Abstract
Mammalian α-defensins are approximately 4- to 5-kDa broad-spectrum antimicrobial peptides and abundant granule constituents of neutrophils and small intestinal Paneth cells. The bactericidal activities of amphipathic α-defensins depend in part on electropositive charge and on hydrophobic amino acids that enable membrane disruption by interactions with phospholipid acyl chains. Alignment of α-defensin primary structures identified conserved hydrophobic residues in the loop formed by the CysIII-CysV disulfide bond, and we have studied their role by testing the effects of mutagenesis on bactericidal activities. Mouse α-defensin 4 (Crp-4) and rhesus myeloid α-defensin 4 (RMAD-4) were selected for these studies, because they are highly bactericidal in vitro and have the same overall electropositive charge. Elimination of hydrophobicity by site-directed mutagenesis at those positions in Crp-4 attenuated bactericidal activity markedly. In contrast to native Crp-4, the (I23/F25/L26/G)-Crp-4 variant lacked bactericidal activity against Salmonella enterica serovar Typhimurium and did not permeabilize Escherichia coli ML35 cells as a result of removing aliphatic side chains by Gly substitutions. Ala replacements in (I23/F25/L26/A)-Crp-4 restored activity, evidence that hydrophobicity contributed by Ala methyl R-groups was sufficient for activity. In macaques, neutrophil α-defensin RMAD-6 is identical to RMAD-4, except for a F28S difference, and (F28S)-RMAD-4 mutagenesis attenuated RMAD-4 bactericidal activity and E. coli permeabilization. Interestingly, (R31/32D)-Crp-4 lacks activity in these assays despite the presence of the Ile23, Phe25, and Leu26 hydrophobic patch. We infer that electrostatic interactions between cationic α-defensin residues and negative charge on bacteria precede interactions between critical hydrophobic residue positions that mediate membrane disruption and bacterial cell killing.
INTRODUCTION
Mammalian α-defensins are ∼4.5-kDa, cationic, broad-spectrum microbicidal peptide effectors of innate immunity in phagocytic leukocytes and in the small intestinal lumen following secretion by Paneth cells (1–3). The canonical α-defensin tertiary structure formed by CI-CVI, CII-CIV, CIII-CV disulfide pairings constrains the peptides into a common triple-stranded, β-sheet topology that brings the N and C termini into proximity (4–9). Despite their shared biochemical features and common overall topology, α-defensin peptides have highly diverse primary structures, and individual molecules may differ in target cell specificities and mechanisms of action. For example, human Paneth cell α-defensins HD5 and HD6 have very different primary structures (10), and HD5 has direct bactericidal effects, but HD6 lacks such activity. Instead, HD6 forms higher-ordered self-assemblies that entrap bacteria in nanonets, a mechanism of action unique to the HD6 molecule (11). Because the variable α-defensin primary structures share the same peptide topology (1), discerning common determinants of bactericidal activity has proven challenging.
Conserved or canonical α-defensin characteristics include the tridisulfide array, an Arg-Glu salt bridge, and an invariant Gly residue in the beta bulge position, but none of these features determines microbicidal activity per se (9, 12–15). For example, disulfide-null mutants of Crp-4 and RMAD-4 are more bactericidal than the native molecules against certain microbes, but disulfide mutagenesis results in susceptibility to proteolysis (15). In addition, Arg-Glu salt bridge variants of HD5 and Crp-4 retain bactericidal activity but fold less efficiently and are sensitive to proteolysis (14, 16). α-Defensin microbicidal activity requires electropositive charge as inferred from Crp-4 loss of function caused by charge reversal or charge neutralization mutations at varied Arg residue positions (15, 17–19). Complete Arg-to-Lys substitutions in both Crp-4 and HNP-1, but not in RMAD-4, attenuate in vitro bactericidal activity and also increase sensitivity to NaCl inhibition in the three peptides (18, 19).
The amphipathicity of α-defensins has long been recognized (20–22), yet the contribution of specific hydrophobic residue positions to antimicrobial activities has not been investigated in diverse α-defensins. An alanine mutagenesis scan of HNP-1 showed that (W26A)-HNP-1 had reduced bactericidal activity against Staphylococcus aureus, and substitutions at Trp26 with aliphatic amino acids of variable chain length restored activity (23). By aligning the primary structures of several α-defensins, Trp26 in HNP-1 was recognized to be one of several aliphatic and aromatic residue positions that occur in conserved hydrophobic region, or “patch,” near the C termini of many α-defensins (Fig. 1). To test whether this cluster of hydrophobicity is a determinant of α-defensin bactericidal activity, hydrophobic residues in the patch were mutagenized in two microbicidal α-defensins: Crp-4, a highly cationic mouse Paneth cell α-defensin, and RMAD-4, a macaque neutrophil α-defensin of equal charge. Gly or Ser substitutions generating (I23/F25/L26/G)-Crp-4, (F25/L26/S)-Crp-4, and (F28S)-RMAD-4 (Fig. 2) resulted in loss of bactericidal activity associated with reduced patch hydrophobicity. Also, the ability to permeabilize Escherichia coli at low peptide concentrations was lost in the less hydrophobic variants, showing that hydrophobicity at specific residue positions is a critical determinant of bactericidal activity in these α-defensin peptides.
FIG 1.
Alignment of α-defensins discloses a conserved hydrophobic patch. The primary structures of human neutrophil α-defensins (HNPs), human Paneth cell α-defensins (HD-5 and HD-6), mouse Paneth cell α-defensins (Crps), rat Paneth cell α-defensin (RD-5), rat neutrophil α-defensins (NP3B, NP-1, NP-2, and NP-6), rabbit neutrophil α-defensins (NP-3A, NP-4, NP-5, and NP-6), rhesus macaque Paneth cell α-defensins (REDs), and rhesus macaque myeloid α-defensins (RMADs) were aligned in single-letter notation using the CysI residue position as the reference point. Dashes (“-”) in the HD-6 and Crp-4, -20, and -21 sequences were introduced to maintain alignment of Cys residues in all sequences. Residue positions with hydrophobic side chains near the peptide C termini between Cys residue positions 4 and 5 are shaded, and asterisks denote the three naturally occurring α-defensins investigated in these studies.
FIG 2.
Primary structures of α-defensins and peptide variants investigated. Crp-4, (R31/R32/D)-Crp-4, (I23/F25/L26/G)-Crp-4, (I23/F25/L26/A)-Crp-4, (F25/L26/S)-Crp-4, proRMAD-4, RMAD-4, and RMAD-6 were aligned in single-letter notation. Amino acid substitutions introduced by mutagenesis (see Materials and Methods) in Crp-4 or RMAD-4 are denoted by boldface italics and underlined.
