ABSTRACT
Infections with high-risk human papillomaviruses (hrHPV) contribute to cervical carcinoma. The cdk inhibitor and tumor suppressor p16INK4A is consistently upregulated in cervical carcinoma cells for reasons that are poorly understood. We report here that downregulation of p16INK4A gene expression in three different cervical carcinoma cell lines reduced expression of the E7 oncogene, suggesting a positive feedback loop involving E7 and p16INK4A. p16INK4A depletion induced cellular senescence in HeLa but not CaSki and MS-751 cervical carcinoma cells.
IMPORTANCE This study demonstrates that the cdk inhibitor p16INK4A, frequently used as surrogate marker for transforming infections by human papillomaviruses of the high-risk group, is required for high-level expression of the E7 oncoproteins of HPV-16, HPV-18, and HPV-45 in cervical carcinoma cells. It is also demonstrated that depletion of p16INK4A induces senescence in HeLa but not CaSki or MS-751 cervical carcinoma cells.
INTRODUCTION
Persistent infections by human papillomaviruses of the high-risk type (hrHPV) are the main etiologic factor for cervical cancer (1). Infections by hrHPV have been detected in virtually all cervical cancers (2), with HPV-16 and HPV-18 being the most prevalent genotypes worldwide in cervical cancer (3). Initial events of cervical carcinogenesis after viral infection by hrHPV types are specific changes that overcome the transcriptional control of viral gene expression in the infected keratinocytes (4). Inactivation of these cellular control functions permits deregulated transcription of the early viral genes E6 and E7, consistent with an increase of the high-risk E7 protein levels during early steps of carcinogenesis in cells of the cervical squamous epithelium (5). High-risk E7, in cooperation with high-risk E6, can efficiently immortalize human primary keratinocytes (6, 7), and the consistent overexpression of these two oncogenes is required to induce and to maintain the transformed phenotype of cervical cancer cells (8); accordingly, silencing E6/E7 gene expression in cervical carcinoma cells induced senescence (9), which was essentially driven by the loss of E7 effects on the pRb pathway (10, 11).
Immortalization of keratinocytes by the E7 oncoprotein involves its ability to bind and thereby functionally inactivate cell cycle regulatory proteins such as the retinoblastoma tumor suppressor protein pRb (5, 12). E7 proteins trigger the release of E2F from pRb, which leads to continuous activation of the cell cycle. Physiologically, E2F activation is mediated by phosphorylation of the Rb protein (13). This pathway is strictly regulated by a set of cyclin-dependent kinase inhibitors, among them p16INK4A, which block cyclin-dependent kinases (cdks) phosphorylating pRb. In cells with hrHPV infections, the regulation of the pRb-E2F pathway is disturbed by E7, and therefore the activation of p16INK4A has no downstream effect. Instead, p16INK4A is strongly overexpressed and accumulates in the cells (14, 15). p16INK4A overexpression has been shown in the vast majority of cervical precancers and cancers, whereas p16INK4A expression is low in normal cervical tissue (15, 16). Therefore, p16INK4A is considered a surrogate marker for persistent hrHPV infection and has been introduced as a diagnostic marker for cervical cancer and precancer (15). However, it is unclear if p16INK4A carries out any relevant function in the context of a persistent hrHPV infection. Whereas current hypotheses state that p16INK4A is highly expressed in cervical carcinoma cells because it is well tolerated in cells with a compromised Rb pathway (reviewed in reference 17), it is unclear whether p16INK4A carries out any relevant function in the cervical carcinoma cells.
MATERIALS AND METHODS
Cell culture.
HeLa, CaSki, MS-751, and U2-OS cells (all cell lines obtained from ATCC, Manassas, VA, USA) were grown in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf serum (FCS; Biochrom AG), 4 mM l-glutamine (Invitrogen), and 1% penicillin streptomycin (Invitrogen). All cells were grown in an atmosphere of 5% CO2 at 37°C and were subcultured by trypsinization with 0.05% trypsin–EDTA (Invitrogen).
Calculation of proliferation rate.
Cumulative population doublings (cPDL) were calculated using the equation cPDL = (log A − log B)/0.301, where A is the number of cells at the end of one passage and B is the number of cells that were seeded at the beginning of one passage.
SA-β-gal staining.
Senescence-associated-β-galactosidase (SA-β-gal) staining was used to determine the senescence status of the cells. To stain for SA-β-gal, cells were grown on 6-well plates and washed three times with phosphate-buffered saline (PBS; Sigma Aldrich, Germany). Afterwards, the cells were fixed with 2% formaldehyde and 0.4% glutaraldehyde in PBS for 5 min at room temperature. Cells were washed three times with PBS and prepared for staining as described previously (18). Cells were covered with staining solution (150 mM NaCl, 2 mM MgCl, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 40 mM citric acid, 12 mM sodium phosphate [pH 6.0], with 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside [X-Gal] added directly before use) and incubated for 24 h at 37°C without light exposure. The reaction was stopped by washing off the staining solution with PBS. Cells were covered with PBS, and blue staining, indicating the presence of SA-β-gal, could be detected under the microscope. To calculate the percentage of SA-β-gal-positive cells, stained cells were counted and compared to the total cell number.
