Abstract
Kingella kingae is a human oral bacterium that can cause diseases of the skeletal system in children and infective endocarditis in children and adults. K. kingae produces a toxin of the RTX group, RtxA. To investigate the role of RtxA in disease pathogenesis in vivo, K. kingae strain PYKK081 and its isogenic RtxA-deficient strain KKNB100 were tested for their virulence and pathological consequences upon intraperitoneal injections in 7-day-postnatal (PN 7) rats. At the doses above 8.0 × 106 cells/animal, PYKK081 was able to cause a fatal illness, resulting in rapid weight loss, bacteremia, and abdominal necrotic lesion formation. Significant histopathology was observed in thymus, spleen, and bone marrow. Strain KKNB100 was less toxic to animals. Neither weight loss, bacteremia, nor histopathological changes were evident. Animals injected with KKNB100 exhibited a significantly elevated circulating white blood cell (WBC) count, whereas animals injected with PYKK081 had a WBC count that resembled that of the uninfected control. This observation parallels the subtleties associated with clinical presentation of K. kingae disease in humans and suggests that the toxin contributes to WBC depletion. Thus, our results demonstrate that RtxA is a key K. kingae virulence factor. Furthermore, our findings suggest that the PN 7 rat can serve as a useful model for understanding disease caused by K. kingae and for elucidating diagnostic parameters in human patients.
INTRODUCTION
Kingella kingae, a Gram-negative bacterium of the Neisseriaceae family, colonizes the posterior pharynges of young children (1, 2). While often carried asymptomatically in the respiratory tract, K. kingae can be associated with invasive infections, including bacteremia, osteoarticular infections, infective endocarditis, meningitis, and infections involving the lower respiratory tract, the central nervous system, and the eyes (2).
Improvements in culture techniques and molecular detection methods have led to the recognition of K. kingae as a frequent cause of osteomyelitis and septic arthritis in pediatric patients younger than 2 years old (3–9). K. kingae is a member of the HACEK (Haemophilus species, Aggregatibacter actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and K. kingae) group of organisms, which are collectively responsible for 3 to 5% of cases of infective endocarditis (10), an infection of the heart valves and lining. Infective endocarditis caused by K. kingae has been diagnosed in otherwise healthy children or adult patients with underlying disease, and unlike that mediated by the other HACEK organisms, K. kingae-mediated disease progresses very rapidly (11–16). K. kingae endocarditis is a severe infection associated with serious complications, including mycotic aneurisms, pulmonary infarctions, meningitis, valvular abscesses, septic embolization, and perforation of the posterolateral leaflet (17–20), and the overall mortality rate is 16% (2). Recent reports describe epidemiological cases of invasive K. kingae infections in day care centers, illustrating that the bacterium is able to cause outbreaks of disease within communities of children (21–23).
K. kingae produces a 100-kDa protein toxin of the RTX group, RtxA, which has been implicated in the organism's virulence (24). Five genes, rtxC, rtxA, rtxB, rtxD, and tolC, are necessary for the production and secretion of RtxA. The genes are organized in a single gene cluster on the chromosome. In in vitro studies, disruption of the structural RTX toxin gene, rtxA, by transposon mutagenesis resulted in a loss of cytotoxicity of K. kingae (24). Consequently, rtxA was used as a specific molecular marker to diagnose K. kingae infections (25–27).
The toxins of the RTX (repeats in toxin) family are large secreted proteins that contain glycine-rich repeats and are secreted from bacterial cells via type I secretion by employing an uncleaved C-terminal recognition signal (28, 29). The repeats are responsible for binding divalent calcium, which is required for the toxin's activity. These toxins are modified with fatty acid moieties attached to internal lysine residues, which is a unique characteristic of this group of toxins (30–32). To date, the role of RtxA in the pathogenesis of K. kingae is not known.
Bacteremia is the common presentation of K. kingae infections, and it is an important component of the pathophysiology of K. kingae-mediated disease (2, 33). In this study, we took advantage of the peritoneal cavity as a niche for inducing bacteremia. The peritoneal cavity has an extensive vascular supply, lined by a thin epithelium; therefore, it served as a conducive environment for engineering K. kingae epithelial breach and subsequent bacteremia (34). This model was used to investigate the pathophysiology of a K. kingae infection and the contribution of RtxA to K. kingae toxicity and virulence in vivo.
MATERIALS AND METHODS
Bacterial strain.
K. kingae strain PYKK081 was isolated in 1991 in Israel from the ankle joint of an 8-month-old boy with septic arthritis. This strain was previously used in our studies (35) and in genome sequencing (36). The bacteria were grown on Columbia agar (CA) (Oxoid Ltd., Hampshire, England) with 5% sheep blood (Hemostat Laboratories, Dixon, CA) or on AAGM agar (37) at 37°C with 10% CO2 and were stored in AAGM broth with 10% dimethyl sulfoxide (DMSO) at −80 °C.
In vitro cell viability assay.
THP-1 human monocytic cells were purchased from ATCC and grown in RPMI medium containing 10% fetal bovine serum (FBS) according to the manufacturer's instructions. For toxicity tests, 1.0 × 105 bacterial cells were added to 0.5 × 106 mammalian cells and incubated for 3 h. Quantitation of cell membrane permeability was done with the trypan blue assay using a Vi-cell cell viability analyzer (Beckman Coulter, Hialeah, FL). ATP production was detected using the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer's instructions. Culture plates were read in a Synergy HT plate reader in the luminescence mode (Bio-Tek, Winooski, VT). All reactions were run in technical duplicate; the assay was performed three independent times.
Immunoassay. (i) RtxA purification.
The bacteria were grown on AAGM plates for 25 h, and biomass was collected and subjected to centrifugation at 150,000 × g for 2 h to separate the bacterial pellet and liquid secreted fraction (35). One milliliter of the resultant supernatant was filtered through a 0.22-μm filter and was then loaded into a G-100 column (bed volume, 45 ml), equilibrated with 20 mM Tris-HCl–250 mM NaCl–2 mM CaCl2 (pH 6.8) buffer, and then eluted with the same buffer in 1-ml fractions. The purified toxin sample was resolved by SDS-PAGE, the RtxA band was excised from the gel, and the protein was isolated by overnight electroelution using an electroeluter (Bio-Rad, Hercules, CA). The procedure was repeated until the desired amount (2.5 mg) of protein was purified.
(ii) Protein assay.
Proteins were resolved by 4 to 20% SDS-PAGE and visualized by staining with GelCode blue stain reagent (Pierce, Rockford, IL). The protein concentration was measured with a bicinchoninic acid assay kit (Pierce, Rockford, IL). Individual 100-kDa protein bands were excised from the SDS-polyacrylamide gel and digested with trypsin, and peptides were extracted for matrix-assisted laser desorption ionization–time of flight/time of flight mass spectrometry (MALDI-TOF/TOF MS) analysis using an ABI 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA). A search for the peptide mass fingerprint was carried out with the nrNCBI nonredundant database.
(iii) Anti-RtxA monoclonal antibody production.
A specific anti-RtxA monoclonal antibody was generated to use in immunoassays (ProMab Biotechnologies, Inc., Richmond, CA). Five BALB/c mice were immunized intraperitoneally (i.p.) with 0.5 mg RtxA by standard immunization protocol. Once an acceptable titer was obtained, hybridoma fusion was prepared using splenocytes from mouse and myeloma cells. Supernatants from the growing hybridoma cells were screened by enzyme-linked immunosorbent assay (ELISA) for sensitivity to RtxA. The 10A7D7 clone was selected for anti-RtxA antibody production.
(iv) Western blot analysis.
The protein samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were subjected to Western blot analysis with 10A7D7 hybridoma supernatants (1:1,000 dilution) and then secondary horseradish peroxidase (HRP)-goat anti-mouse IgG (Fc) (1:10,000) (Pierce, Rockford, IL). For the toxin accumulation experiments, the THP-1 cell culture samples were collected at different time points, and the cells were pelleted by centrifugation at 2,000 × g for 5 min. Ethanol was added to the cell-free supernatants at a ratio of 2.5:1, the mixtures were incubated for 30 min at −20°C, and then the proteins were concentrated by centrifugation (18,000 × g, 10 min).
Creation of an rtxA mutant.
RTX toxin-deficient strain KKNB100 was obtained using the mariner transposon derivative Solo, which carries the aphA3 kanamycin resistance gene, using a previously described protocol (24) with modifications. The rtxA gene was PCR amplified from genomic DNA of PYKK081 using an Expand high-fidelity PCR system (Roche, Mannheim, Germany) and the primers RtxA-EcoRI FW (5′-CGAATTCTGAAAATTGAGGTGAATTATGAAC-3′) and RtxA-EcoRI RV (5′-CGAATTCGTTATTAATGGTATTAATATTTCAG-3′). The ∼3,000-bp product was ligated into EcoRI-digested pUC19 vector (New England BioLabs, Ipswich, MA), creating pUC19-rtxA. The aphA3 gene was PCR amplified from plasmid pFALCON2 using Solo-NsiI FW (5′-CCATGCATCGATACCGTCGACCTCGAGGGGG-3′) and Solo-NsiI RV (5′-CCATGCATCATGTTTGACAGCTTATCATCG-3′) primers. The ∼1,000-bp product was then ligated into NsiI-digested pUC19-rtxA, creating pUC19-rtxA::kan. The rtxA gene containing aphA3 was PCR amplified and used for the transformation of PYKK081. In order to carry this out, the bacteria were grown overnight on CA with 5% sheep blood and resuspended to an A600 of 0.8 in Trypticase soy broth containing 2% yeast extract. After 30 min of incubation, the cells were resuspended in Trypticase soy broth containing 10 mM CaCl2, 12% horse plasma, and DNA. The transformation mixture was incubated for 1 h at 37°C and plated on CA with 5% sheep blood plates containing kanamycin (40 μg/ml). The transformants were grown for 48 h at 37°C in a 10% CO2 incubator. PCR of the rtxA gene was then used to verify that the intended homologous recombination had occurred, disrupting the gene within the selected transformants.