MATERIALS AND METHODS
PCR mutagenesis at α-defensin hydrophobic residue positions.
Crp-4 and RMAD-4 peptides were prepared as described previously (24–28). Briefly, recombinant peptides were expressed in E. coli as His6-tagged fusion proteins using the pET28a expression vector (Novagen, Inc., Madison, WI) at the EcoRI and SalI sites. Methionine codons introduced at the N termini of all expressed peptide products provided unique CNBr cleavage sites for removal of the N-terminal His6 tags after Ni2+ affinity purification. Primers and the PCR-based mutagenesis approach are summarized in Table S1 in the supplemental material.
Recombinant peptide expression and purification.
Recombinant α-defensins were expressed as His6-tagged peptides. E. coli BL21(DE3)-CodonPlus-RIL cells were grown in Terrific broth with shaking at 37°C to an optical density at 600 nm of 0.6, and recombinant protein expression was induced by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacterial cells were harvested by centrifugation after growth at 37°C for 4 to 6 h after induction and stored overnight at −20°C. Bacteria were lysed by resuspending cell pellets in 6 M guanidine-HCl–100 mM Tris-HCl (pH 8.1) and sonicated. Lysates were clarified by centrifugation, and recombinant His6 tag fusion proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA; Qiagen) resin affinity chromatography, dialyzed in 5% acetic acid, and then lyophilized. The Met residue incorporated at the N termini of peptide constructs provided a unique cyanogen bromide (CNBr) site for cleavage and separation of defensin peptides from His6-tagged fusion linkers. Lyophilized peptides were dissolved in 80% formic acid, reaction mixtures were adjusted to 10 mg of CNBr/ml and incubated overnight in the dark at room temperature, and reactions were quenched by the addition of 10 volumes of H2O and lyophilized. Peptides were purified to homogeneity by successive rounds of reversed-phase high-performance liquid chromatography as assessed by acid-urea polyacrylamide gel electrophoresis, a highly sensitive method for resolving correctly folded peptides from misfolded forms. Molecular masses were verified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, and peptides were quantified by using extinction coefficient calculations at 280 nm, obtained at EXPASY (http://ca.expasy.org/tools).
Bactericidal assays.
Recombinant peptides were tested for microbicidal activity against E. coli ML35, Vibrio cholerae O395, S. aureus 710a, S. aureus 502a, Listeria monocytogenes 10403s, Salmonella enterica serovar Typhimurium 14028s, S. enterica serovar Typhimurium CS022, S. enterica serovar Typhimurium JSG210, and S. enterica serovar Typhimurium ΔphoP mutant. Exponentially growing bacteria were deposited by centrifugation at 10,000 × g for 3 min and washed three times with 10 mM PIPES (pH 7.4) supplemented with a 0.01 volume of 1% (vol/vol) Trypticase soy broth (10 mM PIPES-TSB; pH 7.4), and approximately 1 × 106 to 5 × 106 CFU of bacteria/ml were exposed to peptides in 50-μl incubation mixtures. Assay tubes were incubated at 37°C with shaking for 1 h, diluted 1:100 in 10 mM PIPES (pH 7.4), and plated on TSB agar plates using an Autoplate 4000 (Spiral Biotech, Inc., Bethesda, MD). After incubation overnight at 37°C, bacterial cell survival was determined by counting CFU. The bactericidal assays shown below are representative of two or three replicate assays performed on separate days.
Peptide-mediated permeabilization of live E. coli.
α-Defensin-mediated permeabilization of live E. coli was assayed by measuring hydrolysis of ONPG (o-nitrophenyl-β-d-galactopyranoside) by cytosolic β-galactosidase (β-Gal) and colorimetric detection of o-nitrophenol (ONP), the product of ONPG hydrolysis. Permease-deficient, β-Gal-constitutive E. coli ML35 cells allow ONPG to diffuse into cells after peptide-induced membrane disruption, which is then hydrolyzed by intracellular β-Gal. In triplicate, log-phase E. coli ML35 cells were washed and resuspended in 10 mM PIPES-TSB, and the cells (5 × 106 CFU/ml) were exposed to peptides and 2.5 mM ONPG for 2 h at 37°C in 100 μl of PIPES-TSB. The kinetics of ONPG conversion to ONP was measured at A405 using a SpectraMAX plate spectrophotometer (Molecular Devices, Sunnyvale, CA). Results are shown with the standard deviations.
Kyte and Doolittle hydropathy analyses.
Hydropathicity of Crp-4 and Crp-4 mutant primary structures was assessed according to the method of Kyte and Doolittle (29). Peptide amino acid sequences were entered into the ProtScale tool on ExPaSy (30) and exported as both numerical and graphical data based upon the hydropathy parameters of Kyte and Doolittle.
RESULTS
Deletion of hydrophobic side chains attenuates Crp-4 bactericidal activity.
