Abstract
During host colonization, Campylobacter jejuni is exposed to harmful reactive oxygen species (ROS) produced from the host immune system and from the gut microbiota. Consequently, identification and characterization of oxidative stress defenses are important for understanding how C. jejuni survives ROS stress during colonization of the gastrointestinal tract. Previous transcriptomic studies have defined the genes belonging to oxidant stimulons within C. jejuni. We have constructed isogenic deletion mutants of these identified genes to assess their role in oxidative stress survival. Phenotypic screening of 109 isogenic deletion mutants identified 22 genes which were either hypersensitive or hyposensitive to oxidants, demonstrating important roles for these genes in oxidant defense. The significance of these genes in host colonization was also assessed in an in vivo chick model of C. jejuni colonization. Overall, our findings identify an indirect role for motility in resistance to oxidative stress. We found that a nonmotile flagellum mutant, the ΔmotAB mutant, displayed increased sensitivity to oxidants. Restoration of sensitivity to superoxide in the ΔmotAB mutant was achieved by fumarate supplementation or tandem deletion of motAB with ccoQ, suggesting that disruption of the proton gradient across the inner membrane resulted in increased superoxide production in this strain. Furthermore, we have identified genes involved in cation transport and binding, detoxification, and energy metabolism that are also important factors in oxidant defense. This report describes the first isogenic deletion mutant library construction for screening of relevant oxidative stress defense genes within C. jejuni, thus providing a comprehensive analysis of the total set of oxidative stress defenses.
INTRODUCTION
Campylobacter jejuni is a Gram-negative, microaerophilic, human pathogen (1) that is the second most reported cause of food-borne bacterial gastroenteritis in the United States (2) and results in 400 million to 500 million cases of infection worldwide per year (2, 3). Illness caused by C. jejuni typically results in symptoms such as watery or bloody diarrhea, fever, nausea, and abdominal pain (4). Furthermore, C. jejuni infection has also been linked with the development of a rare but serious neuromuscular disorder known as Guillain Barré syndrome (5). As a microaerophilic bacterium, C. jejuni requires low levels of molecular oxygen for proper growth due to its dependence on an oxygen-dependent ribonucleotide reductase (6). However, this dependence on the presence of oxygen for growth inevitably results in the exposure of important biological molecules, such as DNA, proteins, and lipids, to reactive oxygen species (ROS). These ROS originate from several different sources, both within C. jejuni and from its environment. Superoxide radicals (O2·−) and hydrogen peroxide (H2O2) are produced within C. jejuni during normal respiration as a consequence of molecular oxygen nonspecifically oxidizing respiratory chain dehydrogenases (7). In addition, oxidation of cellular ferrous ions by H2O2 results in the production of the particularly powerful oxidizing species hydroxyl radicals (·OH) (8). ROS are also produced by neutrophils, which are recruited to the gut in large numbers as part of the immune response and which catalyze the formation of O2·− as a strategy for killing pathogenic bacteria (9). Finally, the gut microbiota, in particular lactic acid bacteria, also produces exogenous H2O2 in an attempt to eliminate bacteria competing to colonize the same niche (8, 10). Consequently, C. jejuni contains numerous ROS detoxification pathways to survive both endogenously and exogenously produced ROS and colonize its host. The importance of these cellular defenses for C. jejuni survival against ROS has been demonstrated by characterizing ROS detoxification enzymes such as KatA, SodB, AhpC, Tpx, and Bcp (11–14). In addition, these oxidative stress defense enzymes play an important role in host colonization and pathogenesis. Recent work has highlighted this role by demonstrating that in the neonate piglet infectious model a ΔkatA mutant was outcompeted by the wild-type C. jejuni strain (15). Clearly, oxidative stress defenses play an important role in C. jejuni pathogenesis.
In order to identify unforeseen players in ROS defense in C. jejuni, our laboratory previously used genome-wide transcriptome analysis to characterize the oxidant stimulons of C. jejuni. Specifically, our work defined C. jejuni's transcriptomic response to 1 mM H2O2, 1 mM cumene hydroperoxide (CHP), or 1 mM menadione sodium bisulfite (MND) exposure (16). Furthermore, we also characterized the transcriptomic responses in a ΔperR mutant background to identify potential novel oxidative stress defense genes regulated by the PerR peroxide-sensing regulator (16, 17). In this study, we describe the construction of a library of isogenic deletion mutants with mutations in the genes identified by our microarray analysis and their subsequent phenotypic characterization. A total of 109 isogenic deletion mutants were constructed, followed by in vitro phenotypic analysis of oxidant sensitivity and in vivo characterization of selected mutants using chick colonization assays. We have identified 22 mutants that were either hypersensitive or hyposensitive to H2O2, cumene hydroperoxide, and/or menadione sodium bisulfite and thus have revealed important roles for these genes in oxidative stress defense in C. jejuni. The identified genes function in processes such as detoxification, cation transport and binding, energy metabolism, and phosphate transport. We also identified an indirect role for bacterial motility in protecting C. jejuni against oxidative stress. The relevance of the oxidative stress defense mutants in chick colonization was also assessed and revealed important genes required for successful colonization of the chick cecum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α and K-12 strains were cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar plates. LB broth and plates were supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and/or 10 μg/ml chloramphenicol as required. Campylobacter jejuni NCTC11168 was grown under microaerophilic conditions (83% N2, 4%H2, 8% O2, and 5% CO2) at 37°C in a MACS-VA500 workstation (Don Whitley, West Yorkshire, England). C. jejuni was cultured in Mueller-Hinton (MH) broth in biphasic flasks or on MH agar plates. Campylobacter strains containing antibiotic resistance cassettes were grown on MH agar plates supplemented with 10 μg/ml kanamycin and/or 20 μg/ml chloramphenicol as required. The plasmids and bacterial strains used in this study are listed in Table S1 in the supplemental material.