Genetic complementation.
To genetically complement the rtxA gene, the complementation was performed as previously described (38). A fragment of the locus containing the rtxA gene and the upstream rtxC genes (rtxAC) was PCR amplified from PYKK081 genomic DNA using an Expand high-fidelity PCR system (Roche, Mannheim, Germany) and primers rtxAC FW-KpnI (5′-CCGGTACCTTACACTGCGCTAGGTGCTAATAC-3′) and rtxAC RV-BamHI (5′-GGGGATCCTCGCTTTTTTCAACCAATGTTATTCCAGC-3′). The ∼3,500-bp product was cloned into pCR-XL-TOPO vector using the TOPO XL PCR cloning kit (Invitrogen, Carlsbad, CA). The construct was digested with KpnI/BamHI, and the rtxAC fragment was subcloned into a KpnI/BamHI pUC19-based K. kingae complementation vector (pCErm), resulting in pCErm/rtxAC. An ∼1,000-bp sequence upstream of the start codon of the rtxB gene containing the putative promoter region, rtxPro, was PCR amplified with primers BamHI-rtxPro FW (5′-CCGGATCCTTGCCAACTCTTGAACCGTA-3′) and XbaI-rtxPro RV (5′-CCTCTAGAAGACTTTTTCTCTCGTTATCTA-3′). The amplified product was digested with BamHI/XbaI and ligated into BamHI/XbaI-digested pCErm/rtxAC, generating the pCErm/rtxAC/rtxPro construct. This plasmid, containing rtxAC and the putative rtx promoter region, was used to transform KKNB100 as described above. The transformants were selected on CA plates with 5% sheep blood and erythromycin (1 μg/ml). PCR of the rtxA gene was then used to verify that the intended homologous recombination had occurred, introducing rtxAC-rtxPro into the selected transformants.
Animal studies. (i) Ethics statement.
The animal experiments (protocol 11119) were approved by the Animal Care and Use Committee of Rutgers University and were compliant with all institutional policies and federal guidelines.
(ii) Animals.
Nursing Sprague-Dawley rat dams with postnatal day 18 (PN 18) or PN 4 pups were obtained from Charles River Laboratories (Wilmington, MA). The animal cages were supplied with a ventilation unit that maintained a continuous filtered airflow through the cages, preventing aerosolized contamination and keeping the animals in healthy conditions.
(iii) Bacterial injections and postinjection monitoring.
Inocula for animal challenge were prepared by growing bacteria on Columbia agar with 5% sheep blood. The bacterial suspension was made in phosphate-buffered saline (PBS) (pH 7.4) to the desired inoculum concentration and stored no longer than 30 min before injection. CFU were calculated by plating the bacteria on Columbia agar with 5% sheep blood from serial dilutions. On PN 21 or PN 7, rat pups were weaned from mothers and i.p. injected with different doses (from 1.0 × 106 to 1.0 × 109 cells/animal) of K. kingae strains. Vehicle control groups were injected with PBS in volumes of 200 μl and 100 μl for PN 21 and PN 7, respectively. All injections were carried out with a 27 5/8-gauge syringe needle within a laminar flow hood using aseptic techniques. After the injection, PN 7 pups were returned to the mothers and PN 21 animals were separated from the mothers. Each animal group was caged separately. The rats were monitored for mortality and clinical signs of illness twice a day for 14 days or for the duration of each experiment. The animals that were terminally ill were euthanized by CO2 inhalation. Fifty percent lethal doses (LD50) were determined from the 24-h postinjection death rates, corresponding to 4 and 5 serially diluted doses of KKNB100 and PYKK081, respectively.
(iv) Autopsy and histology.
Rats were aseptically dissected, and samples of different organs (heart, lung, kidney, thymus, spleen, hip joint, and tibia bone marrow) were taken for histopathological examination. Tissues were fixed in 10% phosphate-buffered formalin and were embedded in paraffin. The tissue sections were stained with hematoxylin and eosin and subjected to light microscopy and analysis.
(v) Immunohistochemistry.
Skin samples were fixed in 4% neutral buffered formaldehyde and embedded in paraffin. Sections were treated in an automated pressure cooker (Retriever 2100; Electron Microscopy Sciences, Hatfield, PA) in citrate buffer (pH 6.0), rinsed, stained with polyclonal anti-K. kingae outer membrane vesicle (OMV) antibody (35) (1:100 dilution) in Superblock buffer (Pierce, Rockford, IL) for 1 h at room temperature, rinsed with Superblock, and stained with goat anti-rabbit IgG labeled with Alexa 488 (1:200 dilution) for 2 h at room temperature. Nuclei were stained with 7-aminoactinomycin D (7-AAD) (Invitrogen, Carlsbad, CA) for 30 min. Stained sections were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and examined under a microscope (Zeiss LSM 510; Carl Zeiss Inc., Thornwood, NY).
(vi) Blood collection and analysis.
Immediately after euthanasia, intracardiac puncture was done with a sterile 5-ml syringe and a 27 5/8-gauge syringe for blood collection. Up to 300 μl of blood was drawn. Fifty microliters of freshly collected animal blood was processed using a Coulter HmX Hematology Analyzer (BD Biosciences, Franklin Lakes, NJ).
Real-time qPCR analysis.
Total DNA from blood samples was purified using the Qiagen (Valencia, CA) DNeasy blood and tissue kit. Quantitative PCR (qPCR) was carried out in an Roche LightCycler 480 device (Roche, Indianapolis, IN) by using the Kapa Syber Fast Universal qPCR kit (Woburn, MA). The rtxA and rtxB toxin genes were used as a specific target to detect K. kingae as previously described (26). The PCR amplification was carried out in a total volume of 10 μl, containing 4 μl of 2× SYBR green PCR master mix, approximately 25 ng of the DNA sample, and 200 nM each primer. The reaction conditions were 95°C for 3 min followed by 32 cycles of 95°C for 15 s, 61°C for 20 s, and 72°C for 30 s; this was followed by a final 72°C for 3 min. Standard melting curve analysis was performed. All reactions were run in technical duplicate; the assay was performed three independent times. The presence of the rtxA and rtxB genes was analyzed using the software provided by Roche. The second-derivative maximum of the slope of the amplification curves was used to define crossing point (CP) values. Standard curves and minimum detectable CFU were determined using serial dilutions of a purified K. kingae genomic DNA preparation from PYKK081 with a known corresponding CFU value.
Statistical analysis and reproducibility.
Life span evaluations and the determination of LD50 were done from at least three independent experiments. Within each experiment, the dosage- and strain-specific groups were composed of 4 to 8 animals. To limit liter-specific effects, rat pups from different dams were combined in a separate cage before injection and randomly divided into groups. Weight loss, white blood cell (WBC) levels, and in vitro cytotoxicity analysis for each strain group were compared using a one-way analysis of variance (ANOVA) test and post hoc Tukey test. A P value of ≤0.05 was considered statistically significant. A nonlinear regression was used to calculate the LD50. Survival rates were compared by Kaplan-Meier analysis, and dose dependence was assessed using a log rank test. All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). Results are expressed as mean ± standard deviation, unless otherwise indicated.
RESULTS
Generation of an RtxA-deficient strain.
