Abstract
The outgrowth of Porphyromonas gingivalis within the inflammatory subgingival plaque is associated with periodontitis characterized by periodontal tissue destruction, loss of alveolar bone, periodontal pocket formation, and eventually, tooth loss. Potential virulence factors of P. gingivalis are peptidylarginine deiminase (PPAD), an enzyme modifying free or peptide-bound arginine to citrulline, and the bacterial proteases referred to as gingipains (Rgp and Kgp). Chemokines attract leukocytes during inflammation. However, posttranslational modification (PTM) of chemokines by proteases or human peptidylarginine deiminases may alter their biological activities. Since chemokine processing may be important in microbial defense mechanisms, we investigated whether PTM of chemokines by P. gingivalis enzymes occurs. Upon incubation of interleukin-8 (IL-8; CXCL8) with PPAD, only minor enzymatic citrullination was detected. In contrast, Rgp rapidly cleaved CXCL8 in vitro. Subsequently, different P. gingivalis strains were incubated with the chemokine CXCL8 or CXCL10 and their PTMs were investigated. No significant CXCL8 citrullination was detected for the tested strains. Interestingly, although considerable differences in the efficiency of CXCL8 degradation were observed with full cultures of various strains, similar rates of chemokine proteolysis were exerted by cell-free culture supernatants. Sequencing of CXCL8 incubated with supernatant or bacteria showed that CXCL8 is processed into its more potent forms consisting of amino acids 6 to 77 and amino acids 9 to 77 (the 6-77 and 9-77 forms, respectively). In contrast, CXCL10 was entirely and rapidly degraded by P. gingivalis, with no transient chemokine forms being observed. In conclusion, this study demonstrates PTM of CXCL8 and CXCL10 by gingipains of P. gingivalis and that strain differences may particularly affect the activity of these bacterial membrane-associated proteases.
INTRODUCTION
Cytokine and chemokine activity is regulated at multiple levels, including posttranslational modification (PTM) (1–3). Reduced or enhanced receptor affinity/specificity and chemokine activity have been reported, depending on the chemokine and on the type of PTM (4, 5). Most PTMs on inflammatory chemokine ligands depend on proteolytic cleavage, with highly specific proteases mainly affecting the NH2-terminal region of the protein (2, 5–7). Many metalloproteases, such as aminopeptidase N/CD13 and various matrix metalloproteases, and a number of serine proteases, including thrombin, plasmin, cathepsin G, and the dipeptidylpeptidase CD26, were reported to cleave specific chemokines in the NH2-terminal region. The biological effect of such proteolytic processing varies depending on the chemokine and protease involved. First, limited NH2-terminal proteolytic processing of chemokines by proteases can result in enhanced biological activity, e.g., CXCL8 processing by thrombin (8, 9) or matrix metalloproteinase 9 (MMP-9)/gelatinase B (10) and CCL3L1 processing by DPPIV/CD26 (11), or decreased biological activity, e.g., CCL7 processing by MMP-2/gelatinase A (12) and CXCL10 processing by CD26 (13). For some chemokines, like CXCL7 and CCL14, more extensive NH2-terminal truncation is even mandatory to obtain receptor signaling and chemotactic properties (14–16). A detailed description of chemokine-protease interactions and the consequences for the biological functions of chemokines has been published in a number of recent reviews (2, 5, 6, 17). Also, naturally occurring N- and O-glycosylated forms of chemokines with altered lectin binding properties have been identified (18–20). In addition to NH2- and COOH-terminal proteolytic processing and glycosylation, citrullination, i.e., deimination of arginine (Arg) to citrulline (Cit), is a recently discovered PTM on the natural chemokines interleukin-8 (IL-8; CXCL8) and gamma interferon-inducible protein 10 (IP-10; CXCL10) (8, 13, 21–24). CXCL8, a ligand for CXC chemokine receptors CXCR1 and CXCR2, is known to be the major human neutrophil chemoattractant. The CXCR3 ligand CXCL10 predominantly chemoattracts activated lymphocytes. For both chemokines, Arg at position 5 (Arg5) was converted into Cit. The mammalian enzymes responsible for the conversion are peptidylarginine deiminases (PADs) (25). Chemokines were the first identified PAD substrates that signal in a receptor-dependent manner. Most other PAD substrates are structural proteins (e.g., keratin and fillagrin) (26–31). Citrullination may considerably influence the biological activity of proteins, since it may change ionic interactions in macromolecules, resulting in altered protein folding, and may cause altered binding of proteins to other macromolecules, such as DNA, glycosaminoglycans, and other proteins (28, 32).
Currently available in vitro and in vivo data are in line with an anti-inflammatory role for PADs in local acute inflammation by citrullinating and thereby inactivating chemokines, such as CXCL5, CXCL8, CXCL10, CXCL11, and CXCL12, and, consequently, dampening leukocyte migration (33). In addition, chemokine citrullination may be important, in particular, in microbial defense mechanisms, since peptidylarginine deiminating activity has been reported in one prokaryotic organism, Porphyromonas gingivalis (25, 34). It is well-known that an outgrowth of P. gingivalis within the periodontal pocket is associated with periodontitis, an inflammatory disorder characterized by periodontal tissue destruction, loss of alveolar bone, and, eventually, tooth loss. In contrast to other mammalian PADs, PAD of P. gingivalis (PPAD) preferentially citrullinates C-terminal arginine residues and is also able to convert free Arg into Cit. In contrast to mammalian enzymes, PPAD deiminates peptidylarginine residues in a calcium-independent manner (25, 34, 35). Its ammonia-producing properties are well studied as a response to acidic cleansing cycles in the mouth (34, 36, 37). Though the substrates for the action of PPAD are unidentified and the exact role of PPAD in assisting the bacterium in circumventing the host immune defense is unknown, PPAD has been suggested to function as an additional virulence factor (25, 34). In the context of an innate immune response, chemokine citrullination by PPAD may establish a negative feedback on local leukocyte-mediated inflammation and, hence, bacterial clearance. Together with inflammation-associated and PPAD-exerted citrullination of host proteins, this may contribute to the breakdown of immunotolerance to citrullinated epitopes and the eventual development of rheumatoid arthritis (RA) (38).
Gingipains, including arginine-specific gingipains (RgpA and RgpB) and the lysine-specific gingipain (Kgp), are considered to be essential virulence factors of P. gingivalis (39). These proteinases are either cell surface associated or secreted. Gingipains play an important role in the evasion and dysregulation of the host's immune response by the degradation of proinflammatory cytokines, complement factors, antimicrobial peptides, and immunoglobulins at the site of infection (39–41). Since both PPAD and gingipains were found to significantly contribute to P. gingivalis virulence, we investigated whether PTM of chemokines by these enzymes represents a way by which P. gingivalis regulates the inflammatory response.
MATERIALS AND METHODS
Reagents and materials.
The 77 amino acid forms of the chemokines CXCL8, CXCL10, and macrophage inflammatory protein 1α (MIP-1α; CCL3) were obtained from PeproTech (Rocky Hill, NJ).
N-Acetylo-l-Arg, l-Cit, 2,3-butanedione monoxime, thiosemicarbazide, antipyrine, and 2,3-butanedione (>99.4% GC content) were purchased from Sigma-Aldrich (St. Louis, MO).
RgpB was purified as previously described (42, 43). To obtain PPAD, P. gingivalis W83 was engineered to secrete a soluble form of PPAD with a C-terminal hexahistidine affinity tag using the same molecular strategy reported previously for the RgpB protease in the mutant 662i6H (44). Subsequently, PPAD was separated from the culture medium via ion-exchange and gel filtration chromatography, as described for the purification of soluble Kgp from the growth medium (45). Briefly, bacteria were removed by centrifugation, and proteins in the medium were precipitated with acetone, resuspended in phosphate buffer (pH 6.5), dialyzed, and passed through a DE-52 column (Whatman, GE Healthcare, Pittsburgh, PA) to remove excess hemin. The flowthrough was dialyzed against 50 mM Tris-HCl, 0.02% NaN3, pH 8.0, and loaded on a Mono Q column (GE Healthcare). Adsorbed proteins were eluted with an NaCl gradient, and fractions containing PPAD activity were pooled. The final PPAD purification was achieved by gel filtration chromatography using a Superdex 75 column (GE Healthcare). The purity of PPAD was evaluated by SDS-PAGE, followed by silver staining. Gingipain-specific inhibitors KYT-1 and KYT-36 were obtained from PeptaNova (Sandhausen, Germany).
