Abstract
A growing family of virulence factors called serine protease autotransporters of Enterobacteriaceae (SPATEs) are secreted by Shigella, Salmonella, and Escherichia coli pathotypes. SPATEs are subdivided into class 1 and class 2 based on structural features and phylogenetics. Class 1 SPATEs induce cytopathic effects in numerous epithelial cell lines, and several have been shown to cleave the cytoskeletal protein spectrin in vitro. However, to date the in vivo role of class 1 SPATEs in enteric pathogenesis is unknown. Citrobacter rodentium, a natural mouse pathogen, has recently been shown to harbor class 1 and class 2 SPATEs. To better understand the contribution of class 1 SPATEs in enteric infection, we constructed a class 1 SPATE null mutant (Δcrc1) in C. rodentium. Upon infection of C57BL/6 mice, the Δcrc1 mutant exhibited a hypervirulent, hyperinflammatory phenotype compared with its parent, accompanied by greater weight loss and a trend toward increased mortality in young mice; the effect was reversed when the crc1 gene was restored. Using flow cytometry, we observed increased infiltration of T cells, B cells, and neutrophils into the lamina propria of the distal colon in mice fed the Δcrc1 mutant, starting as early as 5 days after infection. No significant difference in epithelial cytotoxicity was observed. Reverse transcription-PCR (RT-PCR) analysis of distal colonic tissue on day 10 postinfection showed significant increases in mRNA encoding cytokines interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), IL-1β, and inducible nitric oxide synthase (iNOS) but not in mRNA encoding IL-17, IL-4, or IL-10 in the Δcrc1 mutant-infected mice. Our data suggest a previously unsuspected role for class 1 SPATEs in enteric infection.
INTRODUCTION
Despite diverse pathogenic strategies, nearly all pathogenic Enterobacteriaceae harbor one or more members of a single family of virulence factors: the serine protease autotransporters of Enterobacteriaceae (SPATEs) (1–3). Importantly, although in vitro phenotypes have been reported for several SPATEs (4–7), none have yet been characterized in whole-animal enteric models. A recent review of the SPATEs published by our group suggested two large phylogenetic clusters, distinguished by structure (3). The class 1 SPATEs are cytopathic toxins that cause cell rounding in vitro, putatively by cleaving the cytoskeletal protein spectrin (8). Class 2 SPATEs, the larger phylogenetic cluster, comprise O-glycoproteases that cleave mucin (9) and other O-glycoproteins on the surface of hematopoietic cells (10); class 2 SPATEs have been associated with roles in colonization and immunomodulation (11). We have previously reported that a Shigella strain deleted for the class 2 SPATE, called Pic, is more inflammatory in the guinea pig keratoconjunctivitis model (10).
A suitable animal model for pathogenic Escherichia coli infection does not yet exist. However, Citrobacter rodentium, a natural mouse pathogen that causes attaching-effacing (A/E) lesions similar to those observed with enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC), is used to model E. coli pathogenesis, given that the microorganism harbors homologs of virulence factors found in several E. coli pathotypes (12–14). C. rodentium causes transmissible murine colonic hyperplasia (TMCH) (15). The infections do not usually result in clinical diarrhea, although the T cell infiltration, cytokine production, and epithelial cell proliferation seen in C. rodentium-infected mice resemble the abnormalities observed in inflammatory bowel disease and infectious enteritis of humans (16, 17).
The most commonly used archetype C. rodentium strain is predicted to have genes encoding three SPATEs: two belonging to class 2 and one belonging to class 1 (3). Here, we address the potential contribution of the class 1 SPATE, called Crc1, to enteric pathogenesis. Our data suggest that interruption of the Crc1 gene results in a phenotype of increased inflammation, suggesting that the protease may play a previously unsuspected immunomodulatory role.
MATERIALS AND METHODS
Construction of a C. rodentium SPATE deletion mutant and a crc1-repaired strain.
Nonpolar deletion mutants in the gene encoding Crc1 were constructed using the λ Red recombinase system (18). PCR fragments containing a kan-sacB insert (10) were generated by PCR using primers described in Table 1 and electroporated into C. rodentium strain ATCC 51459 expressing the λ Red recombinase system encoded by plasmid pKM200 (18). Mutants were selected on LB agar plates containing kanamycin (50 mg/ml). All mutations were confirmed by PCR amplification using primers external to the disrupted gene. The crc1-restored strain was constructed by recombination of the native crc1 gene locus to replace the kanamycin cassette using the λ Red recombinase system. The revertant clones were obtained by screening for kanamycin-sensitive clones.
TABLE 1.
Primers used in this study
| Primer/specificity | Sequence (5′–3′) |
|---|---|
| crc1-kan-sacB/26423-26255; 22157–22328 (pCROD1 plasmid) | AAAATAAAAACTAAATGTAACGATTTATCTTGTGTTAACTGAGTCACAATAAAGCGAGACGTCATTCAATATATTCCCCTCCTTGTGATAACTCCTGGTTATGGTTATCACCAAGGCATTTAGCTGATTTATTTTTCATATTATCTGTGATTTTCATGGAGGGTTTGCTTTTCTTGAAAATTTTTTTTTTGACTCAATAT; AACTCTTATTACCTTAAGCATTTATGGTTGTGCCATTAATAGCAGCATCGTTATCATTTTTCGGTTACTTCGATACCTTTGAAAAATTCTTATTATTCTGATTCAAAGTTTATTCTGACAGCCCTGTTCCCCACAAGAAAAATGAAGCCCCTTTTCAGGGGCCTCTCCCTGTTATTTGTTAACTGTTAATTGTCCTTGTT |
| pVV1/26865–26894; 21763–21794 (pCROD1 plasmid) | TGCTACTCCGCTCGAGCGGGCCGCTGGCAAGGCGAACGTAGTAAATCAC; ACTGCCCAAGCTTGGGTGAAGTTCTACAGAGCGCGAGAAACAGATGGT |
| pVV2/26217–26253; 1540–1579 (pCROD1; pACYC177) | ACTTCGAAATACTAGTTAATACGACTCACTATAGGGAGACCACAACGGTTTCCCTCTAGAAATAATTTTCTTTAACTTTAAGAAGGAGATATACATATGAACAAAATATATTCTCTTAAATATTGCCCTGTTAC; CTGCATGCCGTACATGGTAGTACTAGTTTACTATGTTCCCACTGATGAGGGTGTCAGTGA |
Infection of mice.
The C. rodentium model was used as described previously (19). Briefly, 3- to 4-week-old, C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Five mice per group were inoculated with approximately 1010 CFU of C. rodentium wild type (WT), the Δcrc1 mutant, or the repaired mutant strain (crc1-repaired strain) suspended in 200 μl of phosphate-buffered saline (PBS); organisms were administered by oral gavage using a 20-gauge intubation needle. Control animals received 200 μl of sterile PBS. At specified time points after inoculation, fecal pellets were collected aseptically from each mouse, weighed, and emulsified in PBS. The number of viable bacteria per gram of feces was determined by serial dilutions of the samples on medium containing appropriate selective antibiotics.
Animal studies were carried out in strict compliance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Virginia Animal Care and Use Committee (protocol number 3894). All efforts were made to keep pain and suffering to a minimum.