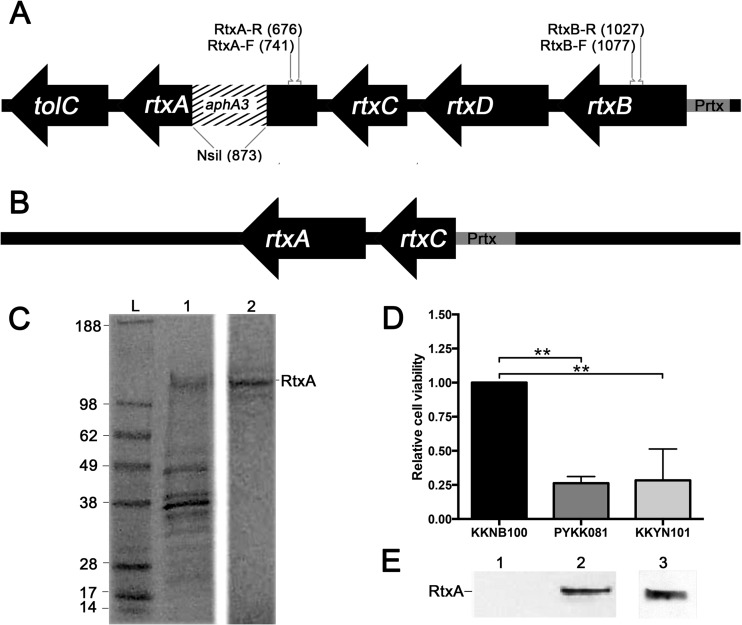
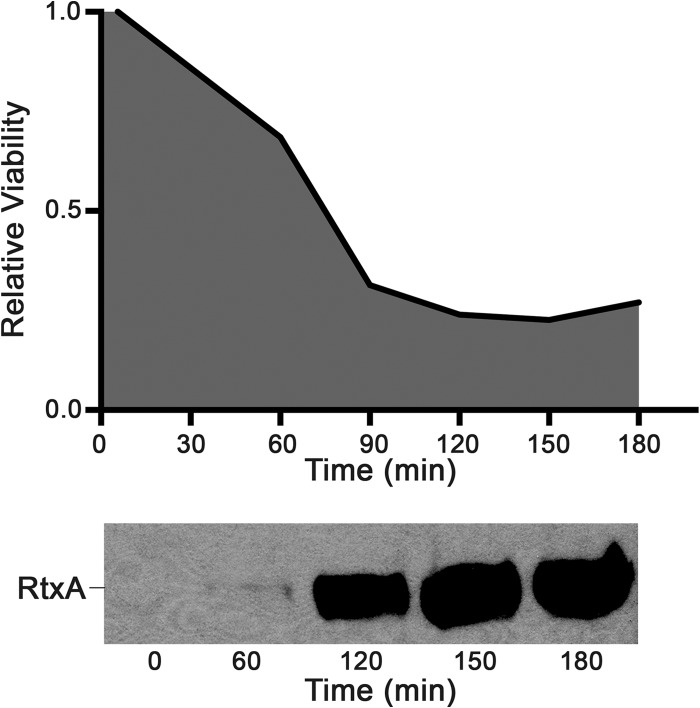
In order to investigate the role of RtxA in K. kingae virulence, we created an isogenic toxin-deficient mutant, designated KKNB100, for K. kingae septic arthritis isolate PYKK081. Only one copy of the rtx toxin gene was found in the genome of PYKK081 (36). The mutation in rtxA was generated by insertion of mariner transposon derivative Solo, which carries the aphA3 kanamycin resistance gene (24, 39, 40) (Fig. 1A). A comparison of growth rates between the wild-type and RTX-toxin-deficient strains did not reveal any statistically significant variations in the growth patterns of these strains (data not shown). In order to detect the production of RtxA by K. kingae, a specific monoclonal anti-RtxA antibody was generated. To obtain the highest-purity antigen, the RtxA band was eluted from semipurified PYKK081 secreted fraction (Fig. 1C). The protein was identified by MALDI-TOF/TOF MS as RtxA toxin with >95% confidence. The production of RtxA in KKNB100 was not detected by Western blotting using the anti-RtxA antibody generated in this study (Fig. 1E). The cytotoxic effect of the bacteria on THP-1 cells was quantified with two independent methods, a trypan blue dye exclusion assay and an ATP quantitation assay, which demonstrated very similar results. The cytotoxicity results from the ATP quantitation assay are shown in Fig. 1D. The results for KKNB100 resembled those for the vehicle control, while PYKK081 was able to kill 74.0% ± 0.5% of the leukocytes after the 3 h of incubation. To confirm that the rtxA gene disruption did not have nonspecific effects on virulence, we performed complementation experiments with strain KKNB100. We found that the leukotoxic activity of RtxA was restored when wild-type rtxA was introduced into KKNB100 by chromosome-based genetic complementation (Fig. 1B). The complemented strain, designated KKYN101, was able to produce RtxA, as identified by Western blotting (Fig. 1E), and was able to lyse 71.7% ± 23.0% of cultured THP-1 cells. The cytotoxic effect of this strain was slightly less than that of strain PPKK081 (Fig. 1D). Thus, we confirmed that the cytotoxicity of PYKK081 on THP-1 cells is due to the RtxA product. We then assayed the accumulation of RtxA versus THP-1 cell killing by PYKK081 over time in vitro and observed the correlation between the accumulated toxin and cell viability (Fig. 2).
FIG 1.
Mutagenesis of rtxA and genetic complementation. (A) The schematic presentation of the rtx operon demonstrates the aphA3 kanamycin cassette insertion in the rtxA gene of KKNB100. The positions of the primers used for qPCR analysis of K. kingae are shown. (B) Schematic presentation of the genetic construct used for chromosome-based genetic complementation of the rtxA gene of KKNB100. (C) Purification of RtxA from the PYKK081 secreted fraction. The PYKK081 secreted fraction was isolated and subjected to G-100 chromatography as described in Materials and Methods. The samples were resolved by SDS-PAGE, and the proteins were stained with Coomassie blue. The 100-kDa protein band was eluted from the gel by electroelution. The purified RtxA was used as an antigen to obtain anti-RtxA monoclonal antibody. The protein ladder is shown in lane L, G-100-purified proteins (125 μg of protein loaded) are shown in lane 1, and the RtxA band (10 μg of protein loaded) after electroelution is shown in lane 2. (D) Determination of cytoxicity for PYKK081 and genetic constructs. For toxicity tests, 1 × 105 K. kingae cells were added to 0.5 × 106 THP-1 cells in RPMI with 10% FBS. Cell viability was estimated by ATP production, which was detected using the CellTiter-Glo luminescence assay. Each condition was assayed in duplicate, and luminescence values were recorded after 3 h. Mean values from three independent experiments are displayed. A one-way ANOVA was preformed among the three conditions; relative cell viability differed significantly (P < 0.05). A post hoc Tukey test revealed that PYKK081 and KKYN101 each differed significantly (P < 0.05) compared to KKNB100. Relative cell viability was defined as the luminescence value divided by the average mean KKNB100 luminescence value. (E) Western blot analysis of RtxA in the cell lysates of PYKK081 (lane 1), KKNB100 (lane 2), and KKYN101 (lane 3) using 10A7D7 antibody. Twenty micrograms of protein was loaded.
FIG 2.
In vitro assay of K. kingae toxicity. For toxicity tests, 1.0 × 105 PYKK081 cells were added to 0.5 × 106 THP-1 cells in RPMI with 10% FBS. Each time point was assayed in duplicate in three independent experiments, and samples were collected every 30 min for 3 h. The data in the figure are representative of the three experiments. The cell viability was estimated by ATP production, which was detected using the CellTiter-Glo luminescence assay. Relative viability refers to the PYKK081 luminescence value divided by the KKNB100 luminescence value at a given time point. THP-1 negative-control wells at each time point were consistently at or slightly below the luminescence of KKNB100. For the toxin accumulation experiment, the protein was precipitated from 200 μl of THP-1 culture medium before loading on the gel. The presence of toxin from PYKK081 was identified by Western blotting using 10A7D7 antibody. The Western blot shown contains the samples taken from the relative viability experiment described above. The correlation is representative of those in the other experiments.
K. kingae toxicity and virulence in PN 21 rats.
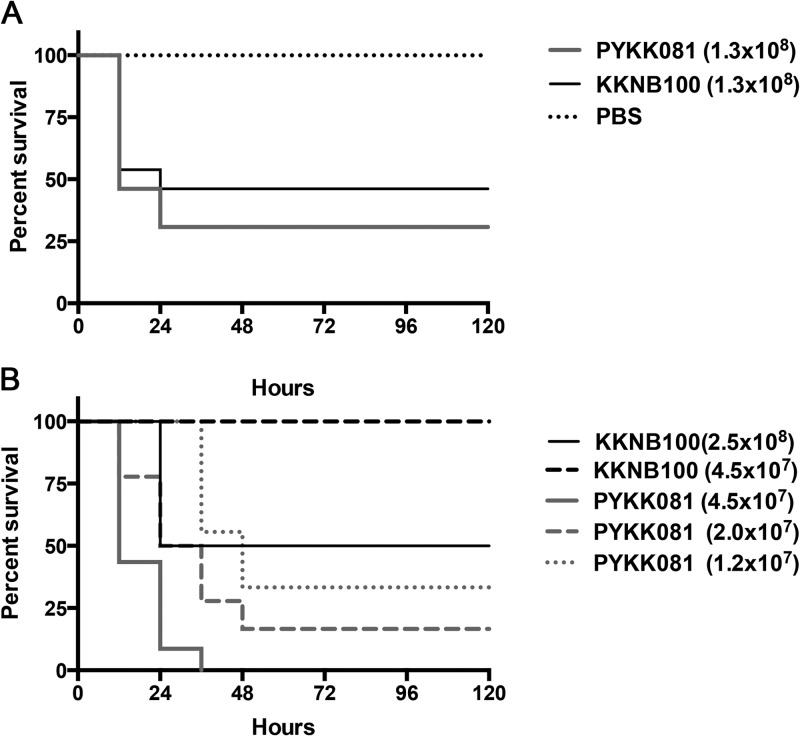
We performed a study to identify the i.p. PYKK081 LD50, the number of bacteria required to cause lethality in half of the exposed rats during the first 24 h. The results showed that the bacterium could cause lethality in PN 21 animals (average weight, 50.56 ± 0.53 g) at the doses above 1.0 × 108 cells/animal; the LD50 was identified as 1.3 × 108 cells/animal (Fig. 3A). Animals that survived after 24 h demonstrated ruffled fur, decreased activity, and hunched posture; however, they fully recovered several days after injection. No weight loss or inflammation in the site of injection was detected. In order to compare wild-type and RtxA-deficient strains in vivo, PYKK081 and KKNB100 LD50s (1.3 × 108 cells/animal) were applied to PN 21 rats. Kaplan-Meier survival analysis indicated there was no significant difference in mortality among the groups (Fig. 3A).
FIG 3.