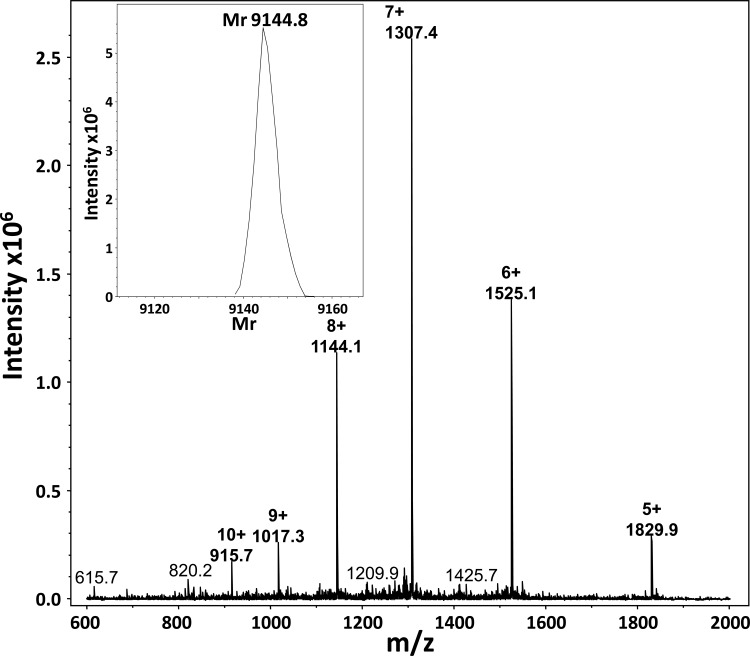
Biotinylated CXCL8 (biotCXCL8) was synthesized, folded, and purified in the Laboratory of Molecular Immunology at the Department of Microbiology and Immunology, Rega Institute. Briefly, CXCL8 was synthesized as previously described (23). After removal of the final 9-fluorenylmethoxy carbonyl (Fmoc) group of the most NH2-terminal amino acid, the free NH2 terminus was biotinylated with biotin-p-nitrophenyl ester (Novabiochem, Darmstadt, Germany) on a 433A solid-phase peptide synthesizer (Applied Biosystems, Foster City, CA) using standard coupling chemistry. The synthetic biotinylated CXCL8 peptide was cleaved from the resin, purified, folded, and repurified as described for intact CXCL8 (23). The purity of biotinylated CXCL8 was confirmed by ion trap mass spectrometry after purification using reverse-phase high-pressure liquid chromatography (RP-HPLC) (Fig. 1).
FIG 1.
Ion trap mass spectra of purified synthetic biotinylated CXCL8. CXCL8 was synthesized using 9-fluorenylmethoxy carbonyl (Fmoc) chemistry and purified using RP-HPLC. After removal of the final Fmoc group of the most NH2-terminal amino acid, the free NH2 terminus was biotinylated with biotin-p-nitrophenyl ester using standard coupling chemistry. The Mr of the purified synthetic biotCXCL8 was determined by deconvolution of the multiple charged ions in the raw spectra. As expected, the Mr of biotCXCL8 differed by almost exactly 226.3 mass units (Mr = 9,144.8) from that of the unmodified CXCL8 (Mr = 8,918.3).
Strains and culture conditions.
P. gingivalis ATCC 33277 was used as a reference strain. Five capsule-typed strains (K1 to K5) (46, 47) and five clinical isolates with nontyped capsule (Pg1 to Pg5) (48) were included in the analysis (Table 1). P. gingivalis strains were grown on blood agar plates (Oxoid, Basingstoke, United Kingdom) supplemented with 5 μg/ml hemin (Sigma-Aldrich), 1 μg/ml menadione, and 5% sterile horse blood (Biotrading, Keerbergen, Belgium). Bacteria were collected from blood agar plates, transferred to 10 ml tryptone soy broth (Oxoid), and incubated overnight at 37°C in an anaerobic atmosphere. The bacterial concentration was adjusted by measuring the optical density at 600 nm to obtain bacterial suspensions with starting concentrations of 5 × 107 CFU/ml. Supernatants were obtained by centrifuging for 3 min at 13,200 rpm.
TABLE 1.
P. gingivalis strains useda
| Strain | Strain code | Disease type | Probing depth (mm) |
|---|---|---|---|
| Laboratory strain ATCC 33277 | NK | NK | NK |
| Capsule-typed strains K1 to K5 | NK | NK | NK |
| Clinical isolates | |||
| Pg1 | C1-IIIa | CAP | 8.3 |
| Pg2 | C7-1 | RPP | 7.7 |
| Pg3 | R33-2 | Res | 4.0 |
| Pg4 | T9-6 | CAP | 8.0 |
| Pg5 | T1-IIIb | CAP | 7.3 |
NK, not known; CAP, chronic adult periodontitis; RPP, rapidly progressive periodontitis; Res, resistance to periodontitis.
PPAD activity assay.
PPAD activity was tested by a colorimetric method for the determination of enzymatically produced l-Cit as described by Knipp and Vasak (49). A standard for l-Cit ranging from 0 to 1,200 μM was prepared in 0.1 M Tris-HCl buffer (pH 7.5), and subsequently, 50 μl was pipetted in a 96-well plate. Ten microliters of each sample was mixed with 40 μl 0.1 M Tris-HCl buffer (pH 7.5) supplemented with 10 mM N-acetylo-l-Arg before pipetting into the 96-well plate. The 96-well plate containing the standard and samples was incubated for 1 h at 37°C. Finally, 150 μl of freshly prepared citrulline detection reagent {1 volume of solution A [80 mM 2,3-butanedione monoxime and 2 mM thiosemicarbazide] and 3 volumes of solution B [3 M H3PO4, 6 M H2SO4, and 2 mM NH4Fe(SO4)2·12H2O]} was added, and the 96-well plate was incubated for 15 min at 95°C. Cit was detected by measuring the absorbance at 535 nm.
In vitro incubation of CXCL8 and CCL3 with PPAD.
CXCL8 or CCL3 was incubated with purified activated PPAD and gingipain-specific inhibitors (10 μM KYT-1 and KYT-36) for 1.5 h at 37°C at an enzyme-substrate (E/S) molar ratio of 1/10 and a substrate concentration of 7.6 μM. Incubations with PPAD were carried out in 50 mM Tris-HCl, 150 mM NaCl (pH 7.8). Enzymatic citrullination was stopped with 0.1% (vol/vol) trifluoroacetic acid (TFA).
In vitro incubation of CXCL8, CXCL10, and [Cit5]CXCL8 with gingipains.
RgpB was first activated for 10 min at 37°C in activation buffer (0.2 M HEPES, 10 mM l-Cys, 10 mM CaCl2, pH 8). CXCL8, CXCL10, or citrullinated CXCL8 ([Cit5]CXCL8) was then incubated with activated RgpB for 1 h at 37°C at an E/S molar ratio of 1/1 and a substrate concentration of 1 μM. Incubations with RgpB were carried out in 20 mM Tris-HCl, 5 mM CaCl2, 0.1 M NaCl (pH 8). Enzymatic processing was stopped with 0.1% (vol/vol) TFA and investigated by Edman degradation on a 491 Procise cLC protein sequencer (Applied Biosystems, Foster City, CA).
PTM of CXCL8 and CXCL10 by different P. gingivalis strains.
Bacterial suspensions with starting concentrations of 5 × 107 CFU/ml were first grown for 24 h in 96-well plates (200 μl/well) at 37°C in an anaerobic atmosphere. Then, the strains were treated with 5 μg/ml CXCL8 or CXCL10 for 2 h, and the supernatants were harvested and stored at −20°C after centrifuging for 3 min at 13,200 rpm.
To investigate the potential binding of CXCL8 to the bacterial membrane, strains were treated with 500 ng/ml biotCXCL8 and protease inhibitor (cOmplete ULTRA, Mini, EDTA-free EASYpack; Roche) supplemented with 5 mM EDTA for 2 h. Supernatants and pellets were harvested and stored at −20°C after centrifuging for 3 min at 13,200 rpm.
Proteolytic processing of CXCL8 and CXCL10 by secreted versus membrane-bound gingipains.