Transfection.
When cells reached 80 to 90% confluence, they were transfected in 6-well plates with oligonucleotides (at a final concentration of 100 to 200 pmol per well) targeting p16INK4A (siGENOME SMARTpool CDKN2a; Thermo Fisher Scientific Inc.) or nontargeting control oligonucleotides (siGENOME ON-TARGET plus; Thermo Fisher Scientific Inc.), using Lipofectamine 2000 (Invitrogen). The transfection procedure was repeated three times at different time points, depending on the proliferation rate, to ensure sustained downregulation of p16INK4A expression. HeLa cells were also stably transfected with a plasmid (pLXSN empty or pLXSN-HPV-18 E6/E7) carrying the HPV-18 E6/E7 oncogenes under the control of the Moloney murine leukemia virus promoter. U2-OS cells were transfected with cytomegalovirus (CMV)-driven expression vectors expressing either p16INK4A or the E7 gene of HPV-16, -18, or -45. p16INK4A small interfering RNAs (siRNAs) or nontargeting control oligonucleotides were cotransfected.
RNA isolation.
Total RNA was isolated with the RNeasy Minikit (Qiagen) according to the manufacturer's protocol. RNA was eluted using 30 μl of RNase-free water. The RNA concentration was quantified by photometric measurement at 260 nm and 280 nm.
Quantitative real-time PCR (qRT-PCR) analysis of mRNAs.
RNA was isolated using the RNeasy Minikit (Qiagen) and quantified as described above. For cDNA synthesis, 1 μg of total RNA was reverse transcribed with the Transcriptor first-strand cDNA synthesis kit (Roche Applied Science) and diluted 1:4. Amplification of bicistronic HPV-16, -18, and -45 E6/E7 mRNAs from HeLa, Caski, and MS-751 cells was carried out with the following primers: HPV-16E6E7 forward, 5′-GTTACTGCGACGTGAGGTATATG-3′; HPV-16E6E7 reverse, 5′-CATTTATCACATACAGCTATGGATTC-3′; HPV-18E6E7 forward, 5′-CAGAGGTATTTGAATTTGCATTT-3′; HPV-18E6E7 reverse, 5′-AATCTATACATTTATGGCATGCAG-3′; HPV-45E7 forward, 5′-TTGCATTTGGAACCTCAGAA-3′; and HPV-45E7 reverse, 5′-TTTCCTCCTCTGACTCGCTT-3′. Amplification of HPV-16, -18, and -45 E7 mRNAs expressed in U2-OS cells was carried out with the following primers: HPV-16E7 forward, 5′-GGAGATACACCTACATTGCATGA-3′; HPV-16E7 reverse, 5′-CATCCTCCTCCTCTGAGCTG-3′; HPV-18E7 forward, 5′-ATGAAATTCCGGTTGACCTT-3′; HPV-18E7 reverse, 5′-GGGCTGGTAAATGTTGATGA-3′; HPV-45E7 forward, 5′-TTGCATTTGGAACCTCAGAA-3′; and HPV-45E7 reverse, 5′-TTTCCTCCTCTGACTCGCTT-3′. Amplification of p16INK4A, p21WAF-1/Cip1, and beta-2-microglobulin (B2M) was carried out with the following primers: CDKN2B (p16INK4A) forward, 5′-GATGAGGACAATGAGGCAAAG-3′; CDKN2B (p16INK4A) reverse, 5′-TGGGAAGAAAAGCAAGACAAC-3′; B2M forward, 5′-GAATTCACCCCCACTGAAAA-3′; and B2M reverse, 5-CTCCATGATGCTGCTTACA-3′).
Protein isolation.
For Western blot samples, whole-cell extracts were prepared from osteosarcoma cells (U2-OS), epithelial cervical cancer cells (HeLa, CaSki, and MS-751), and human diploid fibroblasts (HDF), washed twice with cold PBS, and scraped off on ice in NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% [vol/vol] NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (Complete, EDTA free; Roche Diagnostics, Germany). Cells were deep-frozen in liquid nitrogen and thawed three times and further kept on ice for 30 min. After centrifugation at 20,000 × g for 10 min at 4°C, the supernatant was used to determine the protein concentration using the bicinchoninic acid (BCA) protein assay (Pierce Thermo Scientific).
Western blotting.
Equal amounts of protein were subjected to SDS-PAGE (12.5% acrylamide) and blotted onto a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked with 5% skim milk in PBS–0.1% Tween 20 and incubated overnight with the following primary antibodies: rabbit polyclonal antiserum to HPV-16 E7 (5), monoclonal rabbit anti-HPV-18E7/anti-HPV-45E7 antibodies (clone 143-7 [19]), anti-human p16INK4A antibodies (BD Pharmingen), mouse monoclonal anti-p21WAF1 (556430; BD Pharmingen), mouse monoclonal anti-p53 (sc-126; Santa Cruz), and antitubulin antibodies (Sigma, MO, USA). Proteins of interest were detected after incubation with horseradish peroxidase-conjugated secondary antibodies (Dako Cytomation, Germany) and visualized with ECL Prime Western blotting detection reagent (GE Healthcare, United Kingdom).