Construction of isogenic deletion mutants.
Construction of isogenic deletion mutants was done using the In-fusion Dry-down PCR cloning kit (Clontech). Briefly, target genes plus flanking regions were amplified using Taq polymerase (Invitrogen) with the corresponding gene primers (Invitrogen) listed in Table S2 in the supplemental material. The In-fusion Dry-down cloning kit was used to directionally clone the amplified gene products into BamHI (Invitrogen)-digested pUC19. Subsequently, inverse PCR was used to amplify pUC19 plus the flanking end regions of the target gene. A chloramphenicol or kanamycin antibiotic resistance cassette was directionally cloned into the inverse PCR product, disrupting the target gene. The final construct was sequenced to confirm the absence of point mutations and then naturally transformed into C. jejuni NCTC11168. Clones were selected for on chloramphenicol- or kanamycin-supplemented MH agar plates, and positive colonies were confirmed by PCR.
Double-deletion mutants were constructed by growing mutant strains on MH agar plates supplemented with the appropriate antibiotic for 3 days under microaerophilic conditions at 37°C. The mutant strains were then cultured overnight in biphasic flasks, spotted onto MH agar plates, and allowed to grow overnight. DNA extracted from strains containing the desired secondary gene deletion was naturally transformed into the C. jejuni mutant strains grown on the MH agar plates. Clones were selected for on chloramphenicol- and kanamycin-MH agar plates, and PCR was used to confirm the presence of both mutated genes.
Construction of complemented strains.
Complemented C. jejuni NCTC11168 mutant strains were constructed as described previously (18). Target genes were amplified from extracted C. jejuni genomic DNA using the high-fidelity polymerases Pfx (Invitrogen), Pwo (Roche), or Phusion (Finnzymes). The amplified gene products were subsequently directionally cloned into XhoI (Invitrogen)-digested pRRK (18) using the In-fusion Dry-down cloning kit. The plasmids were sequenced to confirm the absence of mutations in the target genes. The corresponding C. jejuni mutant strains were then naturally transformed with the final construct, and successful transformants were selected for on MH agar plates supplemented with chloramphenicol and kanamycin. Positive colonies were confirmed by PCR using the kanamycin cassette-specific primer AR56 and ribosomal region-specific primers ak233-ak235 (see Table S2 in the supplemental material).
Disc inhibition and motility assays.
C. jejuni NCTC11168 wild-type, mutant, and complemented strains were grown for 3 days under microaerophilic conditions on MH agar plates supplemented with chloramphenicol and/or kanamycin as required. Strains were then cultured in biphasic flasks overnight and subsequently diluted to an optical density at 600 nm (OD600) of 1 in MH broth. For each strain, 100 ml of MH agar (cooled to approximately 45°C and supplemented with 20 mM sodium fumarate when required) was prepared, followed by the addition of 4 ml of the culture at an OD600 of 1.0. The C. jejuni-MH agar mixture was then poured in equal volumes into three petri dishes and allowed to solidify. Paper discs (6-mm diameter) were placed upon the surface of the agar, followed by the addition of 10 μl 3% H2O2, 3% cumene hydroperoxide (CHP), and 90 mM menadione sodium bisulfite (MND) to each paper disc. Next, the MH agar plates were incubated under microaerophilic conditions, and the diameter of growth inhibition (mm) was measured after 28 h. Each mutant and complemented strain was tested in at least biological triplicate. The averages of the clear zones were used to determine if statistically significant differences existed between the mutant, complemented, and wild-type strains, using Bayesian statistical analysis. P values of <0.001 were considered statistically significant.
The motilities of all strains were assayed on 0.4% MH agar plates. Plates were incubated for 28 h under microaerophilic conditions at 37°C, followed by measurement of the diameter of motility (mm). Motility assays were performed in at least biological triplicate, and statistical significance was determined using Bayesian statistical analysis (a P value of <0.001 was considered significant).
Chick colonization model.
The chick colonization model for C. jejuni was employed as described previously (19). Briefly, 1-day-old, specific-pathogen-free layer chicks, raised at Ottawa Laboratory (Fallowfield) (OLF), Canadian Food Inspection Agency (CFIA), were housed in groups of 10 in temperature-controlled isolators (32 to 34°C) and provided with clean water and a commercial, custom-made chicken crumbles feed (Ritchie Feed and Seed, Ottawa, Ontario, Canada). C. jejuni strains were grown on MH agar plates for 3 days under microaerophilic conditions at 37°C. Several colonies from each plate were then selected and transferred to 0.4% MH agar motility plates and allowed to grow for an additional 48 h. The most motile C. jejuni organisms from the motility plates were then subcultured in biphasic flasks and grown overnight. The cultures were then centrifuged, resuspended in fresh MH broth, and diluted to approximately 105 CFU/ml. Each chick was orally inoculated with 0.5 ml of the prepared culture. To confirm that the chicks received approximately the same number of viable C. jejuni organisms for each strain tested, the inoculums were serially diluted and plated onto MH agar plates. The plates were incubated for 1 to 2 days under microaerophilic conditions before enumeration. The chicks were euthanized at 7 days postinoculation, and the ceca were collected and individually weighed. The cecal contents were homogenized, serially diluted, and plated onto selective Karmali agar (Oxoid) supplemented with chloramphenicol and/or kanamycin as required. The Karmali plates were incubated for 2 days under microaerophilic conditions at 42°C, and the resulting colonies were counted. Colonization levels of wild-type, mutant, and complemented strains are expressed as CFU/g cecal content, and statistical significance was analyzed using a nonparametric Mann-Whitney rank sum test. Strains were considered significantly different from the wild-type at a P value of <0.05. Chicks were used in accordance with regulations outlined by the Canadian Council on Animal Care, and experimental procedures were approved by the animal care committee at OLF, CFIA.