Bacterial suspensions of 5 × 107 CFU/ml were cultured for 24 h in 96-well plates (200 μl/well) at 37°C in an anaerobic atmosphere, and supernatants were harvested. To examine the proteolytic processing of chemokines by P. gingivalis proteases, CXCL8 and CXCL10 were incubated with 5 μl of supernatants or with the complete bacterial culture at 37°C. Samples were separated by SDS-PAGE using 14% ProSieve50 gels (Lonza, Basel, Switzerland) with a Tris-Tricine electrode buffer (0.1 M Tris, 0.1 M Tricine, 0.1% SDS). Gels were blotted onto a polyvinylidene difluoride (PVDF) membrane that was subsequently stained using Coomassie brilliant blue. Finally, the NH2-terminal protein sequences of the protein bands visible on the blot were determined by Edman degradation on a 491 Procise cLC protein sequencer (Applied Biosystems).
Detection of total CXCL8, biotCXCL8, and CXCL10.
The levels of human CXCL8 and CXCL10 were quantified by specific sandwich enzyme-linked immunosorbent assays (ELISAs) (50–52).
For human CXCL8, a 96-well plate was coated with goat polyclonal anti-human CXCL8 antibody generated in our laboratory (53), followed by blocking with phosphate-buffered saline (PBS; pH 7.4) containing 0.1% (wt/vol) casein and 0.05% (vol/vol) Tween 20. Human CXCL8 bound to the coating antibody was detected with mouse monoclonal anti-human CXCL8 antibody (R&D Systems, Abingdon, United Kingdom), followed by a secondary peroxidase-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Peroxidase activity was quantified by measuring the conversion of 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma-Aldrich) at 450 nm.
To measure biotCXCL8, plates were coated with goat polyclonal anti-human CXCL8 antibody (see above), and biotCXCL8 was detected with peroxidase-conjugated streptavidin (R&D Systems).
The sandwich ELISA for human CXCL10 consisted of mouse monoclonal anti-human CXCL10 as the coating antibody, biotinylated rabbit polyclonal anti-human CXCL10 as the detecting antibody, and peroxidase-conjugated streptavidin as the secondary reagent (all were from R&D Systems).
Detection of citrullinated chemokines.
The procedure to chemically modify and quantify a citrulline-containing protein of interest was performed as described before (54). Briefly, peptidylcitrulline residues in samples were first chemically modified by addition of 50 mM antipyrine, 16% (vol/vol) TFA, and 12.5 mM 2,3-butanedione (55), followed by incubation for 2 h at 37°C in the dark. Then, the strongly acidic reaction mixture was dialyzed against PBS containing 0.05% Tween 20 (pH 7.4) overnight at room temperature and protected from light using Slide-A-Lyzer Mini dialysis units (Pierce, Rockford, IL).
To detect [Cit5]CXCL8, a specific sandwich ELISA was developed (54). A 96-well plate was coated with mouse monoclonal anti-human CXCL8 antibody (R&D Systems), followed by blocking with PBS containing 0.1% casein and 0.05% Tween 20. Human [Cit5]CXCL8 in chemically modified samples was detected by specific rabbit antibodies against chemically modified citrulline residues and by a secondary peroxidase-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories). Peroxidase activity was quantified by measuring the conversion of TMB (Sigma-Aldrich) at 450 nm. Citrullination of CCL3 was detected in a similar way by using a mouse monoclonal anti-human CCL3 antibody (R&D Systems) instead of the mouse monoclonal anti-human CXCL8 antibody.
In addition, for the in vitro incubation of CXCL8 with PPAD, the NH2-terminal sequences of CXCL8 were determined by Edman degradation on a 491 Procise cLC protein sequencer (Applied Biosystems) to verify if citrullination occurred on the more NH2-terminally located Arg residues.
RESULTS
Enzymatic modification of chemokines by PPAD and gingipains.
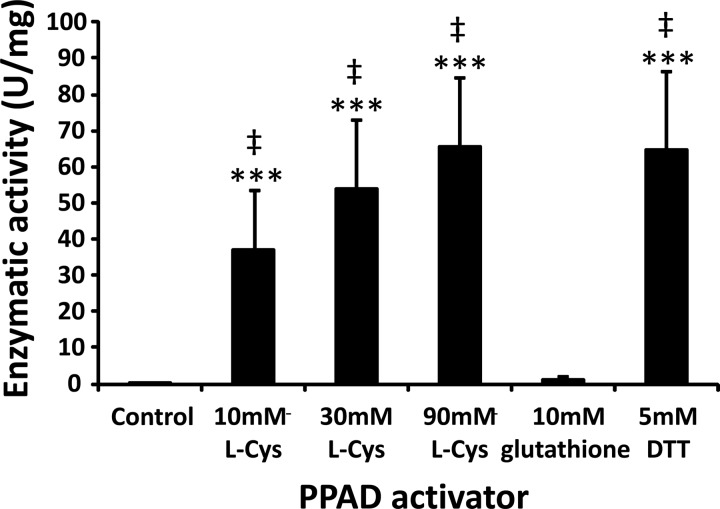
Before incubating chemokines with PPAD in vitro, the activation of the enzyme by different reducing agents was evaluated with a PPAD activity assay (49). In contrast to 10 mM reduced glutathione, both l-cysteine (l-Cys) (10 mM, 30 mM, and 90 mM) and dithiothreitol (DTT; 5 mM) were able to activate PPAD (Fig. 2).
FIG 2.
Activation of PPAD by different reducing agents. Before incubation of CXCL8 with PPAD in vitro, the activation of the enzyme by different reducing agents was evaluated with the PPAD activity assay (49). The mean enzymatic activity (U/mg enzyme) ± SEM is presented for each activator (n = 6). Statistical analysis was performed using the Mann-Whitney U test. ***, P < 0.001 for the comparison with the control; ‡, P < 0.001 for the comparison with glutathione.
Upon incubation of intact CXCL8 with activated PPAD (30 mM l-Cys) at an E/S molar ratio of 1/10 for 1.5 h, no citrullination was detected by the specific ELISA. However, NH2-terminal sequencing revealed about 10% citrullination on the most NH2-terminal Arg but no citrullination of the Arg in the ELR motif of CXCL8. Besides CXCL8, CCL3 is also reported to be an important inflammatory chemokine mediator in periodontitis (56, 57). However, upon incubation of intact CCL3 with activated PPAD (30 mM L-Cys) at an E/S molar ratio of 1/10 for 1.5 h, no citrullination could be detected by the specific ELISA to measure citrullinated CCL3. In contrast to CXCL8, the NH2 terminus of CCL3 contains no Arg, so NH2-terminal sequencing was not performed. Activated RgpB efficiently cleaved CXCL8 and CXCL10 in vitro. After 30 min, no intact CXCL8 was present anymore, as only NH2-terminally processed forms of CXCL8 (e.g., forms of CXCL8 consisting of amino acids 6 to 77, 12 to 77, and 32 to 77 [the 6-77, 12-77, and 32-77 forms, respectively]) were detected. Also for CXCL10, cleavage occurred very rapidly, as after 5 min of incubation, less than 25% intact chemokine could be detected (data not shown). Interestingly, citrullination of CXCL8 prevented this chemokine from being cleaved by RgpB. This confirms the high specificity of RgpB for Arg—X bonds.
Enzymatic citrullination of chemokines by P. gingivalis.
CXCL8 and CXCL10 were incubated with full cultures of different P. gingivalis strains for 2 h. Subsequently, the total amounts of CXCL8 and CXCL10 were determined by standard ELISA and citrullinated chemokine was quantified by a specific sandwich ELISA, as recently described (54).
