Abstract
Prevotella intermedia is an oral bacterium implicated in a variety of oral diseases. Although internalization of this bacterium by nonphagocytic host cells is well established, the molecular players mediating the process are not well known. Here, the properties of a leucine-rich repeat (LRR) domain protein, designated AdpF, are described. This protein contains a leucine-rich region composed of 663 amino acid residues, and molecular modeling shows that it folds into a classical curved solenoid structure. The cell surface localization of recombinant AdpF (rAdpF) was confirmed by electron and confocal microscopy analyses. The recombinant form of this protein bound fibronectin in a dose-dependent manner. Furthermore, the protein was internalized by host cells, with the majority of the process accomplished within 30 min. The internalization of rAdpF was inhibited by nystatin, cytochalasin, latrunculin, nocodazole, and wortmannin, indicating that microtubules, microfilaments, and signal transduction are required for the invasion. It is noteworthy that preincubation of eukaryotic cells with AdpF increased P. intermedia 17 internalization by 5- and 10-fold for HeLa and NIH 3T3 fibroblast cell lines, respectively. The addition of the rAdpF protein was also very effective in inducing bacterial internalization into the oral epithelial cell line HN4, as well as into primary cells, including human oral keratinocytes (HOKs) and human umbilical vein endothelial cells (HUVECs). Finally, cells exposed to P. intermedia 17 internalized the bacteria more readily upon reinfection. Taken together, our data demonstrate that rAdpF plays a role in the internalization of P. intermedia 17 by a variety of host cells.
INTRODUCTION
Leucine-rich repeat (LRR) domain proteins play a major role in host-pathogen interactions (1). These are proteins containing repeats of 20 to 29 residues that form very versatile arc-shaped structural surfaces that are ideal for the formation of protein-protein interactions (2). As such, they are present in a variety of organisms, serving mainly as receptors. Viruses, bacteria, archaea, and eukaryotes have been shown to use LRR domain proteins to mediate immune response, apoptosis, adhesion, invasion, and signal transduction, as well as DNA/RNA processing (2, 3, 4). In eukaryotes, LRR domain proteins form pattern recognition receptors, such as Toll-like receptors (TLRs), which are involved in the immune response to invading pathogens (5, 6). In bacteria, LRR domain proteins have been shown to also mediate a multitude of processes, including the ability of pathogens to attach to and be internalized by host cells (7). However, despite the widespread presence of LRR domain proteins, their roles in host-pathogen interactions remain underinvestigated.
The oral cavity is inhabited by a large number of bacteria of as many as 700 various phylotypes (8). This number may even be higher, as recent studies using high-throughput sequencing, such as 454 pyrosequencing, have revealed a much greater diversity of the oral microbiome; for instance, plaque derived from 98 healthy individuals has been shown to be composed of approximately 10,000 phylotypes (9). Although oral bacteria are mainly believed to be extracellular, it is now well established that many microbial species are also present within gingival epithelial cells (10, 11, 12). The ability of bacteria to be internalized allows them to escape host innate immunity surveillance, provides them with a nutritional niche, and shields them from the action of antibiotics. For these reasons, intracellular pathogens can serve as a microbial reservoir for future reinfections.
We investigated Prevotella intermedia strain 17, a Gram-negative, anaerobic bacterium that is associated with the development and progression of periodontal disease based on its high prevalence in adult periodontitis lesions (13). It is also found at healthy sites (14, 15); however, the virulence of the bacterium may be different at these sites, as it has been shown that the profile of degradative enzymes produced by P. intermedia varies depending on the site at which it is present (16). The primary oral health problems associated with P. intermedia are endodontic infections, including root canal infection, apical periodontitis, and periapical lesions (17). In addition, extraoral diseases, such as tracheitis in children and cancrum oris (also known as NOMA, an infection that destroys oral facial tissues) lesions have been shown to contain P. intermedia (18, 19). The health burden associated with this bacterium may even be higher, as various studies have shown an association between periodontitis and other systemic conditions, such as coronary heart disease and preterm delivery of low-birth-weight infants (20). Indeed, nucleic acid from periodontopathogens, including P. intermedia, has been found in atherosclerotic plaques (20). Also, a significantly higher prevalence of positive fetal IgM to P. intermedia has been demonstrated for preterm compared to full-term infants (21).
P. intermedia 17 is among the oral bacteria that are capable of invading a variety of nonphagocytic eukaryotic cells (22, 23). Previous work has shown that P. intermedia 17 type C fimbriae are required for invasion of epithelial cells (23). Also, our laboratory has identified and characterized a surface protein, AdpB, which binds a variety of host extracellular matrix components (24). The role of the protein in promoting adhesion and invasion of the bacterium is still unknown. Interestingly, we have also identified several genes coding for LRR domain proteins (2). One of the gene products, AdpC, has been shown to be a cell-surface-exposed outer membrane protein that confers an invasive phenotype on Escherichia coli cells expressing the protein (25). Here, we characterized another LRR domain protein, AdpF, encoded by the PI0493 gene (Los Alamos annotation, The Bioinformatics Resource for Oral Pathogens [BROP] at genome.brop.org), that binds fibronectin and promotes the invasion of P. intermedia 17 into a variety of host cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. intermedia 17, Porphyromonas gingivalis strain W83, Streptococcus sanguinis strain SK-36, and Streptococcus gordonii strain DL-1 were grown anaerobically (80% N2, 10% H2, 10% CO2) at 37°C in an anaerobic chamber (Coy Manufacturing, Ann Arbor, MI, USA) on blood agar plates using Trypticase soy agar (TSA II with 5% sheep blood; BBL, Cockeysville, MD) or brain heart infusion (BHI) broth containing hemin (5 μg/ml; Sigma, St. Louis, MO). Escherichia coli TOP10 and BL21(DE3) cells were grown in Luria-Bertani (LB) medium (Gibco, Gaithersburg, MD) with or without 1.5% agar. Transformants carrying either recombinant pCR2.1 vector or the pET30a-adpF plasmid were selected with 50 μg/ml of kanamycin sulfate. For the expression of AdpF in E. coli BL21(DE3), LBM medium (10 g tryptone, 5 g yeast extract, 10 g NaCl, 50 g glycerol, pH 6.9) was used, and E. coli cells were grown at 30°C after induction.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Plasmid | Description | Source |
|---|---|---|---|
| P. intermedia 17 | Kai Leung, U.S. Army Dental Research Detachment, Great Lakes, IL | ||
| P. gingivalis W83 | This study | ||
| S. sanguinis SK-6 | Todd Kitten's collection | ||
| S. gordonii DL-1 | Todd Kitten's collection | ||
| E. coli strains | |||
| One Shot | Chemically competent | Invitrogen | |
| TOP10 strains | |||
| TOP10 parent strain | pCR2.1 | Cloning vector | Invitrogen |
| V2927 | pV2927 | Kanr, pCR2.1 containing the 1.989-kb adpF gene | This study |
| V2928 | pV2828 | Kanr, pCR2.1 containing the truncated adpF gene (−103 bp) | |
| V2720 | pV2720 | Kanr, pET30a | Novagen |
| BL21(DE3)-pLysE strains | Chemically competent | Bioline | |
| V2932 | pV2932 | Kanr, pET30a containing the 1.989-kb adpF gene | This study |
| V2933 | pV2933 | Kanr, pET30a containing the truncated adpF gene (−103 bp) | This study |
Eukaryotic cells and culture conditions.
Primary cultures of human oral keratinocytes (HOKs; ScienCell Research Laboratories, Carlsbad, CA), isolated from human oral mucosa, and human umbilical vein endothelial cells (HUVECs) were cultivated in oral keratinocyte medium (OKM; ScienCell Research Laboratories, Carlsbad, CA) and Vasculife vascular endothelial growth factor (VEGF) cell culture medium (Lifeline Cell Technologies, Frederick, MD), respectively, at 37°C in an atmosphere containing 5% CO2. NH4 cells, which were derived from a primary tongue squamous cell carcinoma (26), were provided by Andrew Yeudall of Virginia Commonwealth University, Richmond, VA. HeLa and NIH 3T3 cells were prepared as previously described (18). The cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 mM sodium pyruvate, and 100 mM HEPES at 37°C in an atmosphere containing 5% CO2.
Expression of the PI0493 gene (encoding AdpF) in E. coli.