Kaplan-Meier survival curves of rat offspring after i.p. injections with K. kingae. PN 21 or PN 7 rats were injected with different doses of K. kingae strains or PBS. The animals were monitored for mortality and clinical signs of illness twice a day for 14 days. After the initial 72 h, no additional mortality was detected. These data were combined from three separate experiments. (A) The survival curve of 21-day-old rats injected with 1.3 × 108 bacterial cells/animal of either PYKK081 or KKNB100 demonstrated no significant difference in mortality (log rank test, P > 0.05). (B) The survival curves of 7-day-old rats injected with 4.5 × 107, 2.0 × 107, and 1.2 × 107 bacterial cells/animal all demonstrated significant difference in mortality when comparing PYKK081 and KKNB100. Additionally, survival curves were significantly dependent upon the concentration of injected PYKK081 (log rank test for trend, P < 0.0001).
K. kingae toxicity and virulence in PN 7 rats.
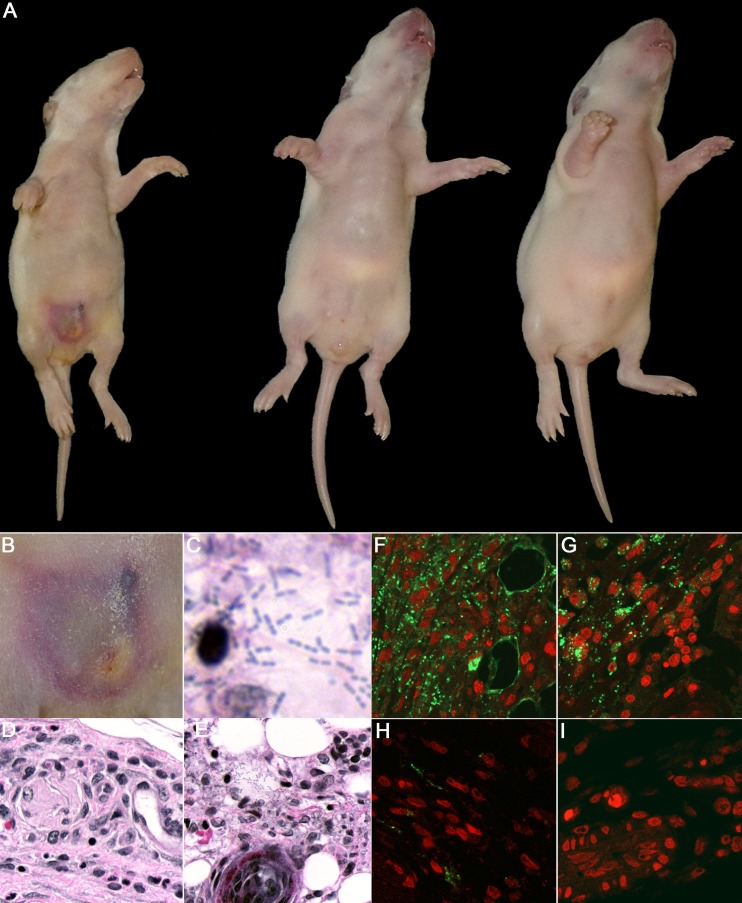
When cohorts of PN 7 rat pups (average weight, 13.44 ± 1.58 g) were injected i.p. with different doses of PYKK081, the LD50 was identified at 1.57 × 107 cells/animal. Interestingly, in contrast to PN 21 animals, PN 7 rat pups that survived after 24 h developed disease after injection of doses at or above 8.0 × 106 cells/animal. The disease presented as necrotic lesion formation at the site of injection (Fig. 4A and B) and a significant lack of age-appropriate weight gain (14.00 ± 0.47 g in sick animals injected with PYKK081 versus 16.83 ± 2.0 g in animals injected with KKNB100 after 48 h), (Fig. 5A). The lesion formation was an indication of the lethal disease; e.g., the animals that developed a lesion did not survive longer than 3 days after the injection. The survival rates are demonstrated in Fig. 3B. We then performed a comparative virulence study using PN 7 rat pups by injections of 4.5 × 107, 2 × 107, and 1.2 × 107cells/animal of PYKK081 or KKNB100. We identified that KKNB100 was not able to cause lethality or disease to PN 7 pups at these doses. The LD50 for KKNB100 was identified at 2.35 × 108 cells/animal. The cumulative survival results are presented in Fig. 3B.
FIG 4.
Gross pathology and histopathological examination of infant rats injected with K. kingae. The animals were i.p. injected with 2.0 × 107 cells of PYKK081 or KKNB100 or with PBS and sacrificed 48 h after injection for tissue analysis. Representative pictures are shown. (A) Ventral view of the animals injected with PYKK081 (left), exhibiting an abdominal necrotic lesion and significant weight loss, KKNB100 (middle), and PBS (right). (B) Magnified view of the skin lesion formed as a result of i.p. injection of PYKK081, which demonstrated considerable necrosis, and associated acute inflammation involving the panniculus carnosus muscularis (PCM), the subdermal tissues, and presumably adherent portions of peritoneum. (C) Skin lesion section from the animal injected with PYKK081, which demonstrates muscle necrosis, some associated acute inflammation, and bacteria. Magnification, ×1,000. (D) Slide from the animal injected with KKNB100, which shows no bacteria or any histopathological changes. Magnification, ×400. (E) Skin lesion section seen in panel C. Magnification, ×400. (F to I) Immunohistochemical analysis of the skin lesion slides was performed using anti-K. kingae OMV antibody and was visualized upon interaction with Alexa 488-conjugated anti-rabbit IgG. Magnification, ×400. Nonspecific fluorescence was not observed in the absence of primary antibody. Cell nuclei were labeled with 7-AAD. An intense green signal, indicating the presence of K. kingae, was clearly observed between the abdominal thin muscle layer underlying the skin and the fatty layer in the samples from animals injected with PYKK081. (F) Skin lesion section from the animal injected with PYKK081. A mixture of intracellular organisms and extensive extracellular K. kingae was detected. (G) Lesion section from the animal injected with PYKK081. The bacteria are predominantly found inside phagocytic cells. (H) In the skin lesion section from the animal injected with KKNB100, the majority of staining was observed in phagocytic cells. The samples show much less staining. (I) Site of injection from the animal injected with PBS. No green signal was detected in these samples.
FIG 5.
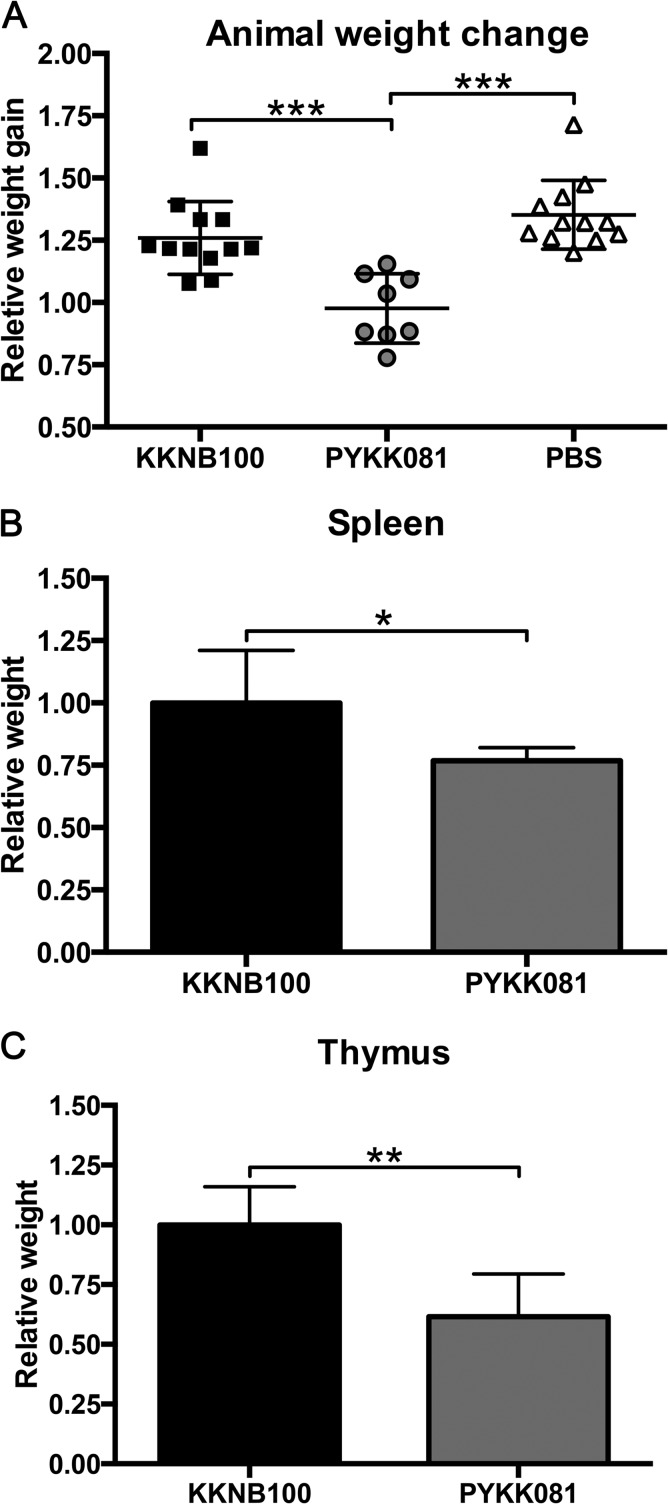
Body and organ weight changes in PN 9 rats 48 h after i.p. injection with K. kingae. Rats were inoculated with 2.0 × 107 CFU of PYKK081, KKNB100, or PBS. Data were combined from three separate experiments. Each symbol represents the value from a single animal, and horizontal lines indicate median values. A one-way ANOVA on the relative body weight gain yielded significant variation between groups (P < 0.0001). A post hoc Tukey test showed that PYKK081 and KKNB100 as well as PYKK081 and the PBS control differed significantly (***, P < 0.05); the PBS control group was not significantly different from the KKNB100 group (P > 0.05). Relative body weights were calculated by dividing an individual animal's PN 9 body weight by the animal's PN 7 preinoculation body weight. Independent unpaired t tests revealed that the means compared differed significantly with respect to both the thymus weight (**, P < 0.0001) and the spleen weight (*, P < 0.022). The thymus and spleen weights were individually normalized by the animal's weight prior to autopsy. The relative organ weights are displayed as values relative to their respective KKNB100-treated average normalized organ weight.