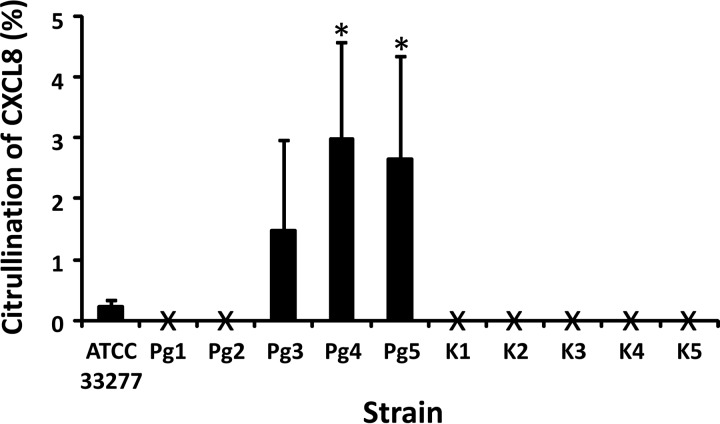
CXCL8 was recovered after incubation with most tested strains; however, only a small amount of CXCL8 (up to 5%) was detected to be citrullinated in samples for all tested strains of P. gingivalis (Fig. 3). At first, an obvious explanation for this could be the NH2-terminal cleavage of CXCL8 at Lys8 by Kgp, which potentially removes the first 8 amino acids, including the Arg in position 5, i.e., the Arg which is most susceptible to citrullination by human PAD2 and PAD4 (8). However, the citrullination levels detected upon incubation with PPAD in vitro were also very low. Therefore, a more plausible reason for the low citrullination of CXCL8 for all tested strains of P. gingivalis is that PPAD citrullinates CXCL8 inefficiently.
FIG 3.
Citrullination of CXCL8 by different clinical isolates of P. gingivalis. Bacterial suspensions with starting concentrations of 5 × 107 CFU/ml were first grown for 24 h at 37°C in an anaerobic atmosphere. Then, the strains were treated with 5 μg/ml CXCL8 for 2 h and the supernatants were harvested. The levels of human CXCL8 were quantified by ELISA. The amount of citrullinated CXCL8 was determined by combined chemical modification and ELISA, as described before (54). The mean percent citrullination of CXCL8 ± SEM is presented for each strain (n = 3). X, citrullination levels which are below the detection limit (0.4 ng/ml). Statistical analysis was performed using the Mann-Whitney U test. *, P < 0.05 for the comparison with laboratory strain ATCC 33277.
To further investigate whether inflammatory chemokines become citrullinated by PPAD, we selected CXCL10, which is a major attractant for natural killer cells and activated T cells and is citrullinated by human PAD2 and PAD4 but which has no NH2-terminal lysine residues (Lys [K]). Thus, the NH2 terminus of CXCL10 (NH2-1VPLSRTVRCT10) is resistant to proteolysis by Kgp, leaving Arg5 and Arg8 available for citrullination by PPAD. In this case, however, only very low levels of CXCL10 were recovered immediately after treatment with P. gingivalis and no CXCL10 was detected for any strain after 2 h of incubation. This suggests that P. gingivalis directly interacts with CXCL10 by binding, degrading, or internalizing the chemokine. When CXCL10 was incubated with supernatants of P. gingivalis, already after 5 min, no intact CXCL10 was present, indicating very fast degradation of CXCL10 by P. gingivalis (see below).
Proteolytic cleavage of chemokines by P. gingivalis.
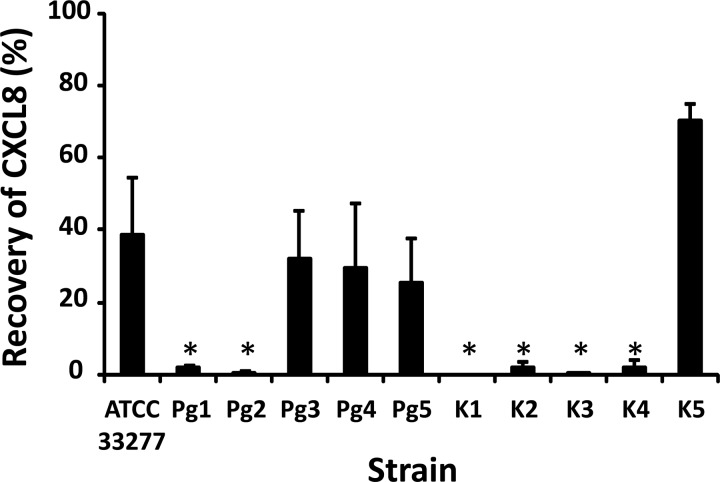
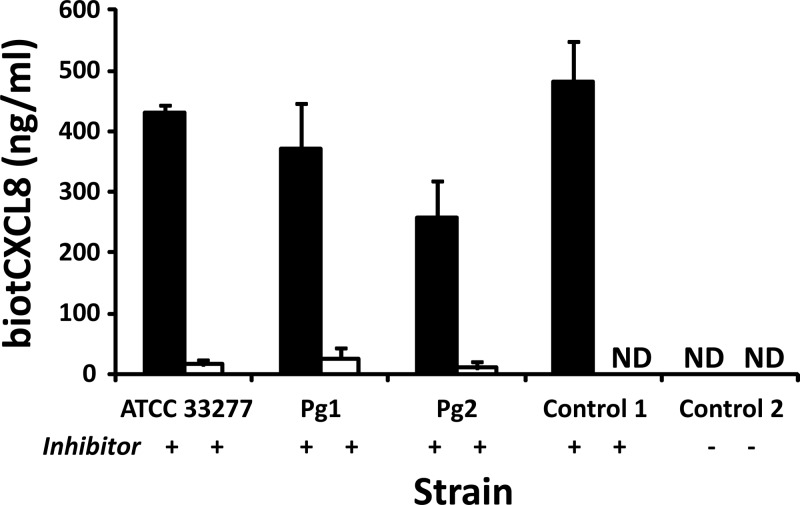
CXCL8 was incubated with different P. gingivalis strains for 2 h, and CXCL8 levels were measured by ELISA. For all strains, the amount of CXCL8 still present after 2 h was lower than the initial quantity added, indicating that clearance of CXCL8 from the bacterial medium occurred (Fig. 4). However, some strains seemed more efficient in degrading CXCL8, since for strains Pg1, Pg2, and K1 to K4, only a very small amount of CXCL8 was detected after 2 h. Samples in which CXCL8 was still present after 2 h were examined by gel electrophoresis and NH2-terminal sequencing (Table 2). Besides intact CXCL8, three NH2-terminally processed forms of CXCL8, i.e., the more potent variants consisting of the CXCL8 from amino acids 6 to 77 [CXCL8(6-77)] and CXCL8(9-77) and the further truncated variant CXCL8(16-77), could be detected, indicating that NH2-terminal truncation by cleavage at Arg5 and Lys8 precedes further degradation of the chemokine. To investigate the potential binding of CXCL8 to the bacterial membrane, biotCXCL8 was incubated with different strains and protease inhibitor for 2 h. CXCL8 was specifically biotinylated at the NH2 terminus by chemical synthesis. Thus, modification of the side chains of Lys was avoided and potential cleavage sites for Kgp remained unaltered. Supernatants and bacteria were harvested, and the biotCXCL8 in both fractions was measured by ELISA (Fig. 5). The amount of biotCXCL8 recovered in the cell pellets was negligible compared to the chemokine levels present in the supernatants. These data confirm the importance of the degradation by proteases (e.g., gingipains) (and not by binding to the bacterial cell surface) in the removal of CXCL8 from the medium.
FIG 4.
Recovery of CXCL8 after incubation with different clinical isolates and capsule-typed strains of P. gingivalis. Bacterial suspensions with starting concentrations of 5 × 107 CFU/ml were first grown for 24 h at 37°C in an anaerobic atmosphere. Then, the strains were treated with 5 μg/ml CXCL8 for 2 h and the supernatants were harvested. The levels of human CXCL8 were quantified by ELISA. The mean CXCL8 recovery ± SEM after 2 h is presented for each strain (n = 3). Statistical analysis was performed using the Mann-Whitney U test. *, P < 0.05 for the comparison with laboratory strain ATCC 33277.
TABLE 2.
NH2-terminally processed forms of CXCL8 after 2 h of incubation with different P. gingivalis strains detected by Edman degradation
| Strain | % of the following NH2-terminally processed forms of CXCL8: |
|||
|---|---|---|---|---|
| 1-77 | 6-77 | 9-77 | 16-77 | |
| Laboratory strain ATCC 33277 | NDa | ND | 50 | 50 |
| Capsule-typed strains | ||||
| K1 | ND | ND | ND | ND |
| K2 | ND | ND | ND | ND |
| K3 | ND | ND | ND | ND |
| K4 | ND | ND | ND | ND |
| K5 | 39 | 32 | 29 | ND |
| Clinical isolates | ||||
| Pg1 | ND | ND | ND | ND |
| Pg2 | ND | ND | ND | ND |
| Pg3 | ND | ND | 100 | ND |
| Pg4 | ND | ND | 100 | ND |
| Pg5 | ND | ND | 100 | ND |
ND, not detectable.