Full-length (1,989 bp) and truncated (starting from base 103 and thus deleting the portion of adpF coding for the signal peptide and ensuring that the gene product will remain intracellular) versions of the PI0493 gene were amplified using the primers shown in Table S1 in the supplemental material. Both amplified products were cloned into bacterial expression vector pET 30a(+) (Novagen), creating pET30-adpF1 (full-length gene) and pET30-adpF2 (truncated gene). These plasmids were transformed into E. coli BL21(DE3)-pLysE cells (Bioline, Taunton, MA), and protein expression was induced by the addition of 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma). Following overnight growth at 30°C, the cells were harvested by centrifugation (6,000 rpm for 10 min), and the recombinant AdpF2 (rAdpF2) protein was purified under native conditions using Ni-nitrilotriacetic acid (NTA) agarose following the manufacturer's instructions (Qiagen, United States). The purity of the rAdpF protein was verified by SDS-PAGE (12% Bis-Tris NuPAGE; Invitrogen). The purified protein was then dialyzed into 25 mM Tris buffer, pH 8.0, and concentrated with an Amicon 10K filter unit (Millipore, MA).
Antibody for rAdpF.
Polyclonal anti-rAdpF antibody was commercially developed in rabbits by Proteintech Group, Inc. (Chicago, IL). Two New Zealand White rabbits were immunized with 200 μg of rAdpF with complete Freund's adjuvant, followed by two booster doses of 100 μg protein in incomplete Freud's adjuvant at 2-week intervals. Sera were prepared from blood collected 2 weeks after the administration of the final booster dose. The antibody was affinity purified by immobilizing rAdpF on a nitrocellulose membrane using standard protocols (27).
Binding of rAdpF to ECM proteins.
Enzyme-linked immunosorbent assays (ELISA) were performed to examine the ability of rAdpF to bind extracellular matrix proteins (ECMs), including fibronectin (from human placenta), fibrinogen fraction I (from human plasma), collagen IV (from human placenta), collagen type I (from human skin), and laminin (from human placenta). All ECMs were purchased from Sigma (St. Louis, MO). Ninety-six-well ELISA plates (Costar, Corning, NY) were coated with 100 μl of ECM protein at a concentration of 20 μg/ml in phosphate-buffered saline (PBS) containing Tween 20 (PBS-T) (0.02%, vol/vol) and immobilized for 2 h at 37°C. Bovine serum albumin (BSA) was used as a control. Following incubation, the ELISA plates were washed with PBS-T buffer and blocked for 2 h in PBS-T containing 3% nonfat milk. The plates were then washed three times, and 100-μl amounts of serial dilutions of rAdpF protein, starting from 250 μg/ml, were added to the wells and incubated at 37°C for 2 h. The plates were then washed and incubated with a 1:2,500 dilution of anti-rAdpF primary antibody for 1 h at 37°C. The plates were washed, and a 1:5,000 dilution of alkaline phosphatase-conjugated anti-rabbit IgG antibody was added. Following incubation, the reaction mixtures were developed using p-nitrophenylphosphate (PNPP) from Bio-Rad, and the absorbance at A405 was monitored with a Thermo Multiskan microplate reader (Fisher Scientific, United States). Competitive ELISA was done using a 96-well plate coated with fibronectin (10 μg/ml). Following blocking, rAdpF (20 μg/ml) preincubated with serial dilutions of fibronectin, starting at a concentration of 10 μg/ml, was added. The bound rAdpF was then detected with anti-rAdpF antibody as described above.
SDS-PAGE and immunoblot analysis.
Protein samples were separated on a 10% NuPAGE Bis-Tris gel (Invitrogen, United States). Gels were stained with 0.2% (wt/vol) Coomassie brilliant blue R-250 (Sigma, St. Louis. MO) in ethanol-acetic acid-water (30:10:60, vol/vol/vol). For Western blot analysis, the proteins were electrophoretically transferred to a nitrocellulose membrane by electroblotting, the membrane was blocked in Hanks balanced salt (HBS) buffer (8 g NaCl, 3.7 g KCl, 1.4 g Na2HPO4, 10 g dextrose, 50 g HEPES, pH 7.0) containing 0.02% Tween 20 with 3% skim milk, and the proteins immunoreacted with a 1:1,000 dilution of mouse anti-HisTag monoclonal antibody (Novagen), followed by incubation with a 1:5,000 dilution of anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (SouthernBiotech, Birmingham, AL). After washing with HBS-T (HBS with 0.02% Tween 20), the protein bands were visualized using a Western Lightning chemiluminescence reagent kit (PerkinElmer LAS, Waltham, MA).
Dot blot analysis.
E. coli cells expressing AdpF, E. coli cells with only the pET30a vector (used as a control), and P. intermedia 17 strains were grown to mid-exponential phase. Cells were centrifuged, washed three times, and suspended in PBS. Two sets of bacterial cells were used, one untreated and one treated with protease for 30 min (to degrade the cell surface proteins). Also, lysates from E. coli cells expressing AdpF and P. intermedia 17 were used for this analysis. In addition, proteins from P. intermedia 17 culture supernatant were concentrated by following the standard ammonium sulfate precipitation technique. Purified rAdpF and BSA were used as controls. Five-microliter aliquots of each sample were spotted on a nitrocellulose membrane and air dried. The membrane was then reacted with anti-rAdpF antibody and developed following the steps described above for Western blot analysis.
Electron microscopy.
P. intermedia 17 was grown in BHI medium to an optical density at 660 nm (OD660) of 0.6. Bacterial cells were harvested by centrifugation (6,000 rpm for 10 min), washed three times with PBS, and suspended in PBS containing 1% skim milk. Anti-rAdpF antibody was added to the bacterial suspension and incubated anaerobically for an additional 2 h. P. intermedia 17 grown without the addition of anti-rAdpF antibody was used as a negative control. After incubation, the bacterial cells were harvested, washed three times with PBS, and suspended in PBS containing 1% skim milk. Anti-rabbit IgG-gold antibody (Sigma-Aldrich, St. Louis, MO) was applied to the bacterial suspension and incubated anaerobically for 2 h. Cells were fixed with 4% paraformaldehyde–0.1% glutaraldehyde in 0.1 M cacodylate buffer. The cells were then dehydrated in serial dilutions of ethanol (50%, 70%, 80%, and 95%) for 5 to 10 min and washed three times with 100% ethanol. The cells were sectioned with an LKB 2128 ultramicrotome. Seven- to 9-μm-thick sections placed on grids and stained with 5% uranyl acetate and Reynold's lead citrate were examined with a transmission electron microscope (Jeol JEM; Jeol USA).
Interaction of rAdpF with host cells using confocal microscopy.
HUVECs, grown as described above, were allowed to adhere to wells of a 6-well tissue culture plate (400,000 cells/well). Both rAdpF and BSA (control protein) were fluorescently labeled using Cy5 (Amersham). Cy5 dye (5 μl at 400 pM) was added to 500 μg of protein solution in PBS (pH 8.5) and incubated on ice in the dark for 1 h. Protein was precipitated using the standard chloroform-methanol protein precipitation method to remove the unbound dye. The dried, labeled protein pellet was dissolved in PBS (pH 7.4). Cy5-labeled rAdpF diluted in cell culture medium was used to replace the old medium, and the plate was incubated in the dark for 2 h. Wells without Cy5-labeled proteins served as controls. The eukaryotic cells were then washed three times with PBS and fixed by incubating in 4% paraformaldehyde for 10 min at room temperature. This solution was replaced with PBS containing 0.05% Tween 20 and incubated again for 10 min. Following the last wash, a coverslip was placed over the glass slide with an antifade reagent containing 4′6-diamidino-2-phenylindole (DAPI) and sealed. The cells were examined using a Zeiss LSM 710 Meta confocal scanning microscope. Wavelengths of 405 nm and 633 nm were used for DAPI and Cy5 analysis, respectively. Z-stack scanning-microscopy images were acquired at steps of 0.1 μm from the bottom to the top of the cell using a 40× objective to determine the position of the red fluorescence of the protein in the cell.
Interaction of P. intermedia 17 with host cells.
All eukaryotic cells were grown as described above. Then, 400,000 cells/well were aliquoted onto a 6-well plate and the plate was incubated for a day to achieve approximately 80% confluence. Next, different concentrations of rAdpF, ranging from 0 to 100 μg/ml, were added to the plate. The plate was then incubated for 2 h before being washed three times with cell medium. The cells were infected with a fresh culture of P. intermedia 17 at a multiplicity of infection (MOI) of 100 and incubated for 30 min. The plate was then washed four times with PBS to remove any unattached bacteria. The total number of interacting bacteria, including all adhered and internalized bacteria, was determined by lysing the eukaryotic cells in BHI medium containing 1% saponin (incubation at 37°C for 10 min), followed by scraping of the lysed cells. The mixture was serially diluted, plated on TSA II blood agar plates, and incubated anaerobically. CFU were determined following 7 days of incubation. The total bacterial interaction (adherence and invasion) was calculated as the percentage of the inoculated bacteria that was recovered from eukaryotic cells. To account for internalized bacteria, the infected eukaryotic cells were incubated in medium containing 300 μg/ml gentamicin and 400 μg/ml metronidazole for 1 h to kill the extracellularly adhered bacteria. Internalized bacteria were determined as described above. Invasion efficiency (%) was also expressed as the percentage of the inoculum of P. intermedia 17 recovered from cells that were treated with antibiotics to kill extracellular bacteria.