Pathology examination in PN 7 rats.
To examine the pathology resulting from K. kingae infections and to identify the role of RtxA in pathogenesis, PN 7 pups were injected with 2.0 × 107 cells/animal PYKK081, KKNB100, or PBS. Animals that received PYKK081 and developed necrotic lesions at 48 h after injection, along with the animals from the other groups, were euthanized, and blood and tissues were collected for analysis.
(i) Analysis of animal blood.
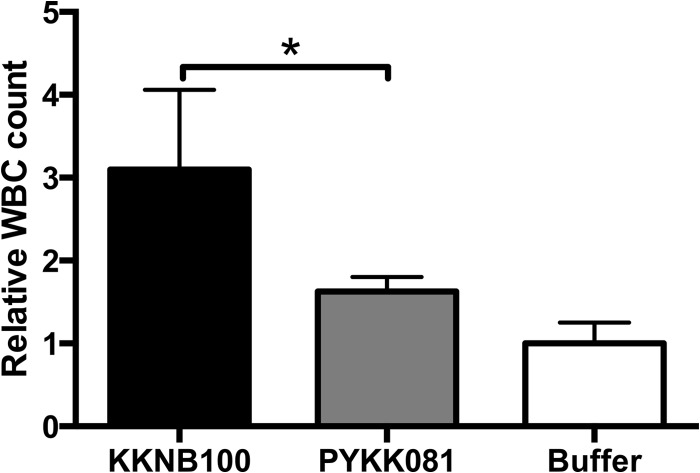
Plating of animal blood on Columbia agar plates with 5% sheep blood did not always lead to recovery of viable K. kingae cells. However, the pathogen was identified in blood samples of animals injected with PYKK081 that developed necrotic lesions using a qPCR assay. The rtxA gene region used for the qPCR amplification was not affected by the transposon insertion in KKNB100. Therefore, this method is appropriate for detection of the gene in this strain (Fig. 1A). The presence of the pathogen was not detected in the blood of animals injected with KKNB100. The CP values for representative animals of each strain group are provided in Table 1, which also demonstrates the correlation between CP values, lesion formation, and the presence of bacteria in the blood (bacteremia). Independently, we analyzed the hematological profiles of the blood collected from each strain group. In animals injected with PYKK081 that developed a lesion, total WBC counts increased by a factor of 1.63 ± 0.18, yet this was not a significant elevation compared to those injected with PBS. In rats injected with KKNB100, the WBC count increased by a factor of 3.10 ± 0.96, which was significant (Fig. 6).
TABLE 1.
Determination of K. kingae in animal blood via detection of rtxA and rtxB by qPCR analysisa
| Inoculum and animal | CP value |
Bacteremia | Lesion | Relative wt gain | |
|---|---|---|---|---|---|
| rtxA | rtxB | ||||
| Buffer | |||||
| 1 | − | − | − | − | 1.22 |
| 2 | − | − | − | − | 1.33 |
| KKNB100 | |||||
| 1 | − | − | − | − | 1.23 |
| 2 | − | − | − | − | 1.29 |
| PYKK081 | |||||
| 1 | 20.63 | 25.78 | + | + | 1.04 |
| 2 | 20.61 | 25.89 | + | + | 1.15 |
| 3 | 18.71 | 25.29 | + | + | 0.88 |
The table lists CP values and the major clinical symptoms for representative animals of each group. CP values are presented as the cycle at which fluorescence is deemed to be detectable above the background during the exponential phase of amplification as described in Materials and Methods. DNA purified from PYKK081 was used to determine the minimum detectable CFU, which was equivalent to 8 to 75 CFU. qPCRs were performed from three PYKK081-, two KKNB100-, and two PBS-injected animals, each in technical duplicate. Bacteremia was defined as a positive call from both the rtxA and rtxB primer sets. The threshold for specific amplification (positive qualitative identification) was set at 26 cycles, below that of the negative control. The major clinical presentations, lesion formation, and weight changes are shown. Mortality is not reflected in the table, since the animals were sacrificed at 48 h postinfection; however, formation of a lesion always correlated with a lethal outcome. The weight gain ratio was determined by dividing the PN 9 (preinjection) by the PN 7 (preinjection) rat weight.
FIG 6.
Effect of K. kingae on WBCs in rat blood. The animals were sacrificed at 48 h after bacterial injections. Fifty microliters of freshly collected animal blood was processed using a Coulter HmX Hematology Analyzer. The average result was calculated from at least three animals. A one-way ANOVA was performed among the three conditions; the relative WBC count differed significantly (P < 0.05). A post hoc Tukey test revealed that PYKK081 and KKNB100 differed significantly (*, P < 0.05); the PBS control group was not significantly different from the PYKK081 group.
(ii) Rat pup autopsy.
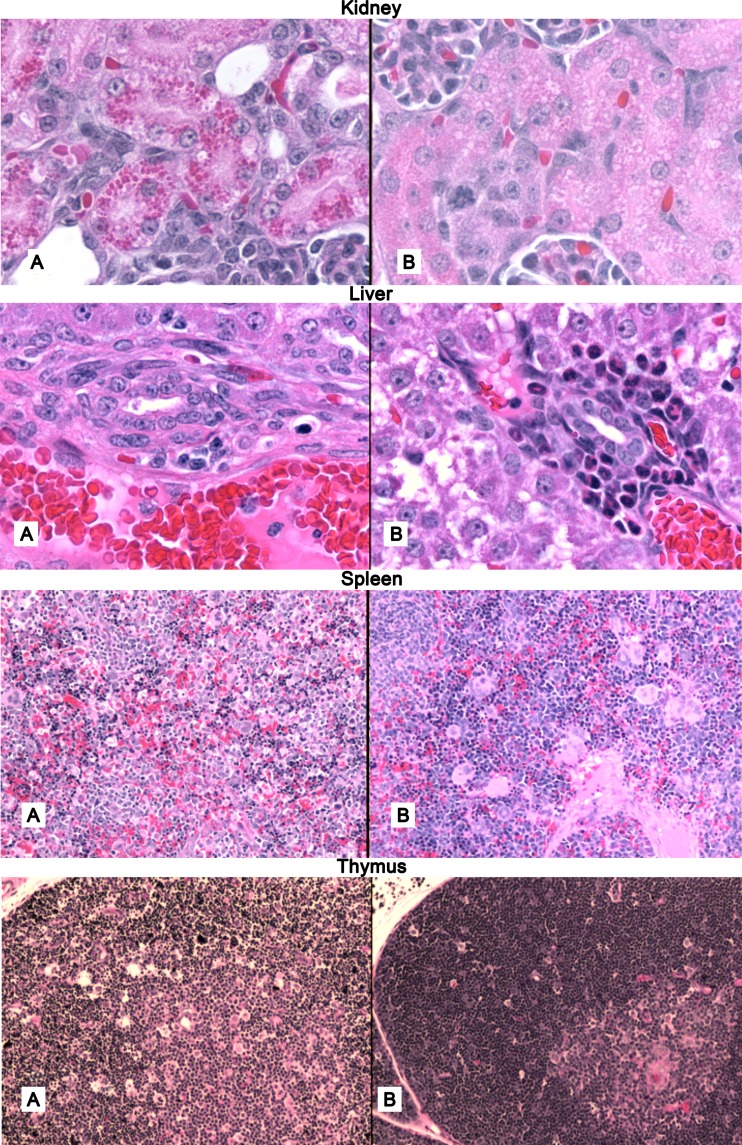
Samples of different organs (heart, lung, hip joint, kidney, thymus, spleen, and tibia bone marrow) were subjected to histopathological examination (Fig. 7). Lungs, joints, and hearts of the pups receiving PYKK081 exhibited no apparent abnormalities. Two-thirds of the pups receiving toxin-producing bacteria had prominent collections of eosinophilic hyaline droplets in the proximal convoluted tubules, which were absent in the pups receiving KKNB100 or PBS. In the livers from animals given PYKK081, accumulations of polymorphonuclear leukocytes (PMNs) around the bile ducts were less prominent than in the livers of animals injected with PBS or KKNB100. In the pups receiving PYKK081, the normalized mean spleen weight was lower (0.77 ± 0.05) than in the other groups (1.0 ± 0.21 and 0.80 ± 0.17 for the KKNB100 and PBS groups, respectively) (Fig. 4), and hematopoiesis was markedly decreased in the red pulp; megakaryocytes were infrequent, were smaller, and contained fewer nuclei. There was a paucity of megakaryocytes, and those that were present were smaller with fewer nuclei. The white pulp in the spleen at this age was insufficiently populated with lymphoid elements to evaluate the effect of toxin on this component. In the bone marrow of the pups receiving PYKK081, hematopoiesis was considerably decreased, and depletion of megakaryocytes was evident. In pups given PYKK081, the thymuses were smaller (0.61 ± 0.18) than in other groups (1.00 ± 0.16 and 0.86 ± 0.40 in the KKNB100 and PBS groups, respectively) (Fig. 4) and on microscopic examination exhibited extensive loss of cortical lymphocytes with widespread karyorrhexis and pyknosis, consistent with apoptosis. The thymuses of pups receiving KKNB100 appeared to be normal. The pathologies observed in spleen, bone marrow, and thymus are consistent with the cytotoxicity that we have demonstrated previously in vitro for PYKK081 with cultured promyoblasts, T lymphoblasts, T lymphocytes, and megakaryoblasts (35).
