Abstract
Borrelia burgdorferi elicits a potent cytokine response through activation of multiple signaling receptors on innate immune cells. Spirochetal lipoproteins initiate expression of NF-κB-dependent cytokines primarily via TLR2, whereas type I interferon (IFN) production is induced through the endosomal receptors TLR7 and TLR9 in human dendritic cells and TLR8 in monocytes. We demonstrate that DNA and RNA are the B. burgdorferi components that initiate a type I IFN response by human peripheral blood mononuclear cells (PBMCs). IFN-α protein and transcripts for IRF7, MX1, and OAS1 were induced by endosomal delivery of B. burgdorferi DNA, RNA, or whole-cell lysate, but not by lysate that had been treated with DNase and RNase. Induction of IFN-α and IFN-λ1, a type III IFN, by B. burgdorferi RNA or live spirochetes required TLR7-dependent signaling and correlated with significantly enhanced transcription and expression of IRF7 but not IRF3. Induction of type I and type III IFNs by B. burgdorferi RNA could be completely abrogated by a TLR7 inhibitor, IRS661. In addition to type I and type III IFNs, B. burgdorferi RNA contributed to the production of the NF-κB-dependent cytokines, IFN-γ, interleukin-10 (IL-10), IL-1β, IL-6, and tumor necrosis factor alpha (TNF-α), by human PBMCs. Collectively, these data indicate that TLR7-dependent recognition of RNA is pivotal for IFN-α and IFN-λ1 production by human PBMCs, and that RNA-initiated signaling contributes to full potentiation of the cytokine response generated during B. burgdorferi infection.
INTRODUCTION
Borrelia burgdorferi is the causative agent of Lyme disease, the most common arthropod-borne disease in the United States (1, 2). Transmission of the spirochete by feeding of an infected tick frequently results in a distinctive skin rash, erythema migrans (EM), which is characterized by an influx of immune cells at the site of inoculation (1, 3). Disseminated infection occurs when spirochetes migrate from the initial site of infection to distal sites in the body, particularly the heart, joints, and central nervous system. Sequelae of disseminated infection are characterized by a robust inflammatory response which results in many of the typical symptoms of Lyme disease, notably carditis, arthritis, and neuroborreliosis (1). B. burgdorferi elicits a variety of pro- and anti-inflammatory cytokines via host recognition of spirochetal pathogen-associated molecular patterns (PAMPs) mediated by various pattern recognition receptors expressed on cells of the innate immune system. Detection of spirochetal lipoproteins by TLR2 has, until recently, been considered to be the key mediator of many B. burgdorferi-induced cytokines (4–9). However, more recent studies have demonstrated that B. burgdorferi, an extracellular pathogen, induces the production of type I interferons (IFNs) and other inflammatory cytokines via endosomal or cytosolic receptors, implicating PAMPs other than lipoproteins as contributors to the innate immune response to B. burgdorferi (10–13).
MyD88-dependent signaling is initiated from within intracellular compartments by the endosomal receptors TLR7, TLR8, and TLR9, as well as by endosomally localized TLR2. MyD88-independent signaling is activated by endosomal TLR3 or by cytosolic receptors (14–16) and mediated through a variety of signal transduction molecules, including TRIF (TLR3/Toll.IL-1R domain-containing adaptor-inducing IFN-β), TBK1 (TANK-binding kinase 1), and IRF3 (interferon regulatory factor 3) (17, 18). Each of these pathways is triggered by detection of nucleic acids and nucleoproteins (15, 16). TLR7 and TLR8 recognize single-stranded RNA motifs; TLR3 and cytosolic RNA helicases, such as RIG-I (retinoic acid-inducible gene 1) and MDA-5 (melanoma differentiation-associated protein 5), detect double-stranded RNA (19); CpG DNA motifs are ligands for TLR9 and cytosolic DNA sensors (17, 18, 20). Detection of RNA or DNA by its cognate receptors promotes a rapid antimicrobial response mediated through transcriptional activation of one or more IRFs. IRF3 is constitutively expressed by almost all cell types (21, 22). Following detection of nucleic acids by TLR3 and cytosolic receptors, IRF3 is phosphorylated and activates transcription of IFNB and IFNA1 (23). This initial type I IFN response then can be amplified through autocrine/paracrine feedback signaling via the type I IFN receptor (IFNAR), which results in transcription of IRF7 and other IFN-responsive genes (21, 23–26). Activation of IRF7 can also occur following ligand recognition by TLR7, TLR8, and TLR9. Some cell types, notably plasmacytoid dendritic cells (pDCs), constitutively express high levels of IRF7 which enable these cells to rapidly produce IFN-α in response to a stimulus without the requirement for IFNAR feedback signaling (24–26).
Published reports describe multiple distinct signaling pathways by which B. burgdorferi elicits a type I IFN response in various human and mouse innate immune cells. We and others have demonstrated that ex vivo stimulation of isolated human monocytes with B. burgdorferi elicits transcription of IFNB through TLR8/IRF7-dependent signaling (10). A more recent report by Cervantes and colleagues identified B. burgdorferi RNA as the ligand that activates this pathway and that also contributes to the production of interleukin-6 (IL-6), IL-10, and tumor necrosis factor alpha (TNF-α) by human monocytes (27). In mouse bone marrow-derived macrophages, transcriptional activation of IFN-responsive genes occurs via an IRF3-dependent but MyD88- and TRIF-independent pathway following recognition of B. burgdorferi RNA and multiple proteins by an unidentified cytosolic receptor (12). Petnicki-Ocwieja and colleagues demonstrated that B. burgdorferi initiates IFNA and IFNB gene transcription in mouse macrophages through activation of endosomally localized TLR2 and contributions from both MyD88- and TRIF-dependent signaling pathways (28). These studies with isolated immune cell populations, while providing crucial mechanistic insights into the production of B. burgdorferi-induced type I IFNs by specific cell types, are limited by the fact that these observations may not fully reflect the immune response in vivo, which results from an intricate network of interactions among multiple cell types.
The present study was designed to identify the B. burgdorferi ligands contributing to the production of IFNs and other cytokines using an experimental system that, while not replicating an in vivo model, more closely approximates the immune response under physiological conditions. We have previously demonstrated that human peripheral blood mononuclear cells (PBMCs), a mixed immune cell population consisting of lymphocytes, natural killer cells, monocytes, and dendritic cells, respond to live B. burgdorferi by transcriptional activation of type I IFNs and IFN-responsive genes, including IRF7, and the secretion of IFN-α protein by dendritic cell populations (11). These responses required spirochete phagocytosis and subsequent activation of TLR7- and TLR9-dependent signaling pathways (11). In the present study, IFN-α protein and transcription of IFN-responsive genes were measured following stimulation of human PBMCs with B. burgdorferi whole-cell lysates and purified DNA and RNA. We also assessed the production of IFN-λ1, a member of the type III IFN family which we have previously implicated in B. burgdorferi pathogenesis (92). In addition, TLR7 was investigated as a receptor for B. burgdorferi RNA and as a contributor to the production of NF-κB-dependent cytokine production by B. burgdorferi-stimulated human PBMCs.
MATERIALS AND METHODS
Isolation of human PBMCs.
Venous blood was obtained from each of four healthy volunteers (1 male, 3 female; 25 to 65 years of age) with no history of Lyme disease as confirmed by serologic testing. Written informed consent was obtained from all subjects before blood collection, in accordance with a protocol approved by the Institutional Review Board of New York Medical College. Blood was collected directly into BD-Vacutainer CPT tubes (BD Biosciences), and PBMCs were obtained by centrifugation per the manufacturer's instructions. PBMCs were washed twice in Hanks' buffered saline solution (HBSS) without calcium, magnesium, or phenol red (Invitrogen) and resuspended in RPMI 1640 without phenol red (Gibco-BRL) but containing 10% (vol/vol) heat-inactivated and endotoxin-free fetal bovine serum (FBS) (HyClone). PBMCs were maintained at 37°C in a humidified incubator containing 5% CO2.
Culture of B. burgdorferi.
B. burgdorferi B515, a human clinical isolate, has been described previously (29). Low-passage (passages 5 to 7) spirochetes were cultured in modified Barbour-Stoenner-Kelly medium at 37°C to the mid- to late-log phase of growth (4 × 107 to 1 × 108/ml) (30). To simulate the temperature shifts that occur during tick-to-mammal transmission and that affect the expression of virulence factors, spirochetes were subcultured and grown at 25°C to the mid-log phase of growth and then subcultured and incubated at 37°C until cultures once again reached the mid- to late-log phase of growth. Spirochetes were enumerated and assessed for motility by dark-field microscopy (31). Prior to coincubation with PBMCs, spirochetes were centrifuged for 10 min at 8,000 × g, washed twice with HBSS, and resuspended at a concentration of 5 × 108/ml in RPMI 1640 containing 10% FBS.
Preparation of B. burgdorferi lysate.