RESULTS
Selection of genes encoding proteins potentially involved in oxidative stress defense and production of a library of isogenic deletion mutants.
The major objective of this study was to systemically identify genes encoding proteins involved in oxidative stress defense in C. jejuni. These genes are commonly induced by oxidants. Based on this premise, we mined the transcriptome data of C. jejuni exposed to menadione (a superoxide generating agent), cumene hydroperoxide (an organic hydroperoxide), or H2O2 (an inorganic peroxide). This analysis led to the selection of 57 gene candidates responsive to one or more of the oxidants tested (Bayesian P value of <10−4) (16). In C. jejuni, the transcriptional regulator PerR is known to repress genes involved in oxidative stress defense (17). In Bacillus subtilis, PerR senses peroxide by Fe2+-catalyzed oxidation of its regulatory binding site (consisting of 2 histidines) leading to gene derepression (20). We previously characterized the PerR regulon by microarray analysis and identified 104 PerR-regulated genes, with 82 of them being PerR repressed (Bayesian P value of <10−4) (16). Based on these transcriptomic analyses, we selected a total of 127 genes as potential candidates for genes of the oxidative stress defense system (see Table S3 in the supplemental material). These genes were upregulated in response to at least one of the three oxidants tested and/or were PerR repressed (fold change of >1.5 with a P value of below 10−4). Genes that were slightly below the cutoff Bayesian P value of 10−4 but encoded proteins of functional interest were also included in the final selection (for a total of 145 targeted genes; see Table S3 in the supplemental material).
Next, the selected genes were targeted for isogenic mutant construction to characterize their role in oxidative stress defense. The mutants were constructed by allelic exchange to introduce a chloramphenicol resistance cassette to disrupt the gene of interest (as described in Materials and Methods). Using this approach, we successfully constructed 109 isogenic deletion mutants (Table 1) from the total 145 gene candidates (see Table S3 in the supplemental material). The construction of the remaining 36 mutants failed after multiple attempts, suggesting an essential role of the targeted genes and/or poor recombinogenic potential. Indeed, of the 36 mutants that were not obtained, 7 of the genes have been identified as essential in C. jejuni, with an additional 10 located next to essential genes (21).
TABLE 1.
C. jejuni isogenic deletion mutant library gene names, functional categories, and annotations
| Functional category and gene name | Annotated gene function |
|---|---|
| Detoxification | |
| cj0020c | Cytochrome C551 peroxidase |
| cj0358 | Putative cytochrome C551 peroxidase |
| rrc | Nonheme iron protein |
| Cation transport/binding | |
| ceuB | Enterobactin uptake permease |
| ceuE | Enterobactin uptake periplasmic binding protein |
| cfbpA | Putative iron uptake ABC transport system, periplasmic iron-binding protein |
| cfbpB | Putative iron uptake ABC transport system permease protein |
| cfbpC | Putative iron uptake ABC transport system ATP-binding protein |
| cfrA | Ferric enterobactin uptake receptor |
| chaN | Putative iron transport protein |
| chuA | Hemin uptake system outer membrane receptor |
| chuB | Putative hemin uptake system permease protein |
| chuC | Putative hemin uptake system ATP-binding protein |
| chuD | Putative hemin uptake system periplasmic hemin-binding protein |
| chuZ | Heme oxygenase |
| cj0045c | Putative iron-binding protein |
| cj0178 | Putative TonB-dependent outer membrane receptor |
| cj1658 | Putative iron permease |
| cj1661 | Possible ABC transport system permease protein |
| cj1663 | Putative ABC transport system ATP-binding protein |
| exbB1 | Biopolymer transport protein |
| exbB2 | Putative ExbB/TolQ family transport protein |
| exbD1 | Biopolymer transport protein |
| exbD2 | Putative ExbD/TolR family transport protein |
| p19 | Periplasmic protein p19 |
| p19 + cj1658 | Periplasmic protein p19/putative iron permease |
| tonB1 | Possible TonB transport protein |
| tonB1+tonB2 | Possible TonB transport protein/putative TonB transport protein |
| tonB2 | Putative tonB transport protein |
| tonB2+tonB3 | Putative tonB transport protein/tonB transport protein |
| tonB3 | TonB transport protein |
| Energy metabolism | |
| acnB | Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase |
| ald | Putative aldehyde dehydrogenase |
| ccoQ | cb-type cytochrome c oxidase subunit IV |
| cj0073c | Nonflavin iron-sulfur-containing oxidoreductase complex subunit |
| cj1207c | Putative lipoprotein thioredoxin |
