Abstract
The VP24 protein plays an essential, albeit poorly understood role in the filovirus life cycle. VP24 is only 30% identical between Marburg virus and the ebolaviruses. Furthermore, VP24 from the ebolaviruses is immunosuppressive, while that of Marburg virus is not. The crystal structure of Marburg virus VP24, presented here, reveals that although the core is similar between the viral genera, Marburg VP24 is distinguished by a projecting β-shelf and an alternate conformation of the N-terminal polypeptide.
TEXT
Marburg virus (MARV) and the ebolaviruses are filamentous, enveloped, negative-sense, single-stranded RNA (ssRNA) viruses that belong to the family Filoviridae and can cause severe hemorrhagic fever in both humans and nonhuman primates. The Marburgvirus genus contains one species, which is eponymously named Marburg virus (1). Marburg virus was the first filovirus to be identified when, in 1967, Marburg virus-infected primates sickened laboratory workers in Germany and Yugoslavia (2). Although early outbreaks were associated with 20 to 40% lethality, more recent outbreaks have been associated with greater pathogenicity and nearly 90% lethality in humans (3, 4). In the Ebolavirus genus are five viruses, termed Ebola virus, Sudan virus (SUDV), Reston virus, Taï Forest virus, and Bundibugyo virus. Among the five ebolaviruses, Reston virus appears to be nonpathogenic to humans, although exposure data are limited (5, 6).
Filoviruses encode just seven genes, encoding NP (nucleoprotein), VP35 (nucleocapsid), VP40 (matrix), GP (glycoprotein), VP24 (nucleocapsid), VP30 (transcription factor), and L (RNA-dependent RNA polymerase). In the ebolaviruses, the nucleocapsid-associated proteins VP24 and VP35 are known to be immunosuppressive (7, 8, 10, 11, 14). In Marburg virus, VP35 and VP40 are immunosuppressive (12), while curiously, Marburg virus VP24 is not. Ebola virus VP24 blocks phosphorylation of p38 mitogen-activated protein kinase (13) and inhibits signaling downstream from both alpha/beta interferon (IFN-α/β) and IFN-γ by sequestering NPI-1 family karyopherin α proteins (α1, α5, and α6) (11, 14). Binding to these proteins prevents them from shuttling activated, phosphorylated STAT1 to the nucleus (11, 14, 15).
Although Marburg virus VP24 is not immunosuppressive, it nonetheless is essential for the virus life cycle. VP24 is bound to the ribonucleoprotein complex in the virion (16), influences the formation of infectious virus particles (17), and like VP24 of Ebola virus, may function in transcription and replication (18–20). No crystal structure of Marburg virus VP24 is yet available. Hence, we set out to determine the structure of this VP24, with its different functional phenotype, in order to provide a 3-dimensional (3-D) template for exploration of the differences between Marburg and ebolavirus and the function(s) of VP24 in the Marburg virus life cycle.
A construct of Marburg virus (strain Musoke) VP24 spanning residues 1 to 241 (MARV VP241-241) in the pET46 Ek/LIC vector was expressed and purified as previously described for ebolavirus VP24 (21). A 12-residue C-terminal truncation improved protein stability, homogeneity, and crystallizability. MARV VP24 was crystallized in 0.1 M N-(2-acetamido)iminodiacetic acid (ADA) (Hampton Research), 0.1 M lithium acetate, 20% glycerol, 2% (vol/vol) polyethylene glycol (PEG) 400, and 8% (vol/vol) PEG 4000 by the hanging-drop vapor diffusion method at 22°C. An amount of 0.2 μl of seed stock was added to the 1.8-μl drop during the initial crystallization set up. Crystals were flash frozen in liquid nitrogen and cryoprotected with unmodified reservoir solution. Diffraction data to 2.65Å were collected at 100 K on Beamline 8.2.2 (Advanced Light Source, Berkeley, CA) and were processed with HKL-2000 (Table 1) (22).
TABLE 1.