Detection of apoptotic cell death.
HeLa cells were transfected with control or p16INK4A siRNA and subsequently washed with PBS, trypsinized, and counted by using a Casy cell counter. Cells (1 × 105) were aliquoted in fluorescence-activated cell sorter (FACS) tubes. Cells were centrifuged for 5 min at 300 × g, washed twice with cold PBS and centrifuged each time. Cell pellets were resuspended in 100 μl 1× binding buffer, 3 μl of annexin V-fluorescein isothiocyanate (FITC) was added, and cells were incubated for 15 min at 25°C. After the incubation, 400 μl of 1× binding buffer was added to each sample, and cells were kept on ice and subsequently analyzed by FACS. The effects of control or p16INK4A siRNA on effector caspase-3 and -7 activities in HeLa cells were detected using the Caspase-Glo 3/7 assay (Promega, Germany). HeLa cells (15,000) were seeded in a 96-well plate in 100 μl DMEM supplemented with 10% heat-inactivated fetal calf serum (FCS). As a positive control, HeLa cells transfected with control siRNA were treated with 1 μM staurosporine for 4.5 h at 37°C. After incubation, 100 μl of caspase-3/7 reagent was added to each well, mixed, and incubated for 1 h at room temperature. Luminescence was measured using the multilabel reader Victor X5 (Perkin-Elmer). Caspase activity was expressed as relative light units (RLU) (counts per second).
RESULTS AND DISCUSSION
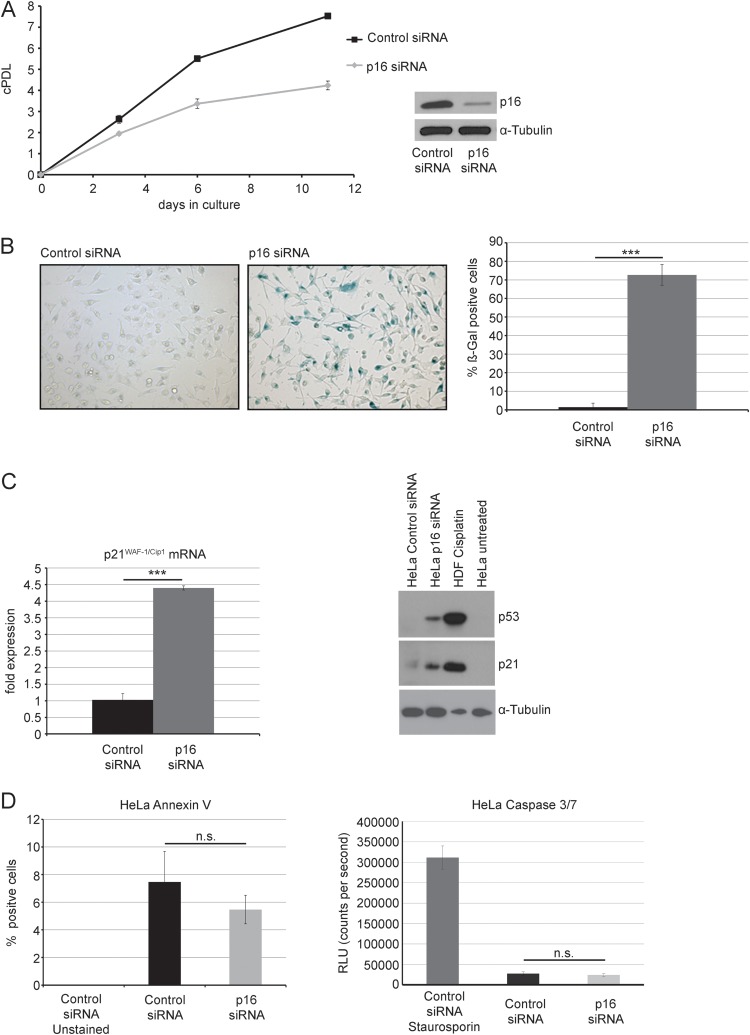
To assess a potential functional role of p16INK4A in hrHPV-positive cervical carcinoma cells, we downregulated p16INK4A by siRNA in HPV-18-positive HeLa cervical carcinoma cells. HeLa cells were transfected in 6-well plates with oligonucleotides targeting p16INK4A or with nontargeting control oligonucleotides. The transfection procedure was repeated after 72 h and after 7 days, to ensure sustained downregulation of p16INK4A expression (Fig. 1A). Seven days after the first transfection, we noted a significant reduction of cell proliferation (Fig. 1A). On day 12, cells were stained in situ for senescence-associated β-galactosidase (SA-β-gal) activity, as described previously (18). Upon p16INK4A depletion, the proportion of SA-β-gal-positive cells was significantly increased, suggesting the appearance of cellular senescence (Fig. 1B). In p16INK4A-depleted HeLa cells, we also observed a significant upregulation of the p53 protein level (Fig. 1C). Accordingly, a strong upregulation in the expression of the cdk inhibitor p21WAF-1/Cip-1, a known downstream target of p53 (20), was observed at both the mRNA and protein levels (Fig. 1C). In contrast, depletion of p16INK4A did not significantly affect the proportion of apoptotic cells, which remained below 10% in all cases (Fig. 1D).