| cj1377c | Putative ferredoxin |
| hypC | Hydrogenase isoenzyme formation protein |
| Surface structures | |
| flaG | Flagellar protein |
| flgD | Flagellar basal body rod modification protein |
| flgE | Flagellar hook protein |
| flgE2 | Flagellar hook protein |
| flgG | Flagellar basal body rod protein |
| flgG2 | Flagellar basal body rod protein |
| flgH | Flagellar basal body L-ring protein |
| flgI | Flagellar basal body P-ring protein |
| flgK | Flagellar hook-associated protein |
| flgL | Flagellar hook-associated protein |
| flgM | Anti-FliA (sigma 28) factor |
| flgP | Putative lipoprotein |
| flgR | Sigma 54-associated transcriptional activator |
| flhB | Flagellar biosynthesis protein |
| fliK | Putative flagellar hook length control protein |
| maf4 | Motility accessory factor |
| maf6 | Motility accessory factor |
| maf7 | Motility accessory factor |
| motAB | Flagellar motor proteins |
| pseB | UDP-GlcNAc-specific C4,6 dehydratase/C5 epimerase |
| Drug efflux | |
| cj0309c | Putative efflux protein |
| cmeA | Periplasmic fusion protein CmeA (multidrug efflux system CmeABC) |
| Membranes, lipoproteins, and porins | |
| cj0385c | Putative integral membrane protein |
| cj0587 | Putative integral membrane protein |
| cj0818 | Putative lipoprotein |
| cj1211 | Putative competence family protein |
| cj1356c | Putative integral membrane protein |
| cj1484c | Hypothetical protein |
| cj0062c | Putative integral membrane protein |
| Miscellaneous | |
| acs | Acetyl coenzyme A synthetase |
| cj0295 | Putative acetyltransferase |
| cj0494 | Putative exporting protein |
| cj0561c | Putative periplasmic protein |
| cj0672 | Putative periplasmic protein |
| cj0947c | Putative carbon-nitrogen hydrolase |
| cj0949c | Putative peptidyl-arginine deiminase family protein |
| cj1036c | Conserved hypothetical protein |
| cj1167 | Putative amino acid metabolism protein |
| cj1209 | Phosphodiesterase |
| cj1241 | Putative major facilitator superfamily transporter protein |
| cj1255 | Putative isomerase |
| cj1340c | Conserved hypothetical protein (1318 family) |
| cj1388 | Putative endoribonuclease L-PSP |
| cj1406c | Putative periplasmic protein |
| cj1623 | Hypothetical protein |
| dprA | DNA-processing protein A |
| folP | Dihydropteroate synthase |
| pstC | Putative phosphate transport system permease protein |
| spoT | Putative guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase |
| trpF | N-(5′-Phosphoribosyl)anthranilate isomerase |
| truB | tRNA pseudouridine synthase B |
| Unknown function | |
| cj0040 | Hypothetical protein |
| cj0044c | Hypothetical protein |
| cj0148c | Hypothetical protein |
| cj0171 | Merged with Cj0170 |
| cj0202c | Hypothetical protein |
| cj0253 | Hypothetical protein |
| cj0260c | Small hydrophobic protein |
| cj0344 | Hypothetical protein |
| cj0416 | Hypothetical protein |
| cj0524 | Hypothetical protein |
| cj0554 | Hypothetical protein |
| cj0741 | Hypothetical protein |
| cj0786 | Small hydrophobic protein |
| cj0814 | Hypothetical protein |
| cj0819 | Hypothetical protein |
| cj0900c | Hypothetical protein |
| cj0977 | Hypothetical protein |
| cj1159c | Small hydrophobic protein |
| cj1242 | Hypothetical protein |
| cj1383c | Hypothetical protein |
| mdaB | MdaB protein homolog |
Phenotypic analysis of the mutant library identifies C. jejuni protective mechanisms against oxidants.
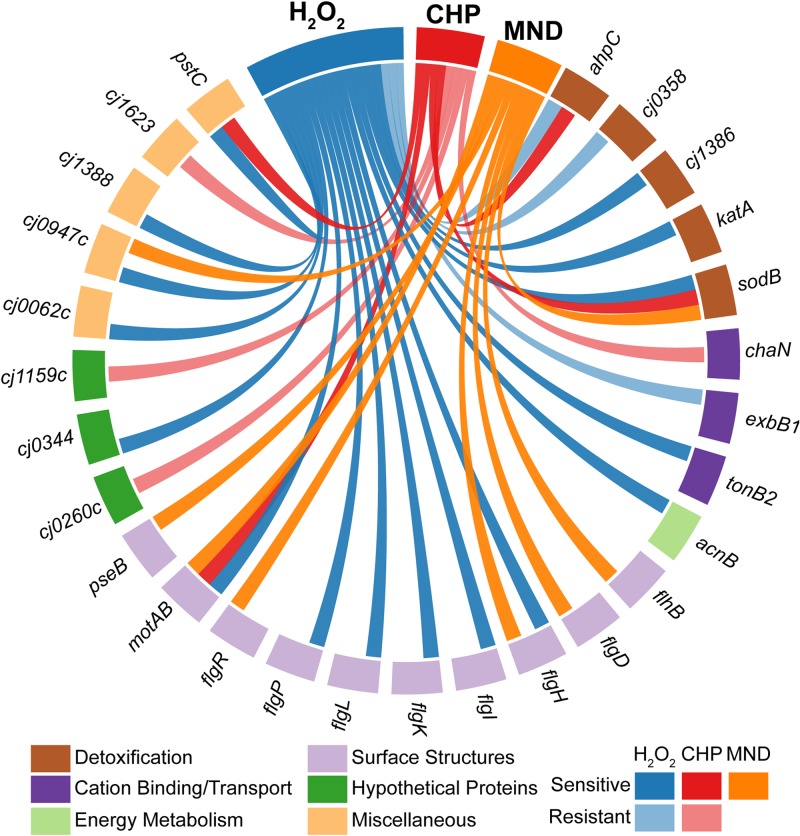
To identify protective mechanisms against oxidants, the entire mutant library was tested for hypersensitivity or resistance to exposure to H2O2, cumene hydroperoxide, and/or menadione bisulfite by disk inhibition assays (Fig. 1; see Table S4 in the supplemental material). Of the 109 mutants tested, 22 were either hypersensitive or more resistant to one or more of the oxidants (Fig. 1; Table 2), with values considered significant at a P value of <0.001 using Bayesian statistical analysis. Of the 22 mutants that had a phenotype, 16 displayed increased sensitivity to one or more of the oxidants. Nine mutants were specifically hypersensitive to H2O2 (ΔflgP, Δcj0062c, Δcj0344, ΔflgI, ΔflgK, ΔflgL, Δcj1388, ΔacnB, and ΔtonB2), 4 were specifically hypersensitive to menadione sodium bisulfite (ΔflgR, ΔflhB, ΔflgD, and ΔpseB), 2 were hypersensitive to both H2O2 and menadione sodium bisulfite (ΔflgH and Δcj0947c), and 1 was hypersensitive to both H2O2 and cumene hydroperoxide (ΔpstC). The remaining 6 mutants displayed increased resistance to the oxidants tested relative to the wild-type C. jejuni. Of these 6 mutants, 2 were specifically more resistant to H2O2 (Δcj0358 and ΔexbB1), and 4 were specifically more resistant to cumene hydroperoxide (Δcj1623, Δcj1159c, ΔchaN, and Δcj0260c).
