicity (7, 13); however, the reversion to an acidic or the original residue is rare in HPAI virus lineages. While a reversion from K to the original E could have occurred on the branch leading to group A, the unusual prevalence of the ancestral state supports repeated reintroduction of the viruses (Fig. 2A).
Outbreaks resulting in high mortality in chickens were reported from Taiwan in 2012 (21). In our study, a variable level of pathogenicity was found in the viruses even though the motifs in the connecting peptides contained multiple basic amino acids. The glycosylation site at the N terminus of the HA protein may interfere with the accessibility of the cleavage enzyme to some extent (10, 11, 41). The contemporary group B viruses, which include Ck/TW/1680/13, have a truncated NS1 protein, while group A viruses have a truncated PB1-F2. This may contribute to the varied pathogenicities, as indicated by the ICPI test. Further investigation of the related molecular basis for this is warranted.
Serological studies revealed that approximately 8.3% and 3.5% of chickens surveyed were seropositive to H5N2 and H6N1 viruses. It could be possible that this reflects prior exposure to circulating viruses. However, the 82.1% seropositive rate to a Hong Kong H9N2 virus, belonging to the Ck/Beijing/94 lineage (38), provided evidence of a broad and recent chicken vaccination program in Taiwan. To date, the Ck/Beijing/94-like H9N2 viruses are highly prevalent mainly in mainland China but have not been reported in Taiwan, nor were they detected in our surveillance. This further supports the possibility that, while apparently prohibited (21), vaccination with an H5N2 virus (potentially a bi- or trivalent vaccine including H6N1 and/or H9N2 viruses) has occurred and that this insufficiently inactivated or inadequately attenuated live-virus vaccine is the source of the H5N2 viruses found in chickens in Taiwan.
Widespread vaccination of poultry is commonly used in several countries to provide protection against avian influenza (50). The quality of the vaccine is critical to the success of such programs, while control measures such as stamping out and localized temporary vaccination upon an outbreak could be an alternative (50, 51). In the case of H5N2 in Taiwan, the reemergence of a North American vaccine strain virus in Asia after approximately 10 years is a strong indication that a vaccine may be the source of this virus. The currently emerging H5N2 variants in Taiwan are suggestive of possible reintroductions from a vaccine source, and some are developing signs of evolving into a highly pathogenic strain. This example provides a strong warning against the use of vaccination in the absence of a present threat and the need for the highest quality of vaccine and strict control of vaccination practice by competent, accountable authorities. Thorough surveillance and a localized response, if needed, may be an effective way to prevent a situation such as the H5N2 influenza virus outbreaks in Taiwan from developing.
ACKNOWLEDGMENTS
This study was supported by the Li Ka Shing Foundation, the National Institutes of Health (NIAID contracts HHSN266200700005C and HHSN266200700006C), and the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06) of the Hong Kong SAR Government.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Alexander DJ. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3–13. 10.1016/S0378-1135(00)00160-7 [DOI] [PubMed] [Google Scholar]
- 2.Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander DJ. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963–973. 10.1007/s007050170128 [DOI] [PubMed] [Google Scholar]
- 3.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36. 10.1016/j.virol.2004.02.015 [DOI] [PubMed] [Google Scholar]
- 4.Kawaoka Y, Webster RG. 1985. Evolution of the A/chicken/Pennsylvania/83 (H5N2) influenza virus. Virology 146:130–137. 10.1016/0042-6822(85)90059-5 [DOI] [PubMed] [Google Scholar]
- 5.Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster RG. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213:223–230. 10.1006/viro.1995.1562 [DOI] [PubMed] [Google Scholar]
- 6.Perdue M, Crawford J, Garcia M, Latimer J, Swayne D. 2003. Occurrence and possible mechanisms of cleavage-site insertions in the avian influenza hemagglutinin gene. Avian Dis. 47(Special Issue):182–193 [Google Scholar]
- 7.Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Suss J, Lipkind M, Kida H, Webster RG. 1996. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40:425–437. 10.2307/1592241 [DOI] [PubMed] [Google Scholar]