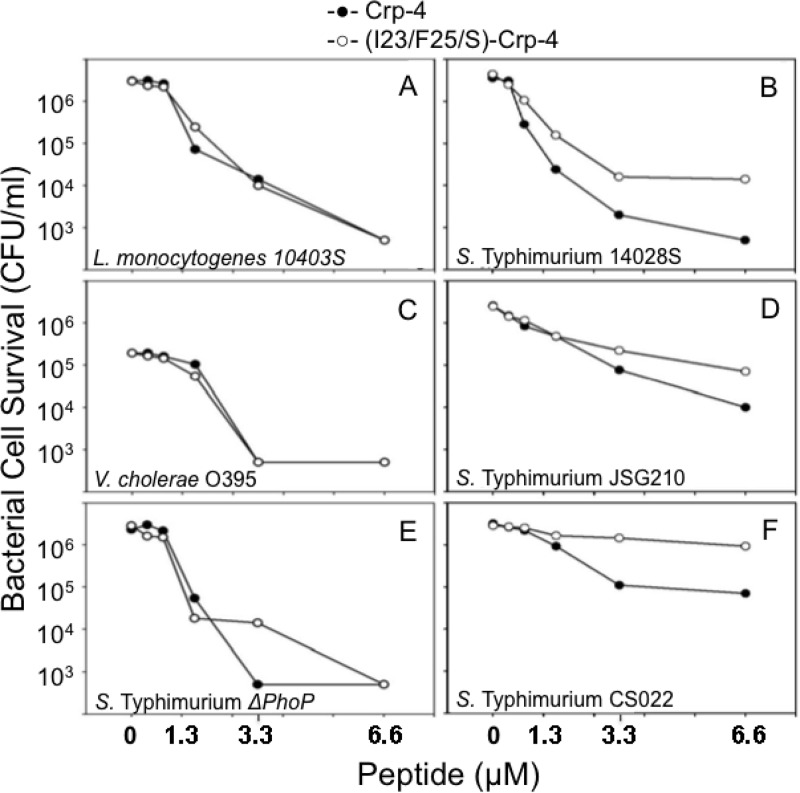
Gly substitutions at three aliphatic/aromatic residues in the Crp-4 hydrophobic patch attenuate bactericidal activity. A W26A substitution in HNP-1 attenuated bactericidal activity against S. aureus (23), and alignment of known α-defensin primary structures showed that HNP-1 Trp26 occurs in a hydrophobic patch conserved in α-defensins in the loop that is formed by the invariant CysIII-CysV disulfide bond (Fig. 1). To test the hypothesis that patch hydrophobic residues are determinants of bactericidal activity, hydrophobic side chains of the patch were ablated by Gly substitutions to produce homogeneous (data not shown) (I23/F25/L26/G)-Crp-4 (Fig. 2) for in vitro bactericidal assays. Under these conditions, (I23/F25/L26/G)-Crp-4 had bactericidal activity that was markedly lower than native Crp-4 against E. coli ML35, S. Typhimurium 14028s, and L. monocytogenes 10403s, representative Gram-positive and Gram-negative bacterial species (Fig. 3). These dose-response assays shown are representative of two or three replicate assays performed on separate days. Because peptide differential activities are more evident when assayed against organisms with low AMP sensitivity, we assayed the peptides against phoP constitutive S. Typhimurium CS022, a strain with low α-defensin sensitivity and found that (I23/F25/L26/G)-Crp-4 lacked activity compared to Crp-4 (Fig. 3D).
FIG 3.
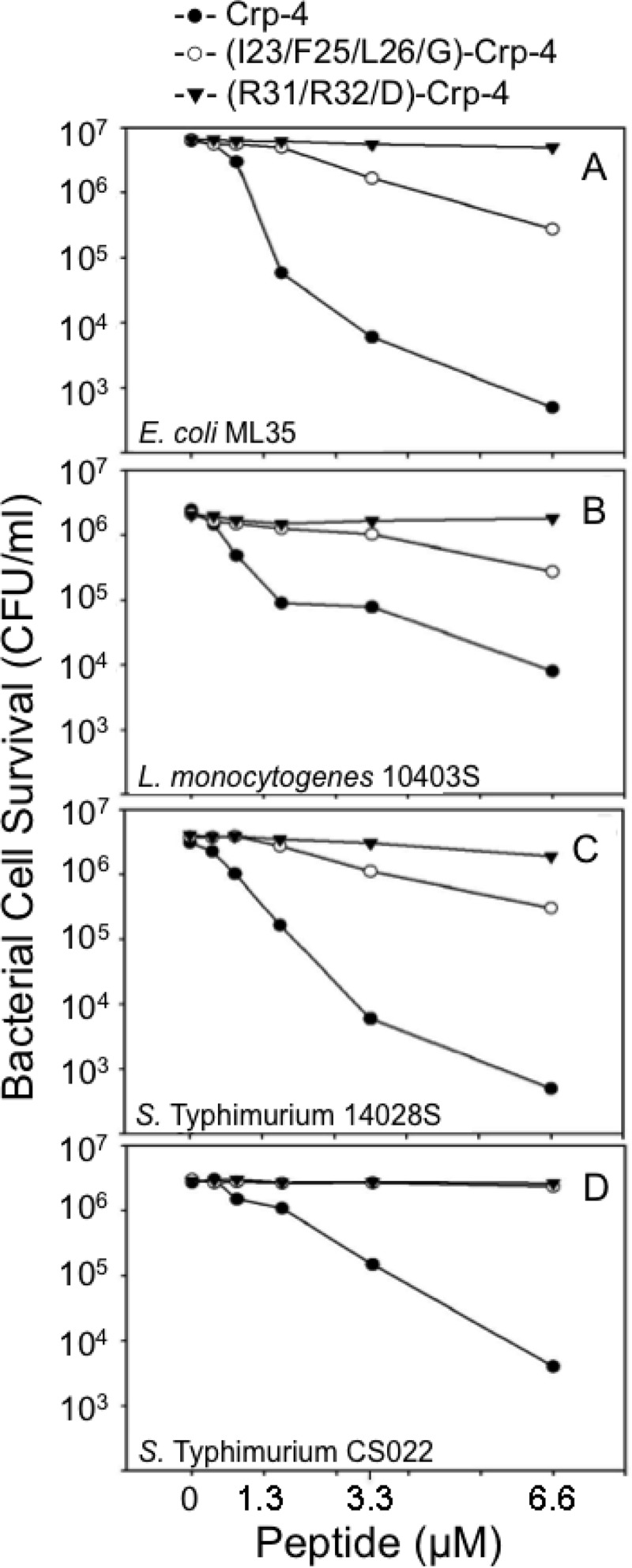
Substitutions in the conserved hydrophobic patch attenuate Crp-4 bactericidal activity. Exponentially growing E. coli ML35 (A), L. monocytogenes 10403s (B), S. Typhimurium 14028s (C), and S. Typhimurium CS022 (D) were exposed to peptides for 1 h at 37°C in 50 μl of PIPES-TSB buffer (see Materials and Methods). Peptide bacterial mixtures were plated on TSB-agar and incubated overnight at 37°C, and surviving bacteria were counted as CFU/ml at each peptide concentration. Assays shown are representative of two or three replicate assays performed on separate days. Values of ≤103 CFU/ml signify that no colonies were detected on plates after overnight growth.
Reducing patch hydrophobicity delays Crp-4 permeabilization of E. coli.