FIG 5.
Levels of biotinylated CXCL8 measured in supernatants and cell pellets of P. gingivalis. Bacterial suspensions with starting concentrations of 5 × 107 CFU/ml were first grown for 24 h at 37°C in an anaerobic atmosphere. Then, the strains were treated with 500 ng/ml biotCXCL8 and protease inhibitor for 2 h. Supernatants (■) and cell pellets (□) were harvested, and the levels of biotCXCL8 were quantified by ELISA. The mean CXCL8 recovery ± SEM after 2 h is presented for each strain (n = 2). Control 1, TSB medium with biotCXCL8 and protease inhibitor incubated for 2 h; control 2, Pg2 treated with 500 ng/ml biotCXCL8 without protease inhibitor for 2 h; ND, not detectable (<2 ng/ml).
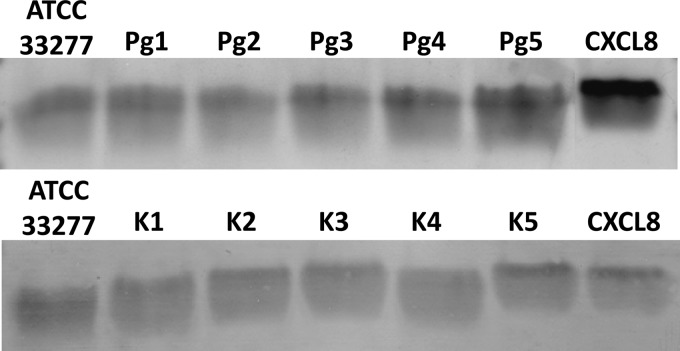
When CXCL8 was incubated for 2 h with supernatants of P. gingivalis, cleavage clearly occurred (Fig. 6). In contrast to the difference in cleavage measured by ELISA for CXCL8 incubated with P. gingivalis cultures (Fig. 4), no quantitative difference in processing of CXCL8 by cell-free culture supernatants of strains ATCC 33277, Pg1 to Pg5, and K1 to K5 could be seen (Fig. 6). Sequencing of the CXCL8 incubated with the supernatant showed that CXCL8 is processed first into its more potent 6-77 and 9-77 forms before being totally degraded. The highly active truncated CXCL8(9-77) was clearly the most dominant form found in all incubations (57% ± 5.4%).
FIG 6.
Cleavage of CXCL8 by the supernatants of different clinical isolates and capsule-typed strains of P. gingivalis. Bacterial suspensions with starting concentrations of 5 × 107 CFU/ml were grown for 24 h at 37°C in an anaerobic atmosphere, and supernatants were harvested. CXCL8 (1 μg) was incubated with 5 μl of the supernatants of ATCC 33277, Pg1 to Pg5, and K1 to K5 for 2 h at 37°C. To examine proteolytic processing of chemokines, samples and untreated CXCL8 (control) were separated by SDS-PAGE, transferred to a PVDF membrane, and stained with Coomassie brilliant blue.
Also, CXCL10 was incubated with different P. gingivalis strains for 2 h, and CXCL10 levels were detected by ELISA. However, none of the CXCL10 was recovered when it was incubated for 2 h with whole bacteria (see above). When CXCL10 was incubated for 5 min, 30 min, or 2 h with supernatants of P. gingivalis, CXCL10 was detectable only after 5 min, as evidenced by gel electrophoresis, blotting, and Edman degradation. However, even after 5 min of incubation, no intact CXCL10 was detected, indicating fast degradation of CXCL10 by P. gingivalis. Two processed forms of CXCL10 (i.e., the 23-77 and 27-77 forms) were detected.
DISCUSSION
Enzymatic citrullination of chemokines by P. gingivalis.
PTMs affect the biological activity of cytokines and chemokines (2, 4, 58). Recently, citrullination, a new PTM of chemokines, has been discovered (8, 21). The conversion of peptidylarginine to peptidylcitrulline is performed by the mammalian PAD enzymes (25). The biological function of chemokines is influenced by citrullination. Citrullination of CXCL8 by PAD protects CXCL8 from NH2-terminal activation by thrombin and plasmin, reduces its glycosaminoglycan binding properties, moderately alters binding of CXCL8 to the CXC chemokine receptors CXCR1 and CXCR2, reduces CXCL8-induced neutrophil extravasation to the peritoneal cavity, and increases the ability of chemokines to mobilize neutrophils to the blood circulation (8, 59). Not only eukaryotes but also the prokaryotic organism P. gingivalis are known to express PAD. Colonization of a biofilm on the tooth surface by P. gingivalis and its subgingival proliferation lead to immune reactions eroding teeth-supporting tissues. Recently, a potential correlation between periodontitis and the autoimmune disease rheumatoid arthritis (RA), which is linked to protein citrullination, has been proposed (32, 60–62). It is suggested that PPAD is responsible for the citrullination of proteins like fibrinogen and α-enolase, leading to the production of anti-citrullinated peptide antibodies (ACPAs) and immune complexes. This results in inflammation and may finally cause chronic RA (63–65). The occurrence of chemokine citrullination by bacterial PPAD, however, was not investigated until now. Therefore, we hypothesized that chemokines are possibly citrullinated by PPAD during P. gingivalis infection. In the gingival crevicular fluid of periodontitis patients, however, CXCL8 is present only in pg/ml concentrations (66) in a milieu containing other proteins and peptides at mg/ml concentrations, which makes detection of [Cit5]CXCL8 with the currently available techniques a great challenge. Future studies should elucidate if CXCL8 is citrullinated by PPAD in vivo by the detection of [Cit5]CXCL8 in gingival crevicular fluid of periodontitis patients.
We observed only a low level of enzymatic citrullination of CXCL8 upon incubation with purified PPAD at an E/S molar ratio of 1/10. In comparison, human PAD2 and PAD4 were able to citrullinate 50% of the NH2-terminal Arg in CXCL8 in 10 min at an E/S molar ratio of 1/200 (8). Also for CCL3, another important inflammatory chemokine mediator in periodontitis (56, 57), no citrullination could be detected. Furthermore, no significant citrullination of CXCL8 could be measured when testing several P. gingivalis strains by a recently developed method for detection of citrullinated proteins (54). In contrast, rapid cleavage of CXCL8 by the gingipains expressed by P. gingivalis occurred. The processing of CXCL8(1-77) into CXCL8(9-77) by secreted gingipains removes the NH2-terminal octapeptide, including the Arg5, which is the residue known to be citrullinated by human PAD (8). Another potential explanation for the failure to detect [Cit5]CXCL8 can be related to the low citrullinating activity of PPAD on Arg5 of CXCL8. Indeed, previous research indicated that PPAD preferentially targets COOH-terminal Arg, in contrast to human PAD, which efficiently citrullinates NH2-terminal or internal Arg (34). However, as only <5% citrullination was detected by ELISA, COOH-terminal citrullination of CXCL8 by PPAD seems to be inefficient.
Proteolytic cleavage of chemokines by P. gingivalis.
As previously reported, CXCL8 was efficiently processed upon incubation with gingipains (RgpB) in vitro (67, 68). Upon incubation with different P. gingivalis strains, cleavage was detected for all strains. However, some strains were more efficient in degrading CXCL8, as for Pg1, Pg2, and K1 to K4, no CXCL8 was detected after 2 h. Clearance of CXCL8 from the medium can be caused by degradation by gingipains or by the binding to the bacterial membrane (and internalization). Our data, however, demonstrate the more important role of enzymatic degradation in removing CXCL8 from the medium compared to the importance of binding to the bacterial membrane. CXCL8 was shown to be first converted to more potent forms by purified gingipains, followed by total degradation (68). This report further demonstrates the consecutive activation and destruction of CXCL8 by incubation with different P. gingivalis strains. Earlier studies have shown the relation between bacterial surface structures and the virulence of different P. gingivalis strains, indicating that bacteria with large hydrophilic capsules are the most virulent ones (69–71). These studies demonstrated that strains K1 (HG66/W83) and K2 (HG184) display a more hydrophilic character than strain ATCC 33277. This might explain the disappearance of CXCL8 from the medium by binding to the bacterial capsule of these strains. Clearance of CXCL8 from the surrounding medium represents a powerful mechanism to downregulate the host's immune response. In this respect, strains Pg1, Pg2, and K1 to K4 can be considered more virulent than the other tested strains, because after 2 h no CXCL8 remained present in the bacterial medium.