- 8.Horimoto T, Kawaoka Y. 1994. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 68:3120–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horimoto T, Kawaoka Y. 1997. Biologic effects of introducing additional basic amino acid residues into the hemagglutinin cleavage site of a virulent avian influenza virus. Virus Res. 50:35–40. 10.1016/S0168-1702(97)00050-6 [DOI] [PubMed] [Google Scholar]
- 10.Deshpande KL, Fried VA, Ando M, Webster RG. 1987. Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc. Natl. Acad. Sci. U. S. A. 84:36–40. 10.1073/pnas.84.1.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaoka Y, Naeve CW, Webster RG. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303–316. 10.1016/0042-6822(84)90376-3 [DOI] [PubMed] [Google Scholar]
- 12.Vey M, Orlich M, Adler S, Klenk HD, Rott R, Garten W. 1992. Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R-X-K/R-R. Virology 188:408–413. 10.1016/0042-6822(92)90775-K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood GW, McCauley JW, Bashiruddin JB, Alexander DJ. 1993. Deduced amino acid sequences at the haemagglutinin cleavage site of avian influenza A viruses of H5 and H7 subtypes. Arch. Virol. 130:209–217. 10.1007/BF01319010 [DOI] [PubMed] [Google Scholar]
- 14.Alexander DJ, Brown IH. 2009. History of highly pathogenic avian influenza. Rev. Sci. Technol. 28:19–38 [DOI] [PubMed] [Google Scholar]
- 15.World Organisation for Animal Health (OIE). 2014, posting date Update on highly pathogenic avian influenza in animals (type H5 and H7). World Organisation for Animal Health (OIE), Paris, France: http://www.oie.int/animal-health-in-the-world/update-on-avian-influenza/2014/ [Google Scholar]
- 16.Becker WB. 1966. The isolation and classification of Tern virus: influenza virus A/Tern/South Africa/1961. J. Hyg. (Lond.) 64:309–320. 10.1017/S0022172400040596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirst M, Astell CR, Griffith M, Coughlin SM, Moksa M, Zeng T, Smailus DE, Holt RA, Jones S, Marra MA, Petric M, Krajden M, Lawrence D, Mak A, Chow R, Skowronski DM, Tweed SA, Goh S, Brunham RC, Robinson J, Bowes V, Sojonky K, Byrne SK, Li Y, Kobasa D, Booth T, Paetzel M. 2004. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 10:2192–2195. 10.3201/eid1012.040743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee CW, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC, Panigrahy B, Rojas H, Spackman E, Alexander DJ. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 10:693–699. 10.3201/eid1004.030396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng MC, Soda K, Lee MS, Lee SH, Sakoda Y, Kida H, Wang CH. 2010. Isolation and characterization of potentially pathogenic H5N2 influenza virus from a chicken in Taiwan in 2008. Avian Dis. 54:885–893. 10.1637/9208-120609-Reg.1 [DOI] [PubMed] [Google Scholar]
- 20.Lee MS, Chang PC, Shien JH, Cheng MC, Chen CL, Shieh HK. 2006. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 50:561–571. 10.1637/7640-050106R.1 [DOI] [PubMed] [Google Scholar]
- 21.World Organisation for Animal Health (OIE). 2012. Update on highly pathogenic avian influenza in animals (type H5 and H7). Chinese Taipei H5N2 14/08/12. Final report. World Organisation for Animal Health (OIE), Paris, France: http://web.oie.int/wahis/reports/en_fup_0000012179_20120814_152200.pdf [Google Scholar]
- 22.Cheung CL, Vijaykrishna D, Smith GJ, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, Poon LL, Peiris JS, Chen H, Guan Y. 2007. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J. Virol. 81:10402–10412. 10.1128/JVI.01157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GJ, Fan XH, Wang J, Li KS, Qin K, Zhang JX, Vijaykrishna D, Cheung CL, Huang K, Rayner JM, Peiris JS, Chen H, Webster RG, Guan Y. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U. S. A. 103:16936–16941. 10.1073/pnas.0608157103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang K, Zhu H, Fan X, Wang J, Cheung CL, Duan L, Hong W, Liu Y, Li L, Smith DK, Chen H, Webster RG, Webby RJ, Peiris M, Guan Y. 2012. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 86:6075–6083. 10.1128/JVI.06389-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 27.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9. 10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- 30.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25:1459–1471. 10.1093/molbev/msn090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terregino C, Capua I. 2009. Conventional diagnosis of Newcastle disease virus infection, p 123–125 In Capua I, Alexander DJ. (ed), Avian influenza and Newcastle disease. Springer, Milan, Italy [Google Scholar]