FIG 1.
Sensitivity of isogenic deletion mutants and complemented strains to H2O2, cumene hydroperoxide, or menadione sodium bisulfite. Mutant and complemented strains are represented along the circle. Lines connecting strains toward 3% H2O2, 3% cumene hydroperoxide (CHP), or 90 mM menadione bisulfite (MND) represent the sensitivity of each strain to the three oxidants assayed relative to wild-type C. jejuni as determined by disc inhibition analysis. Dark and light lines represent hypersensitivity and hyposensitivity to the oxidants, respectively. Each experiment was repeated in quadruplicate. Results for mutants compared to wild-type C. jejuni were considered statistically significant at a P value of <0.001 using Bayesian statistical analysis. Strains are grouped according to functional category.
TABLE 2.
Sensitivities of wild-type C. jejuni, isogenic deletion mutants, and corresponding complemented strains to H2O2, cumene hydroperoxide, or menadione bisulfite
| Functional category and strain | Diam (mm) of zone of inhibition with oxidanta |
||
|---|---|---|---|
| H2O2 | CHP | MND | |
| C. jejuni NCTC11168 | 19.08 ± 0.19 | 24.50 ± 0.22 | 31.63 ± 0.36 |
| Detoxification | |||
| Δcj0358 | 16.44 ± 0.72* | 23.50 ± 0.53 | 29.63 ± 0.80 |
| Δcj0358+cj0358 | 17.40 ± 0.40 | 22.50 ± 0.52 | 31.93 ± 1.13 |
| ΔkatAb | 28.85 ± 0.87* | 26.43 ± 0.47 | 29.85 ± 1.39 |
| ΔkatA+katAb | 10.83 ± 1.24* | 23.83 ± 0.17 | 31.00 ± 1.20 |
| ΔahpCb | 17.23 ± 0.50* | 34.87 ± 0.79* | 29.05 ± 1.81 |
| ΔahpC+ahpCb | 18.45 ± 0.10 | 25.40 ± 0.34 | 28.38 ± 0.39 |
| ΔsodBb | 22.10 ± 0.35* | 30.75 ± 3.53* | 38.28 ± 1.96* |
| ΔsodB+sodBb | 21.78 ± 0.33 | 25.40 ± 0.17 | 37.50 ± 2.62 |
| Δcj1386c | 25.21 ± 0.28* | 25.67 ± 0.76 | 35.04 ± 1.36 |
| Δcj1386+cj1386c | 19.90 ± 0.57 | 24.20 ± 0.51 | 37.30 ± 0.76 |
| Cation transport/binding | |||
| ΔchaN | 17.37 ± 0.30 | 21.47 ± 0.17* | 29.83 ± 0.61 |
| ΔchaN+chaN | 17.26 ± 0.75 | 22.82 ± 0.66 | 34.08 ± 1.97 |
| ΔexbB1 | 16.90 ± 0.31* | 23.13 ± 0.64 | 28.30 ± 0.58 |
| ΔexbB1+exbB1 | 17.33 ± 0.60 | 22.63 ± 0.37 | 32.90 ± 1.16 |
| ΔtonB2 | 21.10 ± 0.46* | 26.47 ± 0.67 | 33.07 ± 1.39 |
| Energy metabolism | |||
| ΔacnB | 21.22 ± 0.45* | 23.28 ± 0.24 | 36.28 ± 0.72 |
| ΔacnB+acnB | 20.27 ± 0.25 | 21.33 ± 0.44 | 33.44 ± 1.49 |
| Surface structures | |||
| ΔflhB | 19.80 ± 0.44 | 23.18 ± 0.32 | 36.53 ± 1.86* |
| ΔflhB+flhB | 18.18 ± 0.76 | 20.86 ± 0.82 | 32.15 ± 0.99 |
| ΔflgD | 20.47 ± 0.47 | 26.27 ± 0.36 | 36.50 ± 0.31* |
| ΔflgD+flgD | 18.40 ± 0.31 | 23.60 ± 0.80 | 31.70 ± 0.58 |
| ΔflgH | 21.00 ± 0.42* | 26.55 ± 0.55 | 37.00 ± 1.30* |
| ΔflgH+flgH | 19.07 ± 0.37 | 23.37 ± 0.23 | 32.33 ± 1.19 |
| ΔflgI | 21.35 ± 0.43* | 26.00 ± 0.30 | 35.38 ± 1.89 |
| ΔflgI+flgI | 18.95 ± 0.34 | 23.95 ± 0.21 | 31.70 ± 1.13 |
| ΔflgK | 21.53 ± 0.13* | 26.13 ± 0.46 | 34.90 ± 2.04 |
| ΔflgK+flgK | 19.67 ± 0.17 | 24.73 ± 0.15 | 31.10 ± 0.49 |
| ΔflgL | 21.00 ± 0.23* | 25.08 ± 0.32 | 34.67 ± 1.73 |
| ΔflgL+flgL | 18.33 ± 1.17 | 21.25 ± 0.12 | 30.67 ± 0.10 |
| ΔpseB | 20.50 ± 0.32 | 25.92 ± 0.53 | 35.95 ± 1.40* |
| ΔpseB+pseB | 18.95 ± 1.13 | 23.67 ± 1.20 | 26.94 ± 2.96 |
| ΔflgP | 21.63 ± 1.24* | 26.25 ± 0.86 | 35.25 ± 2.21 |