Analysis of samples still containing CXCL8 after 2 h of incubation revealed, besides intact CXCL8, three processed forms of CXCL8 (i.e., the 6-77, 9-77, and 16-77 forms). This indicates NH2-terminal cleavage after Arg5 and Lys8, possibly by Lys- and Arg-specific gingipains, before further degradation. Both 6-77 and 9-77 represent forms of CXCL8 more active than intact CXCL8. In the form missing the 15 most NH2-terminal amino acids, the ELR motif is removed, implying a loss of receptor internalization and biological activity. The generation of more potent CXCL8 forms seems to contradict the role of gingipains in the evasion and dysregulation of the host's immune response by the degradation of the host's proinflammatory cytokines at the site of infection (39–41). However, also for other human proteins like the human complement factors, a biphasic effect of gingipains from P. gingivalis has been suggested (72–74). Not only are gingipains able to degrade complement factors, rendering P. gingivalis resistant to the antibacterial activity of complement, but also they initially increase complement activation. The basal level of inflammation caused by this activation is suggested to supply the bacteria with nutrients and might allow colonization (73).
In contrast to clear variations in the ability of full cultures of different strains of P. gingivalis to cleave CXCL8, no such differences were found for cell-free culture supernatants (Fig. 6). A possible explanation is that secreted gingipain activity is modulated by components of the cell surface. Alternatively, the local concentration of enzymes in the microenvironment of the membrane highly surpasses the concentration of soluble gingipains in the medium. The latter is supported by the observation that, except for P. gingivalis strain HG66, all other strains retain the majority of gingipains on the cell surface (75–77). Moreover, CXCL8 was processed into its more active 6-77 and 9-77 forms by the gingipains in the supernatants. CXCL8(9-77) was clearly the most dominant form found in all incubations. These results confirm previous observations that gingipains in the soluble form are able to initially convert CXCL8 to more potent species truncated at the amino terminus, followed by slow degradation and destruction of chemokine biological activity (68).
In contrast to CXCL8, citrullinated CXCL8 is not cleaved by RgpB. Since both human PAD2 and PAD4 together with CXCL8 are present in inflamed periodontal tissue (78), citrullination might protect the chemokine from being processed to more active forms, representing a mechanism to dampen the inflammation. Recently, it has been shown that P. gingivalis can inhibit the production of CXCL10 by epithelial cells, highlighting the immune-disruptive potential of this bacterium (79). Our data show a very fast degradation of CXCL10 by P. gingivalis. Two processed forms of CXCL10 (i.e., the 23-77 and 27-77 frosm) were present after 5 min of incubation with supernatants of the bacteria. These forms might arise from cleavage by secreted gingipains of P. gingivalis present in the supernatants, as residues 22 and 26 are, respectively, Arg and Lys.
At high concentrations (micromolar range) CXCL10 is known to be a chemokine with antimicrobial peptide-like properties for Escherichia coli and Listeria monocytogenes allowing it to bind to cells and induce cell death (80). However, density measurements (data not shown) indicate that the viability of the tested P. gingivalis strains was not influenced by the addition of CXCL10 (5 μg/ml) to bacterial cultures for 2 h. It is possible that CXCL10 is not able to bind to P. gingivalis and, consequently, cause cell death because of its rapid degradation by gingipains secreted in the medium (as shown in Results). In addition, the concentrations CXCL10 that we added were lower than the reported concentrations (80, 81).
In conclusion, this study could not confirm the citrullination of CXCL8 and CXCL10 by PPAD. However, the efficient posttranslational modification of CXCL8 and CXCL10 by gingipains of P. gingivalis was shown.
ACKNOWLEDGMENTS
This work was supported by the Fund for Scientific Research of Flanders (FWO-Vlaanderen), the Concerted Research Actions (GOA) of the Regional Government of Flanders, and the Interuniversity Attraction Poles Programme—Belgian Science Policy (IAP). A.M. is a postdoctoral research fellow of the FWO-Vlaanderen. J.P. is supported by grants from the U.S. NIDR (DE 022597), the European Commission (FP7-HEALTH-2010-261460 [Gums & Joints] and FP7-PEOPLE-2011-ITN-290246 [RAPID]), the Foundation for Polish Science (FNP; TEAM project DPS/424-329/10), the National Science Center (2011/01/B/NZ6/00268, NCN, Krakow, Poland), and the Polish Ministry of Science and Higher Education (project 137/7.PR-EU/2011/2). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of structural funds from the European Union (POIG.02.01.00-12-064/08).
Footnotes
Published ahead of print 31 March 2014
REFERENCES
- 1.Dinarello CA. 1997. Interleukin-1. Cytokine Growth Factor Rev. 8:253–265. 10.1016/S1359-6101(97)00023-3 [DOI] [PubMed] [Google Scholar]
- 2.Mortier A, Van Damme J, Proost P. 2008. Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 120:197–217. 10.1016/j.pharmthera.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 3.Van Damme J, De Ley M, Opdenakker G, Billiau A, De Somer P, Van Beeumen J. 1985. Homogeneous interferon-inducing 22K factor is related to endogenous pyrogen and interleukin-1. Nature 314:266–268. 10.1038/314266a0 [DOI] [PubMed] [Google Scholar]
- 4.Proost P, Struyf S, Schols D, Opdenakker G, Sozzani S, Allavena P, Mantovani A, Augustyns K, Bal G, Haemers A, Lambeir AM, Scharpe S, Van Damme J, De Meester I. 1999. Truncation of macrophage-derived chemokine by CD26/dipeptidyl-peptidase IV beyond its predicted cleavage site affects chemotactic activity and CC chemokine receptor 4 interaction. J. Biol. Chem. 274:3988–3993 [DOI] [PubMed] [Google Scholar]
- 5.Mortier A, Gouwy M, Van Damme J, Proost P. 2010. Effect of posttranslational processing on the in vitro and in vivo activity of chemokines. Exp. Cell Res. 317:642–654. 10.1016/j.yexcr.2010.11.016 [DOI] [PubMed] [Google Scholar]
- 6.Wolf M, Albrecht S, Marki C. 2008. Proteolytic processing of chemokines: implications in physiological and pathological conditions. Int. J. Biochem. Cell Biol. 40:1185–1198. 10.1016/j.biocel.2007.12.009 [DOI] [PubMed] [Google Scholar]
- 7.Ludwig A, Weber C. 2007. Transmembrane chemokines: versatile ‘special agents' in vascular inflammation. Thromb. Haemost. 97:694–703. 10.1160/TH07-01-0035 [DOI] [PubMed] [Google Scholar]
- 8.Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, Dillen C, Ronsse I, Conings R, Struyf S, Opdenakker G, Maudgal PC, Van Damme J. 2008. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J. Exp. Med. 205:2085–2097. 10.1084/jem.20080305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert CA, Luscinskas FW, Kiely JM, Luis EA, Darbonne WC, Bennett GL, Liu CC, Obin MS, Gimbrone MA, Jr, Baker JB. 1990. Endothelial and leukocyte forms of IL-8. Conversion by thrombin and interactions with neutrophils. J. Immunol. 145:3033–3040 [PubMed] [Google Scholar]
- 10.Van den Steen PE, Proost P, Wuyts A, Van Damme J, Opdenakker G. 2000. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood 96:2673–2681 [PubMed] [Google Scholar]
- 11.Proost P, Menten P, Struyf S, Schutyser E, De Meester I, Van Damme J. 2000. Cleavage by CD26/dipeptidyl peptidase IV converts the chemokine LD78beta into a most efficient monocyte attractant and CCR1 agonist. Blood 96:1674–1680 [PubMed] [Google Scholar]
- 12.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. 2000. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 289:1202–1206. 10.1126/science.289.5482.1202 [DOI] [PubMed] [Google Scholar]