Data collection and refinement statistics for MARV VP241-241 crystals
| Parameter | Value(s) |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 42.2, 48.1, 64.1 |
| α, β, γ (°) | 88.4, 78.7, 71.3 |
| Resolution (Å) | 50–2.65 |
| Solvent content (%) | 43 |
| Rsyma (%) | 0.06 |
| I/σ(I) | 12.8 (2.2b) |
| Completeness (%) | 98.1 |
| Redundancy | 2.1 |
| Refinement | |
| Resolution (Å) | 40–2.65 |
| No. of reflections | 13,245 |
| Rwork/Rfree | 18.0/25.5 |
| No. of atoms of: | |
| Protein | 3,685 |
| Water | 48 |
| RMSDc | |
| Bond length (Å) | 0.003 |
| Bond angle (°) | 0.62 |
| Ramachandran plotd | |
| Most favored | 96.2 |
| Additionally allowed | 3.8 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
Rsym = ∑|(I − <I>)|/∑(I).
Value in parentheses refers to the last shell.
RMSD, root mean square deviations.
MolProbity was used to define the indicated regions of the Ramachandran plot.
The structure of MARV VP24 was determined by molecular replacement in Phaser (23) and CCP4 (24), using residues 1 to 233 of Sudan virus VP24 (SUDV VP241-233) as a search model (25). Refinement was performed with Phenix.refine (26, 27), and rebuilding performed in COOT (28). The initial rounds of refinement included TLS parameters (29). The quality of the structure was validated with MolProbity (30) and Procheck (31), with 97% of residues in the most favored region of the Ramachandran plot and no residues in the disallowed regions. The final R and Rfree were 20 and 27%, respectively, with 5% of reflections reserved for Rfree calculations. There are two copies of Marburg virus VP24 in the crystallographic asymmetric unit, termed copy A and copy B.
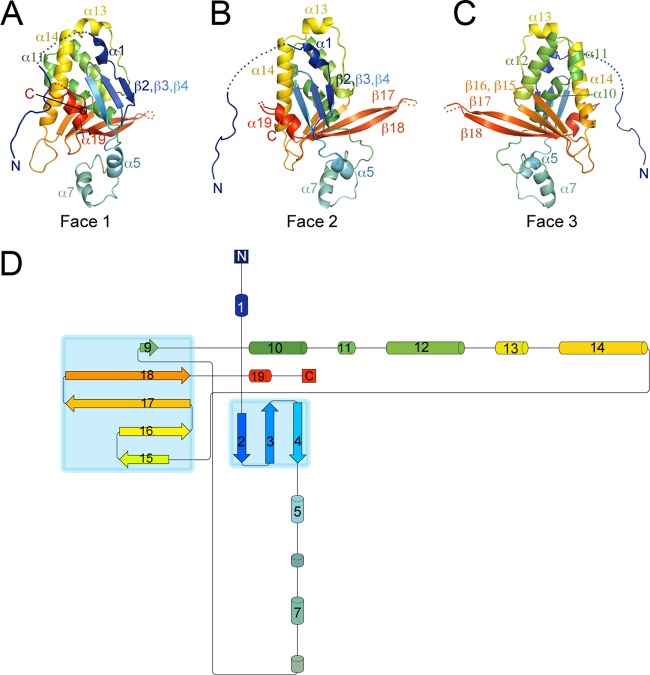
Marburg virus VP24, like our previously determined ebolavirus VP24 structures (from Sudan virus and Reston virus) (25), adopts a single-domain α/β structure with the overall shape resembling a pyramid (Fig. 1). Also like VP24 of the ebolaviruses, two neighboring concave pockets are located at the bottom of the Marburg virus VP24 pyramid. The residues contained inside these pockets are highly conserved across the filovirus family.
FIG 1.
Overall architecture of Marburg virus VP24. (A to C) Faces 1 to 3 of the pyramidal VP24 structure are illustrated in rainbow coloring from the N terminus (navy blue) to the C terminus (red). The extended beta shelf formed by strands β17 and β18 is visible on the right of face 2 and the left of face 3. The descending N terminus of Marburg virus VP24 connects to the protein body via residues 13 to 20, which are disordered. (D) Topology diagram of Marburg virus VP24, with secondary structure elements sequentially numbered and colored from N to C as described for panels A to C. α-Helices are indicated by cylinders, and β-strands by arrows. Panels A to C were produced using Pymol (Delano Scientific) (34), and the topology diagram using Pro-origami (35).
One difference between Marburg virus VP24 and VP24 of the ebolaviruses is found in the bottom platform of the VP24 pyramid, above the conserved pockets. Here, residues 201 to 217, which lie at the interface between face 2 and face 3, appear as long β-strands (β17 and β18) that form part of a greater β-sheet. The two strands jut out from the pyramid to form a shelf (Fig. 1). In Sudan virus VP24, residues 201 to 217 form much shorter β-strands, with the central residues instead adopting a flexible loop structure that contains a small helix (Fig. 2). In Reston virus VP24, these residues are disordered and are not observed in the crystal structure (25).