Fifty-ml cultures of B. burgdorferi B515 were pelleted by centrifugation for 10 min at 8,000 × g and washed three times with PBS that did not contain Ca2+ or Mg2+. The pellet was resuspended in PBS containing 4 μg/ml of lysozyme and incubated at 37°C for 30 min. Acid-washed beads were prepared by transferring 50 g of unwashed glass beads (Sigma) to a 100-ml autoclave-safe bottle, adding 5.8 M HCl to cover the beads and incubating them for 1 h. Beads were washed 10 times with 80 ml of deionized H2O, autoclaved for 20 min, dried overnight at 55°C, and stored in an airtight container. Cells were lysed by the addition of 1.5 g/ml of acid-washed glass beads (425 to 600 nm diameter) followed by 5 cycles of vortexing at maximum speed for 3 min and incubating on ice for 1 min (32–35). Spirochete membrane disruption was confirmed by dark-field microscopy (31). Whole-cell lysate was recovered after the beads had settled. Protein concentration was measured by the Bradford assay and adjusted to 1.0 μg/μl. For some experiments, lysates were treated with RNase A (Qiagen), DNase I (Qiagen), or proteinase K (Sigma) according to the manufacturer's instructions. As a control for Western immunoblotting to assess the presence of lipoproteins in lysate prepared using glass beads, lysate was prepared from separate B515 cultures using BugBuster lysis reagent (Merck Millipore).
Isolation of B. burgdorferi nucleic acids.
Total RNA was prepared from a 50-ml B. burgdorferi B515 culture using the toTALLY RNA kit (Ambion). RNA was resuspended in 30 μl of RNase-free water, and contaminating DNA was removed by treatment with the Turbo DNA-free kit (Ambion). RNA quality and concentration were determined by gel electrophoresis and spectrophotometric readings at 260 and 280 nm. The final concentration was adjusted to 1.0 μg/μl, 0.5 μl of RNase inhibitor (40 U/μl; Promega) was added, and RNA was stored at −80°C in single-use aliquots. Genomic DNA was prepared from 50-ml cultures of B. burgdorferi B515 using the DNeasy kit (Qiagen). DNA was eluted in 100 μl of nuclease-free water; the concentration was determined by spectrophotometry and adjusted to 1.0 μg/μl. DNA was stored at −20°C until use.
DOTAP methosulfate encapsulation of B. burgdorferi lysate and nucleic acids.
DOTAP {N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium} methosulfate (Sigma), a liposomal transfection reagent, was used to deliver B. burgdorferi cellular components to PBMCs via the endosomal pathway (36–40). B. burgdorferi DNA, RNA, or whole-cell lysate were mixed with DOTAP at a 1:1 (vol/vol) ratio and incubated at room temperature for 20 min immediately prior to stimulation of PBMCs.
Coincubation of human PBMCs with B. burgdorferi and spirochetal components.
Freshly isolated human PBMCs were suspended to a final concentration of 5 × 106 viable cells/ml in RPMI 1640 containing 10% heat-inactivated FBS. Live B. burgdorferi (5 × 107 cells; multiplicity of infection [MOI] of 10:1) or 1 μg of B. burgdorferi RNA, DNA, or whole-cell lysate, either alone or complexed with DOTAP, was added to triplicate wells for a final volume of 1.1 ml. Control wells received 100 μl of medium alone. PBMCs were coincubated for 12 h with live B. burgdorferi or spirochetal components in a humidified 37°C incubator containing 5% CO2. Cell-free culture supernatants were stored at −20°C. PBMC pellets were washed twice with PBS and immediately processed for RNA isolation or frozen in aliquots at −20°C for Western immunoblot analysis.
TLR ligands and inhibitors.
Imiquimod (R837) and Pam2CSK4 (InvivoGen) were added to PBMCs at final concentrations of 5 μg/ml and 1 μg/ml, respectively. An oligonucleotide (ODN) inhibitor of TLR7 signaling (IRS661; 5′-TGCTTGCAAGCTTGCAAGCA-3′) and a control ODN sequence (5′-TCCTGCAGGTTAAGT-3′) were synthesized by Integrated DNA Technologies (IDT) and purified by ion-exchange high-performance liquid chromatography (HPLC) (IE-HPLC) (41, 42). Endotoxin levels of all ODNs were <0.1 U/ml, as determined by the Limulus amebocyte lysis assay (LAL; Lonza). ODNs were used at a concentration of 5.6 μM as previously described (11, 41, 42).
Measurement of gene transcripts by real-time RT-PCR.
Total RNA was isolated from PBMCs using the RNeasy kit (Qiagen), and contaminating DNA was removed using the Turbo DNA-free kit (Ambion) per the manufacturer's instructions. RNA was eluted in 30 μl of RNase/DNase-free water, and the concentration was determined by spectrophotometry. RNA samples were stored at −80°C after the addition of 0.8 μl of RNase inhibitor (40 U/μl; Promega). cDNA was synthesized from 1.0 μg of total RNA in 20-μl reaction mixtures containing deoxynucleoside triphosphates (dNTPs) (250 μM each), 0.5 μg random primers, 5 U avian myeloblastosis virus (AMV) reverse transcriptase (RT), 10 U of RNase inhibitor, and 1× RT buffer (all from Promega). Reactions proceeded for 75 min at 42°C and were terminated by heating at 94°C for 5 min. cDNA was stored at −20°C until use.
Real-time RT-PCR was used to determine transcriptional expression of transcription factors and type I IFN-responsive genes. PCR assays were performed in duplicate on 10× diluted cDNA, prepared as described above, using the following predesigned TaqMan human gene expression assays (Applied Biosystems): MX1 (myxovirus [influenza virus] resistance 1, IFN-inducible protein p78 [mouse]), Hs00895608_m1; OAS1 (2′-5′ oligoadenylate synthetase 1), Hs00973637_m1; IRF7 (interferon-responsive factor 7), Hs00185375_m1; and IRF3 (interferon-responsive factor 3), Hs00155574_m1. All assays were performed in 20-μl reaction volumes using the ABI 7900HT SDS sequence detection system (Applied Biosystems) according to the manufacturer's instructions. For each sample, expression of the GAPDH (glyceraldehyde 3-phosphate dehydrogenase) gene was quantified by real-time RT-PCR using the TaqMan human GAPDH endogenous control assay (GenBank accession number NM_002046.3) (Applied Biosystems). A ΔΔCT method was used to calculate the differential expression of each GAPDH-normalized gene in experimental samples relative to unstimulated PBMCs (43).
Quantitation of secreted cytokine proteins.
Protein concentrations of IFN-α and IFN-λ1 in cell-free culture supernatants were quantitated using the human VeriKine IFN-α enzyme-linked immunosorbent assay (ELISA) kit (PBL Biomedical Laboratories) or the human IL-29 Ready-SET-Go ELISA kit (eBioscience), respectively. Concentrations of IFN-γ, TNF-α, IL-6, IL-1β, and IL-10 were measured by cytometric bead array using FlowSimplex kits (eBiosciences). Samples were processed on a MACSQuant Analyzer (Miltenyi Biotec) and analyzed using FlowCytomic 3.0 software (eBioscience).
Western immunoblotting.
Western immunoblotting was performed to detect levels of transcription factors IRF7 and IRF3 in PBMC lysates. Briefly, frozen pellets containing 3.33 × 105 PBMCs were thawed and resuspended in 100 μl of sample buffer containing 40 mM Tris-HCl (pH 6.8), 2% SDS, 1.8 mM EDTA, 10% glycerol, 1% β-mercaptoethanol, and 1× Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Cell suspensions were vortexed for 30 s, heated at 100°C for 10 min, and cooled on ice. Twenty-μl aliquots of lysate (corresponding to 6.7 × 104 PBMCs; 40 μg total protein) were resolved by 12.5% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked for 1 h with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) and incubated at 4°C overnight with rabbit IgG anti-human IRF7 or IRF3 antibody (Cell Signaling) diluted 1:1,000 in TBST containing 5% nonfat milk. Membranes were washed three times for 10 min with TBST, incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (Thermo Scientific) diluted 1:20,000 in TBST containing 3% nonfat milk, and developed using SuperSignal West Pico ECL substrate (Thermo Scientific). Following detection of IRF proteins, polyvinylidene difluoride (PVDF) membranes were stripped and reprobed with an antibody to GAPDH. Briefly, membranes were incubated in stripping buffer (2% SDS, 62.5 mM Tris-HCl, pH 6.8, 1% β-mercaptoethanol) for 30 min at 55°C, washed three times for 10 min in TBST, blocked for 1 h in TBST containing 5% nonfat milk, washed as before, and incubated overnight at 4°C with primary anti-human GAPDH IgG (Cell Signaling) diluted 1:1,000 in TBST-5% nonfat milk. Membranes were incubated with secondary conjugate and developed as described above. Protein levels were quantified by densitometry using ImageJ software (NIH). The average pixel value was calculated for each protein from boxes of equal size, background was automatically subtracted, and pixel values for IRF proteins were divided by pixel values for GAPDH. The IRF/GAPDH density ratio for unstimulated PBMCs was assigned a value of 1. Results from multiple donors were normalized for comparison by designating the smallest and largest densitometry ratios for each donor as baseline values.