- 13.Proost P, Schutyser E, Menten P, Struyf S, Wuyts A, Opdenakker G, Detheux M, Parmentier M, Durinx C, Lambeir AM, Neyts J, Liekens S, Maudgal PC, Billiau A, Van Damme J. 2001. Amino-terminal truncation of CXCR3 agonists impairs receptor signaling and lymphocyte chemotaxis, while preserving antiangiogenic properties. Blood 98:3554–3561. 10.1182/blood.V98.13.3554 [DOI] [PubMed] [Google Scholar]
- 14.Malkowski MG, Lazar JB, Johnson PH, Edwards BF. 1997. The amino-terminal residues in the crystal structure of connective tissue activating peptide-III (des10) block the ELR chemotactic sequence. J. Mol. Biol. 266:367–380. 10.1006/jmbi.1996.0796 [DOI] [PubMed] [Google Scholar]
- 15.Schulz-Knappe P, Magert HJ, Dewald B, Meyer M, Cetin Y, Kubbies M, Tomeczkowski J, Kirchhoff K, Raida M, Adermann K, Kist A, Reinecke M, Sillard R, Pardigol A, Uguccioni M, Baggiolini M, Forssmann WG. 1996. HCC-1, a novel chemokine from human plasma. J. Exp. Med. 183:295–299. 10.1084/jem.183.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Detheux M, Standker L, Vakili J, Munch J, Forssmann U, Adermann K, Pohlmann S, Vassart G, Kirchhoff F, Parmentier M, Forssmann WG. 2000. Natural proteolytic processing of hemofiltrate CC chemokine 1 generates a potent CC chemokine receptor (CCR)1 and CCR5 agonist with anti-HIV properties. J. Exp. Med. 192:1501–1508. 10.1084/jem.192.10.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moelants EA, Mortier A, Van Damme J, Proost P. 2013. In vivo regulation of chemokine activity by post-translational modification. Immunol. Cell Biol. 91:402–407. 10.1038/icb.2013.16 [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Tabak LA, Valente AJ, Graves DT. 1991. Initial characterization of the carbohydrate structure of MCP-1. Biochem. Biophys. Res. Commun. 178:1400–1404. 10.1016/0006-291X(91)91049-I [DOI] [PubMed] [Google Scholar]
- 19.Proost P, Mahieu F, Schutyser E, Van Damme J. 2004. Posttranslational processing of chemokines. Methods Mol. Biol. 239:27–44. 10.1385/1-59259-435-2:27 [DOI] [PubMed] [Google Scholar]
- 20.Durkan AM, Alexander RT, Liu GY, Rui M, Femia G, Robinson LA. 2007. Expression and targeting of CX3CL1 (fractalkine) in renal tubular epithelial cells. J. Am. Soc. Nephrol. 18:74–83. 10.1681/ASN.2006080862 [DOI] [PubMed] [Google Scholar]
- 21.Loos T, Mortier A, Gouwy M, Ronsse I, Put W, Lenaerts JP, Van Damme J, Proost P. 2008. Citrullination of CXCL10 and CXCL11 by peptidylarginine deiminase: a naturally occurring posttranslational modification of chemokines and new dimension of immunoregulation. Blood 112:2648–2656. 10.1182/blood-2008-04-149039 [DOI] [PubMed] [Google Scholar]
- 22.Loos T, Mortier A, Proost P. 2009. Isolation, identification, and production of posttranslationally modified chemokines. Methods Enzymol. 461:3–29. 10.1016/S0076-6879(09)05401-9 [DOI] [PubMed] [Google Scholar]
- 23.Mortier A, Berghmans N, Ronsse I, Grauwen K, Stegen S, Van Damme J, Proost P. 2011. Biological activity of CXCL8 forms generated by alternative cleavage of the signal peptide or by aminopeptidase-mediated truncation. PLoS One 6:e23913. 10.1371/journal.pone.0023913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa H, Hatakeyama S, Ikesue A, Miyai H. 1991. Generation of interleukin-8 by plasmin from AVLPR-interleukin-8, the human fibroblast-derived neutrophil chemotactic factor. FEBS Lett. 282:412–414. 10.1016/0014-5793(91)80526-9 [DOI] [PubMed] [Google Scholar]
- 25.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. 2003. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 25:1106–1118. 10.1002/bies.10357 [DOI] [PubMed] [Google Scholar]
- 26.Senshu T, Kan S, Ogawa H, Manabe M, Asaga H. 1996. Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem. Biophys. Res. Commun. 225:712–719. 10.1006/bbrc.1996.1240 [DOI] [PubMed] [Google Scholar]
- 27.Harding CR, Scott IR. 1983. Histidine-rich proteins (filaggrins): structural and functional heterogeneity during epidermal differentiation. J. Mol. Biol. 170:651–673. 10.1016/S0022-2836(83)80126-0 [DOI] [PubMed] [Google Scholar]
- 28.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. 1996. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 271:30709–30716 [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Hagiwara T, Yamada M. 2002. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 277:49562–49568. 10.1074/jbc.M208795200 [DOI] [PubMed] [Google Scholar]
- 30.Hagiwara T, Hidaka Y, Yamada M. 2005. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry 44:5827–5834. 10.1021/bi047505c [DOI] [PubMed] [Google Scholar]
- 31.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. 2002. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem. Biophys. Res. Commun. 290:979–983. 10.1006/bbrc.2001.6303 [DOI] [PubMed] [Google Scholar]
- 32.Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI. 2006. Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 38:1662–1677. 10.1016/j.biocel.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 33.Moelants EAV, Mortier A, Van Damme J, Proost P, Loos T. 2012. Peptidylarginine deiminases: role in physiology, inflammation and pathology. Drug Discov. Today Technol. 9:e261–e280. 10.1016/j.ddtec.2012.06.002 [DOI] [Google Scholar]
- 34.McGraw WT, Potempa J, Farley D, Travis J. 1999. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67:3248–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Touz MC, Ropolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, Nash TE. 2008. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 121:2930–2938. 10.1242/jcs.026963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquis RE, Bender GR, Murray DR, Wong A. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirai H, Blundell TL, Mizuguchi K. 2001. A novel superfamily of enzymes that catalyze the modification of guanidino groups. Trends Biochem. Sci. 26:465–468. 10.1016/S0968-0004(01)01906-5 [DOI] [PubMed] [Google Scholar]
- 38.Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, Gawron K, Mizgalska D, Marcinska KA, Benedyk M, Pyrc K, Quirke AM, Jonsson R, Alzabin S, Venables PJ, Nguyen KA, Mydel P, Potempa J. 2013. Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD). PLoS Pathog. 9:e1003627. 10.1371/journal.ppat.1003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bostanci N, Belibasakis GN. 2012. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 333:1–9. 10.1111/j.1574-6968.2012.02579.x [DOI] [PubMed] [Google Scholar]
- 40.Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. 2009. The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol. Immunol. 24:11–17. 10.1111/j.1399-302X.2008.00467.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moutsopoulos NM, Kling HM, Angelov N, Jin W, Palmer RJ, Nares S, Osorio M, Wahl SM. 2012. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J. Autoimmun. 39:294–303. 10.1016/j.jaut.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen IB, Enghild JJ, Travis J. 1998. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273:21648–21657. 10.1074/jbc.273.34.21648 [DOI] [PubMed] [Google Scholar]
- 43.Potempa J, Nguyen KA. 2007. Purification and characterization of gingipains. Curr. Protoc. Protein Sci. Chapter 21:Unit 21.20. 10.1002/0471140864.ps2120s49 [DOI] [PubMed] [Google Scholar]
- 44.Zhou XY, Gao JL, Hunter N, Potempa J, Nguyen KA. 2013. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol. Microbiol. 89:903–917. 10.1111/mmi.12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sztukowska M, Veillard F, Potempa B, Bogyo M, Enghild JJ, Thogersen IB, Nguyen KA, Potempa J. 2012. Disruption of gingipain oligomerization into non-covalent cell-surface attached complexes. Biol. Chem. 393:971–977. 10.1515/hsz-2012-0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Winkelhoff AJ, Appelmelk BJ, Kippuw N, De Graaff J. 1993. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol. Immunol. 8:259–265. 10.1111/j.1399-302X.1993.tb00571.x [DOI] [PubMed] [Google Scholar]
- 47.Laine ML, Appelmelk BJ, van Winkelhoff AJ. 1996. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J. Periodontal Res. 31:278–284. 10.1111/j.1600-0765.1996.tb00494.x [DOI] [PubMed] [Google Scholar]