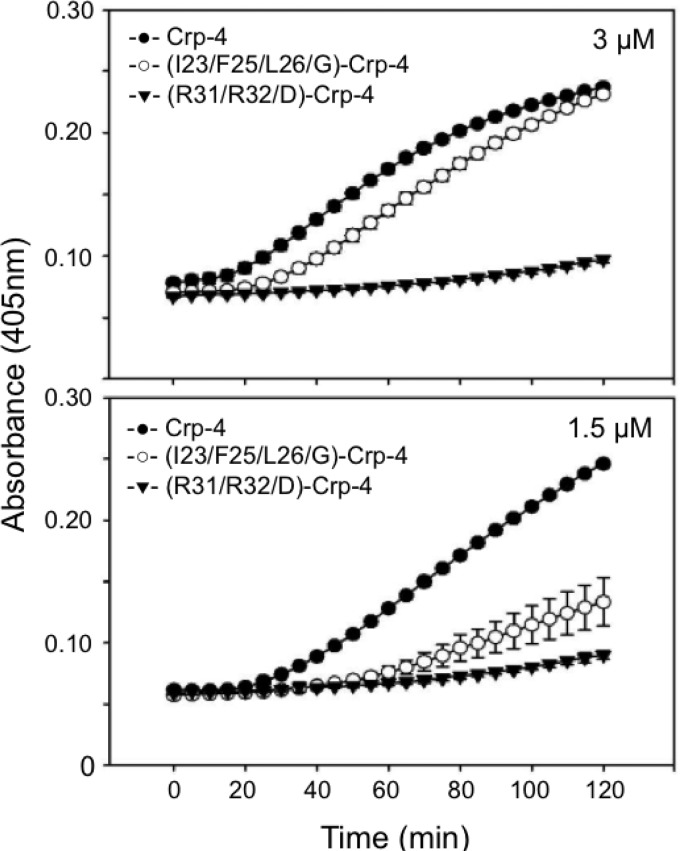
Many α-defensins kill bacteria by selective disruption of cell membranes by forming stable pores or by creating transient defects that lead to a loss of cell integrity (24, 26, 31, 32). To test whether attenuation of (I23/F25/L26/G)-Crp-4 bactericidal activity results from impaired ability to permeabilize bacterial cells, E. coli ML35 cells were exposed to Crp-4, (I23/F25/L26/G)-Crp-4, and (R31/32/D)-Crp-4 in the ONPG conversion assay (32). In this assay, permeabilization or membrane disruption of E. coli enables the lactose analog ONPG to diffuse into the cell cytosol, where constitutively expressed β-Gal converts colorless ONPG to ONP, which is measured based on the absorbance at 450 nm (see Materials and Methods). Crp-4 induces rapid ONPG hydrolysis at low micromolar peptide concentrations (19, 32), and the ONP production by E. coli ML35 exposed to 3 μM Crp-4 or (I23/F25/L26/G)-Crp-4 was similar (Fig. 4A). However, at reduced peptide levels (1.5 μM), ONP production by (I23/F25/L26/G)-Crp-4 was markedly lower than native Crp-4 (Fig. 4B), a finding consistent with peptide levels required for bactericidal activity (Fig. 3A). It should be noted that the kinetics and extent of ONPG-to-ONP conversion in E. coli ML35 cells exposed to defensins is not necessarily coincident with bacterial cell killing. For example, a 1-h exposure to 3.3 μM (I23/F25/L26/G)-Crp-4 reduced ML35 cell survival ∼5-fold (open symbols, Fig. 3A). In the permeabilization assays, ONP production was delayed in cells exposed to Gly-substituted Crp-4, and conversion was lower at the 1-h time point. A correlation exists between cell killing and permeabilization, but no direct relationship between ONP values and cell death has been determined. Thus, Gly substitutions that remove local hydrophobicity in the patch attenuate Crp-4 bactericidal activity and impair E. coli cell permeabilization by membrane disruption, demonstrating a role for these residues in Crp-4 bactericidal activity. The double charge reversal peptide (R31/32D)-Crp-4 lacks bactericidal activity in these assays (15), even though it has a complete Ile23, Phe25, and Leu26 hydrophobic patch (Fig. 3 to 5). In the context of the Crp-4 molecule, it appears that electropositive charge contributes an essential function that precedes bilayer disruption by hydrophobic side chains.
FIG 4.
Delayed permeabilization of E. coli ML35 cells by (I23/F25/L26/G)-Crp-4. Log-phase E. coli ML35 cells were exposed to 3 μM (upper panel) and 1.5 μM (lower panel) concentrations of peptides in the presence of ONPG for 2 h at 37°C (see Materials and Methods). β-Gal-mediated hydrolysis of ONPG was measured by monitoring absorbance at 405 nm; error bars denote the standard deviations.
FIG 5.
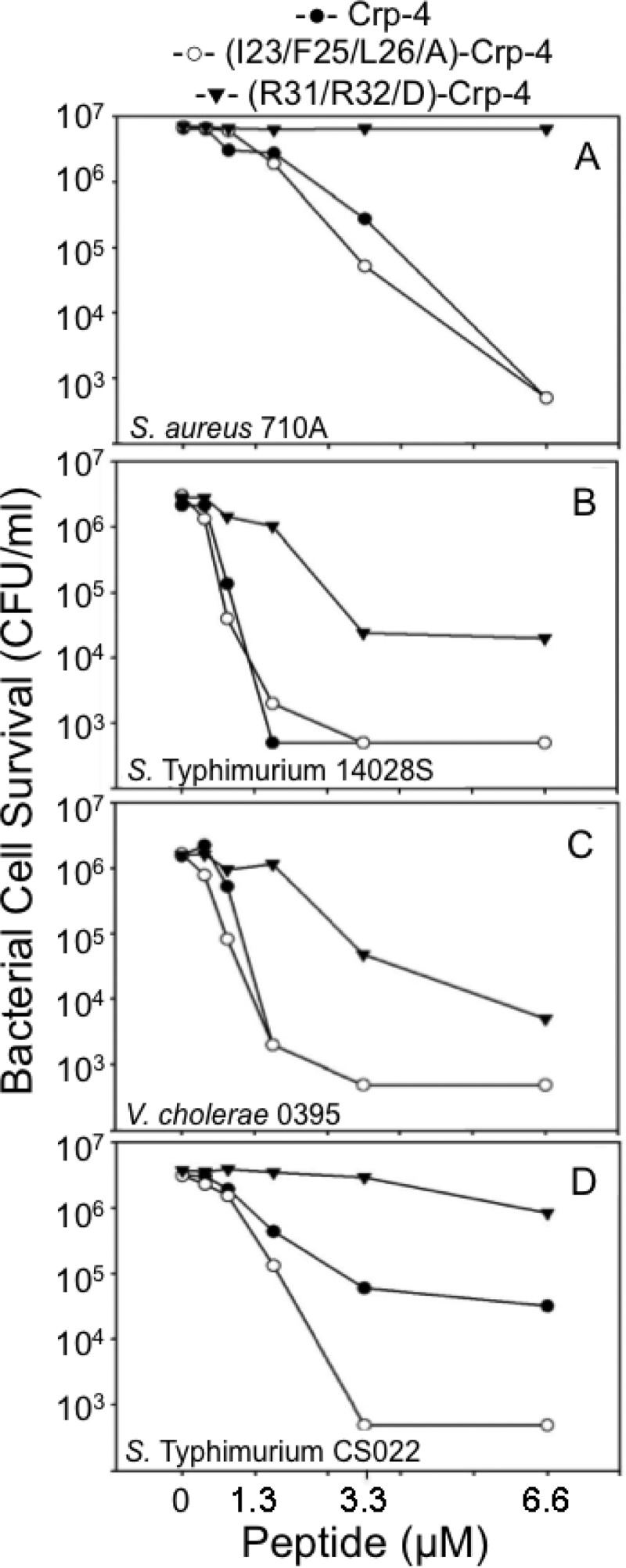
Alanine replacements in the hydrophobic patch restore Crp-4 bactericidal activity. Exponentially grown S. aureus 502A (A), S. Typhimurium 14028s (B), V. cholerae O395 (C), and S. Typhimurium CS022 were exposed to peptides, and bacterial survival was determined as in Fig. 3 (see Materials and Methods). Assays shown are representative of two or three replicate assays performed on separate days.