For detection of OspA and OspC, B. burgdorferi lysates were suspended at a 2:1 (vol/vol) ratio in 3× concentrated sample buffer and prepared as described above. Equal amounts of proteins were resolved by 12.5% SDS-PAGE and transferred onto a PVDF membrane. The membranes were blocked and incubated with a rabbit polyclonal antibody against B. burgdorferi OspC (generously provided by Patricia Rosa, NIH) diluted 1:1,000 in 0.1% TBST containing 5% nonfat milk. Membranes were washed, incubated for 1 h with HRP-conjugated goat anti-rabbit IgG (Thermo Scientific), and developed using SuperSignal ECL substrate (Thermo Scientific). Following detection of OspC, PVDF membranes were stripped, reprobed using a mouse monoclonal antibody against B. burgdorferi OspA (a gift from Doris Bucher, NYMC), and developed using substrate containing 5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt (BCIP) and nitroblue tetrazolium (Thermo Scientific).
Statistics.
Differences in cytokine levels were analyzed by one-way analysis of variance (ANOVA) with Tukey-Kramer's posttest or by an unpaired Mann-Whitney U test as indicated. The fold change was calculated from real-time RT-PCR ΔΔCT values and analyzed for statistical significance using either Student's t test or a one-way ANOVA with Tukey-Kramer's posttest where appropriate (44). Densitometric values of Western blots were analyzed by one-way ANOVA with Tukey-Kramer's posttest. For all tests, P values of less than 0.05 were considered statistically significant. Statistical analyses were performed using GraphPad Prism 5.0 software (GraphPad Prism Software, Inc.).
RESULTS
B. burgdorferi nucleic acids induce expression of IFN-α protein and IFN-responsive genes by human PBMCs.
We have previously shown that B. burgdorferi elicits the production of type I IFN protein and transcription of IFN-responsive genes by human PBMCs; this response requires phagocytosis and signaling via TLR7 and TLR9 (11). In order to identify the specific B. burgdorferi PAMPs that are recognized by these phagosomal receptors, PBMCs from healthy donors were stimulated with live spirochetes (MOI of 10), B. burgdorferi whole-cell lysate, or purified nucleic acids that had been complexed with DOTAP, a liposomal transfection reagent that triggers uptake through the endolysosomal pathway (36–40, 45, 46). After a 12-h coincubation, the type I IFN response was assessed by measurement of IFN-responsive gene transcripts and quantitation of secreted IFN-α protein. Consistent with our previously reported results (11), live B. burgdorferi elicited a significant increase in transcript levels for IRF7 (3.02-fold ± 0.64-fold), MX1 (14.51-fold ± 3.77-fold), and OAS1 (9.56-fold ± 2.6-fold) relative to PBMCs incubated with medium alone (Fig. 1A). Similarly, 1 μg of purified, DOTAP-complexed B. burgdorferi RNA or DNA induced significant transcriptional activation of these genes, with fold change values of 7.04 ± 0.99 and 7.47 ± 1.62 (IRF7), 28.54 ± 4.68 and 22.37 ± 3.35 (MX1), and 19.63 ± 1.23 and 16.09 ± 6.4 (OAS1) for RNA and DNA, respectively, relative to PBMCs stimulated with DOTAP only (Fig. 1A). DOTAP-complexed B. burgdorferi lysate also elicited significant transcriptional activation of these genes, with fold changes of 2.36 ± 0.05 (IRF7), 4.19 ± 0.14 (OAS1), and 7.15 ± 3.08 (MX1) relative to the unstimulated control. However, this response could be abrogated by treatment of the lysate with both DNase I and RNase A prior to DOTAP encapsulation (Fig. 1A). In contrast, treatment of lysate with proteinase K did not significantly reduce transcript levels of IRF7, MX1, or OAS1 (data not shown). In order to verify that spirochetal lipoproteins were not eliminated during preparation or nuclease treatment of the whole-cell lysate, Western immunoblotting for the presence of outer surface protein A (OspA) and OspC, which are well-characterized ligands for TLR2, was performed. Comparable levels of OspA and OspC were detected in B. burgdorferi lysate that had been prepared using either the glass bead or chemical lysis method, as well as in lysates that had been treated with RNase A or DNase I (Fig. 1C).
FIG 1.
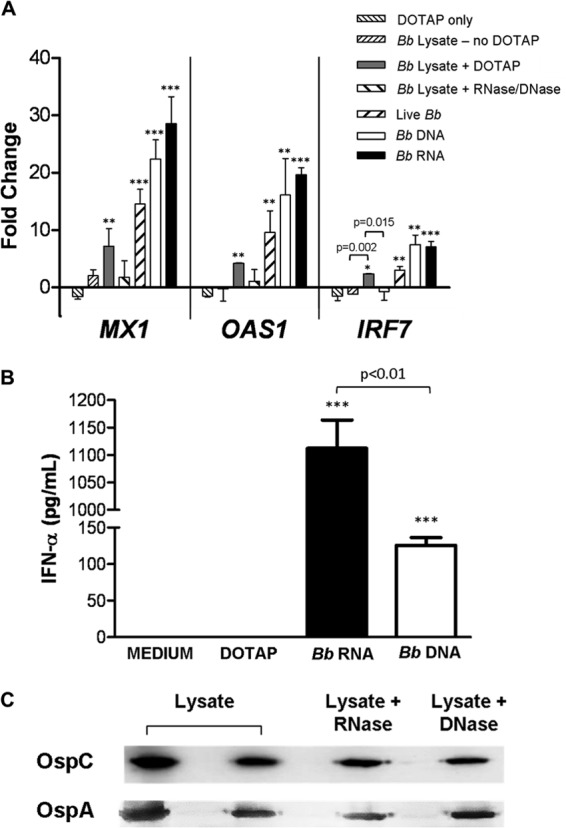
B. burgdorferi nucleic acids induce a type I IFN response in human PBMCs. Human PBMCs (5 × 106) were stimulated for 12 h with 5 × 107 live B. burgdorferi (Bb) spirochetes, DOTAP-complexed B. burgdorferi DNA or RNA (1 μg/ml), or B. burgdorferi whole-cell lysate (1 μg/ml) with or without DOTAP added. (A) Transcriptional expression of MX1, OAS1, and IRF7 was measured by RT-PCR and normalized to transcript levels for GAPDH. Data are presented as the mean fold changes ± standard deviations (SD) relative to PBMCs incubated with medium alone. (B) Protein concentration of IFN-α in cell-free supernatants was measured by ELISA. Data shown are the means ± SD of values from two donors assessed in triplicate in two independent experiments and are representative of one additional donor. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) relative to PBMCs incubated with medium alone. (C) B. burgdorferi B515 lysate was prepared by vortexing in the presence of glass beads until cellular breakage was verified via dark-field microscopy. Whole-cell lysate, or lysate that had been treated with RNase A or DNase I, was used to stimulate PBMCs. In order to confirm the presence of TLR2 ligands in the lysate preparations, volumes containing 10 μg total protein were resolved by 12.5% SDS-PAGE and immunoblotted with specific antibodies for OspC and OspA, spirochetal lipoproteins known to initiate TLR2-mediated signaling. B. burgdorferi B515 lysate prepared from a separate culture using a chemical lysis method was included as a positive control (far left lane).
To confirm the ability of B. burgdorferi nucleic acids to induce type I IFN, levels of IFN-α protein in PBMC culture supernatants were quantitated by ELISA. Significant amounts of IFN-α were secreted by PBMCs in response to purified, DOTAP-complexed B. burgdorferi RNA (1112 pg/ml) or DNA (126.0 pg/ml) (Fig. 1B). Collectively, these data identify B. burgdorferi RNA and DNA, but not protein, as the bacterial PAMPs that elicit the production of IFN-α protein and type I IFN-responsive gene transcripts in human PBMCs.
B. burgdorferi RNA induces expression of IRF7 but not IRF3 in human PBMCs.
Stimulation of human PBMCs with live B. burgdorferi and B. burgdorferi nucleic acids results in significant transcriptional activation of IRF7 (11) (Fig. 1), a critical transcription factor in the TLR7-MyD88-dependent signaling pathway (25). In contrast, IRF3 is utilized by TLR3 and some cytosolic RNA receptors for the generation of type I IFNs (21, 22). Transcript levels for IRF7 and IRF3 were determined by real-time RT-PCR following coincubation of PBMCs for 12 h with either live B. burgdorferi or DOTAP-complexed B. burgdorferi RNA. No significant change in IRF3 transcript level was observed for any of the experimental conditions tested relative to unstimulated PBMCs (Fig. 2). In contrast, transcript levels for IRF7 were significantly induced in PBMCs by live B. burgdorferi (fold change of 7.19) and by B. burgdorferi RNA (fold change of 8.00) (Fig. 2). The induction of IRF7 by B. burgdorferi could be ablated by the prior addition of IRS661, a specific inhibitor of TLR7, whereas a control ODN had no effect.
FIG 2.