Analysis of leukocyte populations in the distal colon.
On days 1, 3, 5, and 10 after inoculation, 3 mice per group were euthanized, and 4 cm of the rectum and colon (extending to the anal verge) was excised and placed on a petri dish containing Hanks balanced salt solution (HBSS) buffer. Mesentery, fat, and feces were removed from the distal colon. The colon was opened lengthwise and cut into 1-cm segments. The tissue sections from 3 mice were pooled in 20 ml of wash buffer (HBSS, 5% fetal bovine serum [FBS], and 120 μl EDTA). The samples were incubated at 37°C with agitation (250 rpm) for 15 min. The tissues were then transferred to 10 ml of C buffer (10 ml HBSS, 5% FBS, and 10 mg collagenase IV) (Sigma C5138; Carlsbad, CA). The intestines were incubated with C buffer at 37°C for 20 min, and then the contents were passed through a 100-μm cell strainer; retentates were gently mashed and washed through the filter using HBSS. The filtrate containing the cells was centrifuged, and the cells were enumerated. The cells were adjusted to 1 million per group using fluorescence-activated cell sorting (FACS) buffer (HBSS plus 0.2% bovine serum albumin [BSA]) and stained for flow cytometry analysis.
Cell suspensions of colonic tissues were first blocked for 15 min with anti-Fc receptor CD16/CD32 blocking antibody (clone 2.4D2) (BD Pharmingen, San Diego, CA) at 4°C, followed by incubation with antibodies to Gr-1 (clone RB6-8C5) (BD Pharmingen), CD3 (clone 17A2) (BD Pharmingen), CD45 (clone 30-F11) (Life Technologies, Carlsbad, CA), and CD19 (clone 1D3) (BD Pharmingen). Aqua stain (Life Technologies) was included to identify viable cells. The analysis was performed on a Beckman Coulter Cyan ADP LX flow cytometer (Brea, CA). Data were analyzed with FlowJo version 4.5 software (TreeStar, Ashland, OR).
Histopathological score.
Distal colonic sections were fixed in Bouin's fixative and embedded in paraffin. Five-micrometer sections were deparaffinized and stained with hematoxylin and eosin. Slides were read by a veterinary pathologist in a blind manner. The criteria for histopathology scoring were neutrophil infiltration, submucosal edema, hyperplasia, and the presence of bacteria (20). The numerical score was as follows: 0, no lesion; 1, minimal; 2, mild; 3, moderate; 4, marked; 5, severe.
Fluorescence assay of HEp-2 cells.
Fluorescent actin staining was performed as previously described (6). Briefly, HEp-2 cells (30,000 cells/well) were treated with recombinant Pet, EspC, Crc1, or Crc1S251A, diluted directly into tissue culture medium without antibiotics or serum at indicated concentrations. The cells were incubated at 37°C for 6 h, washed twice with PBS, and fixed with 2% formalin in PBS for 20 min at room temperature. The fixed cells were permeabilized by adding 0.1% Triton X-100 in PBS for 4 min at room temperature. Actin filaments in the cells were stained by incubation with rhodamine-phalloidin (Life Technologies) for 30 min. Cells were air dried and mounted in Prolong Gold reagent (Life Technologies). Cells were viewed using an Olympus BX51fluorescent microscope.
Cloning strategy for Crc1.
All restriction and ligation enzymes were purchased from New England BioLabs (Ipswich, MA). All genetic manipulations were performed in E. coli K-12 strain DH5α. The crc1 gene was amplified from genomic DNA of C. rodentium using primers described in Table 1 and cloned in pACYC177 vector as an XhoI/HindIII fragment to generate plasmid pVV1. pVV1 was sequenced and verified for the absence of any inadvertent mutations in the crc1 gene. The T7 promoter and the Shine-Dalgarno sequence of the T7 phage were created upstream of crc1 by reverse PCR (pVV2) (Table 1). Vector pVV2 was transformed into T7 Express lysY/Iq competent E. coli (New England BioLabs). Site-directed mutagenesis was performed to modify the predicted catalytic serine (S251A), using the QuikChange site-directed mutagenesis kit from Stratagene (Santa Clara, CA) according to the manufacturer's protocols. The strains were maintained on L agar or L broth containing 100 μg of ampicillin/ml.
Purification of Crc1.
The minimal clone of Crc1 was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to express and secrete Crc1. Total protein in the supernatant was precipitated using 50% ammonium sulfate. The precipitate was centrifuged and resuspended in 50 mM Tris-HCl, pH 8.0. The precipitate was dialyzed three times in a 100× volume of 50 mM Tris-HCl, pH 8.0, using a 50-kDa-pore-size dialysis membrane. After overnight dialysis, the protein precipitate was loaded onto a QA Fast Flow Sepharose (Sigma, St. Louis, MO) anion exchange column; bound protein was eluted using various concentrations of sodium chloride, spanning from 100 mM to 500 mM. The fraction of eluent containing maximum protein was identified by SDS-PAGE, and this fraction was dialyzed with PBS. This purified fraction was quantified and used for in vitro cell culture infections.
Statistical analysis.
Experimental data were expressed as mean ± standard error of the mean (SEM) in each group. The means of groups were combined and analyzed by a two-tailed Student t test for pairwise comparisons. Analysis of variance (ANOVA) with post hoc correction was used to compare means of multiple populations. A P value of ≤0.05 was considered statistically significant.
Real-time PCR.
Distal colon tissue weighing approximately 10 mg (0.5 cm) was collected from the anal verge and homogenized using 1-mm zirconia beads in a Bead-Beater (Fisher Scientific). RNA was extracted using the RNeasy kit (Qiagen). DNA contaminants were removed by DNase digestion (Invitrogen). Approximately 1 μg of RNA was used for cDNA synthesis (iSCRIPT; Bio-Rad). Reverse transcription-PCR (RT-PCR) was performed in a 7500 Real-Time PCR system from Applied Biosystems. Fold induction was calculated using the threshold cycle (ΔΔCT) method. The sense and antisense primer sequences are as follows: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-ATCAACGACCCCTTCATTGACC-3′ and 5′-CCAGTAGACTCCACGACATACTCAGC-3′; tumor necrosis factor alpha (TNF-α), 5′-GACCCTCACACTCAGATCATCTTCT-3′ and 5′-CCACTTGGTGGTTTGCTACGA-3′; gamma interferon (IFN-γ), 5′-CTGCCACGGCACAGTCATTG-3′ and 5′-TGCATCCTTTTTCGCCTTGC-3′; KC, 5′-TGTCAGTGCCTGCAGACCAT-3′ and 5′-GCTATGACTTCGGTTTGGGTG-3′; interleukin-17A (IL-17A), 5′-GCTCCAGAAGGCCCTCAGA-3′ and 5′-CTTTCCCTCCGCATTGACA-3′; IL-22, 5′-TCCGAGGAGTCAGTGCTAAA and 5′-AGAACGTCTTCCAGGGTGAA-3′; IL-6, 5′-TCCAATGCTCTCCTAACAGATAAG-3′ and 5′-CAAGATGAATTGGATGGTCTTG-3′; inducible nitric oxide synthase (iNOS), 5′-GTCTTTGACGCTCGGAACTGT-3′ and 5′-GATGGCCGACCTGATGTTG-3′; IL-1β, 5′-ACAGAATATCAACCAACAAGTGATATTCTC-3′ and 5′-GATTCTTTCCTTTGAGGCCCA-3′; IL-4, 5′-GCATTTTGAACGAGGTCACAGG-3′ and 5′-TATGCGAAGCACCTTGGAAGC-3′; IL-10, 5′-ATTTGAATTCCCTGGGTGAGAAG-3′ and 5′-CACAGGGGAGAAATCGATGACA-3′.