To determine the contribution of AdpF to P. intermedia 17 internalization, bacteria were pretreated with affinity-purified anti-AdpF antibody for 30 min prior to infection. The infection was carried out as described above.
Flow cytometry analysis of internalized P. intermedia 17.
P. intermedia 17 cells from culture grown to mid-exponential phase (OD660 of 0.4 to 0.7) were harvested, washed with PBS, and incubated with 3.5 μl of fluorescein isothiocyanate (FITC) (Fluorescein-5-EX, succinimidyl ester; Invitrogen) for 1 h 40 min at 4°C (28). Following washing, the bacteria were suspended in antibiotic-free eukaryotic cell medium and were used for infection of HUVECs. HUVEC cells, preincubated previously with rAdpF or P. intermedia 17 (MOI of 1:100), were infected with FITC-labeled P. intermedia 17 for 30 min and washed with cell medium to remove extracellular bacteria. The infected cells were then incubated with trypan blue (1:500 dilution; Sigma) to quench the fluorescence from extracellular bacteria. Internalized bacteria were quantified by flow cytometry (using a BD Canto II at the VCU Flow Cytometry Core Laboratory). All flow cytometric analyses were performed on live cells, based on incorporation of propidium iodide and/or forward and side scatter, with BD FACSDiva software. FITC fluorescence was captured (emission wavelength of 518 nm) and plotted against the number of cells. At least 10,000 cells were analyzed in each sample. Healthy cell populations were gated and analyzed. Gates were set to eliminate any unstained material or debris. Since trypan blue has been widely used as an extracellular quencher of FITC, 0.4% trypan blue solution (Sigma-Aldrich) that was diluted 50 times was added to the sample and incubated for 2 min at room temperature before the reading. In addition, crystal violet solution at a final concentration of 500 μg/ml for 10 min was added to the cells before the reading as an intracellular quencher.
Effects of metabolic inhibitors on the association of rAdpF with HUVEC cells.
Metabolic inhibitors were obtained from Sigma-Aldrich, and the following final inhibitor concentrations were used: 10 mM methyl-β-cyclodextrin (MBCD; cholesterol inhibitor), 50 μg/ml nystatin, 2 μM latrunculin A (disrupts microfilament-mediated processes), 25 μM nocodazole (disrupts microtubules), 0.5 μg/ml cytochalasin (actin polymerization inhibitor), and 90 nM wortmannin (phosphatidylinositol 3-kinase [PI3-K] inhibitor). In addition, cells were incubated at 4°C to see if cellular metabolism played a role in mediating the interaction with rAdpF. HUVEC cells were grown on a 10-cm plate for 24 h, and then the cell medium was changed and the culture treated with inhibitors for 30 min at 37°C. The cells were then washed with PBS and incubated with 25 μg of fluorescence-labeled rAdpF for 1 h at 37°C. Washed and trypsinized cells were examined by flow cytometry as described above.
Bioinformatics analysis.
NCBI (www.ncbi.nlm.nih.gov) and ExPASy (www.expasy.org) were used for bioinformatics studies. An AdpF homology model was generated using I-TASSER, which combines the methods of threading, ab initio modeling, and structural refinement. The iterative threading assembly refinement (I-TASSER) server is an integrated platform for automated protein structure/function prediction based on the sequence-to-structure-to-function paradigm (29, 30).
Data analysis.
All experiments were performed at least three times. Data are summarized as the means ± standard deviations (SD) in all graphs. The statistical analysis of the results was performed by one-way analysis of variance (ANOVA) using Graphpad Prism (GraphPad Software, San Diego, CA). A P value of <0.05 was considered significant. Error bars represent the standard deviations between the samples.
RESULTS
Bioinformatic characterization of AdpF.
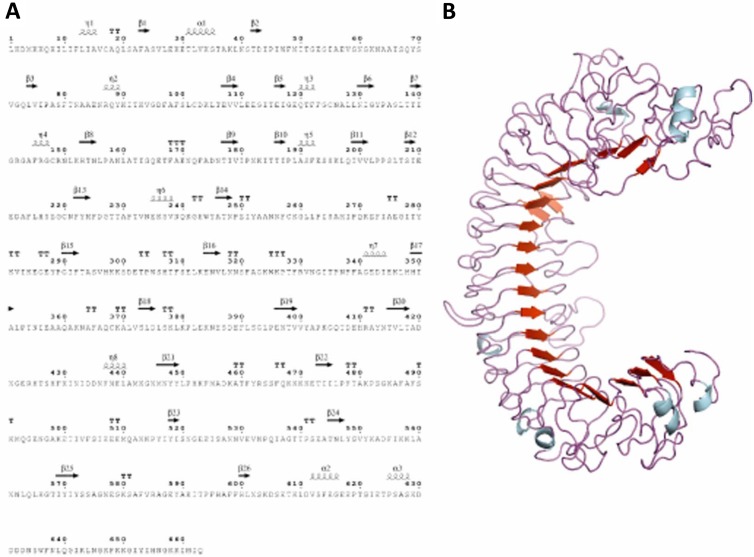
Analysis of the P. intermedia 17 genomic sequence revealed the presence of an open reading frame composed of 1,989 bp encoding a predicted LRR domain-like protein, herein designated AdpF. A BLAST search showed that it had the highest similarity to putative proteins from Prevotella nigrescens, Prevotella sp. oral taxon 472, and Prevotella sp. oral taxon 317. The closest similarity to a protein that has been experimentally verified was to the surface antigen BspA from Tannerella forsythensis (31); the AdpF protein shared 34% identity and 57% similarity to that protein. Bioinformatics analysis of the predicted protein using the TMHMM tool (www.expasy.org) detected the initial 8 N-terminal amino acids as intracellular, the 9th to 18th N-terminal amino acids as comprising a single transmembrane helix (signal peptide), and the remainder of the 663 amino acids of the protein as extracellular (Fig. 1A). Nine full LRRs and three partial LRRs with similarity to the Treponema palladium LRR protein were identified within the protein (see Fig. S1 in the supplemental material). Molecular modeling of the protein structure revealed an arc-shaped structure where the typical parallel LRRs form β-sheets with a concave structure (Fig. 1B) (32).
FIG 1.
Characteristics of P. intermedia 17 AdpF. (A) AdpF is encoded by the PI0493 gene (Los Alamos annotation using BROP Genome Viewer at genome.brop.org). The AdpF amino acid sequence is shown, and the positions of the beta sheets indicated. (B) Predicted structure of AdpF, determined using I-Tasser (30).
AdpF is a cell surface protein.
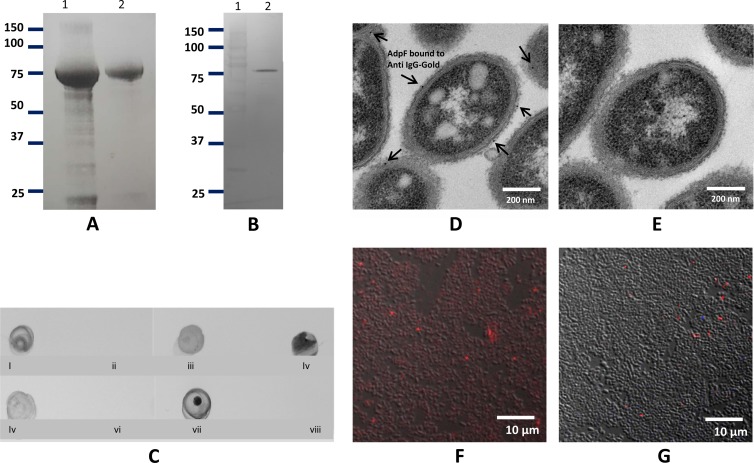
To determine the localization and role of the protein, we prepared two forms of recombinant AdpF (rAdpF), full length and truncated (lacking the predicted mechanisms for protein secretion). Bacterial cell lysates containing the truncated form of rAdpF had higher levels of protein expression than lysates containing full-length protein (data not shown). Thus, the truncated form of the protein was selected to prepare the recombinant form of the protein in this study. The protein remained soluble in bacterial cells and was purified by affinity chromatography using a Ni-NTA resin under native conditions. Following purification, only one protein band with the expected 80-kDa size was observed, and the purity of our protein was confirmed on SDS-PAGE gel (Fig. 2A, lane 2). We further investigated the presence of the protein in P. intermedia 17 using Western blot analysis. First, we examined the presence of AdpF in culture supernatants; a protein band of approximately 80 kDa was detected (Fig. 2B). These data indicated that the protein is present in the culture supernatant. To verify the localization, we performed a dot blot assay. As shown in Fig. 2C, the protein was detected in P. intermedia 17 cells; however, it was not detected in cells that were treated with protease prior to dot blot analysis. As the protease cleaved any accessible proteins on the bacterial cell surface, the data show that AdpF is a cell surface protein. As expected, it was present in lysed P. intermedia 17 cells, as well as in culture supernatant (Fig. 2C). Results for E. coli cells expressing rAdpF and carrying the control vector are shown in Fig. 2C panels v and vi, respectively. Finally, anti-rAdpF antibody reacted with rAdpF but not with BSA (Fig. 2C panels vii and viii respectively). These data demonstrate that AdpF is a P. intermedia 17 cell surface protein that is also secreted into culture medium.