B. burgdorferi RNA induces transcription of IRF7 in human PBMCs via TLR7-dependent signaling. Human PBMCs (5 × 106) were cultured in the presence of medium, a control ODN (5.6 μM), or the TLR7 inhibitor IRS661 (5.6 μM) for 1 h before stimulation with live B. burgdorferi (5 × 107), DOTAP-complexed B. burgdorferi RNA (1 μg/ml), or a synthetic TLR7 agonist, R837 (5 μg/ml). PBMC RNA was isolated 12 h after addition of stimuli, and real-time RT-PCR was used to measure transcriptional expression of signaling mediators. GAPDH-normalized values were used to calculate fold changes in transcript levels for IRF7 or IRF3 relative to PBMCs incubated with medium alone. Columns represent the means ± SD of results obtained using PBMCs from two donors assessed in triplicate in independent experiments. ***, P < 0.001 relative to PBMCs incubated with medium alone; NS, not significantly different.
Expression of IRF7 and IRF3 proteins was assessed by Western immunoblotting using PBMC lysates from 12-h coincubation experiments. Basal levels of IRF7 could not be detected in unstimulated PBMCs (Fig. 3A). Stimulation of PBMCs with live B. burgdorferi or DOTAP-complexed B. burgdorferi RNA resulted in measurable levels of IRF7 that were significantly higher than that of the barely detectable IRF7 protein induced by B. burgdorferi lysate (normalized IRF7/GAPDH density values were the following: 87.7, B. burgdorferi; 82.5, B. burgdorferi RNA) (Fig. 3A). In contrast to IRF7, basal levels of IRF3 could be detected in unstimulated PBMCs, and these levels did not change significantly in response to any stimulus (Fig. 3B). These results suggest that the type I IFN response elicited by live B. burgdorferi and B. burgdorferi RNA in human PBMCs is mediated by IRF7.
FIG 3.

B. burgdorferi RNA promotes expression of IRF7 human PBMCs. Human PBMCs (5 × 106) were incubated for 12 h with 5 × 107 live B. burgdorferi spirochetes, DOTAP-complexed B. burgdorferi RNA (1 μg/ml), or B. burgdorferi whole-cell lysate (1 μg/ml) added without DOTAP. PBMC lysates were resolved by 12.5% SDS-PAGE for Western immunoblotting with rabbit anti-human IRF7 (A) or IRF3 (B). Signals were quantified by densitometry and are expressed as ratios to GAPDH. Columns represent the means ± SD of results from three donors. Western blot images from a single representative donor are shown. ***, P < 0.001 relative to PBMCs incubated with medium alone. NS, not significantly different.
Type I IFN and type III IFNs are induced by TLR7-dependent recognition of B. burgdorferi RNA.
Stimulation of human PBMCs for 12 h with live B. burgdorferi, DOTAP-complexed B. burgdorferi RNA, or the TLR7-specific agonist R837 resulted in the expression of significant levels of IFN-α protein (130.8 pg/ml, 133.3 pg/ml, and 264.6 pg/ml, respectively) (Fig. 4A). In contrast, stimulation of PBMCs with B. burgdorferi whole-cell lysate that had not been complexed with DOTAP or with Pam2CSK4 did not elicit significant IFN-α production. The IFN-α response to B. burgdorferi RNA could be completely ablated by the addition of IRS661 prior to B. burgdorferi RNA stimulation (P < 0.01 relative to the control ODN) or by pretreatment of B. burgdorferi RNA with RNase A (Fig. 4A).
FIG 4.

TLR7-dependent signaling by B. burgdorferi RNA elicits production of type I and type III IFNs by human PBMCs. Human PBMCs (5 × 106) were cultured in the presence of medium, a control ODN (5.6 μM), or the TLR7 inhibitor IRS661 (5.6 μM) for 1 h prior to stimulation with 5 × 107 live B. burgdorferi spirochetes, DOTAP-complexed B. burgdorferi RNA (1 μg/ml), or B. burgdorferi whole-cell lysate (1 μg/ml) added without DOTAP. R837 (5 μg/ml) and Pam2CSK4 (1 μg/ml) were used as positive controls for activation of TLR7 or TLR2, respectively. IFN-α (A) and IFN-λ1 (B) protein concentrations in culture supernatants were quantitated by ELISA. Data represent the means ± SD of values from three donors assessed in triplicate in three independent experiments. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) relative to PBMCs incubated with medium alone, as determined by one-way ANOVA with Tukey-Kramer's posttest, with the exception of the positive-control R837, which was determined by nonparametric Mann-Whitney U test. NS, not significantly different.
We have recently identified the ability of B. burgdorferi to induce the expression of both type I and type III IFNs by human PBMCs (11, 92). Therefore, we sought to determine if recognition of B. burgdorferi RNA by TLR7 also elicits type III IFN production. Significant concentrations of IFN-λ1 were detected by ELISA in the culture supernatants of human PBMCs following stimulation with live B. burgdorferi (15.90 pg/ml) or DOTAP-complexed B. burgdorferi RNA (14.56 pg/ml) (Fig. 4B); similar to the results for IFN-α, addition of IRS661 prior to stimulation completely inhibited the IFN-λ1 response to RNA (Fig. 4B). In contrast, B. burgdorferi lysate that had not been complexed with DOTAP did not induce IFN-λ1.
Transcriptional induction of the IFN-responsive genes MX1 and OAS1 paralleled the expression of IFN-α and IFN-λ1. Stimulation of PBMCs with live spirochetes or B. burgdorferi RNA elicited significant induction of both genes (MX1, 22.16-fold and 47.63-fold, respectively; OAS1, 8.19-fold and 20.78-fold, respectively) (Fig. 5). This induction by B. burgdorferi RNA could be completely ablated by the addition of IRS661 prior to stimulation, whereas a control ODN had no significant effect (Fig. 5). In addition, RNase A treatment completely abolished the ability of B. burgdorferi RNA to induce MX1 transcription (Fig. 5A). Taken together, these results demonstrate that type I and type III IFN production by human PBMCs is mediated, at least in part, by TLR7-dependent recognition of B. burgdorferi RNA.
FIG 5.
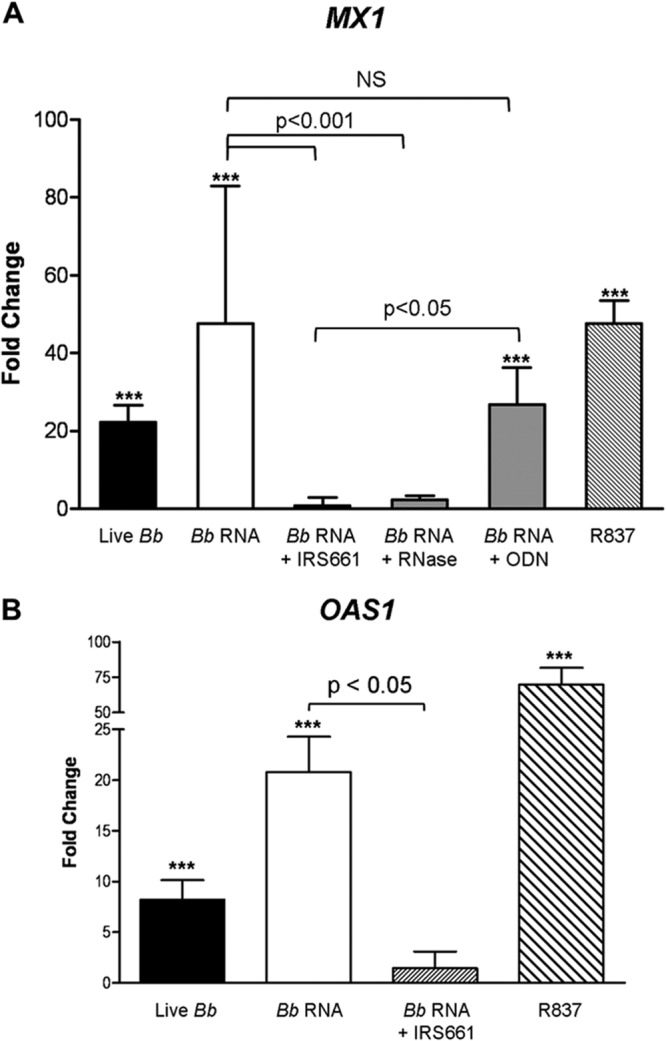
B. burgdorferi RNA induces transcription of IFN-responsive genes in human PBMCs via TLR7. Human PBMCs (5 × 106) were cultured in the presence of medium, a control ODN (5.6 μM), or the TLR7 inhibitor IRS661 (5.6 μM) for 1 h before stimulation with live B. burgdorferi (5 × 107), the synthetic TLR7 agonist R837 (5 μg/ml), or DOTAP-complexed B. burgdorferi RNA (1 μg/ml) that had been treated with RNase A or left untreated. Total RNA was isolated 12 h after addition of stimuli, and transcriptional expression of IFN-responsive genes was measured by real-time RT-PCR. GAPDH-normalized values were used to calculate fold changes in transcript levels for MX1 (A) or OAS1 (B) relative to PBMCs incubated with medium alone. Columns represent the mean fold changes ± SD obtained using PBMCs from two donors assessed in independent experiments. ***, P < 0.001 relative to PBMCs incubated with medium alone. NS, not significantly different.