FIG 2.
Differences between Marburg virus and ebolavirus VP24s. (A) Marburg virus (MARV) and Sudan virus (SUDV) VP24 structures are superimposed and illustrated with face 3 oriented toward the viewer. Conformational differences in α13 between Marburg virus copy B and Sudan virus are highlighted with a black circle. The projecting β17-β18 shelf in Marburg virus is apparent on the left, and the position of the Marburg virus VP24 N terminus on the right. (B) Rotation of the superimposed structures so that differences in structure in β17-β18 are apparent. In the ebolaviruses, the equivalent residues do not make an extended shelf but instead form shorter strands connected by a loop/helical structure. (C) Marburg virus VP24 is illustrated in gray, oriented as described for panel A. VP24 residues conserved across the filovirus family (Marburg and five ebolaviruses) are colored dark blue; those visible in this view are labeled. Conservation in VP24 focuses on face 3, the N terminus, and the pocket at the base connecting faces 1 and 3. Figures were created using PyMol (Delano Scientific) (34).
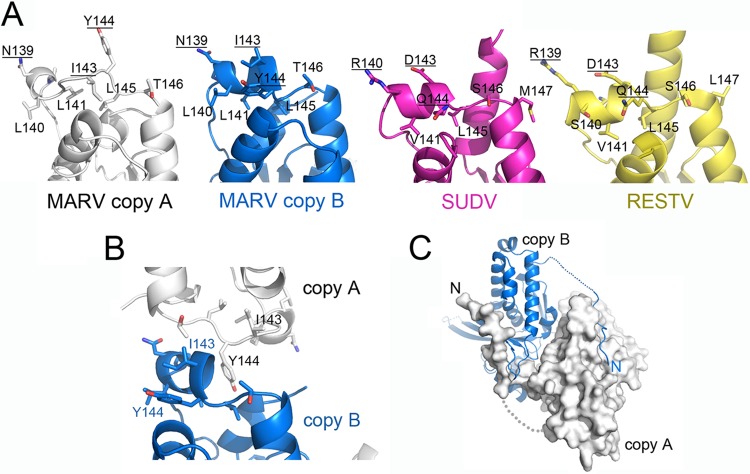
In both ebolavirus VP24s, amino acid residues 142 to 146 form a short α-helix at the top of the pyramid (25) and are proposed to interact with karyopherin α1 (11). These residues also appear as an α-helix in copy B of Marburg VP24 but form a mostly nonhelical loop structure in copy A (Fig. 3A and B). Differences in the structure appear to be dictated by differences in crystal packing, as the central Tyr144 is rotated 180° between the two monomers (Fig. 3B). Both conformers are likely available to VP24 in solution.
FIG 3.
(A) The top of the VP24 pyramids for Marburg virus copy A and copy B and the ebolaviruses (Sudan virus [SUDV] and Reston virus [RESTV]). Residues 142 to 146, which are thought to interact with karyopherin α1 in Ebola virus VP24, differ in sequence from the same residues in the nonimmunosuppressive Marburg virus VP24. Note, for example, the conserved placement of Arg 139/140, Asp 143, and Gln 144 in the two ebolaviruses. These positions are occupied by Asn 139, Ile 143, and Tyr 144 in Marburg virus. This site appears somewhat flexible in Marburg virus, as copies A and B of Marburg virus VP24 adopt different conformations. Copy B forms helix α13 with Tyr 144 rotated downward and Ile 143 upward, but copy A adopts a more extended structure with Tyr 144 rotated upward and Ile 143 downward. (B) Differences in conformation in residues 139 to 143 (α13) appear to be caused by crystal packing interactions. Illustrated here are the crystal packing interactions of copies A and B in the asymmetric unit. Note the alternate rotation of Tyr 144 between the two structures. (C) The N terminus of VP24 may be involved in VP24 oligomerization (33). In the crystal packing, the N termini of the two copies of Marburg virus VP24 reach across to bind into a shallow groove on the surface of the neighboring VP24.