- 32.Lee CW, Senne DA, Suarez DL. 2004. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 78:8372–8381. 10.1128/JVI.78.15.8372-8381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin V, Forman A, Lubroth J. 2009. Preparing for highly pathogenic avian influenza, rev. ed. Food and Agriculture Organization of the United Nations, Rome, Italy: http://www.fao.org/docrep/012/i0808e/i0808e00.htm [Google Scholar]
- 34.Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, Liu YL, Lo YC, Yang CH, Chuang JH, Lin MC, Chung WC, Liao CH, Lee MS, Huang WT, Chen PJ, Liu MT, Chang FY. 2013. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet. Respir. Med. 1:771–778. 10.1016/S2213-2600(13)70221-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan Y, Peiris JS, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. U. S. A. 99:8950–8955. 10.1073/pnas.132268999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Astrahan P, Arkin IT. 2011. Resistance characteristics of influenza to amino-adamantyls. Biochim. Biophys. Acta 1808:547–553. 10.1016/j.bbamem.2010.06.018 [DOI] [PubMed] [Google Scholar]
- 37.Dundon WG, Capua I. 2009. A closer look at the NS1 of influenza virus. Viruses 1:1057–1072. 10.3390/v1031057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. 10.1038/nature12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manz B, Schwemmle M, Brunotte L. 2013. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 87:7200–7209. 10.1128/JVI.00980-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmolke M, Manicassamy B, Pena L, Sutton T, Hai R, Varga ZT, Hale BG, Steel J, Perez DR, Garcia-Sastre A. 2011. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 7:e1002186. 10.1371/journal.ppat.1002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawaoka Y, Webster RG. 1989. Interplay between carbohydrate in the stalk and the length of the connecting peptide determines the cleavability of influenza virus hemagglutinin. J. Virol. 63:3296–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Smith GJ, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, Cheung CL, Xu KM, Duan L, Huang K, Qin K, Leung YH, Wu WL, Lu HR, Chen Y, Xia NS, Naipospos TS, Yuen KY, Hassan SS, Bahri S, Nguyen TD, Webster RG, Peiris JS, Guan Y. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. U. S. A. 103:2845–2850. 10.1073/pnas.0511120103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam TT, Hon CC, Lemey P, Pybus OG, Shi M, Tun HM, Li J, Jiang J, Holmes EC, Leung FC. 2012. Phylodynamics of H5N1 avian influenza virus in Indonesia. Mol. Ecol. 21:3062–3077. 10.1111/j.1365-294X.2012.05577.x [DOI] [PubMed] [Google Scholar]
- 44.Lam TT, Ip HS, Ghedin E, Wentworth DE, Halpin RA, Stockwell TB, Spiro DJ, Dusek RJ, Bortner JB, Hoskins J, Bales BD, Yparraguirre DR, Holmes EC. 2012. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecol. Lett. 15:24–33. 10.1111/j.1461-0248.2011.01703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. 1999. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J. Gen. Virol. 80:3167–3171 [DOI] [PubMed] [Google Scholar]
- 46.Webby RJ, Woolcock PR, Krauss SL, Walker DB, Chin PS, Shortridge KF, Webster RG. 2003. Multiple genotypes of nonpathogenic H6N2 influenza viruses isolated from chickens in California. Avian Dis. 47:905–910. 10.1637/0005-2086-47.s3.905 [DOI] [PubMed] [Google Scholar]
- 47.Wertheim JO. 2010. The re-emergence of H1N1 influenza virus in 1977: a cautionary tale for estimating divergence times using biologically unrealistic sampling dates. PLoS One 5:e11184. 10.1371/journal.pone.0011184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamatsu M, Saito T, Mase M, Tsukamoto K, Yamaguchi S. 2007. Characterization of H5N2 influenza A viruses isolated from chickens in Japan. Avian Dis. 51:474–475. 10.1637/7573-033106R1.1 [DOI] [PubMed] [Google Scholar]
- 49.Food and Health Bureau, Agriculture, Fisheries, and Conservation Department. 2013. Update on the avian influenza vaccination programme in local chicken farms. ACFEH paper 3/2013. Food and Health Bureau, Government of Hong Kong SAR [Google Scholar]
- 50.Swayne DE, Spackman E, Pantin-Jackwood M. 12 September 2013. Success factors for avian influenza vaccine use in poultry and potential impact at the wild bird-agricultural interface, EcoHealth. 10.1007/s10393-013-0861-3 [DOI] [PubMed] [Google Scholar]
- 51.Marangon S, Capua I. 2006. Control of avian influenza in Italy: from stamping out to emergency and prophylactic vaccination. Dev. Biol. (Basel) 124:109–115 [PubMed] [Google Scholar]