FIG 7.
Histopathological examination rat organs. Hematoxylin-eosin stain preparation of organ sections (original magnification, ×100) obtained from autopsied PN 9 rats at 48 h postinjection with PYKK081 (A) or KKNB100/PBS (B). At least four animals from each strain group were sampled for histological analysis. Representative slides are shown.
(iii) Analysis of abdomen.
Culturing of peritoneal fluid samples resulted in recovery of PYKK081 colonies, suggesting the presence of living bacteria in the abdomen. Samples of abdominal skin containing the site of injection and surrounding area were collected and analyzed. We observed the development of inflammation in and around the site of PYKK081 strain injection (Fig. 4B). Histopathological examination of these samples exhibited diffuse distribution of weakly stained short rods, located principally in and around the panniculus carnosus muscularis (PCM) and the subdermal tissue (Fig. 4C and E). Considerable necrosis and associated acute inflammation were present, involving the PCM and the subdermal tissues and probably adherent portions of the peritoneum. Some slides exhibited some local muscle necrosis in the PCM without evident inflammation. Immunohistochemical analysis was used to demonstrate the presence and location of K. kingae in tissue sections; this was accomplished with an antibody which specifically binds to K. kingae OMVs and visualized with Alexa 488 (Fig. 4F to I). An intense presence of K. kingae organisms or bacterial components was found between the thin muscle layer underlying the skin and the fatty layer. No viable bacteria were recovered from the peritoneal cavities of animals infected with KKNB100 by direct culturing. The skin samples from animals injected with KKNB100 did not show any histopathological changes and did not reveal any bacteria, yet they exhibited less abundant staining by immunohistochemical examination (Fig. 4H).
DISCUSSION
Using the rat model, we demonstrated that infant rats are suitable animals to study K. kingae infections and identified pathological conditions that result from injection of the bacteria. While this route of infection is not the natural mode, this model may serve as a K. kingae bacteremia model, which recapitulates what are thought to be the general features of K. kingae pathogenesis, namely, a localized colonization and subsequent epithelial breach resulting in bacteremia, which may enable the formation of distant focal K. kingae sites of infection (2). In our experimental system we demonstrated that injected bacteria could colonize the skin and peritoneal cavity of infected animals and travel from the area of injection into the bloodstream. This model can be used to assay the virulence potential of different bacterial strains and to identify bacterial virulence factors. Indeed, the PN 5 rat model was used previously by another group to compare the toxicity and virulence of K. kingae strains of different sequence types, which were identified by multilocus sequence typing (MLST) (41). Significantly, our model can serve to mimic some aspects of human immunity and consequently pathogen susceptibility. It can also be used to investigate the correlation between immune system development and susceptibility to bacterial infections caused by K. kingae and other organisms.
The majority of K. kingae diseases in humans occur in those between 6 and 24 months of age. This age range may represent the gap between a maternally contributed antibody-mediated defense and the development of a child's own specific immune defenses (42). Indeed, it has been previously found that the peak of K. kingae infections in children correlates with the lowest level of antibodies specific for K. kingae outer membrane proteins (42). We empirically discovered differential symptoms in PN 7 and PN 21 rats injected with strain PYKK081. We observed that younger animals exhibit RtxA-dependent disease symptoms. We believe that in addition to a host's specific immune response to K. kingae, developmental changes may underlie the symptomatic differences that we observed and that these changes are important contributors to disease susceptibility during developmental maturity. Between 6 and 24 months there are many changes occurring in the human WBC population, e.g., fluctuation in the proportion of circulating neutrophils (43). During this early stages of life, neutrophils and monocytes together make up progressively less of the total WBC population, which correlates with susceptibility to Haemophilus influenzae pneumonia and otitis media (44, 45). Interestingly, animal models have demonstrated that depleting neutrophils prior to pathogen inoculation can increase the pathogen burden, which has been shown for a Listeria monocytogenes disease model (46, 47). Additionally, a depletion of monocytes can increase the risk of death from listerial infection (48). It is unknown at present how the human developmental changes are paralleled in the infant rats; however, we hypothesize that rats may share a similar age-dependent modulation of immunity. For instance, our preliminary results demonstrate a difference in the number of various WBCs in PN 7 versus PN 21 rats (data not shown), suggesting that our model may reveal important contributors to disease susceptibility, which may parallel human developmental changes. Our future investigations will involve delineating which factors contribute to our differential observations at 7 and 21 days. These studies may reveal biomarkers for disease susceptibility and aid in the development of targeted therapies.
RTX toxins are produced by both human and animal Gram-negative pathogens (49). The roles of several of these toxins in the bacterial pathogenesis have been investigated. For instance, EhxA (enterohemorrhagic Escherichia coli [EHEC] hemolysin), an RTX toxin produced by enterohemorrhagic Escherichia coli, is an important virulence factor for EHEC-mediated diseases (50, 51). OMV-associated EhxA plays a key role in disease pathogenesis by irreversibly injuring epithelial cells and microvascular endothelial cells (52). Some RTX toxins affect cells in a species-specific manner: the primary biological effect of LktA produced by Mannheimia haemolytica is restricted to ruminant leukocytes and platelets (53), whereas A. actinomycetemcomitans LtxA kills only lymphocytes and granulocytes from humans, the Great Apes, and Old World monkeys (54). To date it remains difficult to appropriately model and evaluate these toxins' contributions to the virulence of the pathogen.
In our study, we used strain PYKK081, which belongs to MLST ST-22, rtxA allele type 1, and porin allele type 9 and is classified by pulse-field gel electrophoresis as clone B (41, 55, 56). This group of K. kingae strains was associated with septic arthritis and bacteremia in different geographic regions, indicating that it is an invasive clone. Our in vitro results show that PYKK081 RtxA affects various cell types but causes a toxic effect primarily on leukocytes from multiple species (35). To investigate the role of RtxA in K. kingae pathogenesis in vivo, PYKK081 and its isogenic RtxA-deficient strain KKNB100 were tested for their toxicity and virulence in PN 7- and PN 21 rat models after i.p. injections. Our in vivo data enabled us to determine i.p. LD50s for these strains.
In the current work, we chose not to test the effect of purified toxin on animals or cells. We believe that the full effect of the toxin is most accurately observed when the toxin is naturally produced and secreted by K. kingae cells. Indeed, we observed the highest cytotoxic effect when RtxA was administered via the introduction of viable K. kingae. We have previously shown that the toxin could exist in soluble and outer membrane vesicle-associated forms, indicating that K. kingae may utilize different mechanisms for toxin delivery (35). Thus, our findings parallel other studies performed on RTX toxins (52). Additionally, the RTX toxins of other organisms, such as that of A. actinomycetemcomitans, which produces LtxA, can be efficiently purified and can maintain a stable cytotoxic effect on WBCs (57). In contrast, our conventional purification of RtxA demonstrated instability in diluted solutions and reduced cytotoxicity after storage. These observations highlight the complex conditions required for RtxA activity. Therefore, future investigations will be carried out to define possible protein and lipid factors among other conditions required to stabilize RtxA activity.
In PN 7 rats, at doses at or above 8.0 × 106 cells/animal, PYKK081 was able to cause a lethal illness, which resulted in weight loss, bacteremia, and necrotic lesion formation at the site of injection when the strain was introduced into the peritoneal cavity. The lesion appearance was a major clinical manifestation of the disease. This process likely contributes to the overall morbidity. However, we also demonstrated that bacteria could enter the bloodstream, severely affecting white blood cells in blood and organs. Thus, we conclude that it is likely that both these events contribute to the lethal outcome of the disease. Histopathological examination of sick animals showed significant immunotoxic effect in rat pup thymuses, spleen, and bone marrow. Strain KKNB100 was less toxic to animals and could not cause lesion formation at any dose tested. Thus, our experiments in the infant rat model demonstrate that the toxin contributes significantly to K. kingae toxicity and plays the key role in the organism's virulence. Interestingly, we observed lethality in PN 7 pups after their injection with the same high dose (3.6 × 108 CFU) of live and heat-killed KKNB100 (data not shown). By using the toxin-deficient strain, we excluded any potential contribution of RtxA to the overall toxicity of the bacteria. These results allow us to speculate regarding the potential toxicity of other bacterial components, such as lipopolysaccharide (LPS), which are known for their ability to induce septic shock in animals (26).