B. burgdorferi RNA contributes to, but is not sufficient for, full production of NF-κB-dependent cytokines.
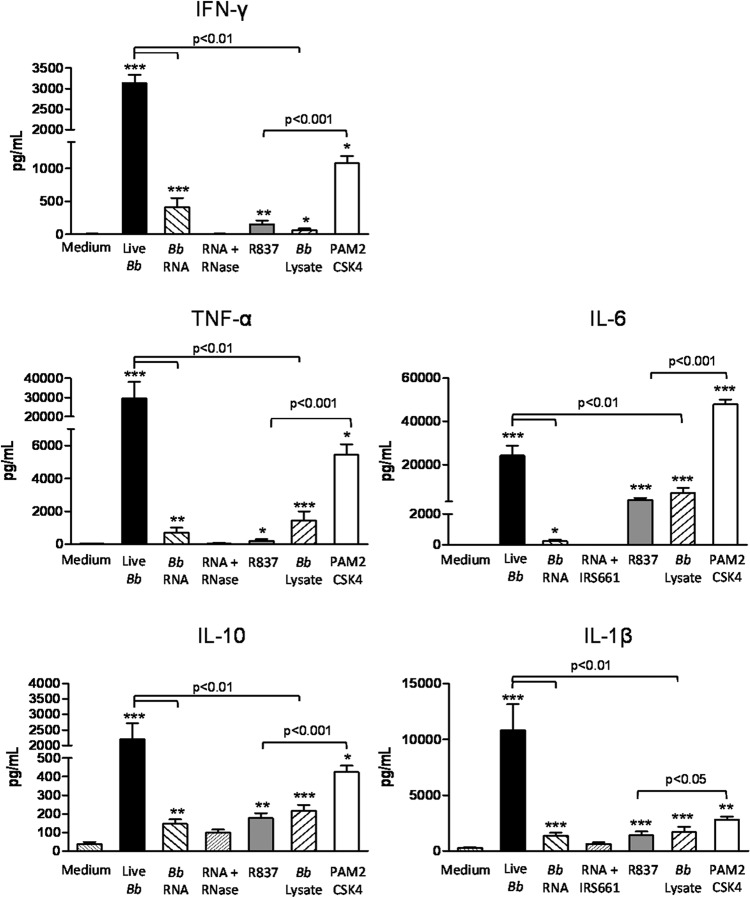
Engagement of TLR7 elicits the production of cytokines and chemokines, including type I IFNs, through MyD88-dependent activation of IRF7 and NF-κB. Therefore, the contribution of RNA to the cytokine profile elicited by B. burgdorferi in human PBMCs was investigated next. A cytometric bead array was employed to measure protein concentrations for IFN-γ, IL-1β, IL-10, IL-6, and TNF-α in the culture supernatants after 12 h of coincubation. Stimulation of PBMCs with live B. burgdorferi resulted in the production of significant levels of IFN-γ (3,100 pg/ml), TNF-α (29,500 pg/ml), IL-10 (2,200 pg/ml), IL-6 (24,100 pg/ml), and IL-1β (10,800 pg/ml) (Fig. 6). Coincubation of PBMCs with B. burgdorferi lysate that had not been complexed with DOTAP, which would be expected to elicit cytokine production predominantly through detection of B. burgdorferi lipoproteins by TLR2, produced modest but significant levels of IFN-γ (63.3 pg/ml), TNF-α (1,312 pg/ml), IL-10 (217.4 pg/ml), IL-6 (6,870 pg/ml), and IL-1β (1,700 pg/ml) (Fig. 6). However, levels of these cytokines were significantly lower than those induced by live B. burgdorferi (P < 0.01 for all cases). Similarly, Pam2CSK4, a synthetic TLR2 ligand, elicited significant production of all cytokines; with the exception of IL-6, these concentrations were significantly lower than those induced by live B. burgdorferi (IFN-γ, P < 0.001; TNF-α, P < 0.05; IL-10, P < 0.01; IL-1β, P < 0.01) (Fig. 6). DOTAP-complexed B. burgdorferi RNA induced significant production of IFN-γ (413.4 pg/ml), TNF-α (715.5 pg/ml), IL-10 (146.1 pg/ml), IL-6 (231.1 pg/ml), and IL-1β (1,374 pg/ml) (Fig. 6); treatment of RNA with RNase A, or preincubation of PBMCs with the TLR7-specific inhibitor IRS661, ablated production of these cytokines. Notably, the induction of cytokines by B. burgdorferi RNA or by the TLR7 ligand (R837) was significantly lower than that elicited in response to live B. burgdorferi. Furthermore, cytokine levels induced by the TLR2-specific ligand Pam2CSK4 were significantly higher than those induced by the TLR7-specific ligand R837 (Fig. 6). Taken together, these data indicate that TLR2 and TLR7 signaling both contribute to the production of NF-κB-dependent cytokines by human PBMCs in response to live B. burgdorferi.
FIG 6.
B. burgdorferi RNA contributes to the induction of NF-κB-dependent cytokines by human PBMCs. Human PBMCs (5 × 106) were incubated for 12 h with 5 × 107 live B. burgdorferi spirochetes, DOTAP-complexed B. burgdorferi RNA (1 μg/ml), or B. burgdorferi whole-cell lysate (1 μg/ml) added without DOTAP. R837 (5 μg/ml) and Pam2CSK4 (1 μg/ml) were used as controls for activation of TLR7 and TLR2, respectively. Protein concentrations of IFN-γ, IL-1β, TNF-α, IL-10, and IL-6 in cell-free culture supernatants were measured by cytometric bead array using a MACSQuant analyzer. Columns depict the mean values ± SD of results from three donors assessed in triplicate in three independent experiments. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) relative to PBMCs incubated with medium alone, as determined by Mann-Whitney U test.
DISCUSSION
Infection with Borrelia burgdorferi leads to a robust inflammatory response that is characterized by the production of IFN-γ, TNF-α, IL-6, IL-10, IL-1β, and type I IFNs (4–7, 9–11, 13, 47). Until recently, it was believed that the key inflammatory mediators consisted of B. burgdorferi lipoproteins that signal through TLR2 heterodimers expressed on the mammalian cell surface (48, 49). Our group has previously shown that live B. burgdorferi induces expression of IFN-α protein and IFN-responsive gene transcripts by pDC and immature mDC populations in human PBMCs following spirochete phagocytosis. This response could be partially but significantly reduced by the addition of synthetic inhibitors of either TLR7 or TLR9 to PBMCs prior to stimulation with live spirochetes, and it could be completely ablated by simultaneous addition of both inhibitors (11). In the present study, we examined the B. burgdorferi PAMPs that may mediate this response. Purified B. burgdorferi DNA or RNA, delivered via the endosomal pathway, elicited expression of IFN-α protein and induction of the IFN-responsive transcripts that had been assessed in our previous study. This result is consistent with our observation that concomitant signaling by TLR7 and TLR9 is required for full induction of IFN-α by live B. burgdorferi in human PBMCs (11). However, RNA induced significantly larger amounts of IFN-α than did the same concentration of DNA (1 μg/ml). Whole B. burgdorferi cell lysate was also able to induce a type I IFN response, but only when delivered to the phagosome using DOTAP methosulfate as a vehicle (36–40). Pretreatment of lysate with RNase A and DNase I abolished its ability to induce type I IFN. These findings demonstrate that B. burgdorferi nucleic acids, but not proteins, are type I IFN-inducing ligands recognized by human PBMCs.
We observed significant increases in IRF7 transcript and protein levels in human PBMCs in response to stimulation by either live B. burgdorferi or B. burgdorferi RNA, but not by extracellular B. burgdorferi lysate that had not been complexed with DOTAP and did not have access to receptors within the phagosome. In contrast, no changes were observed for either IRF3 transcript or protein. This is in contrast to a previous study by Miller et al. which suggested that B. burgdorferi RNA and protein elicit a type I IFN response through a MyD88-independent, IRF3-dependent pathway initiated by an unidentified cytosolic receptor (12). These seemingly contradictory findings are likely attributable to a number of variables, including inherent immunological differences between the species (human versus mouse) and cell types (PBMCs versus macrophages) used in the respective systems (50), as well as methodological differences in the approaches used to deliver spirochetal cellular components and to measure the type I IFN response. We previously identified pDCs as predominant producers of IFN-α in B. burgdorferi-stimulated PBMCs (11). DC populations constitutively express high basal levels of IRF7; thus, they are immediately primed to respond to the appropriate stimuli (24, 25). Although we did not observe any detectable IRF7 in unstimulated PBMCs, this is not surprising, as DCs comprise only 0.1 to 1.0% of peripheral blood cells (51). In contrast to DCs, macrophages produce initial levels of type I IFNs in an IRF3-dependent manner in response to cytosolic sensing of other pathogens (12, 21–23, 52–57). Collectively, the present findings demonstrate that, in an ex vivo human PBMC experimental model, production of type I IFNs in response to live B. burgdorferi, or endosomal delivery of B. burgdorferi RNA, occurs through an IRF7-dependent pathway.