RESULTS
The C. rodentium genome encodes multiple functional SPATE proteases.
Annotation of the genome of C. rodentium strain ATCC 51459 revealed the presence of three full-length predicted SPATE proteases. We here designate Crc1 (C. rodentium class 1; accession number YP_003368469) and Crc2 (C. rodentium class 2; accession number YP_003368482) (3). The third SPATE was not recognized as a member of this family but was previously designated AdcA (CBG90828) (19). Each featured a full beta translocation domain, a predicted beta-helix backbone occupying the C terminus of the secreted domain, and a globular passenger N-terminal domain including the catalytic motif (GDSGSP) (2). We analyzed this strain for the ability to express three SPATEs under laboratory conditions. SDS-PAGE analysis revealed the presence of Crc1 and Crc2 predicted proteases migrating at the molecular mass expected for full-length expression (Fig. 1B). The inability to detect AdcA in vitro may be due to the fact that this SPATE was reported to be regulated by the activator RegA (19).
FIG 1.
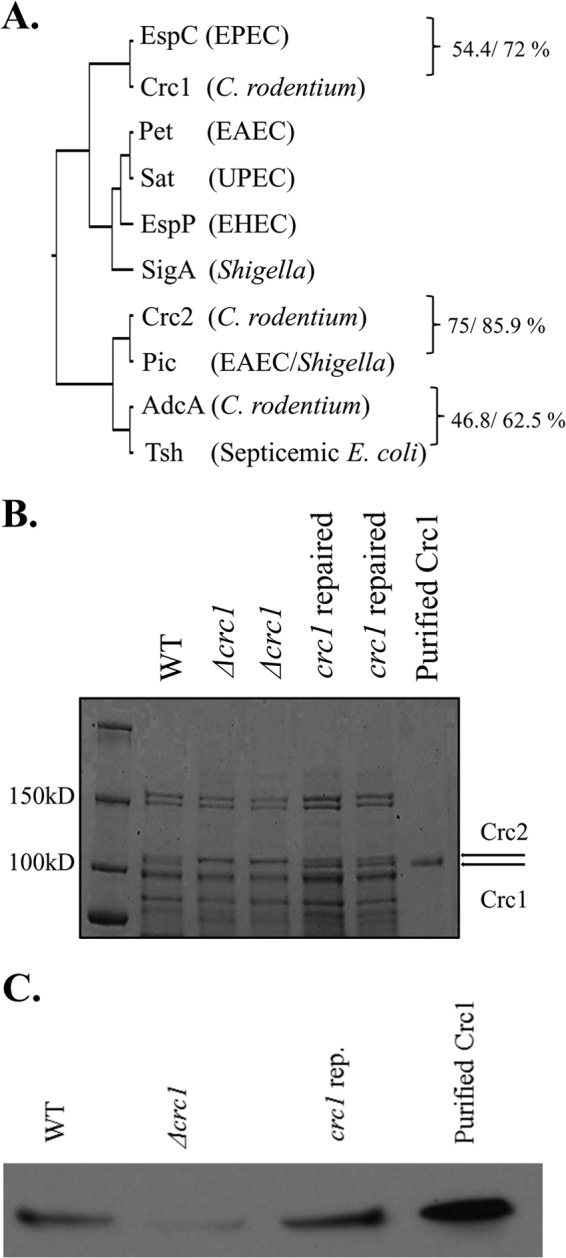
(A) Phylogenetic analysis of C. rodentium SPATEs. Alignment of sequences was built with ClustalW, and the phylogenetic tree was constructed using UPGMA (unweighted pair group method using arithmetic mean). C. rodentium SPATEs and their closest homologs were aligned using the BLOSUM50 alignment matrix, and identity/similarity was calculated using the Matrix Global Alignment Tool. (B) Secretion of Crc1 and Crc2 into the culture supernatant by C. rodentium ICC168. Supernatants from C. rodentium WT, Δcrc1, and crc1-repaired strain bacterial cultures were collected. The supernatant was concentrated 1,000-fold using Amicon filters with a 70-kDa cutoff size. The supernatants were analyzed by SDS-PAGE. Crc2 (predicted molecular mass, 109.3 kDa) and Crc1 (predicted molecular mass, 104.9 kDa) appeared above the 100-kDa band of the molecular mass ladder (shown by arrows). Crc1 purification is illustrated in Fig. S1 in the supplemental material. (C) Confirmation of Crc1 deletion. Supernatants from C. rodentium WT, Δcrc1, and crc1-repaired strain bacterial cultures were collected. The supernatant was concentrated 1,000-fold using Amicon filters with a 70-kDa cutoff size. The supernatants and the purified Crc1 toxin were analyzed by immunoblotting using anti-Crc1 rabbit antibody.
To ascertain the phylogenetic relationships of C. rodentium SPATEs, we performed multiple sequence alignments of the passenger domain sequences of the three C. rodentium SPATEs and represent them along with their closest homologs from human pathogens (Fig. 1). The predicted class 1 SPATE in the ATCC 51459 genome, Crc1, is most closely related to EspC from enteropathogenic E. coli (EPEC). By using the Matrix Global Alignment Tool (MATGAT) (21), the passenger domains of EspC and Crc1 exhibited 54.4% amino acid identity and 72% similarity with minimal gapping. Class 2 SPATE Crc2 was shown to be most closely related to Pic; the previously described class 2 SPATE called AdcA (19) was most closely related to the Tsh protease (22) of avian-pathogenic and extraintestinal E. coli (Fig. 1).
Cytotoxic effects of the C. rodentium class 1 SPATE.