With the exception of endocarditis, K. kingae infections in children are frequently characterized by a mild clinical presentation, low-grade fever, mildly elevated acute-phase reactants, a normal to slightly elevated leukocyte count, and a paucity of leukocyte content in the synovial fluid of children with joint infections caused by the organism (58). The WBC cutoff employed by pediatricians to distinguish between viral infections, characterized by lower counts, and those caused by bacteria, which usually exhibit high WBC counts, is 15,000/mm3 (59). Our results suggest that the RTX toxin may play a role in the low inflammatory response observed in these patients. These findings may have significance for diagnostics of K. kingae infections. In fact, many events of invasive K. kingae infections are probably unsuspected and undiagnosed because, in the absence of fever or leukocytosis, patient specimens are not cultured and nucleic acid amplification assays are not performed (2).
K. kingae has been previously reported to be difficult to recover directly from infected human blood or synovial fluid. The application of PCR analysis has significantly increased the number of identified K. kingae infection cases (60). Thus, the poor recovery of the organism from the blood of infected animals observed in our study correlates well with the analogous clinical observation. We used qPCR analysis to confirm the presence of bacteria in the blood. The rtxA and rtxB toxin genes are used as a specific target to diagnose K. kingae infection in clinical samples (61). We adopted this molecular diagnostic tool for our study. The bacteria were detected in neither the peritoneal cavity nor the blood of animals infected with KKNB100, suggesting that RtxA is required for bacterial survival in an animal's body. As a cytotoxic agent, RtxA may contribute to host immune response evasion that allows bacteria to survive in the host's blood; additionally, RtxA may facilitate an epithelial breach that would precede its entrance into the bloodstream. Evaluating the rtxA transcriptional levels in vivo was problematic due to the small size of PN 9 rats; additionally, the animals that developed a lesion lost weight and in turn had lower blood collection yields. Therefore, the collection of blood to perform a sequential time point assessment of rtxA expression was not an available option. Alternatively, we chose to assay the accumulation of RtxA versus cytotoxicity over time, in vitro, using THP-1 cell culture. The accumulation of RtxA correlated with increasing cytotoxicity on the mammalian cells. This finding further supports the role of RtxA in K. kingae disease pathogenesis.
In this study, we have further established an infant rat model for disease caused by K. kingae. We also demonstrated that RtxA is a significant contributing factor to the organism's toxicity and is specifically a key virulence factor for K. kingae. Notably, other Kingella species (K. denitrificans and K. oralis), which do not produce RtxA, have rarely been implicated in human systemic infections. The infant rat model is a powerful tool for future mechanistic investigations of disease pathogenesis and virulence and may reveal molecular markers of disease susceptibility, as well as aid in the development of targeted therapeutics.
ACKNOWLEDGMENTS
We are thankful to Pablo Yagupsky for providing K. kingae isolate PYKK081 and for discussion of the results. We thank David Lagunoff and Scott Kachlany for suggestions during the project. We thank Joseph St. Geme III and Eric Porsch for providing the mariner transposon mutagenesis system and the vector for genetic complementation in K. kingae.
This work was supported by American Heart Association grant 09SDG2310194 and Foundation of UMDNJ grant PC15-11 to N.V.B., as well as NIH grant R01GM098052 to E.Z.
Footnotes
Published ahead of print 24 March 2014
REFERENCES
- 1. Yagupsky P, Peled N, Katz O. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 40:4180–4184. 10.1128/JCM.40.11.4180-4184.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yagupsky P, Porsch E, St Geme JW., 3rd 2011. Kingella kingae: an emerging pathogen in young children. Pediatrics 127:557–565. 10.1542/peds.2010-1867 [DOI] [PubMed] [Google Scholar]
- 3. Yagupsky P, Dagan R, Prajgrod F, Merires M. 1995. Respiratory carriage of Kingella kingae among healthy children. Pediatr. Infect. Dis. J. 14:673–678. 10.1097/00006454-199508000-00005 [DOI] [PubMed] [Google Scholar]
- 4. Yagupsky P, Dagan R, Howard CB, Einhorn M, Kassis I, Simu A. 1993. Clinical features and epidemiology of invasive Kingella kingae infections in southern Israel. Pediatrics 92:800–804 [PubMed] [Google Scholar]
- 5. Verdier I, Gayet-Ageron A, Ploton C, Taylor P, Benito Y, Freydiere AM, Chotel F, Berard J, Vanhems P, Vandenesch F. 2005. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr. Infect. Dis. J. 24:692–696. 10.1097/01.inf.0000172153.10569.dc [DOI] [PubMed] [Google Scholar]
- 6. Li X, Kolltveit KM, Tronstad L, Olsen I. 2000. Systemic diseases caused by oral infection. Clin. Microbiol. Rev. 13:547–558. 10.1128/CMR.13.4.547-558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ilharreborde B, Bidet P, Lorrot M, Even J, Mariani-Kurkdjian P, Liguori S, Vitoux C, Lefevre Y, Doit C, Fitoussi F, Pennecot G, Bingen E, Mazda K, Bonacorsi S. 2009. New real-time PCR-based method for Kingella kingae DNA detection: application to samples collected from 89 children with acute arthritis. J. Clin. Microbiol. 47:1837–1841. 10.1128/JCM.00144-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gene A, Garcia-Garcia JJ, Sala P, Sierra M, Huguet R. 2004. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr. Infect. Dis. J. 23:886–888. 10.1097/01.inf.0000137591.76624.82 [DOI] [PubMed] [Google Scholar]
- 9. Chometon S, Benito Y, Chaker M, Boisset S, Ploton C, Berard J, Vandenesch F, Freydiere AM. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26:377–381. 10.1097/01.inf.0000259954.88139.f4 [DOI] [PubMed] [Google Scholar]
- 10. Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48:25–33. 10.1146/annurev.med.48.1.25 [DOI] [PubMed] [Google Scholar]
- 11. Verbruggen AM, Hauglustaine D, Schildermans F, van der Hauwaert L, Rombouts JJ, Wauters G, Vandepitte J. 1986. Infections caused by Kingella kingae: report of four cases and review. J. Infect. 13:133–142. 10.1016/S0163-4453(86)92841-0 [DOI] [PubMed] [Google Scholar]
- 12. Lewis MB, Bamford JM. 2000. Global aphasia without hemiparesis secondary to Kingella kingae endocarditis. Arch. Neurol. 57:1774–1775. 10.1001/archneur.57.12.1774 [DOI] [PubMed] [Google Scholar]
- 13. Korach A, Olshtain-Pops K, Schwartz D, Moses A. 2009. Kingella kingae prosthetic valve endocarditis complicated by a paravalvular abscess. Isr. Med. Assoc. J. 11:251–253 [PubMed] [Google Scholar]
- 14. Goutzmanis JJ, Gonis G, Gilbert GL. 1991. Kingella kingae infection in children: ten cases and a review of the literature. Pediatr. Infect. Dis. J. 10:677–683. 10.1097/00006454-199109000-00011 [DOI] [PubMed] [Google Scholar]
- 15. Elyes B, Mehdi G, Kamel BH, Hela Z, Imen BS. 2006. Kingella kingae septic arthritis with endocarditis in an adult. Joint Bone Spine 73:472–473. 10.1016/j.jbspin.2005.10.021 [DOI] [PubMed] [Google Scholar]
- 16. Brachlow A, Chatterjee A, Stamato T. 2004. Endocarditis due to Kingella kingae: a patient report. Clin. Pediatr. 43:283–286. 10.1177/000992280404300311 [DOI] [PubMed] [Google Scholar]
- 17. Rotstein A, Konstantinov IE, Penny DJ. 2010. Kingella-infective endocarditis resulting in a perforated aortic root abscess and fistulous connection between the sinus of Valsalva and the left atrium in a child. Cardiol. Young 20:332–333. 10.1017/S1047951110000314 [DOI] [PubMed] [Google Scholar]
- 18. Lee WL, Dooling EC. 1984. Acute Kingella kingae endocarditis with recurrent cerebral emboli in a child with mitral prolapse. Ann. Neurol. 16:88–89. 10.1002/ana.410160117 [DOI] [PubMed] [Google Scholar]
- 19. Holmes AA, Hung T, Human DG, Campbell AI. 2011. Kingella kingae endocarditis: a rare case of mitral valve perforation. Ann. Pediatr. Cardiol. 4:210–212. 10.4103/0974-2069.84664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berkun Y, Brand A, Klar A, Halperin E, Hurvitz H. 2004. Kingella kingae endocarditis and sepsis in an infant. Eur. J. Pediatr. 163:687–688. 10.1007/s00431-004-1520-z [DOI] [PubMed] [Google Scholar]
- 21. Kiang KM, Ogunmodede F, Juni BA, Boxrud DJ, Glennen A, Bartkus JM, Cebelinski EA, Harriman K, Koop S, Faville R, Danila R, Lynfield R. 2005. Outbreak of osteomyelitis/septic arthritis caused by Kingella kingae among child care center attendees. Pediatrics 116:e206–213. 10.1542/peds.2004-2051 [DOI] [PubMed] [Google Scholar]
- 22. Sena AC, Seed P, Nicholson B, Joyce M, Cunningham CK. 2010. Kingella kingae endocarditis and a cluster investigation among daycare attendees. Pediatr. Infect. Dis. J. 29:86–88. 10.1097/INF.0b013e3181b48cc3 [DOI] [PubMed] [Google Scholar]