| ΔflgP+flgP | 19.94 ± 0.59 | 25.74 ± 0.62 | 33.08 ± 1.26 |
| ΔflgR | 20.47 ± 0.75 | 25.63 ± 0.34 | 36.78 ± 1.35* |
| ΔflgR+flgR | 17.90 ± 0.61 | 22.17 ± 0.35 | 32.83 ± 0.87 |
| Hypothetical unknown proteins | |||
| Δcj0260c | 17.42 ± 0.19 | 21.67 ± 0.44* | 30.87 ± 0.75 |
| Δcj0260c+cj0260c | 17.50 ± 0.35 | 22.83 ± 0.57 | 30.85 ± 0.35 |
| Δcj0344 | 20.92 ± 0.14* | 23.92 ± 0.45 | 33.33 ± 1.01 |
| Δcj0344+cj0344 | 18.12 ± 0.73 | 21.25 ± 1.24 | 30.36 ± 1.33 |
| Δcj1159c | 19.71 ± 0.90 | 22.25 ± 0.40* | 30.42 ± 0.72 |
| Miscellaneous | |||
| Δcj0062c | 21.73 ± 0.21* | 26.00 ± 0.39 | 35.67 ± 0.33 |
| Δcj0062c+cj0062c | 19.44 ± 0.39 | 25.20 ± 0.50 | 33.63 ± 1.80 |
| Δcj0947c | 23.94 ± 1.20* | 26.44 ± 0.80 | 43.00 ± 3.76* |
| Δcj0947c+cj0947c | 20.66 ± 1.50 | 25.55 ± 0.50 | 32.33 ± 0.19 |
| Δcj1388 | 21.17 ± 0.41* | 25.38 ± 0.61 | 35.33 ± 2.20 |
| Δcj1623 | 18.33 ± 0.50 | 21.95 ± 0.19* | 34.78 ± 2.50 |
| Δcj1623+cj1623 | 17.44 ± 0.49 | 22.70 ± 0.39 | 32.63 ± 1.44 |
| ΔpstC | 20.75 ± 0.31* | 27.25 ± 1.00* | 32.33 ± 1.24 |
| ΔpstC+pstC | 20.60 ± 0.25 | 25.30 ± 0.41 | 36.84 ± 1.79 |
The diameter of the zone of inhibition is represented as the mean clear zone ± standard error for each strain after exposure to 10 μl of 3% H2O2, 3% cumene hydroperoxide (CHP), or 90 mM menadione bisulfite (MND). Each experiment was repeated in quadruplicate. Values were considered significant (*) at a P value of <0.001 using Bayesian statistical analysis.
Data are from reference 16.
Data are from reference 15.
These 22 genes group into a wide range of functional categories, including several that are not often identified as having an important role in oxidative stress defense. The categories include detoxification (Δcj0358), cation transport/binding proteins (ΔchaN, ΔexbB1, and ΔtonB2), energy metabolism (ΔacnB), surface structures (ΔflhB, ΔflgD, ΔflgH, ΔflgI, ΔflgK, ΔflgL, and ΔpseB), membranes, lipoproteins, and porins (Δcj1623 and ΔflgP), hypothetical proteins (Δcj0062c, Δcj0260c, Δcj0344, Δcj1388, and Δcj1159c), and miscellaneous functions (ΔpstC, ΔflgR, and Δcj0947c).
To confirm the observed phenotypes and rule out the possibility of secondary mutations or polar effects on neighboring genes, complemented strains were constructed for 19 of 22 of the mutant strains as described previously (18) (see Materials and Methods). Complementation of the mutants either partially or completely restored the wild-type phenotype for oxidant resistance (Table 2), indicating that the specific genes mutated were responsible for the observed phenotypic differences. The partial but statistically significant restoration of the phenotype may be due to the differences in the level of gene expression between the complemented strain and wild-type C. jejuni (as gene transcription is driven from the kanamycin resistance cassette promoter in the complemented strains). Of the 109 mutant strains, 87 mutants showed no detectable phenotype toward oxidants (see Table S4 in the supplemental material), suggesting that these genes are not functionally important for cell protection against oxidative stress under the assay conditions.