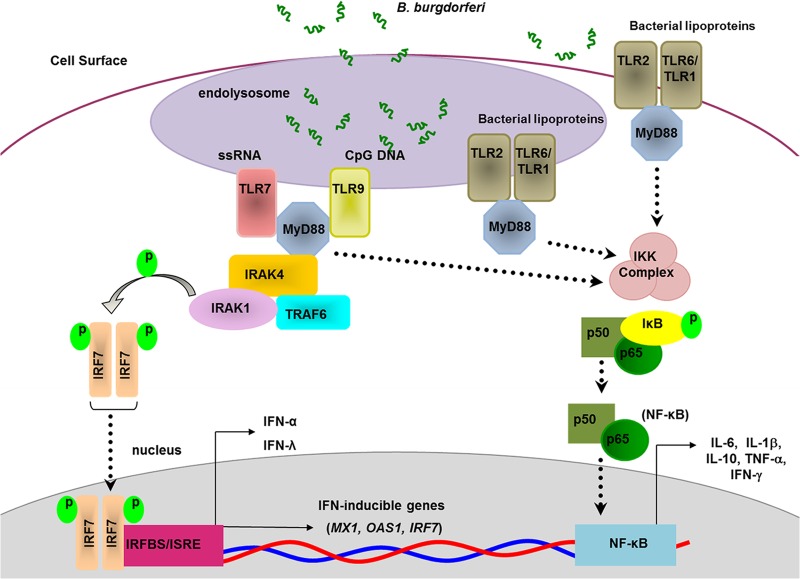
TLR2 has been implicated in the production of many proinflammatory cytokines associated with Lyme disease through recognition of B. burgdorferi lipoproteins (4–6, 8, 48, 49, 58). However, TLR2 has been shown to traffic to the endosomal membrane upon recognition of B. burgdorferi lipoproteins and contributes to MyD88/TRIF-dependent transcriptional activation of type I IFN gene products (28, 48). Therefore, it was important to establish the potential contribution of TLR2-specific ligands to the type I IFN response we observed. We identified the presence of lipoproteins within the B. burgdorferi lysates utilized for PBMC stimulation, specifically the presence of OspA and OspC. However, only B. burgdorferi lysate delivered endosomally via DOTAP was able to induce a type I IFN response, and treatment with RNase and DNase abolished this response. This allows us to conclude that in human PBMCs, recognition of TLR2 ligands, namely, B. burgdorferi lipoproteins, does not contribute to the type I IFN response observed. Based on these results and those from our previous study using live B. burgdorferi (11), we infer that phagocytosis of B. burgdorferi by human PBMCs results in the release of spirochetal nucleic acids and the subsequent activation of TLR7 and TLR9 (Fig. 7).
FIG 7.
Proposed model of B. burgdorferi-induced cytokine production by human dendritic cells. Following phagocytosis of B. burgdorferi, spirochetal RNA is detected by TLR7 and initiates MyD88-dependent signaling that leads to activation of IRF7 and production of IFN-α and IFN-λ1. TLR7-MyD88 signaling contributes to the production of NF-κB-dependent cytokines, including IL-6, IL-1β, TNF-α, IFN-γ, and IL-10. Additional activation of NF-κB by B. burgdorferi occurs via TLR2-dependent sensing of spirochetal lipoproteins. The figure is based on Petzke et al. (11) and adapted to include data from the present study.
Convergent signaling by multiple receptors results in the production of a maximal inflammatory response to a variety of pathogens, including Francisella tularensis and Streptococcus pyogenes (6, 53, 59–61). Phagocytosis is required for full induction of several NF-κB-dependent cytokines that are elicited in response to B. burgdorferi (10, 47). Several recent reports describe the contributions of TLR8-MyD88 signaling to the production of inflammatory cytokines following recognition of self RNA or microbial RNA, including the requirement for TLR2-TLR8 cooperativity in the generation of TNF-α, IL-6, IL-1β, and IL-10 by B. burgdorferi RNA-stimulated human monocytes (27). In the present study, we observed that isolated B. burgdorferi cellular constituents, as well as synthetic agonists specific for TLR7 and TLR2, elicited the production of the NF-κB-dependent cytokines implicated in the pathogenesis of B. burgdorferi. However, levels of these inflammatory proteins generally were lower than those induced by stimulation with the live spirochete. Both TLR7 and TLR8 are able to recognize single-stranded RNA motifs and thereby induce the production of type I IFNs (14, 21, 62), although there are distinct differences in the cytokine responses elicited by each of these receptors (63, 64). TLR7 signaling preferentially leads to IFN-α production by plasmacytoid DCs, whereas TLR8 signaling is more commonly associated with monocytes and IFN-β production (63, 65). During DC maturation, TLR7 and TLR8 agonists have differential biological effects on the expression of maturation markers and the production of proinflammatory cytokines, including IL-6 and IL-12 (66). We have previously reported that in the human PBMC system employed here, the predominant IFN-α producers in response to B. burgdorferi are plasmacytoid DCs and myeloid DC precursors (11). We now identify B. burgdorferi RNA as a TLR7 signaling ligand not only for the IFN-α response but also as a contributor to the NF-κB inflammatory profile.
The production of type I IFNs in response to bacterial pathogens can have a variety of effects. Classically thought of as antiviral mechanisms, the potential role of type I IFNs in response to bacterial pathogens is less well defined (14, 62). Type I IFNs constitute pivotal components of the host defense system that limit the pathogenesis of Salmonella enterica serovar Typhimurium, group B streptococci, and Escherichia coli (39, 67, 68). Conversely, the production of type I IFNs by the host in response to pathogens such as Listeria monocytogenes, uropathogenic E. coli (UPEC), and Mycobacterium tuberculosis enhances the pathogenic potential of these bacteria (69–71). Of greater relevance to the current study, type I IFN signaling was shown to be a crucial factor in the development and severity of Lyme arthritis in susceptible mouse strains following infection with B. burgdorferi (72, 73). Therefore, evaluating the signaling pathways that lead to type I IFN production in response to B. burgdorferi is crucial to elucidating the mechanisms that play a role in the pathogenesis of Lyme disease.
Interestingly, we also observed a novel type III IFN response that paralleled the production of IFN-α and confirms our report of B. burgdorferi-mediated type III IFN production. Type III IFNs, which include IL-29/IFN-λ1, IL-28A/IFN-λ2, and IL-28B/IFN-λ3, are recently described inflammatory cytokines that can be induced through recognition of double-stranded RNA motifs via IRF3-, IRF7-, and NF-κB-mediated signaling (74–80). IFN-λs are expressed by monocytes, macrophages, and dendritic cells and can be produced concurrently with type I IFNs (75, 81, 82). Type I and type III IFNs share a variety of biological activities that contribute to antiviral responses, including induction of major histocompatibility complex class I antigen presentation, promotion of NK cell and T cell cytotoxic effects, and transcriptional induction of a subset of interferon-stimulated genes (75, 77, 83–85). Although typically associated with antiviral responses, type III IFNs are able to attenuate the T-helper 2 (Th2) response by limiting inflammation during immune challenge, leading to tolerance; in these instances, DCs were identified to be the predominant cell populations producing IFN-λ (86, 87). Recent studies have established a role for type III IFNs in the pathogenesis of certain bacteria, including Staphylococcus aureus, Pseudomonas aeruginosa, and L. monocytogenes (88–91). L. monocytogenes infection is promoted by type I IFNs and further enhanced by the production of type III IFNs (89). IFN-λR is expressed solely in epithelial tissues, indicating that the immune-modulatory effects are important for pathogens interacting with epithelial surfaces, whereas type I IFNs exert systemic effects (76, 85). These findings suggest that the type III IFN production that was observed in response to B. burgdorferi RNA contributes to the overall progression of infection and development of Lyme disease.
Differential transcription factor activation and coordinated signaling between pathogen recognition receptors of innate immune cells can dramatically alter the inflammatory response to B. burgdorferi. Moreover, the signaling cascade that leads to a type I IFN response is distinct from that of other inflammatory mediators of Lyme disease, as B. burgdorferi RNA contributed only modestly to the levels of NF-κB-mediated cytokines we measured. This provides further evidence that full potentiation of the immune response to B. burgdorferi requires a combination of IFN and NF-κB signaling mediated though multiple PRRs (Fig. 7) (10). Collectively, these data indicate that B. burgdorferi DNA and RNA are pivotal for IFN-α and IFN-λ1 production by human PBMCs, and that RNA-initiated TLR7 signaling contributes to full potentiation of the cytokine response generated during B. burgdorferi infection (Fig. 7). These findings suggest that the magnitude of TLR7 signaling mediated by spirochetal RNA plays a pivotal role in the ability of B. burgdorferi to cause disseminated disease, either by direct effects exerted by type I and III IFNs or through activation of dendritic cells via TLR7 signaling.
ACKNOWLEDGMENTS
We gratefully acknowledge the outstanding phlebotomy services of Diane Holmgren and other staff members in the Division of Infectious Diseases, New York Medical College. We also thank Giacomo Signorino and Michelle Krupna-Gaylord for providing technical expertise.
This work was supported by ARRA supplement 3R01-AR41511 from the National Institutes of Health and grant 5U01CK000153 from the Centers for Disease Control and Prevention (to I.S.) and by a faculty recruitment grant from New York Medical College (to M.M.P.).