FIG 2.
Purification and cellular localization of AdpF. (A) SDS-PAGE analysis of cell extract after IPTG induction (lane 1) and of purified recombinant AdpF from E. coli (rAdpF) (lane 2). Molecular weight (in thousands) is shown. (B) Western blot analysis of concentrated P. intermedia 17 culture supernatant. Anti-rAdpF antibody was used to identify the protein. Lanes contain molecular weight marker (in thousands) (lane 1) and P. intermedia 17 culture supernatant proteins (lane 2). (C) Dot blot assay. Five-microliter aliquots were spotted on a nitrocellulose membrane and developed with anti-rAdpF antibody. Blots are as follows: P. intermedia 17 cells (i), P. intermedia 17 cells treated with protease (ii), P. intermedia 17 cell lysate (iii), P. intermedia 17 culture supernatant (iv), lysate of E. coli expressing rAdpF (v), lysate of E. coli carrying no insert vector, as a control (vi), purified rAdpF (vii), BSA (viii). (D and E) Surface localization of anti-AdpF antibody on P. intermedia 17 was shown by immunoelectron microscopy. The anti-AdpF antibodies, labeled with gold particle (arrows), were localized in the outer membrane of P. intermedia 17 (D), and there were no detectable anti-AdpF antibodies in P. intermedia 17 after protease treatment (E). (F and G) Localization of Cy5-labeled anti-rAdpF antibody in P. intermedia 17 cells without treatment with protease (F) and following treatment with protease (G). Bacteria binding Cy5-labeled anti-AdpF antibody appear red in the image.
To further confirm the surface localization of AdpF, we also performed immunogold electron microscopy studies using polyclonal antibodies generated against AdpF protein as the primary antibody and goat anti-mouse antibody conjugated with gold particles as the secondary antibody. As shown in Fig. 2D, gold-labeled antibody was found only in the outer membrane of P. intermedia 17 when antibodies against rAdpF were used as the primary antibody. In contrast, there were no gold antibodies detected on the surface of P. intermedia 17 pretreated with protease K (Fig. 2E).
For confocal microscopy analysis, anti-rAdpF antibodies were fluorescently labeled using Cy5 dye and applied to P. intermedia 17 cells. The results shown in Fig. 2F indicated that fluorescently labeled anti-rAdpF antibodies were bound to the surface of the bacteria. On the other hand, there were no anti-rAdpF antibodies attached to the membrane of P. intermedia 17 cells pretreated with protease K (Fig. 2G). Based on the results described above, we conclude that AdpF is located on the surface of P. intermedia 17.
AdpF binds fibronectin.
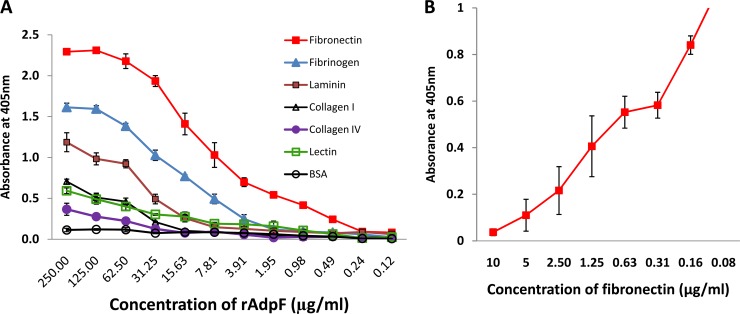
P. intermedia 17 binds a variety of ECM proteins. Previously, we showed that it binds fibrinogen (1). In this study, we have verified that it binds fibronectin. The binding activity is concentrated in the membrane fraction of the cell. To examine the ability of rAdpF to bind ECM proteins, 96-well ELISA plates were coated with ECM proteins, and different concentrations of rAdpF were added and incubated to allow binding. From the results shown in Fig. 3A, rAdpF was found to primarily bind fibronectin. The binding was concentration dependent, and the interactions involved in the binding were specific. We also found that both fibrinogen and laminin showed binding in this assay, thus indicating that AdpF may bind multiple ECMs, although with different affinities. Other ECM proteins, such as collagen I and collagen IV, showed lower levels of binding with rAdpF (Fig. 3A). Also, the control protein, BSA, did not bind rAdpF. To further confirm the AdpF binding specificity, a competitive inhibition assay was performed to inhibit the binding of rAdpF to fibronectin (10 μg/ml) immobilized on the plate. The same steps as outlined above for the ELISA were followed, except that rAdpF was mixed with various concentrations of fibronectin prior to deposition in the 96-well plates. As shown by the results in Fig. 3B, soluble fibronectin inhibited AdpF binding in a dose-dependent manner, thus indicating that the fibronectin binding to rAdpF is specific.
FIG 3.
Binding characteristics of rAdpF to extracellular matrix (ECM) proteins. (A) Results of ELISA analysis of rAdpF binding to various ECM proteins. Absorbance at 405 nm (detection of secondary antibody binding to anti-rAdpF antibody) is plotted on the y axis, and serial dilutions of rAdpF (starting with 250 μg/ml) and immobilization of each ECM protein (20 μg/ml) on the 96-well plate are plotted on the x axis. (B) Competitive inhibition of fibronectin binding to immobilized rAdpF (20 μg/ml) using various amounts of fibronectin (from 10 μg/ml). All experiments were performed in triplicate.
AdpF is internalized by eukaryotic cells.
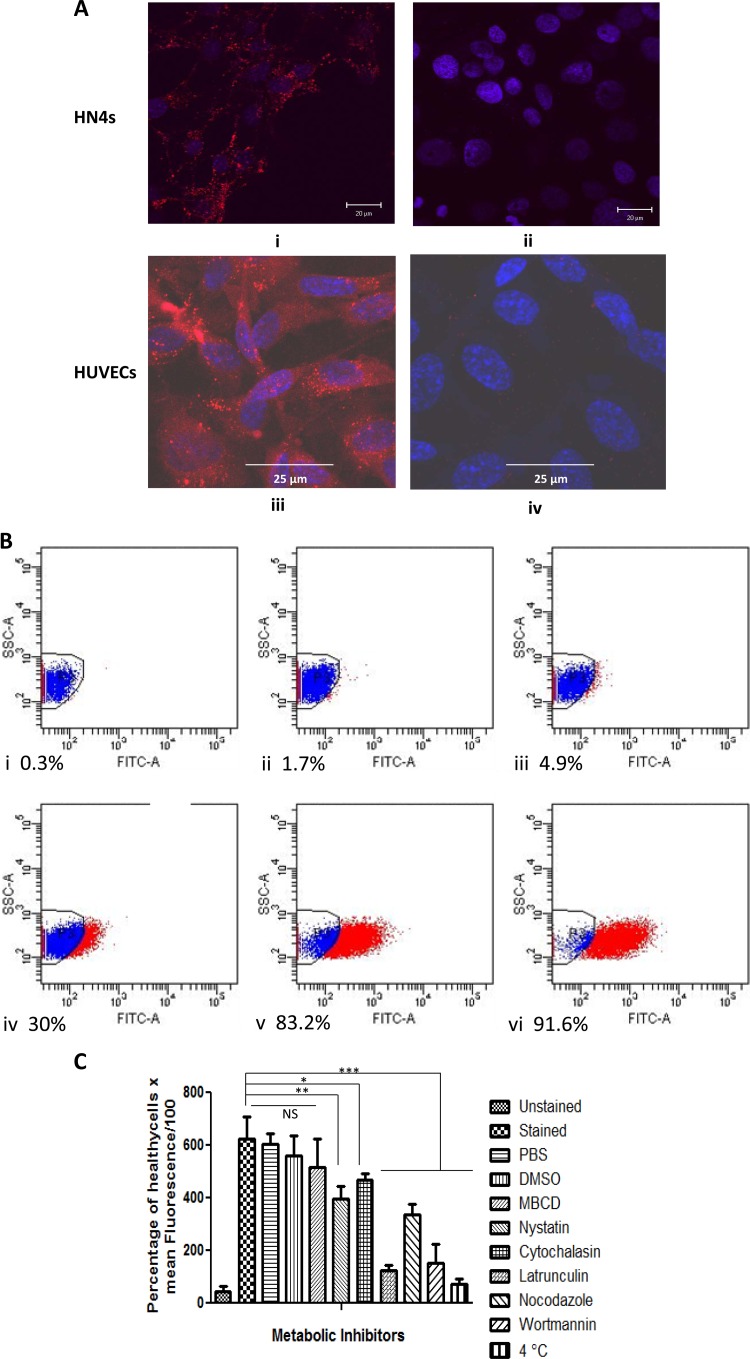
To gain some insight into the biological significance of AdpF, we examined the interaction of rAdpF with eukaryotic cells by using confocal microscopy. The HN4 cells and HUVECs preincubated with Cy5-labeled rAdpF protein showed an intense red signal inside the cells, indicating that rAdpF was internalized by the cells (Fig. 4Ai and Aiii). However, no red signal was detected in cells preincubated with BSA (Fig. 4Aii and Aiv). These results show that rAdpF is internalized by eukaryotic cells and that the protein internalization is specific for rAdpF. The presence of DAPI (blue signal) indicated the nucleus, and the accumulation of red fluorescence surrounding the blue-labeled nucleus for all cell lines suggested that most of the proteins are incorporated in the cytoplasm (Fig. 4).