In vivo chick colonization assays of oxidant-sensitive mutants reveal genes with important roles in colonization of chick ceca.
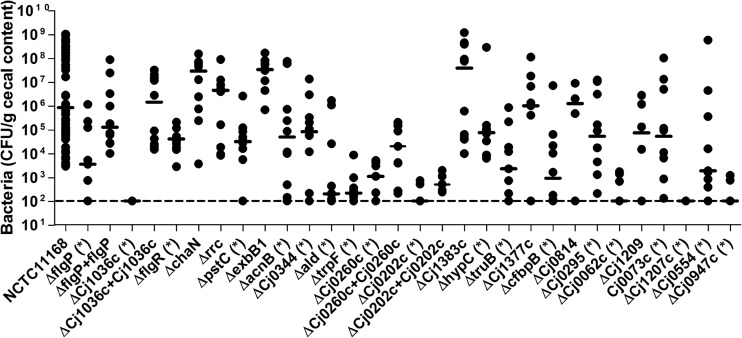
To investigate the biological significance of important oxidative stress defense genes in an in vivo setting, chick colonization assays were performed using the constructed isogenic deletion mutants. Mutants that were identified as being hypersensitive or more resistant to oxidants using the disc inhibition assay were prioritized for testing in the chick colonization assay. Mutants were also selected based upon their functional category (flagellar mutants with sensitivity to oxidants were not tested due to the motility defects associated with these strains). A total of 30 mutant and complemented strains were tested for their colonization ability. Of the 26 mutant strains, 19 (ΔflgP, ΔflgR, ΔpstC, Δcj1036c, ΔacnB, Δcj0344, Δald, ΔtrpF, Δcj0260c, Δcj0202c, ΔcfbpB, Δcj0062c, Δcj0295, Δcj0947c, Δcj0073c, ΔhypC, ΔtruB, Δcj1207c, and Δcj0554) had a significant reduction in their ability to colonize chick ceca relative to the wild-type C. jejuni strain, as shown in Fig. 2. The remaining 7 strains (Δcj1383c, ΔchaN, Δrrc, ΔexbB1, Δcj1209, Δcj1377c, and Δcj0814) were not significantly affected in their colonization levels relative to wild-type C. jejuni. Four complemented strains were tested (ΔflgP+flgP, Δcj1036c+cj1036c, Δcj0260c+cj0260c, and Δcj0202c+cj0202c), with the ΔflgP+flgP, Δcj1036c+cj1036c, and Δcj0260c+cj0260c complements showing statistically significant restoration of the phenotype compared to their respective mutants (Fig. 2), confirming the observed in vivo phenotype. Importantly, the mutant strains assayed in the chicks did not display any in vitro growth defects, except for the flgR and cj0947c mutants (see Fig. S1 in the supplemental material). Thus, it cannot be ruled out that the attenuated colonization observed in these strains is due to general growth defects in addition to increased susceptibility to oxidants.
FIG 2.
C. jejuni wild-type, mutant, and complemented strain colonization levels in the chick cecum. Each data point represents the CFU per gram of cecal content recovered for each strain tested. Bars represent the median colonization of each strain, and the dashed line indicates the detection limit of the assay. An asterisk denotes statistical significance (P < 0.05) using a nonparametric Mann-Whitney rank sum test.
Fumarate supplementation restores the menadione sensitivity of flagellar biogenesis mutants to wild-type levels.
Of the 22 oxidant-sensitive mutants identified, 10 mutants, constructed with mutations in genes encoding components of the flagellar apparatus (FlhB, FlgD, FlgI, FlgK, FlgL, FlgH, and FlgP), proteins involved in flagellar modification (PseB and FlgP), and the flagellar regulatory protein FlgR, exhibited significant hypersensitivity to one or more of the three oxidants tested. Importantly, the complemented strains for these mutants displayed sensitivity to oxidants to levels comparable to those of the wild-type parental C. jejuni strain (Table 2). Although several flagellum gene mutants were not affected in oxidative stress resistance (flgG, flgG2, flgE, flgE2, flaG, fliK, flgM, maf4, maf6, and maf7), the present results indicate an important role for the flagella in oxidant sensitivity within C. jejuni. Interestingly, all the flagellum mutants that were sensitive to oxidants also displayed reduced motility (Table 3). Conversely, the flagellum mutants that remained motile (flgG, flgG2, flgE, flgE2, flaG, fliK, flgM, maf4, maf6, and maf7) showed no defect toward oxidant sensitivity (see Table S4 in the supplemental material). Moreover, mutation of the flgP gene, which encodes a lipoprotein required for motility but not flagellum biogenesis (22), resulted in significant hypersensitivity toward oxidants. These observations suggest that it is bacterial motility as opposed to the flagellum apparatus itself that is key for oxidant resistance.
TABLE 3.