FIG 1.
Knockdown of p16INK4A induces senescence of HeLa cells. (A) HeLa cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 4, and day 7). p16INK4A knockdown was verified by Western blotting (inset). Cells were counted, and a growth curve was established. Shown are the results (mean ± standard deviation [SD]) from three independent experiments. cPDL, cumulated population doublings. (B) At day 12, cells were assessed for presence of SA-β-gal activity. Bars represent the relative percentage of SA-β-gal-positive cells (mean ± SD) observed within three visual fields from three independent experiments. Two representative micrographs are also shown. (C) HeLa cells were treated as described for panel A and harvested at day 10 posttransfection. Left panel, extraction of total RNA was followed by subsequent cDNA synthesis, and mRNA levels of B2M and p21WAF-1/Cip-1 were assessed by qRT-PCR. p21WAF-1/Cip-1 mRNA levels were normalized to B2M mRNA. Shown are the results (mean ± SD) of three independent experiments. Right panel, cell extracts were subjected to SDS-PAGE and analyzed by Western blotting using antibodies for the detection of p21WAF-1/Cip-1, p53, and tubulin, as indicated. Extracts of cisplatin-treated human fibroblasts were added as a positive control for the expression of p21WAF-1/Cip-1 and p53; as a further control, extracts of untreated HeLa cells were used. Three independent experiments were performed, and results of one representative experiment are shown. (D) Left panel, HeLa cells were treated as for panel A, stained with annexin V, and analyzed by flow cytometry, and the relative percentage of annexin V-positive cells was determined. Shown are the results (mean ± SD) of three independent experiments. Right panel, HeLa cells were treated as for panel A, and caspase-3/7 activity was determined and displayed as relative luminescence units (RLU). Staurosporine-treated cells were used as a positive control for the assay. Shown are the results (mean ± SD) of three independent experiments. n.s., not significant.
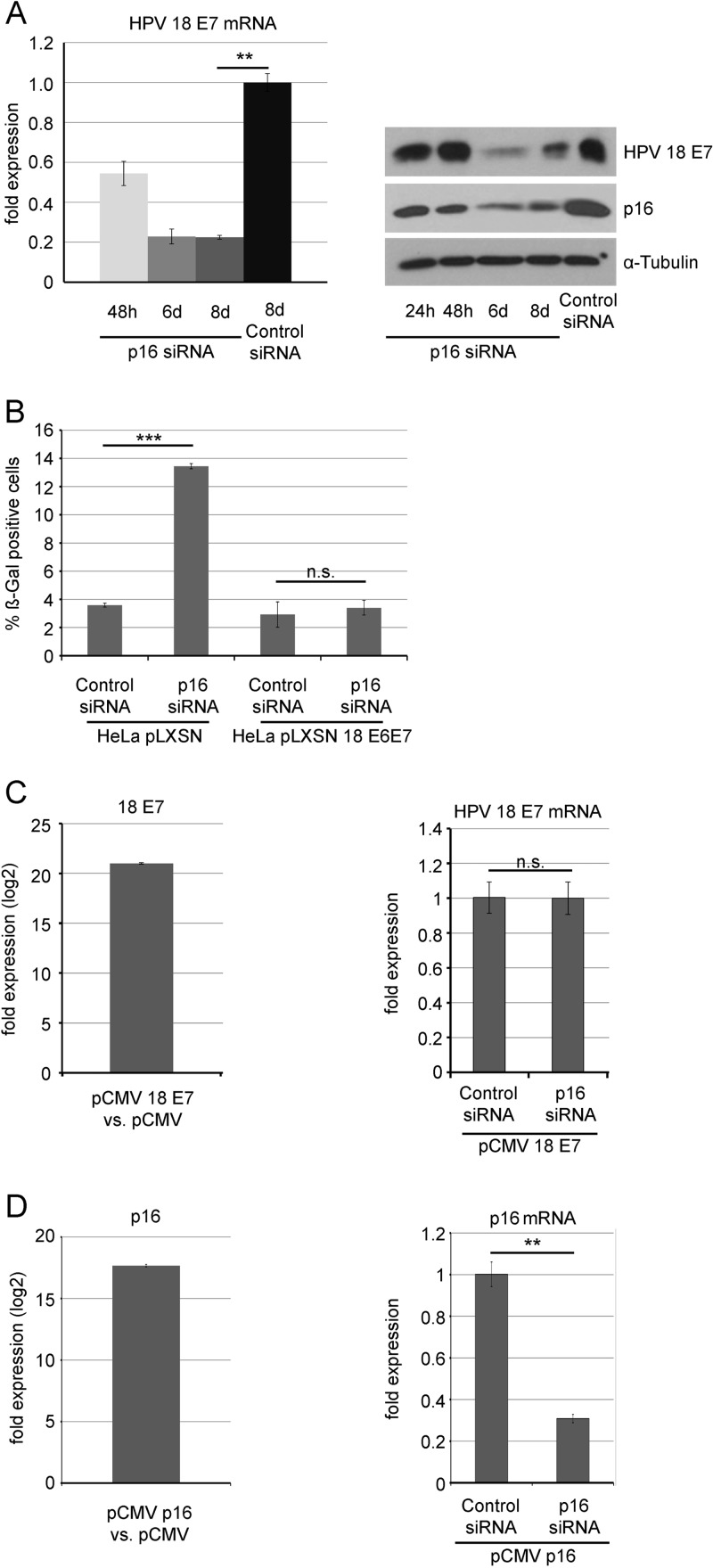
To address the reason for senescence-associated growth arrest observed in p16INK4A-depleted HeLa cells, we analyzed the expression of HPV-18 oncogenes. In HeLa cells, HPV-18 DNA is integrated and the bicistronic E6/E7 open reading frame is transcribed from the viral p97 promoter. The E6/E7 transcript is differentially spliced (E6/E7 and E6*/E7), leading to E7 mRNA as the major transcript (21, 22). In fact, over 97% of the total spliced transcript is E6*/E7, which encodes the E7 oncoprotein (23). Using qRT-PCR, we found that the level of the E7 transcript was significantly reduced 48 h after transfection and was further reduced (roughly 5-fold) at day 8 (Fig. 2A).
FIG 2.