Ala patch substitutions retain Crp-4 bactericidal activity.
To test the effect on bactericidal activity of reducing, rather than ablating, hydrophobicity in Crp-4 at Ile23, Phe25, and Leu26, (I23/F25/L26A)-Crp-4 and Crp-4 were compared in in vitro bactericidal assays. (I23/F25/L26A)-Crp-4 and native Crp-4 were equally active against S. aureus 502A, S. Typhimurium 14028S, and V. cholerae O395, in contrast to the loss of (I23/F25/L26/G)-Crp-4 activity caused by hydrophobic side chain deletions (Fig. 5). Thus, the Ala methyl R-group contributes sufficient hydrophobicity at these patch positions to retain bactericidal activity against these bacterial species. However, against phoP constitutive S. Typhimurium CS022, among the least sensitive Salmonella strains to Crp-4 and defensins in general (33–35), Ala replacements did not restore adequate hydrophobicity for (I23/F25/L26A)-Crp-4 to kill CS022 as effectively as native Crp-4 (Fig. 5D). Because complex modifications occur to the S. Typhimurium cell envelope in the phoP constitutive state, (I23/F25/L26A)-Crp-4 has an intermediate activity against this strain but a wild-type activity against S. aureus, S. Typhimurium 14028S, and V. cholerae O395, all species that are more sensitive to Crp-4.
Polar substitutions of the Crp-4 hydrophobic patch yield intermediate bactericidal activities.
To investigate the role of the Crp-4 hydrophobic patch further, we determined the bactericidal activity of (F25/L26S)-Crp-4, leaving Ile23 unchanged and substituting with polar side chains rather than ablating or minimizing hydrophobicity at Phe25 and Leu26. Against L. monocytogenes, V. cholerae and S. Typhimurium ΔphoP mutant, which are very sensitive to Crp-4, the (F25/L26S)-Crp-4 activity was similar to that of native Crp-4 (Fig. 6A, C, and E). However, as determined for (I23/F25/L26/G)-Crp-4 (Fig. 3D), phoP-constitutive strains JSG210 and CS022 of S. Typhimurium and wild-type S. Typhimurium 14028S, all with low sensitivity to Crp-4, are less susceptible to (F25/L26S)-Crp-4 than to the native peptide (Fig. 6B, D, and F). Also, hydropathy plots show that bactericidal activity of Crp-4 and its variants corresponded directly to local patch hydrophobicity (Fig. 7), supporting the conclusion that the patch is required for Crp-4 bactericidal activity.
FIG 6.
Serine substitutions in the hydrophobic patch attenuate Crp-4 bactericidal activity. The effects of peptide exposure on survival of L. monocytogenes 10403s (A), S. Typhimurium 14028s (B), V. cholerae O395 (C), S. Typhimurium JSG210 (D), S. Typhimurium ΔphoP mutant (E), and S. Typhimurium CS022 (F) were determined as in Fig. 3 (see Materials and Methods).
FIG 7.
Hydropathy plot analyses of Crp-4 and Crp-4 peptide variants. The hydrophobicity at peptide residue positions was measured using the hydropathy plot algorithm of Kyte and Doolittle (see Materials and Methods). The Crp-4 primary structure is shown aligned in register below the hydropathy plots. A direct correspondence exists between diminishing hydrophobicity of Crp-4 variants and their impaired bactericidal activities.
Hydrophobic determinants of RMAD-4 bactericidal activity.
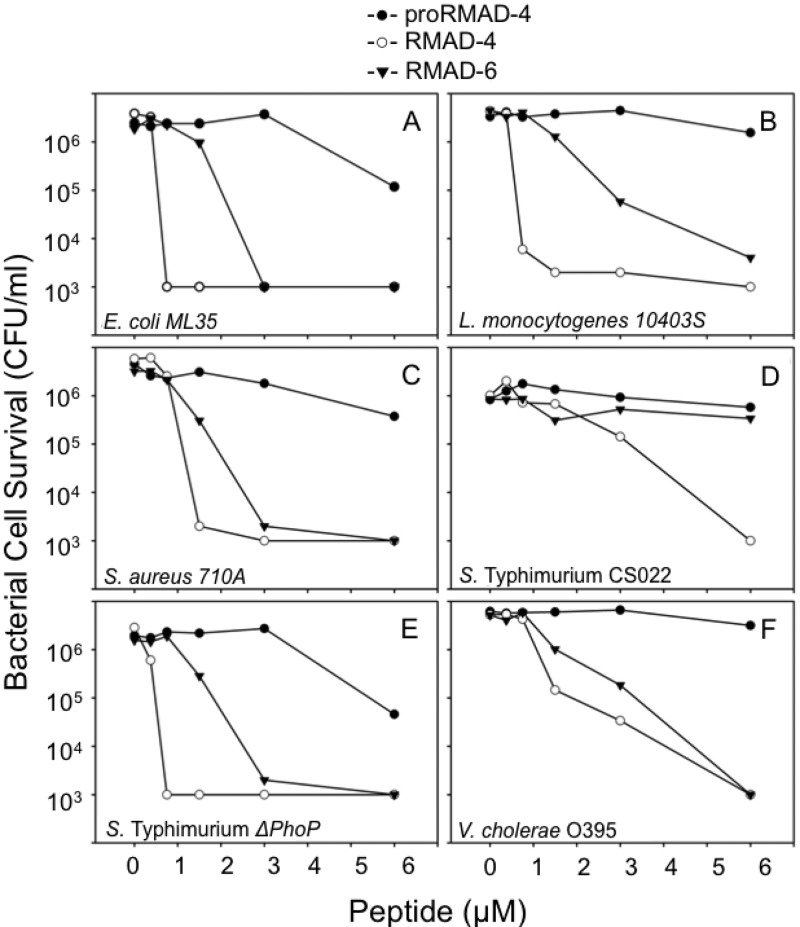
To test whether positioning of hydrophobic residues as shown for Crp-4 is a general requirement for α-defensin bactericidal activity, we tested the effect of modifying hydrophobicity in RMAD-4. RMAD-4 was selected for study, because it has the same overall bactericidal activity (19, 28, 36) and cationic charge as Crp-4 (+8), but RMAD-4 differs from Crp-4 in primary structure and in the distribution of peptide surface charge (Fig. 2) (19). Interestingly, RMAD-6 is a naturally occurring macaque neutrophil α-defensin that has the same primary structure as RMAD-4, except that Phe28 in the RMAD-4 patch is replaced by Ser (36). Accordingly, RMAD-4 and RMAD-6 were tested against the same species assayed with Crp-4 and its hydrophobic variants (Fig. 8). Against E. coli ML35, L. monocytogenes 10403s, S. aureus 710A, S. Typhimurium CS022, S. Typhimurium ΔphoP mutant, and V. cholerae O395, RMAD-6 was less active than RMAD-4, requiring 2- to 4-fold-higher peptide levels to achieve killing activity comparable to that of RMAD-4 (Fig. 8). Thus, the reduced activity of RMAD-6 [(F28S)-RMAD-4] illustrates a role for Phe28 in RMAD-4 bactericidal activity. Thus, naturally occurring RMAD-4 and RMAD-6, nearly identical macaque neutrophil α-defensins, differ substantially in their in vitro bactericidal activities as a consequence of substituting one hydrophobic patch residue with a polar side chain.