The N termini of both Ebola (Zaire) virus and Marburg virus VP24 are thought to be important for nucleocapsid formation and oligomerization (17, 32, 33). In each copy of VP24 crystallized for an ebolavirus (Sudan virus and Reston virus), the N terminus forms a rigid α-helix that extends from the apex of the pyramid to bind into the conserved hydrophobic pocket in face 3. In contrast, in both copies of Marburg virus VP24, the N terminus (residues 1 to 23) does not form a rigid helix but instead forms an extended flexible strand that reaches across to a neighboring copy of Marburg virus VP24, with residues 1 to 10 binding into a groove along the base of the pyramid (Fig. 3C). Approximately 770 Å2 of molecular surface is buried by the 10 residues participating in this interaction. Contact here is mediated by the main chain atoms of the N-terminal peptide, as well as side chain atoms of Leu 4, Arg 7, Tyr 8, Asn 9, and Leu 10. Each of these residues, except Leu 4, is completely conserved across the filovirus family (Fig. 2C). Leu 4 is replaced by Ala 4 in the ebolaviruses. These residues were deleted from constructs used to determine the Reston virus (4D9O) and the 2.1-Å Sudan virus (3VNF) VP24 structures to limit aggregation. They were included in material used to generate the subsequent 2.0-Å Sudan virus structure (3VNE), but residues 1 to 8 are disordered in the resulting electron density maps (25).
In summary, the overall structural conservation observed between VP24 of Marburg virus and the ebolaviruses supports their common essential functions in viral assembly and function. The reasons why Marburg virus VP24 is not immunosuppressive remain elusive, however. Marburg virus and Ebola virus VP24 are 70% different in sequence, and the precise residues responsible for the difference in immunosuppression are unknown, as are the precise role(s) of Ebola virus VP24 in immunosuppression. The crystal structure of Marburg virus VP24 presented here now provides the 3-D template for directed functional exploration of the multiple roles of VP24 in the Marburg virus life cycle and key differences between Marburg virus and the ebolaviruses in immunosuppression.
Protein structure accession number.
The atomic coordinates and structure factors identified in this study have been deposited in the Protein Data Bank under accession number 4OR8.
ACKNOWLEDGMENTS
We thank Stephan Becker (Philipps-Universität Marburg) for his gift of cDNA to express MARV Musoke VP24, Timothy Booth and Daniel Benias of the University of Manitoba and Public Health Agency Canada for helpful discussions about the position of VP24 in cryoelectron microscopy reconstructions of the filovirus nucleocapsid, and Xiaoping Dai of TSRI and the staff of Beamline 8.2.2 of the Advanced Light Source and 23-ID-D of the Advanced Photon Source for assistance with data collection.
E.O.S. was supported by The Skaggs Institute for Chemical Biology and an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. A.P.P.Z. was supported by NIH postdoctoral training grant 5P32 AI007244 to the TSRI Department of Immunology and Microbial Science.
Footnotes
Published ahead of print 26 February 2014
This is manuscript no. 26039 from The Scripps Research Institute.
REFERENCES
- 1.Bukreyev AA, Chandran K, Dolnik O, Dye JM, Ebihara H, Leroy EM, Mühlberger E, Netesov SV, Patterson JL, Paweska JT, Saphire EO, Smither SJ, Takada A, Towner JS, Volchkov VE, Warren TK, Kuhn JH. 13 October 2013. Discussions and decisions of the 2012-2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012-June 2013. Arch. Virol. 10.1007/s00705-013-1846-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slenczka W, Klenk HD. 2007. Forty years of Marburg virus. J. Infect. Dis. 196(Suppl 2):S131–S135. 10.1086/520551 [DOI] [PubMed] [Google Scholar]
- 3.Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, Hartman AL, Comer JA, Zaki SR, Ströher U, Gomes da Silva F, del Castillo F, Rollin PE, Ksiazek TG, Nichol ST. 2006. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 80:6497–6516. 10.1128/JVI.00069-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bausch DG, Nichol ST, Muyembe-Tamfum JJ, Borchert M, Rollin PE, Sleurs H, Campbell P, Tshioko FK, Roth C, Colebunders R, Pirard P, Mardel S, Olinda LA, Zeller H, Tshomba A, Kulidri A, Libande ML, Mulangu S, Formenty P, Grein T, Leirs H, Braack L, Ksiazek T, Zaki S, Bowen MD, Smit SB, Leman PA, Burt FJ, Kemp A, Swanepoel R, International Scientific and Technical Committee for Marburg Hemorrhagic Fever Control in the Democratic Republic of the Congo 2006. Marburg hemorrhagic fever associated with multiple genetic lineages of virus. N. Engl. J. Med. 355:909–919. 10.1056/NEJMoa051465 [DOI] [PubMed] [Google Scholar]