Knockdown of p16INK4A leads to reduced expression of the E7 oncogene in HeLa cells (A) Left panel, HeLa cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 4, and day 7) and harvested at different time points after initial transfection (24 h, 6 days, and 8 days). Extraction of total RNA was followed by subsequent cDNA synthesis, and HPV-18 E7 mRNA levels were assessed by qRT-PCR. Shown are the results (mean ± SD) of three independent experiments. Right panel, HeLa cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 4, and day 7) and harvested at different time points after initial transfection (24 h, 48 h, 6 days, and 8 days). Cell extracts were subjected to SDS-PAGE and analyzed by Western blotting using antibodies for the detection of p16INK4A, HPV-18 E7 protein, and alpha-tubulin, as indicated. Three independent experiments were performed, and results from one representative experiment are shown. (B) HeLa cells were stably transfected with pLSXN and pLSXN-HPV-18 E6/E7, as indicated. p16INK4A was silenced by siRNA as for panel A, and the percentage of senescent cells was determined after SA-β-gal staining. Shown are the results (mean ± SD) of three independent experiments. (C) Left panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-18 E7 or the empty vector pCMV. HPV-18 E7 mRNA was analyzed by qRT-PCR in both cases; the fold difference of expression is shown on a logarithmic scale. Right panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-18 E7 and cotransfected by p16INK4A siRNA or control siRNA, as indicated. HPV-18 E7 mRNA was analyzed by qRT-PCR in both cases; shown are the results (mean ± SD) of three independent experiments. (D) Left panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for p16INK4A or the empty vector pCMV. p16INK4A mRNA was analyzed by qRT-PCR in both cases; the fold difference of expression is shown on a logarithmic scale. Right panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for p16INK4A and cotransfected by p16INK4A siRNA or control siRNA, as indicated. p16INK4A mRNA was analyzed by qRT-PCR in both cases; shown are the results (mean ± SD) of three independent experiments.
HPV-18 E7 oncoprotein expression was determined by Western blotting, using monoclonal rabbit anti-HPV-18 E7 antibodies (clone 143-7) (19). These experiments revealed that depletion of p16INK4A significantly decreased the level of HPV-18 E7 protein in HeLa cells (Fig. 2A). Together, these data suggest that p16INK4A is required to maintain E7 oncogene expression in cervical carcinoma cells. When HeLa cells were stably transfected with a plasmid carrying the HPV-18 E6/E7 oncogenes under the control of the Moloney murine leukemia virus promoter, depletion of p16INK4A did not elicit a senescence response in HeLa cells (Fig. 2B), suggesting that reduced expression of the E6/E7 oncogenes is the cause for senescence induced by p16 depletion in HeLa cells. To control for potential direct off-target effects of p16INK4A siRNAs on E7 transcripts, U2-OS cells were transfected with CMV-driven expression vectors expressing either the E7 gene of HPV-18 (Fig. 2C) or p16INK4A (Fig. 2D), which yielded a high level of the respective mRNAs, as shown by qRT-PCR. When p16INK4A siRNAs were cotransfected, this led to a considerable reduction of p16INK4A mRNA, whereas the E7 mRNA level remained unchanged (Fig. 2C and D), ruling out direct off-target effects of p16INK4A siRNAs on E7 transcripts.