- 23. Yagupsky P, Erlich Y, Ariela S, Trefler R, Porat N. 2006. Outbreak of Kingella kingae skeletal system infections in children in daycare. Pediatr. Infect. Dis. J. 25:526–532. 10.1097/01.inf.0000215243.42501.4f [DOI] [PubMed] [Google Scholar]
- 24. Kehl-Fie TE, St Geme JW., III 2007. Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J. Bacteriol. 189:430–436. 10.1128/JB.01319-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ceroni D, Cherkaoui A, Ferey S, Kaelin A, Schrenzel J. 2010. Kingella kingae osteoarticular infections in young children: clinical features and contribution of a new specific real-time PCR assay to the diagnosis. J. Pediatr. Orthop. 30:301–304. 10.1097/BPO.0b013e3181d4732f [DOI] [PubMed] [Google Scholar]
- 26. Cherkaoui A, Ceroni D, Emonet S, Lefevre Y, Schrenzel J. 2009. Molecular diagnosis of Kingella kingae osteoarticular infections by specific real-time PCR assay. J. Med. Microbiol. 58:65–68. 10.1099/jmm.0.47707-0 [DOI] [PubMed] [Google Scholar]
- 27. Lehours P, Freydiere AM, Richer O, Burucoa C, Boisset S, Lanotte P, Prere MF, Ferroni A, Lafuente C, Vandenesch F, Megraud F, Menard A. 2011. The rtxA toxin gene of Kingella kingae: a pertinent target for molecular diagnosis of osteoarticular infections. J. Clin. Microbiol. 49:1245–1250. 10.1128/JCM.01657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goni FM, Ostolaza H. 1998. E. coli alpha-hemolysin: a membrane-active protein toxin. Braz. J. Med. Biol. Res. 31:1019–1034 [DOI] [PubMed] [Google Scholar]
- 29. Ostolaza H, Soloaga A, Goni FM. 1995. The binding of divalent cations to Escherichia coli alpha-haemolysin. Eur. J. Biochem. 228:39–44. 10.1111/j.1432-1033.1995.tb20225.x, [DOI] [PubMed] [Google Scholar]
- 30. Balashova NV, Shah C, Patel JK, Megalla S, Kachlany SC. 2009. Aggregatibacter actinomycetemcomitans LtxC is required for leukotoxin activity and initial interaction between toxin and host cells. Gene 443:42–47. 10.1016/j.gene.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 31. Stanley P, Koronakis V, Hughes C. 1998. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. Rev. 62:309–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stanley P, Packman LC, Koronakis V, Hughes C. 1994. Fatty acylation of two internal lysine residues required for the toxic activity of Escherichia coli hemolysin. Science 266:1992–1996. 10.1126/science.7801126 [DOI] [PubMed] [Google Scholar]
- 33. Gutierrez K. 2005. Bone and joint infections in children. Pediatr. Clin. N. Am. 52:779–794. 10.1016/j.pcl.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 34. Verger C, Luger A, Moore HL, Nolph KD. 1983. Acute changes in peritoneal morphology and transport properties with infectious peritonitis and mechanical injury. Kidney Int. 23:823–831. 10.1038/ki.1983.101 [DOI] [PubMed] [Google Scholar]
- 35. Maldonado R, Wei R, Kachlany SC, Kazi M, Balashova NV. 2011. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microb. Pathog. 51:22–30. 10.1016/j.micpath.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaplan JB, Lo C, Xie G, Johnson SL, Chain PS, Donnelly R, Kachlany SC, Balashova NV. 2012. Genome sequence of Kingella kingae septic arthritis isolate PYKK081. J. Bacteriol. 194:3017. 10.1128/JB.00421-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063–1076. 10.1016/S0003-9969(99)00089-8 [DOI] [PubMed] [Google Scholar]
- 38. Porsch EA, Kehl-Fie TE, Geme JW., III 2012. Modulation of Kingella kingae adherence to human epithelial cells by type IV pili, capsule, and a novel trimeric autotransporter. mBio 3(5):e00372–12. 10.1128/mBio.00372-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kehl-Fie TE, Porsch EA, Yagupsky P, Grass EA, Obert C, Benjamin DK, Jr, St Geme JW., III 2010. Examination of type IV pilus expression and pilus-associated phenotypes in Kingella kingae clinical isolates. Infect. Immun. 78:1692–1699. 10.1128/IAI.00908-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kehl-Fie TE, Miller SE, St Geme JW., III 2008. Kingella kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells. J. Bacteriol. 190:7157–7163. 10.1128/JB.00884-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amit U, Porat N, Basmaci R, Bidet P, Bonacorsi S, Dagan R, Yagupsky P. 2012. Genotyping of invasive Kingella kingae isolates reveals predominant clones and association with specific clinical syndromes. Clin. Infect. Dis. 55:1074–1079. 10.1093/cid/cis622 [DOI] [PubMed] [Google Scholar]
- 42. Slonim A, Steiner M, Yagupsky P. 2003. Immune response to invasive Kingella kingae infections, age-related incidence of disease, and levels of antibody to outer-membrane proteins. Clin. Infect. Dis. 37:521–527. 10.1086/376913 [DOI] [PubMed] [Google Scholar]
- 43. Koenig JM, Yoder MC. 2004. Neonatal neutrophils: the good, the bad, and the ugly. Clin. Perinatol. 31:39–51. 10.1016/j.clp.2004.03.013 [DOI] [PubMed] [Google Scholar]
- 44. Bamberger EE, Ben-Shimol S, Abu Raya B, Katz A, Givon-Lavi N, Dagan R, Srugo I. 20 January 2014. Pediatric invasive Haemophilus influenzae infections in Israel in the era of Haemophilus influenzae type b vaccine: a nationwide prospective study. Pediatr. Infect. Dis. J. 10.1097/INF.0000000000000193 [DOI] [PubMed] [Google Scholar]
- 45. Teele DW, Klein JO, Rosner B. 1989. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J. Infect. Dis. 160:83–94. 10.1093/infdis/160.1.83 [DOI] [PubMed] [Google Scholar]
- 46. Carr KD, Sieve AN, Indramohan M, Break TJ, Lee S, Berg RE. 2011. Specific depletion reveals a novel role for neutrophil-mediated protection in the liver during Listeria monocytogenes infection. Eur. J. Immunol. 41:2666–2676. 10.1002/eji.201041363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sherrid AM, Kollmann TR. 2013. Age-dependent differences in systemic and cell-autonomous immunity to L. monocytogenes. Clin. Dev. Immunol. 2013:917198. 10.1155/2013/917198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. 2003. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity 19:59–70. 10.1016/S1074-7613(03)00171-7 [DOI] [PubMed] [Google Scholar]
- 49. Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34:1076–1112. 10.1111/j.1574-6976.2010.00231.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bauer ME, Welch RA. 1996. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 64:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmidt H, Beutin L, Karch H. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bielaszewska M, Ruter C, Kunsmann L, Greune L, Bauwens A, Zhang W, Kuczius T, Kim KS, Mellmann A, Schmidt MA, Karch H. 2013. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog. 9:e1003797. 10.1371/journal.ppat.1003797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown JF, Leite F, Czuprynski CJ. 1997. Binding of Pasteurella haemolytica leukotoxin to bovine leukocytes. Infect. Immun. 65:3719–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taichman NS, Simpson DL, Sakurada S, Cranfield M, DiRienzo J, Slots J. 1987. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol. Immunol. 2:97–104. 10.1111/j.1399-302X.1987.tb00270.x [DOI] [PubMed] [Google Scholar]
- 55. Amit U, Dagan R, Porat N, Trefler R, Yagupsky P. 2012. Epidemiology of invasive Kingella kingae infections in 2 distinct pediatric populations cohabiting in one geographic area. Pediatr. Infect. Dis. J. 31:415–417. 10.1097/INF.0b013e318240cf8a [DOI] [PubMed] [Google Scholar]
- 56. Banerjee A, Kaplan JB, Soherwardy A, Nudell Y, Mackenzie GA, Johnson S, Balashova NV. 2013. Characterization of TEM-1 beta-lactamase producing Kingella kingae clinical isolates. Antimicrob. Agents Chemother. 57:4300–4306. 10.1128/AAC.00318-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DiFranco KM, Kaswala RH, Patel C, Kasinathan C, Kachlany SC. 2013. Leukotoxin kills rodent WBC by targeting leukocyte function associated antigen 1. Comp. Med. 63:331–337 [PMC free article] [PubMed] [Google Scholar]
- 58. Dubnov-Raz G, Ephros M, Garty BZ, Schlesinger Y, Maayan-Metzger A, Hasson J, Kassis I, Schwartz-Harari O, Yagupsky P. 2010. Invasive pediatric Kingella kingae Infections: a nationwide collaborative study. Pediatr. Infect. Dis. J. 29:639–643. 10.1097/INF.0b013e3181d57a6c [DOI] [PubMed] [Google Scholar]
- 59. Baraff LJ, Bass JW, Fleisher GR, Klein JO, McCracken GH, Jr, Powell KR, Schriger DL. 1993. Practice guideline for the management of infants and children 0 to 36 months of age with fever without source. Ann. Emerg. Med. 22:1198–1210. 10.1016/S0196-0644(05)80991-6 [DOI] [PubMed] [Google Scholar]
- 60. Levy PY, Fournier PE, Fenollar F, Raoult D. 2013. Systematic PCR detection in culture-negative osteoarticular infections. Am. J. Med. 126:1143.e25–e333. 10.1016/j.amjmed.2013.04.027 [DOI] [PubMed] [Google Scholar]
- 61. Ceroni D, Cherkaoui A, Kaelin A, Schrenzel J. 2010. Kingella kingae spondylodiscitis in young children: toward a new approach for bacteriological investigations? A preliminary report. J. Child. Orthopaed. 4:173–175. 10.1007/s11832-009-0233-2 [DOI] [PMC free article] [PubMed] [Google Scholar]