- 48.Papaioannou W, van Steenberghe D, Cassiman JJ, Dierickx K, Quirynen M. 2003. Adhesion of Porphyromonas gingivalis to cultured pocket epithelium: mono- and multi-layered. Clin. Oral Investig. 7:162–166. 10.1007/s00784-003-0217-4 [DOI] [PubMed] [Google Scholar]
- 49.Knipp M, Vasak M. 2000. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal. Biochem. 286:257–264. 10.1006/abio.2000.4805 [DOI] [PubMed] [Google Scholar]
- 50.Wuyts A, Struyf S, Gijsbers K, Schutyser E, Put W, Conings R, Lenaerts JP, Geboes K, Opdenakker G, Menten P, Proost P, Van Damme J. 2003. The CXC chemokine GCP-2/CXCL6 is predominantly induced in mesenchymal cells by interleukin-1β and is down-regulated by interferon-γ: comparison with interleukin-8/CXCL8. Lab. Invest. 83:23–34. 10.1097/01.LAB.0000048719.53282.00 [DOI] [PubMed] [Google Scholar]
- 51.Proost P, Vynckier AK, Mahieu F, Put W, Grillet B, Struyf S, Wuyts A, Opdenakker G, Van Damme J. 2003. Microbial Toll-like receptor ligands differentially regulate CXCL10/IP-10 expression in fibroblasts and mononuclear leukocytes in synergy with IFN-γ and provide a mechanism for enhanced synovial chemokine levels in septic arthritis. Eur. J. Immunol. 33:3146–3153. 10.1002/eji.200324136 [DOI] [PubMed] [Google Scholar]
- 52.Proost P, Wuyts A, Conings R, Lenaerts JP, Put W, Van Damme J. 1996. Purification and identification of natural chemokines. Methods 10:82–92. 10.1006/meth.1996.0082 [DOI] [PubMed] [Google Scholar]
- 53.Van Damme J, Decock B, Conings R, Lenaerts JP, Opdenakker G, Billiau A. 1989. The chemotactic activity for granulocytes produced by virally infected fibroblasts is identical to monocyte-derived interleukin 8. Eur. J. Immunol. 19:1189–1194. 10.1002/eji.1830190706 [DOI] [PubMed] [Google Scholar]
- 54.Moelants EAV, Van Damme J, Proost P. 2011. Detection and quantification of citrullinated chemokines. PLoS One 6:e28976. 10.1371/journal.pone.0028976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm A, Rise F, Sessler N, Sollid LM, Undheim K, Fleckenstein B. 2006. Specific modification of peptide-bound citrulline residues. Anal. Biochem. 352:68–76. 10.1016/j.ab.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 56.Taylor JJ. 2010. Cytokine regulation of immune responses to Porphyromonas gingivalis. Periodontol. 2000 54:160–194. 10.1111/j.1600-0757.2009.00344.x [DOI] [PubMed] [Google Scholar]
- 57.Yucel-Lindberg T, Bage T. 2013. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev. Mol. Med. 15:e7. 10.1017/erm.2013.8 [DOI] [PubMed] [Google Scholar]
- 58.Mortier A, Loos T, Gouwy M, Ronsse I, Van Damme J, Proost P. 2010. Posttranslational modification of the NH2-terminal region of CXCL5 by proteases or peptidylarginine deiminases (PAD) differently affects its biological activity. J. Biol. Chem. 285:29750–29759. 10.1074/jbc.M110.119388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loos T, Opdenakker G, Van Damme J, Proost P. 2009. Citrullination of CXCL8 increases this chemokine's ability to mobilize neutrophils into the blood circulation. Haematologica 94:1346–1353. 10.3324/haematol.2009.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alivernini S, Fedele AL, Cuoghi I, Tolusso B, Ferraccioli G. 2008. Citrullination: the loss of tolerance and development of autoimmunity in rheumatoid arthritis. Reumatismo 60:85–94. 10.4081/reumatismo.2008.85 [DOI] [PubMed] [Google Scholar]
- 61.Detert J, Pischon N, Burmester GR, Buttgereit F. 2010. The association between rheumatoid arthritis and periodontal disease. Arthritis Res. Ther. 12:218. 10.1186/ar3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Persson RG. 2012. Rheumatoid arthritis and periodontitis—inflammatory and infectious connections. Review of the literature. J. Oral Microbiol. 4:11829. 10.3402/jom.v4i0.11829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Routsias JG, Goules JD, Goules A, Charalampakis G, Pikazis D. 2011. Autopathogenic correlation of periodontitis and rheumatoid arthritis. Rheumatology (Oxford) 50:1189–1193. 10.1093/rheumatology/ker090 [DOI] [PubMed] [Google Scholar]
- 64.Mangat P, Wegner N, Venables PJ, Potempa J. 2010. Bacterial and human peptidylarginine deiminases: targets for inhibiting the autoimmune response in rheumatoid arthritis? Arthritis Res. Ther. 12:209. 10.1186/ar3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, Kinloch A, Culshaw S, Potempa J, Venables PJ. 2010. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62:2662–2672. 10.1002/art.27552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamonal J, Acevedo A, Bascones A, Jorge O, Silva A. 2000. Levels of interleukin-1β, -8, and -10 and RANTES in gingival crevicular fluid and cell populations in adult periodontitis patients and the effect of periodontal treatment. J. Periodontol. 71:1535–1545. 10.1902/jop.2000.71.10.1535 [DOI] [PubMed] [Google Scholar]
- 67.Dias IH, Marshall L, Lambert PA, Chapple IL, Matthews JB, Griffiths HR. 2008. Gingipains from Porphyromonas gingivalis increase the chemotactic and respiratory burst-priming properties of the 77-amino-acid interleukin-8 variant. Infect. Immun. 76:317–323. 10.1128/IAI.00618-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mikolajczyk-Pawlinska J, Travis J, Potempa J. 1998. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 440:282–286. 10.1016/S0014-5793(98)01461-6 [DOI] [PubMed] [Google Scholar]
- 69.Sundqvist G, Figdor D, Hanstrom L, Sorlin S, Sandstrom G. 1991. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand. J. Dent. Res. 99:117–129 [DOI] [PubMed] [Google Scholar]
- 70.Van Steenbergen TJ, Delemarre FG, Namavar F, De Graaff J. 1987. Differences in virulence within the species Bacteroides gingivalis. Antonie Van Leeuwenhoek 53:233–244. 10.1007/BF00393930 [DOI] [PubMed] [Google Scholar]
- 71.Laine ML, van Winkelhoff AJ. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322–325. 10.1111/j.1399-302X.1998.tb00714.x [DOI] [PubMed] [Google Scholar]
- 72.Imamura T, Travis J, Potempa J. 2003. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr. Protein Pept. Sci. 4:443–450. 10.2174/1389203033487027 [DOI] [PubMed] [Google Scholar]
- 73.Popadiak K, Potempa J, Riesbeck K, Blom AM. 2007. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J. Immunol. 178:7242–7250 [DOI] [PubMed] [Google Scholar]
- 74.Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. 1992. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 267:18902–18907 [PubMed] [Google Scholar]
- 75.Smith AJ, Minhas T, Greenman J, Embery G. 1993. The distribution and properties of some hydrolytic enzymes from Porphyromonas gingivalis W50. Microbios 73:185–197 [PubMed] [Google Scholar]
- 76.O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409–426. 10.2174/1389203033487009 [DOI] [PubMed] [Google Scholar]
- 77.Minhas T, Greenman J. 1989. Production of cell-bound and vesicle-associated trypsin-like protease, alkaline phosphatase and N-acetyl-beta-glucosaminidase by Bacteroides gingivalis strain W50. J. Gen. Microbiol. 135:557–564 [DOI] [PubMed] [Google Scholar]
- 78.Harvey GP, Fitzsimmons TR, Dhamarpatni AA, Marchant C, Haynes DR, Bartold PM. 2013. Expression of peptidylarginine deiminase-2 and -4, citrullinated proteins and anti-citrullinated protein antibodies in human gingiva. J. Periodontal Res. 48:252–261. 10.1111/jre.12002 [DOI] [PubMed] [Google Scholar]
- 79.Jauregui CE, Wang Q, Wright CJ, Takeuchi H, Uriarte SM, Lamont RJ. 2013. Suppression of T-cell chemokines by Porphyromonas gingivalis. Infect. Immun. 81:2288–2295. 10.1128/IAI.00264-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cole AM, Ganz T, Liese AM, Burdick MD, Liu L, Strieter RM. 2001. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623–627 [DOI] [PubMed] [Google Scholar]
- 81.Yung SC, Murphy PM. 2012. Antimicrobial chemokines. Front. Immunol. 3:276. 10.3389/fimmu.2012.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]