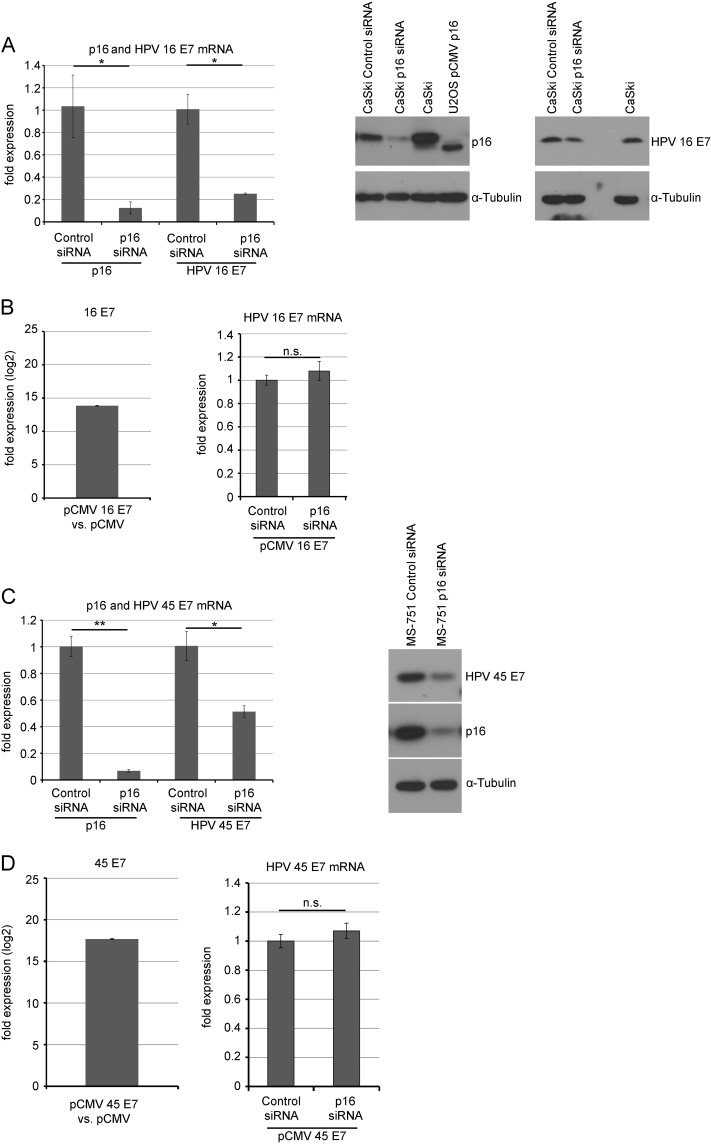
When p16INK4A was depleted by siRNA in CaSki cells and MS-751 cells, established cervical carcinoma cells harboring the genomes of HPV-16 and HPV-45, respectively, the mRNA and protein (Fig. 3A and C) levels of the respective E7 oncoprotein were significantly reduced, indicating that p16INK4A is required for high-level expression of the E7 oncogenes of HPV-16 and HPV-45 as well. As shown for HPV-18 E7 before, ectopic expression of the E7 genes of HPV-16 and HPV-45 from a CMV promoter in U2-OS cells was insensitive to cotransfection of p16INK4A siRNA (Fig. 3B and D), indicating that downregulation of E7 gene expression in cervical cancer cells is not due to off-target effects of p16INK4A siRNAs on E7 transcripts.
FIG 3.
p16INK4A knockdown reduces E7 levels in CaSki and MS-751 cells. (A) CaSki cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day, 3 and day 6). Left panel, at day 8, RNA was prepared and the mRNA levels for p16INK4A and HPV-16 E7 were analyzed by qRT-PCR. Shown are the results (mean ± SD) of three independent experiments. Right panel, at day 8, cells were lysed and protein levels of p16INK4Aand HPV-16 E7 were analyzed by Western blotting. One representative Western blot is shown. (B) Left panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-16 E7 or the empty vector pCMV. HPV-16 E7 mRNA was analyzed by qRT-PCR in both cases; the fold difference of expression is shown on a logarithmic scale. Right panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-16 E7 and cotransfected by p16INK4A siRNA or control siRNA, as indicated. HPV-16 E7 mRNA was analyzed by qRT-PCR in both cases; shown are the results (mean ± SD) of three independent experiments. (C) MS-751 cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 4, and day 8). Left panel, at day 8 RNA was prepared and the mRNA levels for p16INK4A and HPV-45 E7 were analyzed by qRT-PCR. Shown are the results (mean ± SD) of three independent experiments. Right panel, at day 10, cells were lysed and protein levels of p16INK4A and HPV-45 E7 were analyzed by Western blotting. One representative Western blot is shown. (D) Left panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-45 E7 or the empty vector pCMV. HPV-45 E7 mRNA was analyzed by qRT-PCR in both cases; the fold difference of expression is shown on a logarithmic scale. Right panel, U2-OS cells were transiently transfected with a CMV-driven expression vector for HPV-45 E7 and cotransfected by p16INK4A siRNA or control siRNA, as indicated. HPV-45 E7 mRNA was analyzed by qRT-PCR in both cases; Shown are the results (mean ± SD) of three independent experiments.