FIG 4.
Internalization of AdpF by eukaryotic cells. (A) Internalization of rAdpF by various eukaryotic cells. Cy5-labeled rAdpF proteins were incubated with eukaryotic cells as described in the text, and the cells were examined using confocal microscopy. A 100× oil immersion objective lens was used to collect images in the x, y, and z planes. Ratiometric confocal image slices of HN4 cells treated with labeled rAdpF were taken at 0.32-μm intervals. rAdpF (i and iii) and BSA as a control (ii and iv) on HN4 cells and HUVECs were used for this study. The image shows the presence of labeled rAdpF proteins as red spots, and the nuclei of the eukaryotic cells are visualized in blue (stained with DAPI). The images of a cell section at 1 μm below the cell surface are shown. (B) Flow cytometry analysis of time-dependent internalization of fluorescein-labeled AdpF in HN4 cells at 0 min (i), 2 min (ii), 5 min (iii), 12 min (iv), 30 min (v), and 60 min (vi) postexposure. The percentage of cells which were positive and entered the next gate is indicated for each time point. (C) Effects of various inhibitors on internalization of AdpF by HUVEC cells. HUVECs were preincubated with various inhibitors as described in Materials and Methods and were exposed to labeled rAdpF. The ability of rAdpF to be internalized by the host cells was determined by flow cytometry. The inhibitor concentrations resulted in 90% of the cells remaining viable. The relation used in the plot is as follows: (percentage of healthy cells × mean fluorescence)/100. Error bars are described in Materials and Methods. *, P < 0.05; **, P < 0.01; ***, P < 0.0001; NS, not significant.
We then examined the time-dependent entry of rAdpF into epithelial cells by flow cytometry. As shown by the results in Fig. 4B, significant amounts of rAdpF were detected within the first 12 min of incubation, concomitant with a rapid transfer into HN4 cells. With steady transfer, it was nearly complete after a 30-min incubation period.
To investigate possible cellular mechanisms involved in the internalization of rAdpF, we examined the effects of several inhibitors on invasion of the protein. As shown by the results in Fig. 4C, there was no significant inhibition of rAdpF internalization in cells treated with PBS, dimethyl sulfoxide (DMSO), or MBCD. Interestingly, nystatin, a sterol-binding agent shown to increase cell permeability and disrupt the later organization of plasma membrane, resulted in 30% inhibition of rAdpF internalization. Cytochalasin, an inhibitor of actin polymerization, reduced the accumulation of rAdpF inside HUVECs by 20%, while latrunculin, an agent that also alters the state of actin polymerization, as well as disrupting microfilament-mediating processes, inhibited the intracellular accumulation of rAdpF by almost 90% (the largest inhibition observed in our study). Cytochalasin binds to G-actin (actin monomers), while latrunculin binds F-actin (filamentous actin), and thus, while both reagents alter the state of actin polymerization, they do so using different mechanisms. Nocodazole, a microtubule-disrupting agent, inhibited the internalization of rAdpF into HUVECs by 50%. These results indicate that the cell cytoskeleton, as well as both microfilament and microtubule activity, may play a role in internalization of rAdpF by host cells. In cells preincubated with wortmannin, an inhibitor affecting phosphatidylinositol 3-kinase (PI3-K) activity, an 80% reduction in internalization of rAdpF was observed. This result shows that signal transduction mechanisms play a role in the internalization of rAdpF. Finally, metabolically inactive cells, prepared by incubation at 4°C, were not able to internalize rAdpF, suggesting that the internalization of rAdpF requires energy and, thus, is an active process.
AdpF promotes invasion of eukaryotic cells by P. intermedia 17.
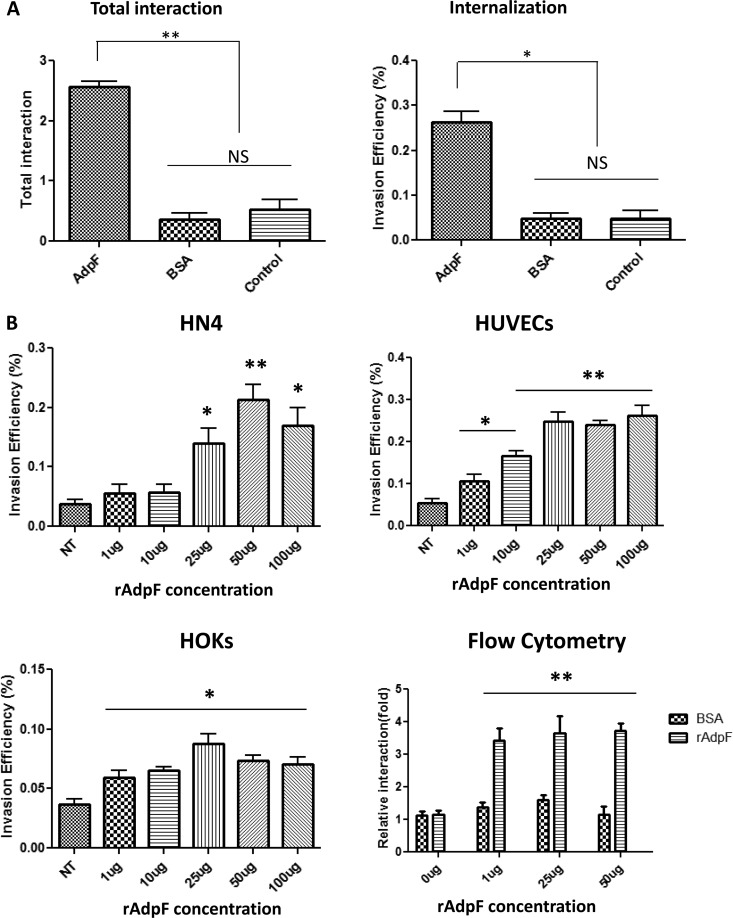
We then speculated that the protein may have an effect on the adhesion/invasion of P. intermedia 17 into nonphagocytic eukaryotic cells. As shown by the results in Fig. 5A, the addition of rAdpF drastically increased the ability of P. intermedia 17 to interact with host cells. The total bacterial interaction (number of bacteria attached and internalized by host cells) with HN4 cells preincubated with rAdpF increased approximately 5-fold compared to the bacterial interaction with HN4 cells preincubated with BSA or not exposed to the protein (control) (Fig. 5A). Similarly, bacterial invasion rates were elevated by 5-fold in HN4 cells exposed to rAdpF compared to the rates in BSA-pretreated and control cells (Fig. 5A). To determine whether the changes were cell specific, we performed the same experiments using HeLa and NIH 3T3 murine fibroblast cell lines. Elevated total bacterial adherence was observed in HeLa cells exposed to rAdpF compared to that in control or BSA-exposed cells. However, there were no significant changes in NIH 3T3 cells (see Fig. S2A in the supplemental material). On the other hand, in HeLa and NIH 3T3 cells pretreated with rAdpF, the rates of invasion increased significantly compared to the rates in cells exposed to BSA or control cells (see Fig. S2B).
FIG 5.