- 5.Retuya TJA, Miranda MEG, Ksiazek TG, Khan AS, Sanchez A, et al. 1997. Is the ebola-reston virus a threat to occupationally exposed humans? J. Clin. Epidemiol. 50(Suppl 1):S32. 10.1016/S0895-4356(97)87270-8 [DOI] [Google Scholar]
- 6.Sanchez A, Khan AS, Zaki SR, Nabel GJ, Ksiazek TG, Peters CJ. 2001. Filoviridae: Marburg and Ebola viruses, p 1279–1304 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 7.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, García-Sastre A, Palese P. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. 97:12289–12294. 10.1073/pnas.220398297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebihara H, Takada A, Kobasa D, Jones S, Neumann G, Theriault S, Bray M, Feldmann H, Kawaoka Y. 2006. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2:e73. 10.1371/journal.ppat.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10.Mateo M, Carbonnelle C, Reynard O, Kolesnikova L, Nemirov K, Page A, Volchkova VA, Volchkov VE. 2011. VP24 is a molecular determinant of Ebola virus virulence in guinea pigs. J. Infect. Dis. 204(Suppl 3):S1011–S1020. 10.1093/infdis/jir338 [DOI] [PubMed] [Google Scholar]
- 11.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 84:1169–1175. 10.1128/JVI.01372-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krähling V, Basler CF, Mühlberger E. 2010. Marburg virus evades interferon responses by a mechanism distinct from Ebola virus. PLoS Pathog. 6:e1000721. 10.1371/journal.ppat.1000721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halfmann P, Neumann G, Kawaoka Y. 2011. The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J. Infect. Dis. 204(Suppl 3):S953–S956. 10.1093/infdis/jir325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156–5167. 10.1128/JVI.02349-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469–13477. 10.1128/JVI.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bharat TA, Riches JD, Kolesnikova L, Welsch S, Krahling V, Davey N, Parsy ML, Becker S, Briggs JA. 2011. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 9:e1001196. 10.1371/journal.pbio.1001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bamberg S, Kolesnikova L, Moller P, Klenk HD, Becker S. 2005. VP24 of Marburg virus influences formation of infectious particles. J. Virol. 79:13421–13433. 10.1128/JVI.79.21.13421-13433.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. 2007. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J. Infect. Dis. 196(Suppl 2):S284–S290. 10.1086/520582 [DOI] [PubMed] [Google Scholar]
- 19.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403:56–66. 10.1016/j.virol.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 20.Iwasa A, Halfmann P, Noda T, Oyama M, Kozuka-Hata H, Watanabe S, Shimojima M, Watanabe T, Kawaoka Y. 2011. Contribution of Sec61alpha to the life cycle of Ebola virus. J. Infect. Dis. 204(Suppl 3):S919–926. 10.1093/infdis/jir324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang AP, Bornholdt ZA, Liu T, Abelson DM, Lee DE, Li S, Woods VL, Jr, Saphire EO. 2012. The Ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 8:e1002550. 10.1371/journal.ppat.1002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Academic Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40(Pt 4):658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 25.Zhang AP, Bornholdt ZA, Liu T, Abelson DM, Lee DE, Li S, Woods VL, Jr, Saphire EO. 2012. The Ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 8:e1002550. 10.1371/journal.ppat.1002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afonine PV, Grosse-Kunstleve RW, Adams PD. 17 July 2005. The Phenix refinement framework. CCP4 Newsl. 42, Contribution 8 [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 29.Painter J, Merritt EA. 2006. TLSMD web server for the generation of multi-group TLS models. J. Appl. Cryst. 39:109–111. 10.1107/S0021889805038987 [DOI] [Google Scholar]
- 30.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383. 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- 32.Noda T, Halfmann P, Sagara H, Kawaoka Y. 2007. Regions in Ebola virus VP24 that are important for nucleocapsid formation. J. Infect. Dis. 196(Suppl 2):S247–S250. 10.1086/520596 [DOI] [PubMed] [Google Scholar]
- 33.Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793–1800. 10.1128/JVI.77.3.1793-1800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 35.Stivala A, Wybrow M, Wirth A, Whisstock JC, Stuckey PJ. 2011. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics 27:3315–3316. 10.1093/bioinformatics/btr575 [DOI] [PubMed] [Google Scholar]