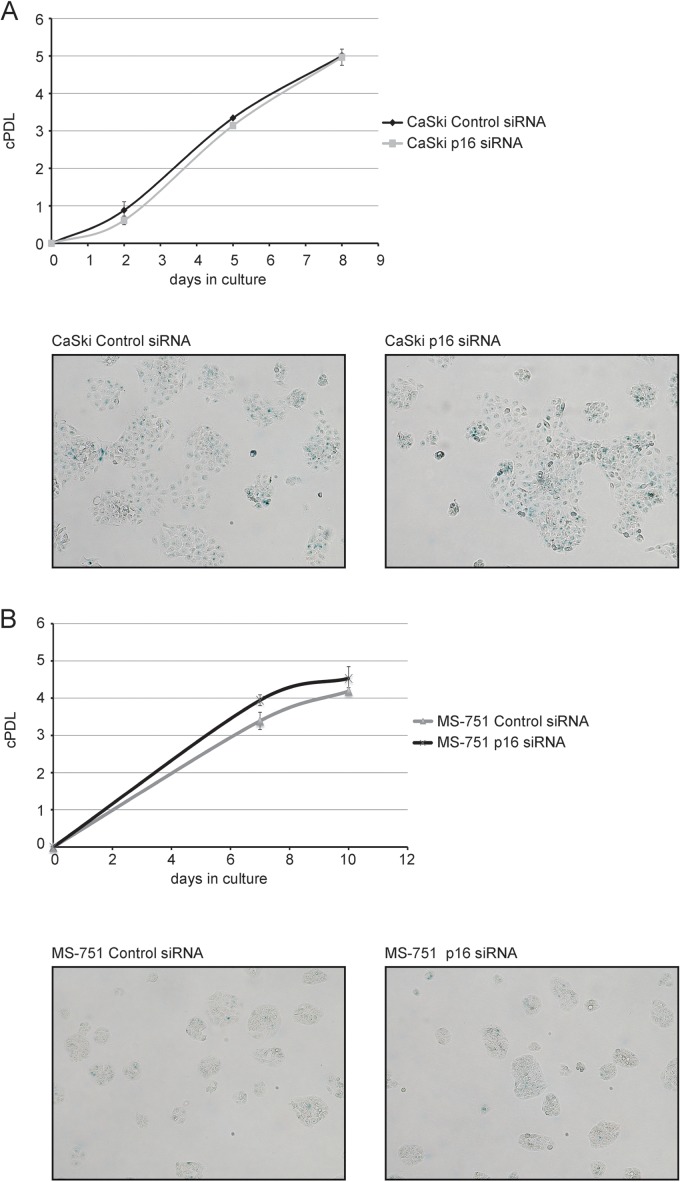
We found that, in contrast to our observations with p16INK4A-depleted HeLa cells, the rate of cell proliferation was not reduced in both CaSki cells and MS-751 cells (Fig. 4), and p16INK4A depletion did not induce a senescence response in these cell types (Fig. 4). We conclude that, whereas downregulation of E7 gene and protein expression reflects a conserved mechanism by which cervical carcinoma cells respond to p16INK4A depletion. The effect of reduced E7 protein levels on the cellular phenotype seems to vary between cell lines.
FIG 4.
p16INK4A knockdown does not affect cell proliferation in CaSki and MS-751 cells. (A) CaSki cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 3, and day 6). Upper panel, cells were counted, and a growth curve was established. Shown are the results (mean ± SD) of three independent experiments. Lower panel, at day 9, cells were assessed for presence of SA-β-gal activity. Two representative micrographs are shown. (B) MS-751 cells were transfected with p16INK4A siRNA or control siRNA (200 pmol per well) for three times in total (day 1, day 4, and day 8). Upper panel, cells were counted, and a growth curve was established. Shown are the results (mean ± SD) of three independent experiments. Lower panel, at day 11, cells were assessed for presence of SA-β-gal activity. Two representative micrographs are shown.
We hypothesize that the threshold level of E7 protein required for cell proliferation may be different for different cervical carcinoma cell lines; it is also possible that CaSki and MS-751 cells have acquired mutations and/or epigenetic alterations that loosen their dependence on high levels of the respective E7 oncoprotein. Others have shown that HPV-16 E7 triggers p16INK4A expression through a mechanism involving the histone demethylase KDM6B and epigenetic reprogramming. They also reported that KDM6B is required for the viability of CaSki cells and hypothesized that E7-expressing cervical carcinoma cells are “addicted” to KDM6B activity (24). The same group recently reported that indeed p16INK4A is necessary for the survival of several cervical carcinoma cell lines and that this is due to the ability of p16INK4A to inhibit cdk4, probably preventing the phosphorylation of so-far-unidentified cdk4 substrates. Of interest, this mechanism was not operative in HeLa cells, the viability of which was not reduced upon p16INK4A depletion (25).