FIG 8.
Attenuation of RMAD-6 bactericidal activity relative to RMAD-4. Log-phase E. coli ML35 (A), L. monocytogenes 10403s (B), S. aureus 710A (C), S. Typhimurium CS022 (D), S. Typhimurium ΔphoP mutant (E), and V. cholerae O395 (F) were exposed to peptides and peptide-mediated bacterial cell killing was measured as in Fig. 3 (see Materials and Methods).
Diminished ability of RMAD-6 to permeabilize E. coli.
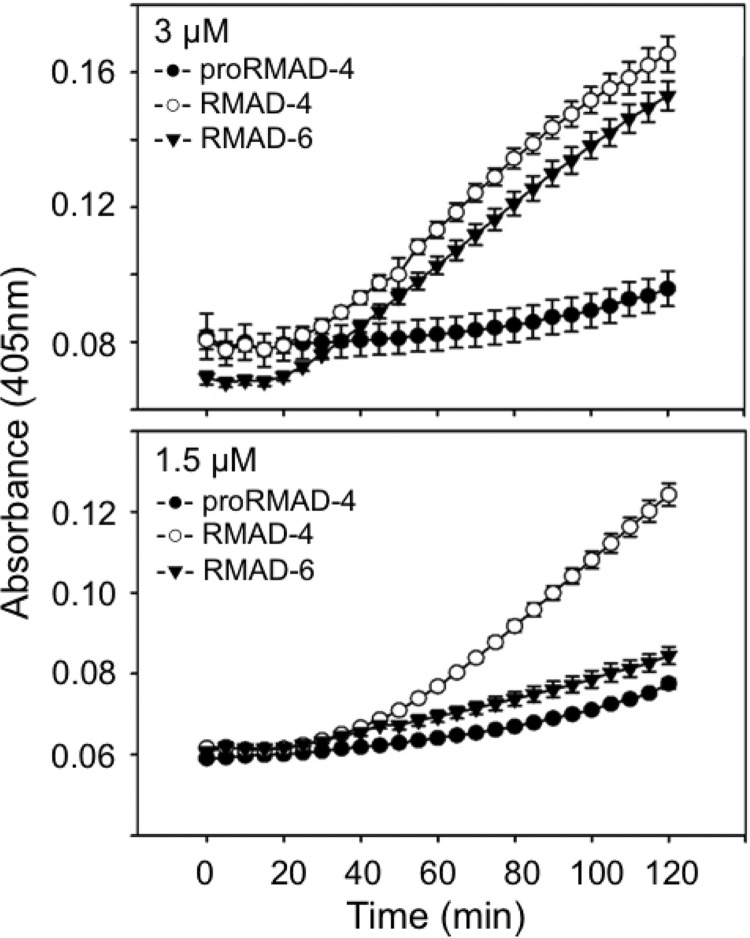
At 3 μM peptide concentrations, RMAD-6 and RMAD-4 killed 99.9% of exposed E. coli ML35 in in vitro bactericidal assays (Fig. 8A), but only RMAD-4 was active at lower peptide levels. To test whether the concentration-dependent differences in bactericidal activity are due to a diminished ability of <3 μM RMAD-6 to permeabilize E. coli, RMAD-6 and RMAD-4 were compared in ONPG conversion assays (Fig. 9). At 3 μM, RMAD-6 and RMAD-4 had equivalent bactericidal activities and permeabilized E. coli ML35 similarly (Fig. 9A). In contrast, the E. coli cells were not permeabilized above baseline levels when exposed to 1.5 μM RMAD-6, but RMAD-4 was membrane disruptive at the same concentration (Fig. 9B). Therefore, the F28S mutation in RMAD-4 caused a loss of bactericidal activity and impaired peptide-mediated cell permeabilization as a result of reducing local hydrophobicity.
FIG 9.
Delayed permeabilization of E. coli ML35 cells RMAD-6 relative to RMAD-6. Permeabilization of log-phase E. coli ML35 cells by exposure for 2 h at 37°C to 3 μM (upper panel) or 1.5 μM (lower panel) concentrations of RMAD-4 and RMAD-6 was measured in the presence of ONPG as in Fig. 4 (Materials and Methods).
DISCUSSION
Cathelicidins and defensins are two major AMP gene families in mammals, and with few exceptions, these diverse peptides are amphipathic, having cationic and hydrophobic peptide surfaces (37–39). The evolutionary conservation of amphipathicity among the AMP families suggests that both cationic and hydrophobic residues are important for biological function. Among the linear, non-disulfide-constrained cathelicidins and other linear AMPs, amphipathicity generally is induced as the disordered peptides adopt α-helical structures in hydrophobic solvents or environments (37). Studies of α-defensins have focused more on the role of electropositive charge on bactericidal activity than on the contributions of hydrophobic residues (15, 19). As noted previously, alanine scanning mutagenesis of HNP-1 and HD5 implicated single hydrophobic residues near the C termini of both peptides as determinants of bactericidal activity (14, 17, 18, 23). At micromolar levels, α-defensins disrupt microbial membranes selectively by inducing either stable or transient defects, as shown by studies of model membranes composed mainly of microbial phospholipids (40, 41). Peptide-membrane interactions cause rapid K+ efflux and lead to target cell permeabilization, depolarization, dissipation of electrochemical gradients, and eventual cell death and leakage (24, 26, 31, 32, 42). On the other hand, at lower peptide concentrations, defensins also may inhibit bacterial peptidoglycan synthesis by lipid II binding (43, 44), and defensins may use a lipid II binding mechanism to exert antimicrobial effects (44). Here, reducing hydrophobicity in a conserved patch in two diverse α-defensins, Crp-4 and RMAD-4, attenuated bactericidal activity and E. coli permeabilization, showing that it is the former mechanism that was affected by mutations at hydrophobic positions.