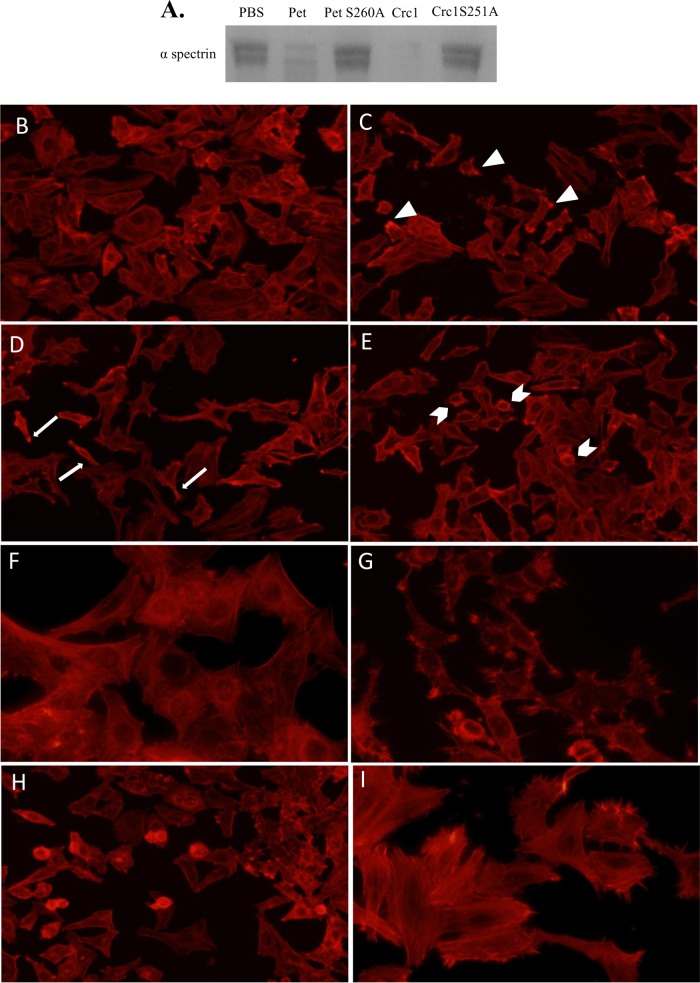
Most class 1 SPATEs characterized to date are cytotoxins in vitro and cleave the cytoskeletal protein spectrin. We showed that 1.2 μg of Crc1 (see Fig. S1 in the supplemental material) cleaved 3 μg of spectrin after overnight incubation, with catalytic activity similar to that observed for the class 1 SPATEs EspC and Pet from EPEC and enteroaggregative E. coli (EAEC) strains, respectively (8). We performed site-directed mutagenesis to abolish the catalytic serine residue of Crc1, predicted by homology with the catalytic serines of EspC and Pet. As expected, the Crc1 mutant S251A was unable to cleave spectrin (Fig. 2A). We did not study the site of cleavage of spectrin by Crc1 as this has been already extensively characterized (23).
FIG 2.
(A) Crc1 cleaves spectrin. Three micrograms of spectrin from erythrocytes was treated with 1 μg of purified Crc1, Crc1S251A mutant, Pet, or Pet S260A mutant overnight at 37°C. The proteins were analyzed by SDS-PAGE. (B to I) Effect of Crc1 on HEp-2 cells. HEp-2 cells were treated with increasing concentrations of purified CrC1—60 μg/ml (B), 120 μg/ml (C), 180 μg/ml (D), and 240 μg/ml (E)—for 6 h, and actin was stained with rhodamine-phalloidin. (C) Formation of punctate actin structures (arrowheads). (D) Spindle formation due to loss of actin stress fibers (arrows). (E) Cell rounding at highest concentration (arrow pins). (F) Cytotoxicity was not observed in the Crc1S251A mutant. (G and H) Pet at 37 μg/ml (G) and EspC at 120 μg/ml (H) were used as positive controls. (I) Untreated HEp-2 cells. Images represent typical fields selected from reproducible experiments.
We also assessed the effect of Crc1 on HEp-2 cells, which are susceptible to the Pet protease. Fifty micrograms per ml of purified Crc1 was incubated with semiconfluent HEp-2 monolayers for 6 h at 37°C. Cellular actin was stained using rhodamine-phalloidin, and cells were visualized under epifluorescence microscopy. Pet (37 μg/ml) was used as a positive control. While extensive cell rounding and loss of actin stress fibers were seen after Pet intoxication (Fig. 2G), few or no signs of cytotoxicity were seen in Crc1-treated cells. Previous studies have shown that the minimum dose of EspC required to cause cytotoxic effects was 120 μg/ml for 6 h (Fig. 2H) (23). To ascertain if a similar effect could be observed upon intoxication with Crc1, we used increasing concentrations of Crc1 from 60 μg/ml through 240 μg/ml, applied for 6 h (Fig. 2B to E). We observed a dose-dependent increase in cytotoxicity, although Crc1 was overall not as potent as Pet or EspC in this system. The cytotoxicity was completely abolished in the Crc1S251A mutant (Fig. 2F).
Role of Crc1 in C. rodentium pathogenesis.
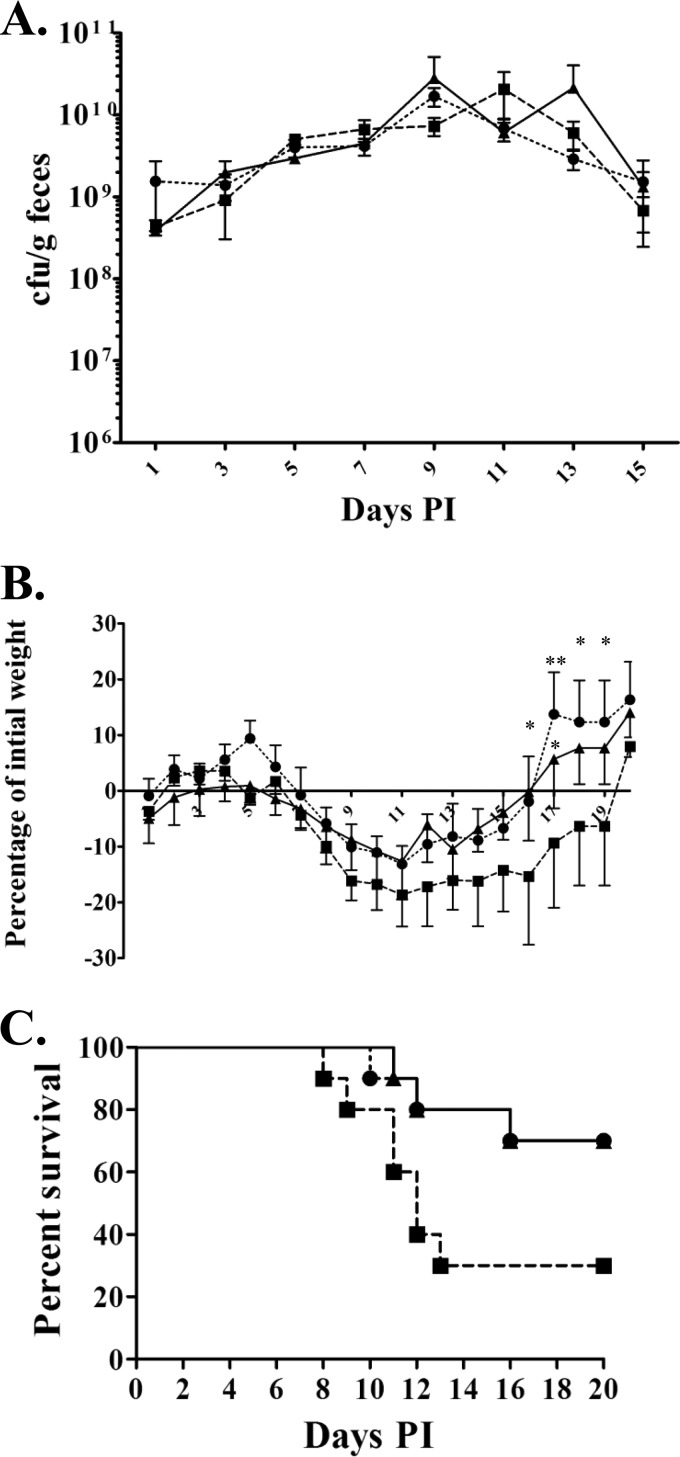
In order to determine the role of Crc1 in C. rodentium pathogenesis, we deleted the crc1 gene in the ATCC 51459 strain. We rescued the crc1 null mutant by repairing the crc1 gene with the native allele and assessed the Crc1 expression by SDS-PAGE and immunoblotting (Fig. 1C). C. rodentium wild-type (WT), Δcrc1, and crc1-repaired strains were fed individually to 4-week-old C57BL/6 mice. We observed no significant difference in fecal shedding among the different strains over the course of infection. All infected mice survived through 14 days with peak shedding at days 7 through 10. After day 14 postinoculation, all mice began to clear the infection with similar kinetics (Fig. 3A). As shown in Fig. 3B, mice began to lose weight on day 7, peaking at day 11, after which they begin to recover their weight. While all the infected groups showed loss of weight during infection, we noticed that the Δcrc1 mutant-infected group lost a significantly greater percentage of initial weight. The Δcrc1 mutant-infected mice showed delayed recovery of weight compared to the group infected with the wild type or the crc1-repaired strain (P < 0.05 on days 16, 17, and 18).