Whereas the existing data suggest that p16INK4A may favor proliferation and survival through more than one molecular mechanism, e.g., inhibition of cdk4 activity versus regulation of E7 expression, future studies are warranted to more precisely define potential interactions between the two pathways. In our view, the most likely scenario is the existence of a positive feedback loop where E7 triggers expression of p16INK4A and p16INK4A in turn helps to maintain sufficiently high expression of E7. Finally, it is interesting to note that p16INK4A, which has been considered a surrogate marker for persistent hrHPV infection (26), now turns out to contribute in several ways to survival and proliferation of cervical carcinoma cells. This observation supports the use of p16INK4A as a functional marker of transforming infection with high-risk human papillomaviruses.
ACKNOWLEDGMENT
This work was supported by a grant from the Austrian Science Funds (FWF P21853) to P.J.-D.
Footnotes
Published ahead of print 5 March 2014
REFERENCES
- 1.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19. [DOI] [PubMed] [Google Scholar]
- 3.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527. 10.1056/NEJMoa021641 [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. 1999. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin. Cancer Biol. 9:405–411. 10.1006/scbi.1999.0144 [DOI] [PubMed] [Google Scholar]
- 5.Fiedler M, Muller-Holzner E, Viertler HP, Widschwendter A, Laich A, Pfister G, Spoden GA, Jansen-Durr P, Zwerschke W. 2004. High level HPV-16 E7 oncoprotein expression correlates with reduced pRb-levels in cervical biopsies. FASEB J. 18:1120–1122. 10.1096/fj.03-1332fje [DOI] [PubMed] [Google Scholar]
- 6.Hudson JB, Bedell MA, McCance DJ, Laiminis LA. 1990. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J. Virol. 64:519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Knebel Doeberitz M, Rittmuller C, Aengeneyndt F, Jansen-Durr P, Spitkovsky D. 1994. Reversible repression of papillomavirus oncogene expression in cervical carcinoma cells: consequences for the phenotype and E6-p53 and E7-pRB interactions. J. Virol. 68:2811–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 97:10978–10983. 10.1073/pnas.97.20.10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFilippis RA, Goodwin EC, Wu L, DiMaio D. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 77:1551–1563. 10.1128/JVI.77.2.1551-1563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johung K, Goodwin EC, DiMaio D. 2007. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 81:2102–2116. 10.1128/JVI.02348-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson N, Howley PM, Munger K, Harlow E. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937. 10.1126/science.2537532 [DOI] [PubMed] [Google Scholar]
- 13.DeGregori J. 2004. The Rb network. J. Cell Sci. 117:3411–3413. 10.1242/jcs.01189 [DOI] [PubMed] [Google Scholar]
- 14.Khleif SN, DeGregori J, Yee CL, Otterson GA, Kaye FJ, Nevins JR, Howley PM. 1996. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc. Natl. Acad. Sci. U. S. A. 93:4350–4354. 10.1073/pnas.93.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. 1998. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am. J. Pathol. 153:1741–1748. 10.1016/S0002-9440(10)65689-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Keyserling H, Kuhn W, Schneider A, Bergmann T, Kaufmann AM. 2012. p16INK(4)a and p14ARF mRNA expression in Pap smears is age-related. Mod Pathol. 25:465–470. 10.1038/modpathol.2011.179 [DOI] [PubMed] [Google Scholar]
- 17.Cuschieri K, Wentzensen N. 2008. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol. Biomarkers Prev. 17:2536–2545. 10.1158/1055-9965.EPI-08-0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unterluggauer H, Hampel B, Zwerschke W, Jansen-Durr P. 2003. Senescence-associated cell death of human endothelial cells: the role of oxidative stress. Exp. Gerontol. 38:1149–1160. 10.1016/j.exger.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 19.Ehehalt D, Lener B, Pircher H, Dreier K, Pfister H, Kaufmann AM, Frangini S, Ressler S, Muller-Holzner E, Schmitt M, Hofler D, Rostek U, Kaiser A, Widschwendter A, Zwerschke W, Jansen-Durr P. 2012. Detection of human papillomavirus type 18 E7 oncoprotein in cervical smears: a feasibility study. J. Clin. Microbiol. 50:246–257. 10.1128/JCM.01108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. 10.1016/0092-8674(93)90500-P [DOI] [PubMed] [Google Scholar]
- 21.Schneider-Gadicke A, Schwarz E. 1986. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 5:2285–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smotkin D, Wettstein FO. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. U. S. A. 83:4680–4684. 10.1073/pnas.83.13.4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu EM, McNicol PJ. 1992. Characterization of HPV-16 E6/E7 transcription in CaSki cells by quantitative PCR. Mol. Cell. Probes 6:459–466. 10.1016/0890-8508(92)90042-V [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin-Drubin ME, Crum CP, Munger K. 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. U. S. A. 108:2130–2135. 10.1073/pnas.1009933108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaughlin-Drubin ME, Park D, Munger K. 2013. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc. Natl. Acad. Sci. U. S. A. 110:16175–16180. 10.1073/pnas.1310432110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Knebel Doeberitz M, Reuschenbach M, Schmidt D, Bergeron C. 2012. Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev. Proteomics 9:149–163. 10.1586/epr.12.13 [DOI] [PubMed] [Google Scholar]