Role of rAdpF in the interaction of Prevotella intermedia 17 with eukaryotic cells. (A) Eukaryotic HN4 cells were exposed to 0.5 ng/ml of rAdpF and infected with P. intermedia 17 as described in Materials and Methods. The total association and internalization of P. intermedia 17 into HN4 cells was determined by the quantification of viable bacteria. The efficiencies of total interaction and internalization are expressed as percentages of the recovered inoculum. *, P < 0.05; **, P < 0.01; NS, not significant. (B) Effect of AdpF concentration on P. intermedia 17 internalization. Various concentrations of rAdpF were used to pretreat HN4 cells, HUVECs, and HOKs prior to infection with P. intermedia 17. The numbers of viable bacteria recovered from eukaryotic cells were determined as described above. These results were further confirmed using flow cytometry analysis. HUVECs were pretreated with various concentrations of AdpF and incubated with fluorescently labeled P. intermedia 17. *, P < 0.05; **, P < 0.01.
We also investigated the dose-dependent effect of rAdpF on the internalization of P. intermedia 17. Three types of eukaryotic cells, HN4 cells, HOKs, and HUVECs, were used for this study. Different concentrations of rAdpF protein (ranging from 1 μg/ml to 100 μg/ml) were preincubated with the eukaryotic cells. For HN4 cells, the invasion efficiency was significantly increased (at least 3 times) by the use of 25 μg of rAdpF (Fig. 5B). Interestingly, using primary cells, i.e., HUVECs and HOKs, we observed that a dose as low as 1 μg of rAdpF (final concentration, 0.5 ng/ml) resulted in significant increases in P. intermedia 17 internalization (Fig. 5B).
To further confirm the induction of internalization of P. intermedia 17 by AdpF, we used flow cytometry. FITC-labeled P. intermedia 17 was added to HUVECs pretreated with different doses of rAdpF and BSA. As shown by the results in Fig. 5A and B, the bacterial internalization was elevated at least 3 times by the addition of rAdpF compared to the bacterial internalization in cells pretreated with BSA (control experiment), thus further verifying the importance of rAdpF for bacterial internalization. Finally, to investigate the contribution of AdpF to P. intermedia 17 invasion, we performed the internalization studies with bacteria preincubated with anti-AdpF antibody. However, as shown by the results in Fig. 6, anti-AdpF antibody did not have any effect on the internalization of P. intermedia 17. Collectively, these results demonstrate that rAdpF enhances the interaction of P. intermedia 17 with host cells.
FIG 6.
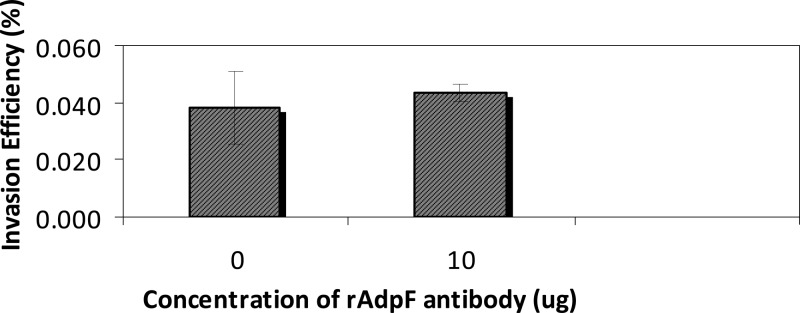
Effect of anti-AdpF antibody on P. intermedia 17 internalization. P. intermedia 17 was prepared as described in Materials and Methods and preincubated with anti-AdpF antibody for 30 min prior to infection of HUVECs. The HUVECs were then infected with the bacteria at an MOI of 1:100 (HUVECs/bacteria) for 30 min. The internalized bacteria were quantified by counting viable bacteria. Invasion efficiency is expressed as the percentage of the inoculum recovered from eukaryotic cells as determined using plating. Values are the means of triplicate samples and are representative of similar results from several experiments.
Infection of eukaryotic cells with P. intermedia 17 increases subsequent internalization of bacteria.
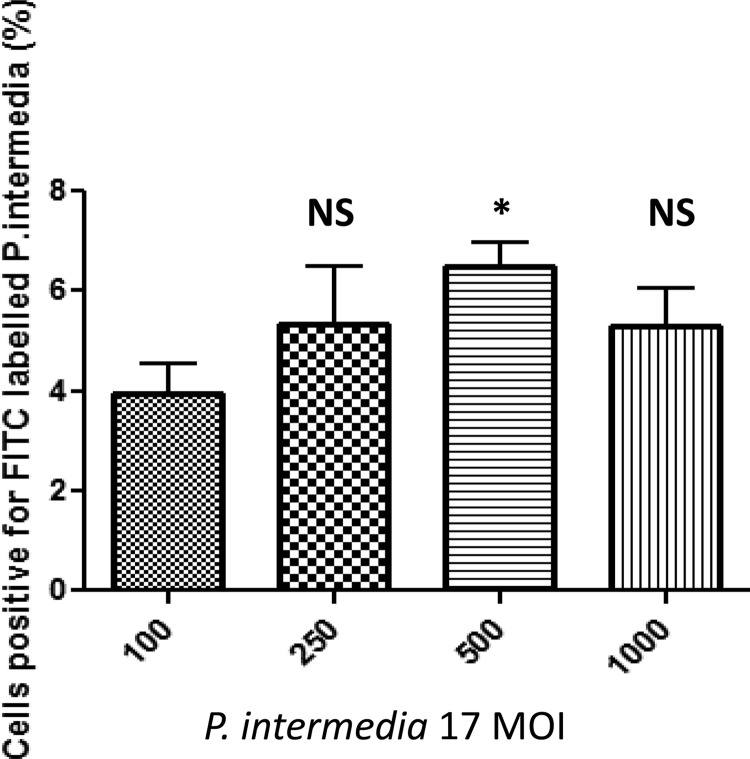
Since rAdpF is a P. intermedia 17 cell surface protein, we reasoned that infection with the bacterium may also have an effect on bacterial internalization. We thus examined the effect of host cell pretreatment with whole bacterial cells. Significant differences in subsequent microbial invasion were noted when using an MOI of 1:500 (Fig. 7). These results demonstrated that exposure of epithelial cells to P. intermedia 17 leads to cellular changes that result in increased uptake of bacteria.
FIG 7.
Effect of P. intermedia 17 challenge on subsequent microbial invasion. P. intermedia 17 was grown as described in Materials and Methods. HN4 cells were infected with different MOIs of P. intermedia 17 for 30 min, washed, and reinfected with fluorescently labeled bacteria. The internalized bacteria were detected by flow cytometry. *, P < 0.05; NS, not significant.
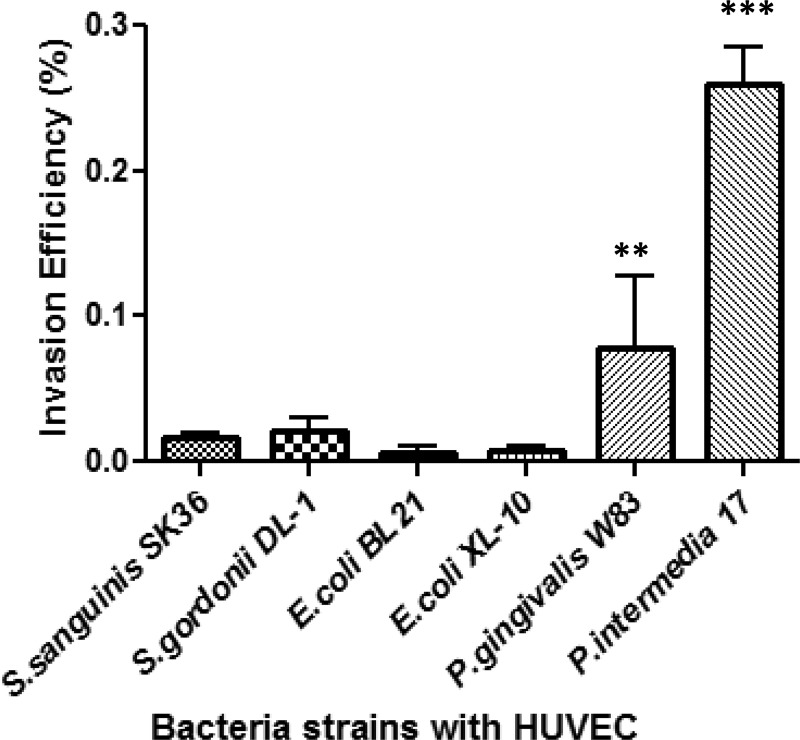
We also examined whether the effect of AdpF on microbial internalization was bacterium specific. P. intermedia 17, Porphyromonas gingivalis W83, Streptococcus sanguinis SK-36, Streptococcus gordonii DL-1, E. coli BL21(DE3), and E. coli XL-10 were used for our study. All of these strains were used to infect HUVEC cells preincubated with 25 μg/ml of rAdpF. As shown by the results in Fig. 8, only P. intermedia 17 was highly invasive, and its invasion efficiency increased significantly upon treatment of the eukaryotic cells with rAdpF, whereas P. gingivalis W83 was only slightly invasive, and no bacterial invasion was observed for S. sanguinis SK-36, S. gordonii DL-1, E. coli BL21(DE3), and E. coli XL-10 (the latter two were used as controls for our study). These results indicate that rAdpF specifically induces the internalization of P. intermedia 17 by nonphagocytic eukaryotic cells.