FIG 3.
Role of Crc1 in C. rodentium pathogenesis. Mice received (via oral gavage) 1010 CFU of C. rodentium wild-type strain (●), Δcrc1 deletion mutant (■), or crc1-repaired strain (▲). (A) Fecal excretion of C57BL/6 mice by derivatives of C. rodentium. The data are means and standard errors of the CFU/gram of feces from at least five individual mice at selected time points after inoculation. (B) Mean percent change in body weight of C57BL/6 mice infected with C. rodentium and its derivatives. The data shown are mean and standard error of the mean percent change in initial body weight of at least 5 mice per group. (*, P < 0.05; **, P < 0.01, using two-way ANOVA with multiple comparisons). (C) Ten mice per group were monitored for survival during the course of infection. P value = 0.0715 using log-rank test. PI, postinfection.
In an effort to observe more subtle differences in virulence, we performed similar challenge experiments in younger C57BL/6 mice (3 weeks old). We found that these mice often died when inoculated with WT C. rodentium (30% mortality); however, the Δcrc1 mutant-infected mice showed increased mortality (70%), manifested earlier in the infection, than did mice infected with the WT or the crc1-repaired strain (Fig. 3C). We observed a trend toward more rapid death in the Δcrc1 mutant-infected group, but the difference in the survival curves was not statistically significant (P value = 0.07 using log-rank test).
Differential colitis severity in distal colons of mice infected with C. rodentium derivatives.
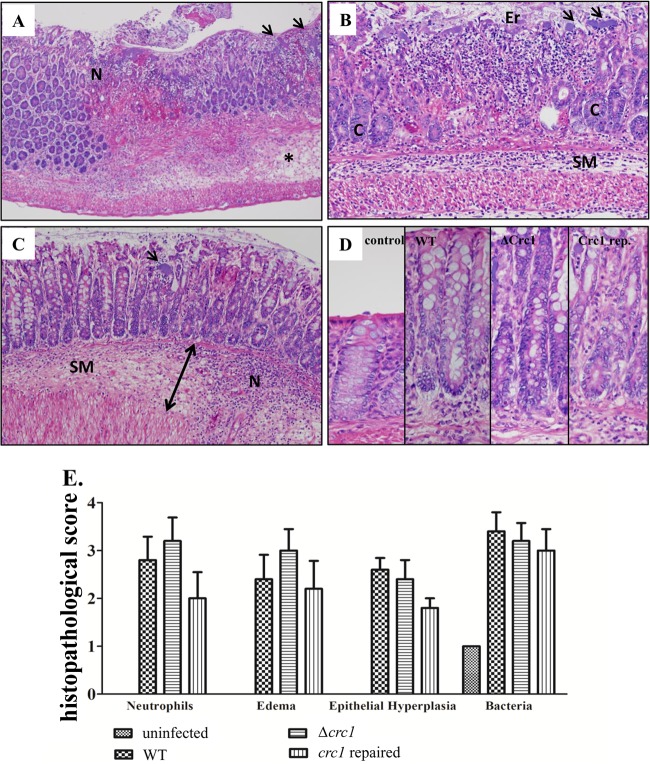
All mice that received a strain of C. rodentium developed acute colitis, which varied in severity from minimal to marked. The colitis was characterized by edema, neutrophilic infiltrate, epithelial hyperplasia, and large numbers of adherent bacteria (Fig. 4A to D). Epithelial erosions with hemorrhage and fibrin were present in more severe cases. The severity of colitis was highest in the group fed the Δcrc1 mutant, in which three of five mice developed colitis with marked edema and moderate to marked neutrophil infiltrate (Fig. 4E). In the WT group, two of five mice developed marked edema, but the neutrophil infiltrate was less abundant. The group that received the crc1-repaired strain had only one animal with marked edema and neutrophil infiltrate. Levels of epithelial hyperplasia were similar among the three infected groups (Fig. 4D). Notably, despite cytotoxicity to epithelial cells exhibited by purified Crc1 and other class 1 SPATEs in vitro, we observed no difference in epithelial cytotoxicity between WT ATCC 51459 and its crc1 mutant. Despite this difference in polymorphonuclear leukocyte (PMN) infiltration and edema by microscopic analysis, the pathological scores between the different infected groups were not statistically significant. Hence, we resorted to a quantitative analysis of neutrophil infiltration by flow cytometry.
FIG 4.
Histopathology on day 8 postinfection. (A) Colonic sections of mice infected with WT C. rodentium, demonstrating large colonies of bacteria (arrows), many neutrophils (N), and the submucosa expanded by edema (asterisk). (B) Colonic sections of mice infected with C. rodentium crc1 deletion mutant demonstrating an area of erosion (Er) in which crypt epithelium (C) has been replaced by neutrophils and cellular debris. The neutrophilic infiltrate extends from the mucosa into the submucosa (SM) and the smooth muscle layers. Large colonies of bacteria are present on the surface of the mucosa (arrows). (C) A mouse infected with the crc1-repaired strain demonstrated neutrophilic colitis (N), colonies of bacteria (arrow), and submucosal (SM) edema (double arrow). (D) Epithelial hyperplasia; the mucosa in infected groups was nearly twice the thickness of the saline control. Hematoxylin and eosin staining; magnification, ×20 (A to C) and ×400 (D). (E) Pathological scoring indicated more neutrophil infiltration and edema in the crc1 mutant-infected group than in other groups. Levels of epithelial hyperplasia were comparable between infected groups.
Deletion of Crc1 causes increased leukocyte infiltration into the submucosa.