Alignment of varied α-defensin primary structures disclosed a conserved patch of aliphatic and aromatic residues between CysIV and CysV, a patch shared by all representative peptides (Fig. 1). Three residues with bulky R-groups within this region were identified in Crp-4, and mutagenesis to glycine [(I23/F25/L26/G)-Crp-4, Fig. 2] eliminated hydrophobicity from this patch, attenuated bactericidal activity, and disabled peptide-mediated membrane permeabilization of E. coli ML35. Possibly, other hydrophobic residues may overcome the loss of patch hydrophobicity at higher peptide concentrations to induce membrane permeabilization. By introducing Ala substitutions at Ile23, Phe25, and Leu26 residue positions, sufficient partial hydrophobicity at those Crp-4 positions to restore bactericidal activity under these assay conditions, although that result had not been anticipated. (F25/L26S)-Crp-4 substitutions introduced polar side chains at two patch residue positions, which had a partial attenuating effect on Crp-4 bactericidal activity. Also, hydropathy plot comparisons of Crp-4 and Crp-4 peptide variants confirmed that a correlation exists between peptide hydrophobicity, bactericidal activity, and induced E. coli permeabilization for these Crp-4 peptides.
To test whether the effects of Crp-4 patch mutagenesis represented a general role for the patch in α-defensins, we compared RMAD-6, an F28/S variant of RMAD-4 in macaque neutrophils, with native RMAD-4 in bactericidal and E. coli permeabilization assays. In those experiments, RMAD-6 had less bactericidal activity than RMAD-4 at concentrations <3 μM, and RMAD-6 did not mediate E. coli cell permeabilization at a 1.5 μM peptide concentration. Thus, reducing patch hydrophobicity, even at a single residue position in these bactericidal α-defensins of equal cationic charge, caused reduced function or loss of function, supporting the conclusion that the patch partly determines α-defensin bactericidal activity.
Reducing overall cationicity of Crp-4 attenuates bactericidal activity, as shown by the loss of function induced in Crp-4 by charge reversal or charge neutralization substitutions at varied Arg residue positions (15). The requirement is specific for Arg, because Arg→Lys substitutions diminish the activity of Crp-4 but not of RMAD-4. In addition, the bactericidal activities of Lys substituted Crp-4 and RMAD-4 are both more sensitive to inhibition by NaCl than the native peptides (19). As noted previously, the double charge reversal variant (R31/32D)-Crp-4 lacks activity (15) despite having a complete Ile23, Phe25, and Leu26 hydrophobic patch (Fig. 2 to 4). In a previous study (45), (R31G/R32G)-Crp-4 and (R31V/R32V)-Crp-4 exhibited attenuated activities but to a lower extent, showing that reduced cationicity and introduction of anionic amino acids both contributed to loss of function in (R31D/R32D)-Crp-4. Also, three additional variants, (R16D/R18D)-Crp-4, (R16D/R24D)-Crp-4, and (R18D/R24D)-Crp-4, all with complete hydrophobic patches, also lacked activity. This implies that cationic charge interactions with bacteria precede the hydrophobic interactions that mediate membrane disruption and cell killing. This is consistent with the early α-defensin literature, which revealed a role for electrostatic interactions in bacterial cell killing (20, 40, 41) and with a model in which early electrostatic events attract α-defensins to the cell envelope with the hydrophobic patch mediating membrane disruption after proximity to the bilayer has been established.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health grants DK044632, AI059346 (A.J.O.), AI022931, and AI058129 and the Southern California Clinical Translational Science Institute UL1RR031986 (M.E.S.). This study was also supported in part by a USC Norris Cancer Center Support Grant P30CA014089 from the National Cancer Institute.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Published ahead of print 10 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01414-13.
REFERENCES
- 1. Selsted ME, Ouellette AJ. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551–557. 10.1038/ni1206 [DOI] [PubMed] [Google Scholar]
- 2. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720. 10.1038/nri1180 [DOI] [PubMed] [Google Scholar]
- 3. Rehaume LM, Hancock RE. 2008. Neutrophil-derived defensins as modulators of innate immune function. Crit. Rev. Immunol. 28:185–200. 10.1615/CritRevImmunol.v28.i3.10 [DOI] [PubMed] [Google Scholar]
- 4. Jing W, Hunter HN, Tanabe H, Ouellette AJ, Vogel HJ. 2004. Solution structure of cryptdin-4, a mouse paneth cell alpha-defensin. Biochemistry 43:15759–15766. 10.1021/bi048645p [DOI] [PubMed] [Google Scholar]
- 5. Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. 2006. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 15:2749–2760. 10.1110/ps.062336606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pardi A, Hare DR, Selsted ME, Morrison RD, Bassolino DA, Bach AC., II 1988. Solution structures of the rabbit neutrophil defensin NP-5. J. Mol. Biol. 201:625–636. 10.1016/0022-2836(88)90643-2 [DOI] [PubMed] [Google Scholar]
- 7. Pardi A, Zhang XL, Selsted ME, Skalicky JJ, Yip PF. 1992. NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry 31:11357–11364 [DOI] [PubMed] [Google Scholar]
- 8. McManus AM, Dawson NF, Wade JD, Carrington LE, Winzor DJ, Craik DJ. 2000. Three-dimensional structure of RK-1: a novel alpha-defensin peptide. Biochemistry 39:15757–15764. 10.1021/bi000457l [DOI] [PubMed] [Google Scholar]
- 9. Rosengren KJ, Daly NL, Fornander LM, Jonsson LM, Shirafuji Y, Qu X, Vogel HJ, Ouellette AJ, Craik DJ. 2006. Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse Cryptdin-4 and (E15D)-Cryptdin-4. J. Biol. Chem. 281:28068–28078. 10.1074/jbc.M604992200 [DOI] [PubMed] [Google Scholar]
- 10. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2009. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76–83. 10.1038/ni.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu W, Lehrer RI, Baumler AJ, Bevins CL. 2012. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337:477–481. 10.1126/science.1218831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie C, Prahl A, Ericksen B, Wu Z, Zeng P, Li X, Lu WY, Lubkowski J, Lu W. 2005. Reconstruction of the conserved beta-bulge in mammalian defensins using d-amino acids. J. Biol. Chem. 280:32921–32929. 10.1074/jbc.M503084200 [DOI] [PubMed] [Google Scholar]