Footnotes
Published ahead of print 24 March 2014
REFERENCES
- 1. Steere AC. 2001. Lyme disease. N. Engl. J. Med. 345:115–125. 10.1056/NEJM200107123450207 [DOI] [PubMed] [Google Scholar]
- 2. Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093–1101. 10.1172/JCI21681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nadelman RB, Nowakowski J, Forseter G, Goldberg NS, Bittker S, Cooper D, Mguero-Rosenfeld Wormser GP. 1996. The clinical spectrum of early Lyme borreliosis in patients with culture-confirmed erythema migrans. Am. J. Med. 100:502–508. 10.1016/S0002-9343(95)99915-9 [DOI] [PubMed] [Google Scholar]
- 4. Ma Y, Seiler KP, Tai KF, Yang L, Woods M, Weis JJ. 1994. Outer surface lipoproteins of Borrelia burgdorferi stimulate nitric oxide production by the cytokine-inducible pathway. Infect. Immun. 62:3663–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Radolf JD, Arndt LL, Akins DR, Curetty LL, Levi ME, Shen Y, Davis LS, Norgard MV. 1995. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytes/macrophages. J. Immunol. 154:2866–2877 [PubMed] [Google Scholar]
- 6. Udalova IA, Vidal V, Scragg IG, Kwiatkowski D. 2000. Direct evidence for involvement of NF-kappaB in transcriptional activation of tumor necrosis factor by a spirochetal lipoprotein. Infect. Immun. 68:5447–5449. 10.1128/IAI.68.9.5447-5449.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sellati TJ, Bouis DA, Kitchens RL, Darveau RP, Pugin J, Ulevitch RJ, Gangloff SC, Goyert SM, Norgard MV, Radolf JD. 1998. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytic cells via a CD14-dependent pathway distinct from that used by lipopolysaccharide. J. Immunol. 160:5455–5464 [PubMed] [Google Scholar]
- 8. Shin OS, Isberg RR, Akira S, Uematsu S, Behera AK, Hu LT. 2008. Distinct roles for MyD88 and Toll-like receptors 2, 5, and 9 in phagocytosis of Borrelia burgdorferi and cytokine induction. Infect. Immun. 76:2341–2351. 10.1128/IAI.01600-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sellati TJ, Bouis DA, Caimano MJ, Feulner JA, Ayers C, Lien E, Radolf JD. 1999. Activation of human monocytic cells by Borrelia burgdorferi and Treponema pallidum is facilitated by CD14 and correlates with surface exposure of spirochetal lipoproteins. J. Immunol. 163:2049–2056 [PubMed] [Google Scholar]
- 10. Cervantes JL, Dunham-Ems SM, La Vake CJ, Petzke MM, Sahay B, Sellati TJ, Radolf JD, Salazar JC. 2011. Phagosomal signaling by Borrelia burgdorferi in human monocytes involves Toll-like receptor (TLR) 2 and TLR8 cooperativity and TLR8-mediated induction of IFN-beta. Proc. Natl. Acad. Sci. U. S. A. 108:3683–3688. 10.1073/pnas.1013776108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petzke MM, Brooks A, Krupna MA, Mordue D, Schwartz I. 2009. Recognition of Borrelia burgdorferi, the Lyme disease spirochete, by TLR7 and TLR9 induces a type I IFN response by human immune cells. J. Immunol. 183:5279–5292. 10.4049/jimmunol.0901390 [DOI] [PubMed] [Google Scholar]
- 12. Miller JC, Maylor-Hagen H, Ma Y, Weis JH, Weis JJ. 2010. The Lyme disease spirochete Borrelia burgdorferi utilizes multiple ligands, including RNA, for interferon regulatory factor 3-dependent induction of type I interferon-responsive genes. Infect. Immun. 78:3144–3153. 10.1128/IAI.01070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salazar JC, Duhnam-Ems S, La VC, Cruz AR, Moore MW, Caimano MJ, Velez-Climent L, Shupe J, Krueger W, Radolf JD. 2009. Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFN-beta. PLoS Pathog. 5:e1000444. 10.1371/journal.ppat.1000444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalliolias GD, Ivashkiv LB. 2010. Overview of the biology of type I interferons. Arthritis Res. Ther. 12(Suppl 1):S1. 10.1186/ar2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 16. Stetson DB, Medzhitov R. 2006. Type I interferons in host defense. Immunity 25:373–381. 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 17. Keating SE, Baran M, Bowie AG. 2011. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32:574–581. 10.1016/j.it.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa H, Barber GN. 2011. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol. Life Sci. 68:1157–1165. 10.1007/s00018-010-0605-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilkins C, Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47. 10.1016/j.coi.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nockher WA, Scherberich JE. 1998. Expanded CD14+ CD16+ monocyte subpopulation in patients with acute and chronic infections undergoing hemodialysis. Infect. Immun. 66:2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 22. Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548. 10.1016/S1074-7613(00)00053-4 [DOI] [PubMed] [Google Scholar]
- 23. Solis M, Goubau D, Romieu-Mourez R, Genin P, Civas A, Hiscott J. 2006. Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages. Biochem. Pharmacol. 72:1469–1476. 10.1016/j.bcp.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 24. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- 25. Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434:1035–1040. 10.1038/nature03547 [DOI] [PubMed] [Google Scholar]
- 26. Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun. 12:399–414. 10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cervantes JL, La Vake CJ, Weinerman B, Luu S, O'Connell C, Verardi PH, Salazar JC. 2013. Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J. Leukoc. Biol. 94:1231–1241. 10.1189/jlb.0413206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petnicki-Ocwieja T, Chung E, Acosta DI, Ramos LT, Shin OS, Ghosh S, Kobzik L, Li X, Hu LT. 2013. TRIF mediates Toll-like receptor 2-dependent inflammatory responses to Borrelia burgdorferi. Infect. Immun. 81:402–410. 10.1128/IAI.00890-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, Liveris D, McClain SA, Wormser GP, Schwartz I. 2002. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis. 186:782–791. 10.1086/343043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang G, Iyer R, Bittker S, Cooper D, Small J, Wormser GP, Schwartz I. 2004. Variations in Barbour-Stoenner-Kelly culture medium modulate infectivity and pathogenicity of Borrelia burgdorferi clinical isolates. Infect. Immun. 72:6702–6706. 10.1128/IAI.72.11.6702-6706.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwartz I, Wormser GP, Schwartz JJ, Cooper D, Weissensee P, Gazumyan A, Zimmermann E, Goldberg NS, Bittker S, Campbell GL, Pavia CS. 1992. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J. Clin. Microbiol. 30:3082–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benov L, Al-Ibraheem J. 2002. Disrupting Escherichia coli: a comparison of methods. J. Biochem. Mol. Biol. 35:428–431. 10.5483/BMBRep.2002.35.4.428 [DOI] [PubMed] [Google Scholar]
- 33. Ranhand JM. 1974. Simple, inexpensive procedure for the disruption of bacteria. Appl. Microbiol. 28:66–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramanan RN, Ling TC, Ariff AB. 2008. The performance of a glass bead shaking technique for the disruption of Escherichia coli cells. Biotechnol. Bioprocess Eng. 13:613–623. 10.1007/s12257-008-0047-y [DOI] [Google Scholar]
- 35. Song DD, Jacques NA. 1997. Cell disruption of Escherichia coli by glass bead stirring for the recovery of recombinant proteins. Anal. Biochem. 248:300–301. 10.1006/abio.1997.2149 [DOI] [PubMed] [Google Scholar]
- 36. Dalpke A, Frank J, Peter M, Heeg K. 2006. Activation of toll-like receptor 9 by DNA from different bacterial species. Infect. Immun. 74:940–946. 10.1128/IAI.74.2.940-946.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sasai M, Linehan MM, Iwasaki A. 2010. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329:1530–1534. 10.1126/science.1187029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer J. 2008. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 180:3729–3738 [DOI] [PubMed] [Google Scholar]
- 39. Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, Beninati C. 2009. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 10:587–594. 10.1038/ni.1733 [DOI] [PubMed] [Google Scholar]
- 40. Kronenwett R, Steidl U, Kirsch M, Sczakiel G, Haas R. 1998. Oligodeoxyribonucleotide uptake in primary human hematopoietic cells is enhanced by cationic lipids and depends on the hematopoietic cell subset. Blood 91:852–862 [PubMed] [Google Scholar]
- 41. Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202:1131–1139. 10.1084/jem.20050914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duramad O, Fearon KL, Chang B, Chan JH, Gregorio J, Coffman RL, Barrat FJ. 2005. Inhibitors of TLR-9 act on multiple cell subsets in mouse and man in vitro and prevent death in vivo from systemic inflammation. J. Immunol. 174:5193–5200 [DOI] [PubMed] [Google Scholar]
- 43. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 44. Yuan JS, Reed A, Chen F, Stewart CN., Jr 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics; 7:85. 10.1186/1471-2105-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wrobel I, Collins D. 1995. Fusion of cationic liposomes with mammalian cells occurs after endocytosis. Biochim. Biophys. Acta 1235:296–304. 10.1016/0005-2736(95)80017-A [DOI] [PubMed] [Google Scholar]
- 46. Litzinger DC, Brown JM, Wala I, Kaufman SA, Van GY, Farrell CL, Collins D. 1996. Fate of cationic liposomes and their complex with oligonucleotide in vivo. Biochim. Biophys. Acta 1281:139–149. 10.1016/0005-2736(95)00268-5 [DOI] [PubMed] [Google Scholar]