FIG 8.
Effect of AdpF on invasion of HUVECs by different bacterial strains. HUVEC cells were pretreated with 25 μg of rAdpF protein (2.5 ng/ml final protein concentration) and then infected with various bacteria as described in Materials and Methods. Invasion efficiency is expressed as the percentage of the inoculum recovered from eukaryotic cells as determined using plating. **, P < 0.01; ***, P < 0.0001.
DISCUSSION
We identified and characterized the LRR domain cell surface protein AdpF from P. intermedia 17. This is the second LRR domain protein identified in P. intermedia 17 that plays a role in the internalization of the bacterium (25, 33). Unlike the previously characterized AdpC protein, which acts on the bacterium and confers an invasive phenotype when expressed heterologously in E. coli, AdpF alters the host cells to more readily take up invasive bacteria. Such an outer membrane protein would be expected to play a significant role in altering host-pathogen interactions, considering that it can be secreted into the extracellular environment via outer membrane vesicles produced by P. intermedia 17 (34). Being an LRR domain protein, it can have an effect both through binding to ECM molecules and through signaling mechanisms (2).
AdpF is an LRR domain protein; it is larger than the previously identified internalin-like protein AdpC, but it is predicted to form the characteristic arc-shaped structure. The LRR domain structure is characteristic of those of other microbial internalins (1); thus, it may be a characteristic feature of proteins that promote the internalization of bacteria. AdpF contains the typical signal peptide that targets it for secretion through microbial membranes. A major portion of the protein is predicted to be extracellular, and indeed, our work verified the cell surface location. However, we also showed that AdpF was present in the culture supernatant. The protein size detected in our study was 80 kDa, which is 10 kDa shorter than the full-length protein. Therefore, it is probable that the protein is cleaved from the bacterial cell surface and released into the culture medium. Another source of AdpF, as already mentioned above, could be from vesicles that are secreted by P. intermedia 17 (34).
As a cell surface-located protein, AdpF can interact with the environment. Since it is associated with a human pathogen, such a protein would be expected to play a role in host-pathogen interactions. Our work showed that AdpF binds to fibronectin; thus, its specificity is similar to that of AdpB, previously characterized by our laboratory (25). The mechanism by which fibronectin, a 440-kDa mosaic glycoprotein, stimulates the internalization of bacteria by mammalian cells is not clear. However, fibronectin-binding proteins present in streptococci and staphylococci have been reported to mediate bacterial adhesion to and invasion of host cells.
Based on the ability of P. intermedia 17 AdpF to bind to fibronectin, we speculated that it may also bind to proteins located on the surface of eukaryotic cells and, thus, serve as an adhesin. As so far the mutagenesis of P. intermedia 17 is not feasible, we reasoned that preincubation of the host cells with rAdpF would saturate the eukaryotic cell surface receptors, thus preventing them from binding to AdpF located on the P. intermedia 17 surface and ultimately reducing bacterial attachment. However, surprisingly, preincubation of eukaryotic cells with rAdpF increased the bacterial interaction with these cells. Both the total interaction and internalization were enhanced. Considering that the total interaction was 10-fold greater than the rate of P. intermedia 17 internalization, we reasoned that the difference was due to high bacterial adherence to the HN4 cells. Thus, we confirmed that both adhesion and invasion were elevated in cells pretreated with AdpF. Since no similar enhancement was observed for the interaction of eukaryotic cells with other oral bacteria or with E. coli, we conclude that the AdpF effect is specific for P. intermedia 17.
The enhanced internalization effect seen upon exposure of mammalian cells to AdpF was observed using two types of primary cells and three different cell lines, i.e., HUVECs, HOKs, HN4, HeLa, and NIH 3T3, respectively. These results suggest that the effect may not be cell specific. Previously, we have shown that the internalin-like P. intermedia 17 protein AdpC confers an invasive phenotype on E. coli cells expressing the protein (25). However, this effect was observed only using endothelial HUVECs and NIH 3T3 cells and was not detected with the oral epithelial HN4 cell line. Thus, it is probable that P. intermedia 17 has cell-specific mechanisms for gaining entry.
The ability of bacteria to gain entry into host cells is mediated by two well-investigated mechanisms, a “zipper” mechanism, in which the bacteria bind to a specific cellular receptor and induce cellular rearrangements, ultimately leading to microbial internalization, and a “trigger” mechanism, in which the bacteria inject effector proteins into the host cell, thus triggering the cellular actin rearrangements required for bacterial entry (35). P. intermedia cells have been shown to require receptor-mediated endocytosis (RME) for bacterial uptake (23), which is consistent with the “zipper” mechanism. However, the role of AdpF in altering the host cell to take up more bacteria has some resemblance to the triggering mechanism, thus showing that the two mechanisms of bacterial uptake may be combined in the case of P. intermedia 17 entry into host cells.
Triggering mechanisms have been shown to be present in paradigm bacteria, such as Salmonella and Shigella, that inject a variety of effectors (including SopE, SopE2, and SopB in the case of Salmonella and IpgD and IpaH in the case of Shigella) into the host cell cytoplasm using the type III secretion system. However, since the type III secretion system is unlikely to play a role in the cell entry of P. intermedia, the entry mechanism for AdpF must differ from the paradigm microbial trigger effectors (36).
Since eukaryotic membranes are hydrophobic, protein delivery across membranes, as well as across membrane-bound compartments, is very limited. Active uptake involving active, energy-requiring protein transporters usually plays a major role. As AdpF is a large protein, we also speculated that cellular mechanisms involving cytoskeletal rearrangement or involvement of lipid rafts would be engaged, similar to the situation seen in cells that internalize bacteria. Indeed, our work showed that cytoskeletal rearrangement and lipid rafts may play a role; however, the inhibition of rAdpF internalization was only 30%, thus showing that a significant amount of protein could still be internalized by other mechanisms. One possible uptake mechanism could involve RME, which is commonly used in bacterial toxin entry (37). Such a mechanism is dependent on a signal transduction cascade involving PI3-K, and indeed, our study showed that AdpF internalization was significantly inhibited by the PI3-K inhibitor wortmannin. However, as this inhibitor may also affect other pathways, such as mammalian target of rapamycin (mTOR), myosin light-chain kinase (MLCK), and mitogen-activated protein kinase (MAPK), at the concentration used (90 nM), it is possible that the signaling cascade is more complex. Thus, further studies are needed to elucidate the details of the signaling mechanisms involved in the internalization of AdpF. Inhibition by cytochalasin, nocodazole, and latrunculin was shown to be 20%, 50%, and 90%, respectively. This evidence shows that the mechanisms of cytoskeletal rearrangement and actin polymerization are very critical for the internalization of AdpF protein.
As epithelial cells are the first physical barrier and, thus, the first line of defense against invading bacteria, the results of their interaction with oral bacteria will ultimately have an effect on the initiation and progression of oral disease. The ability of bacteria to enter host cells has numerous advantages, with the most immediate being escape from the host's immune surveillance and the action of antibiotics (35). Here, we showed that the cell surface of AdpF promotes P. intermedia 17 adhesion/internalization. As P. intermedia 17 is a medically significant bacterium, anything that promotes its adhesion/internalization is of importance, because it may lead to the identification of mechanisms interfering with the action of that protein and ultimately allow reduction of the level of internalized bacteria that can serve as a reservoir for future infections. Our study showed that blocking AdpF using an anti-AdpF antibody had no effect on P. intermedia 17 internalization. This result may be due to the fact that AdpF must be internalized by eukaryotic cells in order to have effect on microbial invasion. In our study, the antibody most probably neutralized cell-associated AdpF. Furthermore, it is also possible that the antibody did not block a portion of the protein playing a role in interaction with the eukaryotic cell. This possibly could be due to the fact that the antibody was raised against denatured AdpF. Thus, further analysis using antibody against native AdpF is needed, as well as direct verification of the contribution of AdpF to P. intermedia 17 invasion using bacterial mutants deficient in AdpF. However, so far, genetic manipulation of this strain has been difficult. It is noteworthy that many pathogens previously considered to be extracellular have now been shown to be able to enter eukaryotic cells. Studies of oral epithelial cells have also shown the presence of oral pathogens, including P. gingivalis, Tanerella forsythensis, Aggregatibacter actinomycetemcomitans, and streptococci (12, 38). Although our study showed that AdpF does not play a role in promoting the internalization of other bacteria, we believe that our findings will provide novel insights into the molecular mechanisms mediating microbial internalization.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by USPHS grants R01DE016124 and R01DE018039 from the National Institute of Dental and Craniofacial Research, awarded to J.P.L.