- 13. Wu Z, Li X, de Leeuw E, Ericksen B, Lu W. 2005. Why is the Arg5-Glu13 salt bridge conserved in mammalian alpha-defensins? J. Biol. Chem. 280:43039–43047. 10.1074/jbc.M510562200 [DOI] [PubMed] [Google Scholar]
- 14. Rajabi M, de Leeuw E, Pazgier M, Li J, Lubkowski J, Lu W. 2008. The conserved salt bridge in human alpha-defensin 5 is required for its precursor processing and proteolytic stability. J. Biol. Chem. 283:21509–21518. 10.1074/jbc.M801851200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maemoto A, Qu X, Rosengren KJ, Tanabe H, Henschen-Edman A, Craik DJ, Ouellette AJ. 2004. Functional analysis of the α-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279:44188–44196. 10.1074/jbc.M406154200 [DOI] [PubMed] [Google Scholar]
- 16. Andersson HS, Figueredo SM, Haugaard-Kedstrom LM, Bengtsson E, Daly NL, Qu X, Craik DJ, Ouellette AJ, Rosengren KJ. 2012. The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance. Amino Acids 43:1471–1483. 10.1007/s00726-012-1220-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Leeuw E, Rajabi M, Zou G, Pazgier M, Lu W. 2009. Selective arginines are important for the antibacterial activity and host cell interaction of human alpha-defensin 5. FEBS Lett. 583:2507–2512. 10.1016/j.febslet.2009.06.051 [DOI] [PubMed] [Google Scholar]
- 18. Zou G, de Leeuw E, Li C, Pazgier M, Zeng P, Lu WY, Lubkowski J, Lu W. 2007. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282:19653–19665. 10.1074/jbc.M611003200 [DOI] [PubMed] [Google Scholar]
- 19. Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. 2009. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect. Immun. 77:5035–5043. 10.1128/IAI.00695-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. 1985. Defensins: natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76:1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Selsted ME, Brown DM, DeLange RJ, Harwig SS, Lehrer RI. 1985. Primary structures of six antimicrobial peptides of rabbit peritoneal neutrophils. J. Biol. Chem. 260:4579–4584 [PubMed] [Google Scholar]
- 22. Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. 1985. Primary structures of three human neutrophil defensins. J. Clin. Invest. 76:1436–1439. 10.1172/JCI112121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei G, Pazgier M, de Leeuw E, Rajabi M, Li J, Zou G, Jung G, Yuan W, Lu WY, Lehrer RI, Lu W. 2010. Trp-26 imparts functional versatility to human alpha-defensin HNP1. J. Biol. Chem. 285:16275–16285. 10.1074/jbc.M110.102749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Satchell DP, Sheynis T, Kolusheva S, Cummings JE, Vanderlick TK, Jelinek R, Selsted ME, Ouellette AJ. 2003. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides 24:1793–1803. 10.1016/j.peptides.2003.08.020 [DOI] [PubMed] [Google Scholar]
- 25. Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. 2003. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J. Biol. Chem. 278:7910–7919. 10.1074/jbc.M210600200 [DOI] [PubMed] [Google Scholar]
- 26. Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. 2003. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes: prosegment inhibition of peptide association with biomimetic membranes. J. Biol. Chem. 278:13838–13846. 10.1074/jbc.M212115200 [DOI] [PubMed] [Google Scholar]
- 27. Figueredo S, Mastroianni JR, Tai KP, Ouellette AJ. 2010. Expression and purification of recombinant alpha-defensins and alpha-defensin precursors in Escherichia coli. Methods Mol. Biol. 618:47–60. 10.1007/978-1-60761-594-1_4 [DOI] [PubMed] [Google Scholar]
- 28. Kamdar K, Maemoto A, Qu X, Young SK, Ouellette AJ. 2008. In vitro activation of the rhesus macaque myeloid alpha-defensin precursor proRMAD-4 by neutrophil serine proteinases. J. Biol. Chem. 283:32361–32368. 10.1074/jbc.M805296200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 30. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lehrer RI, Barton A, Ganz T. 1988. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods 108:153–158. 10.1016/0022-1759(88)90414-0 [DOI] [PubMed] [Google Scholar]
- 32. Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. 2008. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by Cryptdin-4 and its disulfide-null analogue. Biochemistry 47:12626–12634. 10.1021/bi800335e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054–5058. 10.1073/pnas.86.13.5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Groisman EA, Chiao E, Lipps CJ, Heffron F. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. U. S. A. 86:7077–7081. 10.1073/pnas.86.18.7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fields PI, Groisman EA, Heffron F. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059–1062. 10.1126/science.2646710 [DOI] [PubMed] [Google Scholar]
- 36. Tang YQ, Yuan J, Miller CJ, Selsted ME. 1999. Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of alpha-defensins from rhesus macaque leukocytes. Infect. Immun. 67:6139–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 38. Zanetti M. 2005. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 7:179–196 [PubMed] [Google Scholar]
- 39. Ganz T. 2004. Defensins: antimicrobial peptides of vertebrates. C. R. Biol. 327:539–549. 10.1016/j.crvi.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 40. Wimley WC, Selsted ME, White SH. 1994. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 3:1362–1373. 10.1002/pro.5560030902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hristova K, Selsted ME, White SH. 1996. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry 35:11888–11894. 10.1021/bi961100d [DOI] [PubMed] [Google Scholar]
- 42. Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GC. 2011. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133:6720–6727. 10.1021/ja200079a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328:1168–1172. 10.1126/science.1185723 [DOI] [PubMed] [Google Scholar]
- 44. de Leeuw E, Li C, Zeng P, Diepeveen-de Buin M, Lu WY, Breukink E, Lu W. 2010. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 584:1543–1548. 10.1016/j.febslet.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanabe H, Qu X, Weeks CS, Cummings JE, Kolusheva S, Walsh KB, Jelinek R, Vanderlick TK, Selsted ME, Ouellette AJ. 2004. Structure-activity determinants in paneth cell alpha-defensins: loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J. Biol. Chem. 279:11976–11983. 10.1074/jbc.M310251200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.