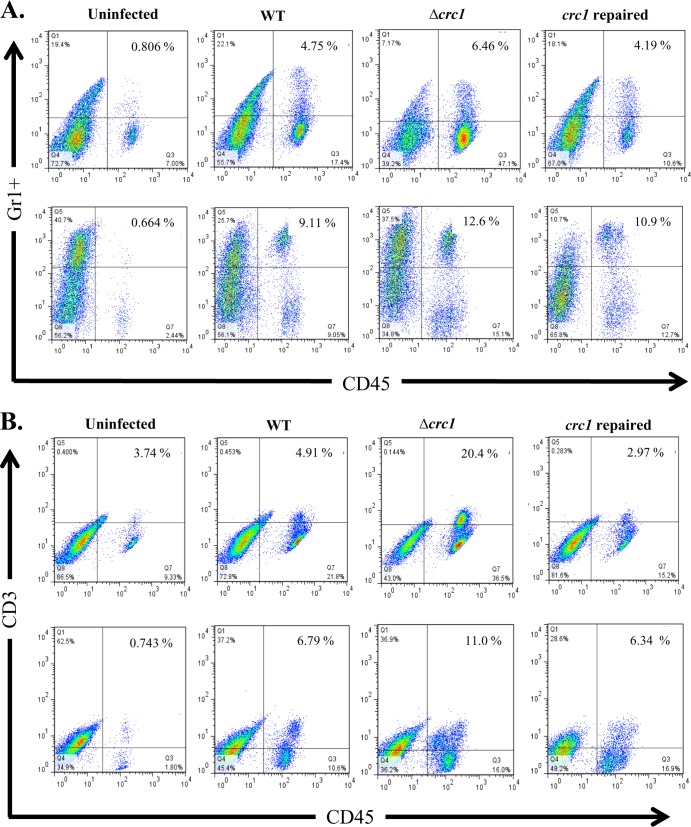
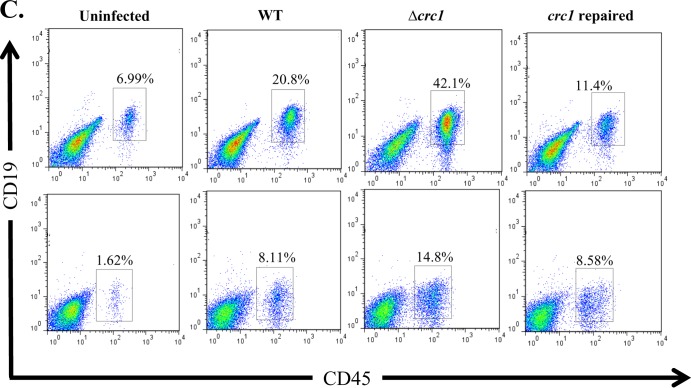
WT C. rodentium pathology features massive infiltration of PMNs, activated lymphocytes, and macrophages (24–27). We sought to study the populations of these immune cell types infiltrating the colon during the course of infection with the wild-type strain or the crc1 mutant. On days 1, 3, 5, and 10 postinfection, we euthanized 3 mice per group and analyzed the tissue extract from distal colon sections by flow cytometry. We observed that on days 1 and 3 there were few T cells, neutrophils, or B cells infiltrating the distal colon (data not shown) and no differences among the three challenge strains. However, on days 5 and 10 we observed a significant increase in the numbers of these cells present in infected submucosa (Fig. 5), which correlated with the extent of bacterial colonization (Fig. 1). Using flow cytometry, we observed a higher percentage of neutrophils, T cells, and B cells in the tissues of the crc1 mutant-infected group on days 5 and 10. Infiltrating T cells were found to be predominantly of the CD4+ T-helper cell subset (data not shown). No difference was observed for macrophage populations.
FIG 5.
C. rodentium Δcrc1 mutant infection is associated with elevated influx of CD45+/Gr-1+ neutrophils (A), CD45+/CD3+ T cells (B), and CD45+/CD19+ B cells (C) in the colons of C57BL/6 mice. Mice were infected orally with C. rodentium WT, Δcrc1 mutant, and crc1-repaired strains and sacrificed on days 5 (top panels) and 10 (bottom panels) postinfection. Single-cell suspensions from the colons of infected and uninfected mice were subjected to flow cytometry staining using CD45 as the marker to differentiate leukocytes from epithelial cells and cell-specific markers to look for infiltration of neutrophils, B cells, and T cells. FACS plots were gated on live cells with Aqua-positive dead cells excluded from analysis. Numbers indicate the percentages of cells in each quadrant. (Statistical analysis showed a P value of 0.03 for PMNs on day 10 and a P value of 0.01 for T cells and 0.0065 for B cells on day 10 using repeated-measure, one-way ANOVA and Greenhouse-Geisser correction). FACS plots are representative of three independent experiments on day 10 with a total of nine mice from each group.
Deletion of Crc1 causes increased transcript levels for proinflammatory cytokines in distal colonic tissue on day 10.
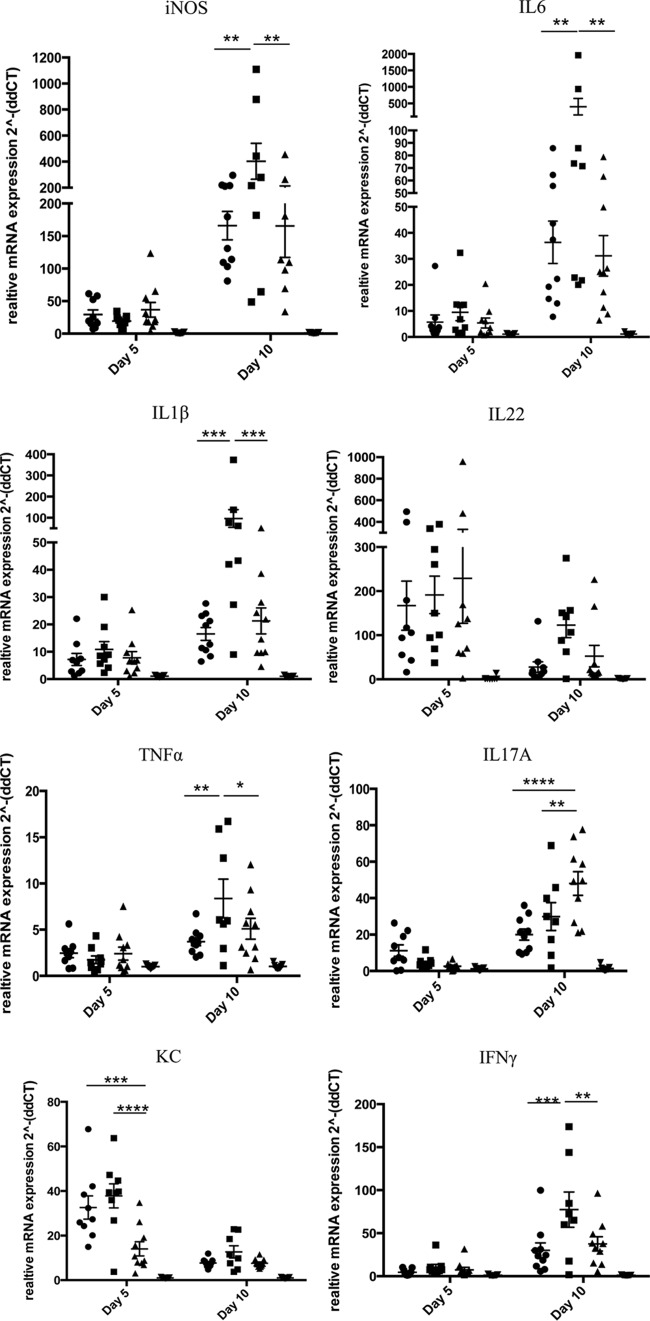
To characterize tissue cytokine expression, we performed real-time RT-PCR on homogenized tissue from the distal colon on days 5 and 10; transcripts encoding proinflammatory cytokines associated with C. rodentium colitis, such as IFN-γ, TNF-α, IL-1β, iNOS, IL-6, IL-17, KC, RANTES, and IL-22, as well as anti-inflammatory cytokines IL-4 and IL-10, were measured (Fig. 6). We observed that all cytokines except KC and IL-22 were upregulated severalfold on day 10 compared with day 5. On day 5, KC and IL-22 were significantly upregulated in all the infected groups in comparison to the uninfected groups but did not exhibit any statistically significant differences among the groups infected with the wild-type, Δcrc1 mutant, or crc1-repaired strain. In the day 10 group, 2 mice in the Δcrc1 mutant-infected group died on day 9. Upon cytokine analysis on day 10, we observed severalfold induction of cytokines IL-6, iNOS, TNF-α, IL-1β, IFN-γ, and IL-17. We observed significant increases in levels of IL-6, iNOS, TNF-α, IL-1β, and IFN-γ in the Δcrc1 mutant-infected mice compared with the other two infected groups. RANTES, IL-4, and IL-10 displayed less than 10-fold induction (data not shown).