The genomic sequence of P. intermedia 17 was obtained from The J. Craig Venter Institute (formerly TIGR) (http://www.jcvi.org) and Los Alamos Oral Pathogen Sequence Database (BROP Genome Viewer at genome.brop.org). The confocal laser-scanning fluorescence microscopy experiments were performed by the VCU Flow Cytometry—Department of Neurobiology & Anatomy Microscopy facility, supported, in part, with funding from NIH-NINDS center core grant 5P30NS047463, Imaging Shared Resource Facility, and NIH grant P30CA16059.
We thank Divya Iyer for her help with the preparation of constructs for AdpF expression, David Svintradze for generating the structural model of AdpF, and Christopher Wunsch for examination of the role of anti-AdpF antibody in P. intermedia 17 internalization. Also, we thank the members of Todd Kitten's laboratory for their assistance with laboratory equipment (the molecular imager and the Fluostar Galaxy reader) and for the streptococcal strains.
Footnotes
Published ahead of print 7 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01361-13.
REFERENCES
- 1.Kedzierski L, Montgomery J, Curtis J, Handman E. 2004. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Ther. Exp. (Warsz.) 52:104–112 http://www2.iitd.pan.wroc.pl/journals/AITEFullText/5239.pdf [PubMed] [Google Scholar]
- 2.Kobe B, Kajava AV. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725–732. 10.1016/S0959-440X(01)00266-4 [DOI] [PubMed] [Google Scholar]
- 3.Braun L, Cossart P. 2000. Interactions between Listeria monocytogenes and host mammalian cells. Microbes Infect. 2:803–811. 10.1016/S1286-4579(00)90365-4 [DOI] [PubMed] [Google Scholar]
- 4.Kobe B, Deisenhofer J. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415–421. 10.1016/0968-0004(94)90090-6 [DOI] [PubMed] [Google Scholar]
- 5.Ng AC, Eisenberg JM, Heath RJ, Huett A, Robinson CM, Nau GJ, Xavier RJ. 2011. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4631–4638. 10.1073/pnas.1000093107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda K, Akira S. 2005. Toll-like receptors in innate immunity. Int. Immunol. 17:1–14. 10.1093/intimm/dxh186 [DOI] [PubMed] [Google Scholar]
- 7.Bierne H, Sabet C, Personnic N, Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156–1166. 10.1016/j.micinf.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 8.Paster BJ, Olsen I, Aas JA, Dewhirst FE. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80–87. 10.1111/j.1600-0757.2006.00174.x [DOI] [PubMed] [Google Scholar]
- 9.Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016–1020. 10.1177/154405910808701104 [DOI] [PubMed] [Google Scholar]
- 10.Colombo AV, da Silva CM, Haffajee A, Colombo AP. 2007. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J. Periodontal Res. 42:236–243. 10.1111/j.1600-0765.2006.00938.x [DOI] [PubMed] [Google Scholar]
- 11.Rudney JD, Chen R, Sedgewick GJ. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700–2707. 10.1128/IAI.69.4.2700-2707.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudney JD, Chen R, Sedgewick GJ. 2005. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J. Dent. Res. 84:59–63. 10.1177/154405910508400110 [DOI] [PubMed] [Google Scholar]
- 13.Lopez NJ. 2000. Occurrence of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Prevotella intermedia in progressive adult periodontitis. J. Periodontol. 71:948–954. 10.1902/jop.2000.71.6.948 [DOI] [PubMed] [Google Scholar]
- 14.Lo Bue AM, Nicoletti G, Toscano MA, Rossetti B, Cali G, Condorelli F. 1999. Porphyromonas gingivalis prevalence related to other micro-organisms in adult refractory periodontitis. New Microbiol. 22:209–218 [PubMed] [Google Scholar]
- 15.Teanpaisan R, Douglas CW, Walsh TF. 1995. Characterisation of black-pigmented anaerobes isolated from diseased and healthy periodontal sites. J. Periodontal Res. 30:245–251. 10.1111/j.1600-0765.1995.tb02129.x [DOI] [PubMed] [Google Scholar]
- 16.Maeda N, Okamoto M, Kondo K, Ishikawa H, Osada R, Tsurumoto A, Fujita H. 1998. Incidence of Prevotella intermedia and Prevotella nigrescens in periodontal health and disease. Microbiol. Immunol. 42:583–589. 10.1111/j.1348-0421.1998.tb02328.x [DOI] [PubMed] [Google Scholar]
- 17.Baumgartner JC, Watkins BJ, Bae KS, Xia T. 1999. Association of black-pigmented bacteria with endodontic infections. J. Endod. 25:413–415. 10.1016/S0099-2399(99)80268-4 [DOI] [PubMed] [Google Scholar]
- 18.Enwonwu CO, Falkler WA, Idigbe EO. 2000. Oro-facial gangrene (noma/cancrum oris): pathogenetic mechanisms. Crit. Rev. Oral Biol. Med. 11:159–171. 10.1177/10454411000110020201 [DOI] [PubMed] [Google Scholar]
- 19.Falkler WA, Jr, Enwonwu CO, Idigbe EO. 1999. Microbiological understandings and mysteries of noma (cancrum oris). Oral Dis. 5:150–155 [DOI] [PubMed] [Google Scholar]
- 20.Haraszthy VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71:1554–1560. 10.1902/jop.2000.71.10.1554 [DOI] [PubMed] [Google Scholar]
- 21.Madianos PN, Lieff S, Murtha AP, Boggess KA, Auten RL, Jr, Beck JD, Offenbacher S. 2001. Maternal periodontitis and prematurity. II. Maternal infection and fetal exposure. Ann. Periodontol. 6:175–182. 10.1902/annals.2001.6.1.175 [DOI] [PubMed] [Google Scholar]
- 22.Dorn BR, Dunn WA, Jr, Progulske-Fox A. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67:5792–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorn BR, Leung KL, Progulske-Fox A. 1998. Invasion of human oral epithelial cells by Prevotella intermedia. Infect. Immun. 66:6054–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F, Iyer D, Anaya C, Lewis JP. 2006. Identification and characterization of a cell surface protein of Prevotella intermedia 17 with broad-spectrum binding activity for extracellular matrix proteins. Proteomics 6:6023–6032. 10.1002/pmic.200600177 [DOI] [PubMed] [Google Scholar]
- 25.Iyer D, Anaya-Bergman C, Jones K, Yanamandra S, Sengupta D, Miyazaki H, Lewis JP. 2010. AdpC is a Prevotella intermedia 17 leucine-rich repeat internalin-like protein. Infect. Immun. 78:2385–2396. 10.1128/IAI.00510-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyazaki H, Patel V, Wang H, Ensley JF, Gutkind JS, Yeudall WA. 2006. Growth factor-sensitive molecular targets identified in primary and metastatic head and neck squamous cell carcinoma using microarray analysis. Oral Oncol. 42:240–256. 10.1016/j.oraloncology.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 27.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658–1666. 10.1128/JB.186.6.1658-1666.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, Curtis MA, Lewis JP. 2011. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect. Immun. 79:4533–4542. 10.1128/IAI.05016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66:5703–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchenko DV, Akins DR, Robinson E, Li M, Popova TG, Cox DL, Radolf JD. 1997. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 179:3188–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizarro-Cerda J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. 10.1016/j.cell.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 34.Stubbs S, Hobot JA, Waddington RJ, Embery G, Lewis MA. 1999. Effect of environmental haemin upon the physiology and biochemistry of Prevotella intermedia R78. Lett. Appl. Microbiol. 29:31–36. 10.1046/j.1365-2672.1999.00570.x [DOI] [PubMed] [Google Scholar]
- 35.Cossart P, Sansonetti PJ. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248. 10.1126/science.1090124 [DOI] [PubMed] [Google Scholar]
- 36.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. 2008. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat. Struct. Mol. Biol. 15:65–70. 10.1038/nsmb1346 [DOI] [PubMed] [Google Scholar]
- 37.Lord JM, Smith DC, Roberts LM. 1999. Toxin entry: how bacterial proteins get into mammalian cells. Cell Microbiol. 1:85–91. 10.1046/j.1462-5822.1999.00015.x [DOI] [PubMed] [Google Scholar]
- 38.Rudney JD, Chen R, Zhang G. 2005. Streptococci dominate the diverse flora within buccal cells. J. Dent. Res. 84:1165–1171. 10.1177/154405910508401214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.