- 47. Moore MW, Cruz AR, LaVake CJ, Marzo AL, Eggers CH, Salazar JC, Radolf JD. 2007. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun. 75:2046–2062. 10.1128/IAI.01666-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marre ML, Petnicki-Ocwieja T, DeFrancesco AS, Darcy CT, Hu LT. 2010. Human integrin alpha(3)beta(1) regulates TLR2 recognition of lipopeptides from endosomal compartments. PLoS. One. 5:e12871. 10.1371/journal.pone.0012871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hirschfeld M, Kirschning CJ, Schwandner R, Wesche H, Weis JH, Wooten RM, Weis JJ. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 163:2382–2386 [PubMed] [Google Scholar]
- 50. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 110:3507–3512. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fearnley DB, Whyte LF, Carnoutsos SA, Cook AH, Hart DN. 1999. Monitoring human blood dendritic cell numbers in normal individuals and in stem cell transplantation. Blood 93:728–736 [PubMed] [Google Scholar]
- 52. Takeuchi O. 2012. IRF3: a molecular switch in pathogen responses. Nat. Immunol. 13:634–635. 10.1038/ni.2346 [DOI] [PubMed] [Google Scholar]
- 53. Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin HS, Valentine C, Hellman J. 2007. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J. Immunol. 178:1164–1171 [DOI] [PubMed] [Google Scholar]
- 54. Nakaya T, Sato M, Hata N, Asagiri M, Suemori H, Noguchi S, Tanaka N, Taniguchi T. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150–1156. 10.1006/bbrc.2001.4913 [DOI] [PubMed] [Google Scholar]
- 55. Prantner D, Darville T, Nagarajan UM. 2010. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J. Immunol. 184:2551–2560. 10.4049/jimmunol.0903704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li XD, Knapp S, Kovarik P. 2011. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathog. 7:e1001345. 10.1371/journal.ppat.1001345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gratz N, Siller M, Schaljo B, Pirzada ZA, Gattermeier I, Vojtek I, Kirschning CJ, Wagner H, Akira S, Charpentier E, Kovarik P. 2008. Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J. Biol. Chem. 283:19879–19887. 10.1074/jbc.M802848200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dennis VA, Dixit S, O'Brien SM, Alvarez X, Pahar B, Philipp MT. 2009. Live Borrelia burgdorferi spirochetes elicit inflammatory mediators from human monocytes via the Toll-like receptor signaling pathway. Infect. Immun. 77:1238–1245. 10.1128/IAI.01078-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abraham S, Nagaraj AS, Basak S, Manjunath R. 2010. Japanese encephalitis virus utilizes the canonical pathway to activate NF-kappaB but it utilizes the type I interferon pathway to induce major histocompatibility complex class I expression in mouse embryonic fibroblasts. J. Virol. 84:5485–5493. 10.1128/JVI.02250-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cole LE, Santiago A, Barry E, Kang TJ, Shirey KA, Roberts ZJ, Elkins KL, Cross AS, Vogel SN. 2008. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 180:6885–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loof TG, Goldmann O, Medina E. 2008. Immune recognition of Streptococcus pyogenes by dendritic cells. Infect. Immun. 76:2785–2792. 10.1128/IAI.01680-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Decker T, Muller M, Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687. 10.1038/nri1684 [DOI] [PubMed] [Google Scholar]
- 63. Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu X, Tomai MA, Alkan SS, Vasilakos JP. 2005. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J. Immunol. 174:1259–1268 [DOI] [PubMed] [Google Scholar]
- 64. Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, Rothenfusser S, Endres S, Hartmann G, Hornung V. 2009. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J. Immunol. 182:6824–6833. 10.4049/jimmunol.0803001 [DOI] [PubMed] [Google Scholar]
- 65. Krieg AM, Vollmer J. 2007. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol. Rev. 220:251–269. 10.1111/j.1600-065X.2007.00572.x [DOI] [PubMed] [Google Scholar]
- 66. Larange A, Antonios D, Pallardy M, Kerdine-Romer S. 2009. TLR7 and TLR8 agonists trigger different signaling pathways for human dendritic cell maturation. J. Leukoc. Biol. 85:673–683. 10.1189/jlb.0808504 [DOI] [PubMed] [Google Scholar]
- 67. Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. 2007. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 178:3126–3133 [DOI] [PubMed] [Google Scholar]
- 68. Hess CB, Niesel DW, Klimpel GR. 1989. The induction of interferon production in fibroblasts by invasive bacteria: a comparison of Salmonella and Shigella species. Microb. Pathog. 7:111–120. 10.1016/0882-4010(89)90030-2 [DOI] [PubMed] [Google Scholar]
- 69. Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200:527–533. 10.1084/jem.20040976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, III, Freedman VH, Kaplan G. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. U. S. A. 98:5752–5757. 10.1073/pnas.091096998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Loughman JA, Hunstad DA. 2012. Induction of indoleamine 2,3-dioxygenase by uropathogenic bacteria attenuates innate responses to epithelial infection. J. Infect. Dis. 205:1830–1839. 10.1093/infdis/jis280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Miller JC, Ma Y, Bian J, Sheehan KC, Zachary JF, Weis JH, Schreiber RD, Weis JJ. 2008. A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J. Immunol. 181:8492–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Crandall H, Dunn DM, Ma Y, Wooten RM, Zachary JF, Weis JH, Weiss RB, Weis JJ. 2006. Gene expression profiling reveals unique pathways associated with differential severity of Lyme arthritis. J. Immunol. 177:7930–7942 [DOI] [PubMed] [Google Scholar]
- 74. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68. 10.1038/ni873 [DOI] [PubMed] [Google Scholar]
- 75. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77. 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- 76. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 179:3434–3442 [DOI] [PubMed] [Google Scholar]
- 77. Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, Lewis-Antes A, Amrute SB, Garrigues U, Doyle S, Donnelly RP, Kotenko SV, Fitzgerald-Bocarsly P. 2012. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J. Immunol. 189:2735–2745. 10.4049/jimmunol.1102038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 282:7576–7581. 10.1074/jbc.M608618200 [DOI] [PubMed] [Google Scholar]
- 79. Decker T, Stockinger S, Karaghiosoff M, Muller M, Kovarik P. 2002. IFNs and STATs in innate immunity to microorganisms. J. Clin. Investig. 109:1271–1277. 10.1172/JCI15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669. 10.1093/emboj/17.22.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uze G. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796–805. 10.1002/eji.200324610 [DOI] [PubMed] [Google Scholar]
- 82. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80:4501–4509. 10.1128/JVI.80.9.4501-4509.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, Okada M, Iwakura Y, Aiba S, Yamaya M. 2007. IL-28 elicits antitumor responses against murine fibrosarcoma. J. Immunol. 178:5086–5098 [DOI] [PubMed] [Google Scholar]
- 84. Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M. 2005. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine 31:109–118. 10.1016/j.cyto.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 85. Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. 2007. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 81:7749–7758. 10.1128/JVI.02438-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gallagher G, Megjugorac NJ, Yu RY, Eskdale J, Gallagher GE, Siegel R, Tollar E. 2010. The lambda interferons: guardians of the immune-epithelial interface and the T-helper 2 response. J. Interferon Cytokine Res. 30:603–615. 10.1089/jir.2010.0081 [DOI] [PubMed] [Google Scholar]
- 87. Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, Kotenko SV, Sideras P, Lehr HA, Tepe M, Klucher KM, Doyle SE, Neurath MF, Finotto S, Andreakos E. 2011. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 3:348–361. 10.1002/emmm.201100142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pietila TE, Latvala S, Osterlund P, Julkunen I. 2010. Inhibition of dynamin-dependent endocytosis interferes with type III IFN expression in bacteria-infected human monocyte-derived DCs. J. Leukoc. Biol. 88:665–674. 10.1189/jlb.1009651 [DOI] [PubMed] [Google Scholar]
- 89. Lebreton A, Lakisic G, Job V, Fritsch L, Tham TN, Camejo A, Mattei PJ, Regnault B, Nahori MA, Cabanes D, Gautreau A, Ait-Si-Ali S, Dessen A, Cossart P, Bierne H. 2011. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331:1319–1321. 10.1126/science.1200120 [DOI] [PubMed] [Google Scholar]
- 90. Bierne H, Travier L, Mahlakoiv T, Tailleux L, Subtil A, Lebreton A, Paliwal A, Gicquel B, Staeheli P, Lecuit M, Cossart P. 2012. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One 7:e39080. 10.1371/journal.pone.0039080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cohen TS, Prince AS. 2013. Bacterial pathogens activate a common inflammatory pathway through IFNlambda regulation of PDCD4. PLoS Pathog. 9:e1003682. 10.1371/journal.ppat.1003682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Krupna M, Wormser G, Schwartz I, Petzke M. 2011. Borrelia burgdorferi pathogenic potential correlates with type I and type III IFN production by human immune cells. J. Immunol. 186:56.18 [Google Scholar]