FIG 6.
RT-PCR analysis of cytokine expression from distal colonic tissue homogenate of infected mice. Groups of 10 animals were sacrificed at indicated time points and analyzed independently. Mice were infected with the C. rodentium wild-type (●), Δcrc1 mutant (■), or crc1-repaired (▲) strain or not infected (▼). Data are means ± standard errors of the means of all 10 mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-way ANOVA with multiple comparisons).
DISCUSSION
SPATEs are encoded in the genomes of a wide variety of enteric and nonenteric pathogens of the family Enterobacteriaceae (3). Despite their broad prevalence, however, little is known with regard to their roles in pathogenesis. For the first time, the presence of SPATEs in C. rodentium has allowed us to address the effect of a class 1 SPATE using a whole-animal model.
Most class 1 SPATEs cleave the cytoskeletal protein spectrin (fodrin) in vitro, leading to cell rounding and cytotoxicity (8). Intoxicated cells display loss of stress fibers, disruption of the actin cytoskeleton, cell rounding, cell detachment, and opening of cell tight junctions in epithelial cells (23, 28–30). However, most of these studies were performed in an in vitro cell culture system employing HEp-2 cells, Vero cells, or HeLa cell lines. Whereas Pet elicits cytotoxicity at a concentration of 37 μg/ml within an hour of treatment on HEp-2 and HT-29 colorectal cancer cells (6), other class 1 SPATEs require longer incubation times and higher concentrations to cause cytoskeletal changes. For example, Sat is cytotoxic to human bladder and kidney epithelial cells at a concentration of 100 μg/ml (5), EspP is cytotoxic to HeLa cells at 200 μg/ml (38), and EspC causes cytotoxicity on HEp-2 cells at 120 μg/ml (23).
Like other class 1 SPATEs, Crc1 cleaves spectrin and exhibits cytotoxicity to HEp-2 cells. However, we observed morphological alterations of intoxicated HEp-2 cells only upon incubation with very high concentrations of Crc1. Given the low efficiency of spectrin cleavage, we doubt that cleavage of spectrin in epithelial cells is pathogenically relevant. It is possible that Crc1 requires injection by a type III secretion system (T3SS) in order to access its target, as suggested previously for EspC (31), and that important in vivo cytopathic effects may not be observable using light microscopic histopathology. It is also possible that the physiologic activity of Crc1 occurs via cleavage of spectrin in leukocytes, and this possibility is under investigation.
EspP encoded by EHEC has been shown to contribute to bacterial colonization in calves (32). However, no effect on colonization has been reported for other class 1 SPATEs. We did not observe an effect of Crc1 on C. rodentium colonization.
A growing number of virulence factors effect a sometimes subtle modulation of the innate immune response, leading to decreased inflammation (33, 34). The C. rodentium model of inflammatory colitis is a fitting model to study such a phenotype. We first showed that deletion of crc1 causes exacerbation of disease, as mice showed an increase in weight loss when infected with this strain compared to infection with the wild-type or crc1-repaired strain. In younger mice, we observed a trend toward increased mortality, although this was not statistically significant. In our studies, the increased virulence of the Δcrc1 mutant was directly correlated with an increase in leukocyte infiltration, but we are unable to infer that the two effects are related causally.
We have shown for the first time a putative role for class 1 SPATEs in immunomodulation. Flow cytometry revealed higher numbers of T cells, B cells, PMNs, and macrophages in mice fed the Δcrc1 mutant than in those fed the wild type or the reconstructed mutant. Increased leukocyte infiltration was associated with increased levels of proinflammatory cytokines during day 10 but not on day 5 in the Δcrc1 mutant-infected group. On day 5, the only cytokines that were significantly upregulated in the infected groups versus uninfected mice were IL-22 and KC. KC is a neutrophil chemoattractant produced by epithelial cells in response to infection during early stages of C. rodentium infection (35). IL-22 expression has been shown to be upregulated during early stages of C. rodentium infection by another study (36). Interestingly, the main source of IL-22 in C. rodentium infection was previously shown to be innate lymphoid cells (ILCs) and dendritic cells (36, 37). However, there was no differential fold induction in IL-22 or KC between the groups fed the C. rodentium WT, Δcrc1 mutant, or crc1-repaired strain. The differential fold induction of cytokines on day 10 with significantly more expression of IFN-γ, TNF-α, IL-1β, iNOS, and IL-6 in the Δcrc1 mutant-infected group correlated with the increased leukocyte infiltration. However, the source of these cytokines remains undetermined.
Notably, the class 2 SPATE Pic was reported to suppress inflammatory responses in the guinea pig keratoconjunctivitis model and to act via cleavage of leukocyte surface signaling glycoproteins (10). Interestingly, we have not observed cleavage of O-glycoproteins by class 1 SPATEs, but they could produce similar phenotypes via cleavage of other protein targets. Of note, the SPATE proteolytic pocket structurally resembles those seen in the chymotrypsin family of serine proteases, among which are several leukocyte immunomodulators (38–40).
From the perspective of natural selection, the immunosuppressive effect of Crc1 could be adaptive to the enteric pathogen by improving survival and thereby providing increased opportunities for disease transmission. This potential contribution is under investigation in our laboratories. It will also be necessary to ascertain whether the phenotype associated with Crc1 pertains to the SPATEs of human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by supported in part by NIH grants AI-033096 and U19AI090873 to J.P.N.
Footnotes
Published ahead of print 7 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01518-13.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 2.Dautin N. 2010. Serine protease autotransporters of Enterobacteriaceae (SPATEs): biogenesis and function. Toxins (Basel) 2:1179–1206. 10.3390/toxins2061179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Perez F, Nataro JP. 2014. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell. Mol. Life Sci. 71:745–770. 10.1007/s00018-013-1355-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson IR, Hicks S, Navarro-Garcia F, Elias WP, Philips AD, Nataro JP. 1999. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect. Immun. 67:5338–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maroncle NM, Sivick KE, Brady R, Stokes FE, Mobley HL. 2006. Protease activity, secretion, cell entry, cytotoxicity, and cellular targets of secreted autotransporter toxin of uropathogenic Escherichia coli. Infect. Immun. 74:6124–6134. 10.1128/IAI.01086-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro-Garcia F, Sears C, Eslava C, Cravioto A, Nataro JP. 1999. Cytoskeletal effects induced by pet, the serine protease enterotoxin of enteroaggregative Escherichia coli. Infect. Immun. 67:2184–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellies JL, Navarro-Garcia F, Okeke I, Frederickson J, Nataro JP, Kaper JB. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315–324. 10.1128/IAI.69.1.315-324.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta PR, Cappello R, Navarro-Garcia F, Nataro JP. 2002. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect. Immun. 70:7105–7113. 10.1128/IAI.70.12.7105-7113.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro-Garcia F, Gutierrez-Jimenez J, Garcia-Tovar C, Castro LA, Salazar-Gonzalez H, Cordova V. 2010. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect. Immun. 78:4101–4109. 10.1128/IAI.00523-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, Murphy E, Cross A, Sztein MB, Nataro JP. 2011. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 108:12881–12886. 10.1073/pnas.1101006108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. 2009. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect. Immun. 77:2465–2473. 10.1128/IAI.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty NK, Feltwell T, Pickard D, Clare S, Toribio AL, Fookes M, Roberts K, Monson R, Nair S, Kingsley RA, Bulgin R, Wiles S, Goulding D, Keane T, Corton C, Lennard N, Harris D, Willey D, Rance R, Yu L, Choudhary JS, Churcher C, Quail MA, Parkhill J, Frankel G, Dougan G, Salmond GP, Thomson NR. 2011. Citrobacter rodentium is an unstable pathogen showing evidence of significant genomic flux. PLoS Pathog. 7:e1002018. 10.1371/journal.ppat.1002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng W, Li Y, Vallance BA, Finlay BB. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323–6335. 10.1128/IAI.69.10.6323-6335.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192:525–538. 10.1128/JB.01144-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luperchio SA, Schauer DB. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:333–340. 10.1016/S1286-4579(01)01387-9 [DOI] [PubMed] [Google Scholar]
- 16.MacDonald TT, Frankel G, Dougan G, Goncalves NS, Simmons C. 2003. Host defences to Citrobacter rodentium. Int. J. Med. Microbiol. 293:87–93. 10.1078/1438-4221-00247 [DOI] [PubMed] [Google Scholar]
- 17.Bhinder G, Sham HP, Chan JM, Morampudi V, Jacobson K, Vallance BA. 2013. The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. J. Vis. Exp.. (72):e50222. 10.3791/50222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy KC, Campellone KG. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. 10.1186/1471-2199-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. 2008. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 76:5247–5256. 10.1128/IAI.00770-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia TA, Ventura CL, Smith MA, Merrell DS, O'Brien AD. 2013. Cytotoxic necrotizing factor 1 and hemolysin from uropathogenic Escherichia coli elicit different host responses in the murine bladder. Infect. Immun. 81:99–109. 10.1128/IAI.00605-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. 10.1186/1471-2105-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stathopoulos C, Provence DL, Curtiss R., III 1999. Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect. Immun. 67:772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro-Garcia F, Canizalez-Roman A, Sui BQ, Nataro JP, Azamar Y. 2004. The serine protease motif of EspC from enteropathogenic Escherichia coli produces epithelial damage by a mechanism different from that of Pet toxin from enteroaggregative E. coli. Infect. Immun. 72:3609–3621. 10.1128/IAI.72.6.3609-3621.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, Jonas AM. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889–894 [PubMed] [Google Scholar]
- 25.Barthold SW, Coleman GL, Jacoby RO, Livestone EM, Jonas AM. 1978. Transmissible murine colonic hyperplasia. Vet. Pathol. 15:223–236. 10.1177/030098587801500209 [DOI] [PubMed] [Google Scholar]
- 26.Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. 1999. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 67:3031–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schauer DB, Falkow S. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djafari S, Ebel F, Deibel C, Kramer S, Hudel M, Chakraborty T. 1997. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol. Microbiol. 25:771–784. 10.1046/j.1365-2958.1997.5141874.x [DOI] [PubMed] [Google Scholar]
- 29.Guignot J, Chaplais C, Coconnier-Polter MH, Servin AL. 2007. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell. Microbiol. 9:204–221. 10.1111/j.1462-5822.2006.00782.x [DOI] [PubMed] [Google Scholar]
- 30.Navarro-Garcia F, Canizalez-Roman A, Luna J, Sears C, Nataro JP. 2001. Plasmid-encoded toxin of enteroaggregative Escherichia coli is internalized by epithelial cells. Infect. Immun. 69:1053–1060. 10.1128/IAI.69.2.1053-1060.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidal JE, Navarro-Garcia F. 2008. EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell. Microbiol. 10:1975–1986. 10.1111/j.1462-5822.2008.01181.x [DOI] [PubMed] [Google Scholar]
- 32.Dziva F, Mahajan A, Cameron P, Currie C, McKendrick IJ, Wallis TS, Smith DG, Stevens MP. 2007. EspP, a type V-secreted serine protease of enterohaemorrhagic Escherichia coli O157:H7, influences intestinal colonization of calves and adherence to bovine primary intestinal epithelial cells. FEMS Microbiol. Lett. 271:258–264. 10.1111/j.1574-6968.2007.00724.x [DOI] [PubMed] [Google Scholar]
- 33.Chavakis T, Hussain M, Kanse SM, Peters G, Bretzel RG, Flock JI, Herrmann M, Preissner KT. 2002. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 8:687–693. 10.1038/nm728 [DOI] [PubMed] [Google Scholar]
- 34.McGuirk P, Mills KH. 2000. Direct anti-inflammatory effect of a bacterial virulence factor: IL-10-dependent suppression of IL-12 production by filamentous hemagglutinin from Bordetella pertussis. Eur. J. Immunol. 30:415–422. [DOI] [PubMed] [Google Scholar]
- 35.Lebeis SL, Bommarius B, Parkos CA, Sherman MA, Kalman D. 2007. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 179:566–577 [DOI] [PubMed] [Google Scholar]
- 36.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- 37.Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, Kc W, Wu X, Thomas SR, Lee WL, Turkoz M, McDonald KG, Meredith MM, Song C, Guidos CJ, Newberry RD, Ouyang W, Murphy TL, Stappenbeck TS, Gommerman JL, Nussenzweig MC, Colonna M, Kopan R, Murphy KM. 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14:937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockmeyer J, Spelten S, Kuczius T, Bielaszewska M, Karch H. 2009. Structure and function relationship of the autotransport and proteolytic activity of EspP from Shiga toxin-producing Escherichia coli. PLoS One 4:e6100. 10.1371/journal.pone.0006100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, Park SY, Tame JR. 2005. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280:17339–17345. 10.1074/jbc.M412885200 [DOI] [PubMed] [Google Scholar]
- 40.Johnson TA, Qiu J, Plaut AG, Holyoak T. 2009. Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease: the structure of Haemophilus influenzae immunoglobulin A1 protease. J. Mol. Biol. 389:559–574. 10.1016/j.jmb.2009.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.