Motilities of wild-type C. jejuni, flagellum gene mutants, and corresponding complemented strains
| Strain | Motility (mm)a |
|---|---|
| NCTC11168 | 33.1 ± 0.4 |
| ΔflhB | 8.0 ± 1.2* |
| ΔflhB+flhB | 31.3 ± 3.6 |
| ΔflgD | 8.0 ± 0.0* |
| ΔflgD+flgD | 33.2 ± 1.2 |
| ΔflgE | 48.4 ± 3.9* |
| ΔflgE+flgE | 43.5 ± 3.3 |
| ΔflgH | 18.3 ± 2.2* |
| ΔflgH+flgH | 28.8 ± 1.5 |
| ΔflgI | 24.0 ± 3.4* |
| ΔflgI+flgI | 38.3 ± 2.7 |
| ΔflgK | 11.2 ± 4.2* |
| ΔflgK+flgK | 11.0 ± 0.6* |
| ΔflgL | 10.5 ± 6.4* |
| ΔpseB | 6.3 ± 0.4* |
| ΔflgG | 34.8 ± 12.4 |
| ΔflgG2 | 28.4 ± 7.5 |
| ΔflgP | 6.0 ± 0.0* |
| ΔflgP+flgP | 10.0 ± 0.8 |
| ΔflgR | 17.1 ± 4.5* |
| ΔflgR+flgR | 29.8 ± 0.2 |
| ΔflgM | 31.5 ± 0.5 |
| ΔflgM+flgM | 29.0 ± 0.8 |
| ΔfliK | 27.2 ± 1.2 |
| ΔfliK+fliK | 34.0 ± 2.8 |
| ΔflaG | 27.8 ± 7.4 |
| ΔflgE2 | 50.3 ± 2.4* |
| Δmaf4 | 41.5 ± 3.5* |
| Δmaf6 | 29.3 ± 1.3 |
| Δmaf7 | 33.5 ± 2.0 |
| ΔmotAB | 6.0 ± 0.0* |
| ΔmotAB+motAB | 23.5 ± 2.6* |
Motility was assayed on 0.4% MH agar after 24 h of incubation under microaerophilic conditions. Experiments were performed in at least biological duplicate. Values (mean ± standard error) were considered significant (*) at a P value of <0.001 using Bayesian statistical analysis.
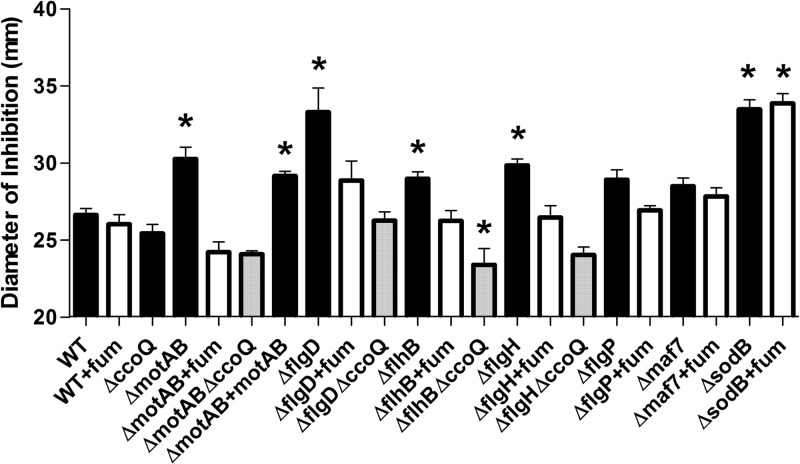
To further investigate this finding, we constructed a ΔmotAB double mutant. The motA and motB genes encode the flagellar motor apparatus, which utilizes the proton motive force across the inner membrane to drive flagellar rotation (23, 24). Previous studies using a ΔmotAB mutant have found that this mutant produces a full-length flagellum but is nonmotile (25). Consequently, we tested our ΔmotAB mutant for sensitivity to oxidants. Interestingly, the ΔmotAB mutant displayed significantly increased sensitivity to all 3 oxidants (Fig. 1; Fig. 3), providing further evidence for a link between motility and oxidant sensitivity in C. jejuni. Complementation of ΔmotAB restored the motility of the ΔmotAB strain (Table 3) and sensitivity to H2O2 and cumene hydroperoxide to wild-type levels (see Table S5 in the supplemental material). Restoration of the phenotype in the presence of menadione was, however, not found to be statistically significant (Fig. 3).
FIG 3.
The use of an alternative electron acceptor or tandem deletion of ccoQ in flagellar mutants restores sensitivity to menadione to parental C. jejuni levels. Wild-type, isogenic single- and double-deletion mutants, and complemented C. jejuni strains were assessed for sensitivity to 90 mM menadione using disc inhibition analysis. When required, the MH agar was supplemented with 20 mM sodium fumarate (fum). The diameter of the zone of inhibition is represented as the mean clear zone ± standard error (in mm) for each strain after exposure to 10 μl of 90 mM menadione bisulfite. Each experiment was repeated in at least quadruplicate. Values were considered significant (*) at a P value of <0.05 using one-way analysis of variance (ANOVA). Black bars, wild-type and mutant C. jejuni strains; white bars, strains plus fumarate; gray bars, double-deletion mutant strains.
The flagellar motor utilizes the proton potential of the inner cell membrane to generate torque for rotation (24). Consequently, flagellum mutants may conceivably exhibit a disturbed proton potential, which in turn may perturb the activity of the electron transport chain (ETC). The ETC is known to be the main source of endogenous ROS production (8). Therefore, ETC disturbance may cause increased production of ROS through electron leakage, leading to the observed hypersensitive phenotype of the flagellum mutants. It has been well documented that electron leakage at complexes I and III of the ETC lead to the endogenous production of superoxide (26). Disruption of proton/electron flow could also lead to the production of harmful oxy-intermediates and free radicals at complex IV (cytochrome c oxidase), which is the only ETC complex that directly interacts with molecular oxygen (26). To test this hypothesis, we assessed the oxidant sensitivity of the flagellar mutants in the presence of an alternative terminal electron acceptor, fumarate. The use of fumarate allows cellular respiration to occur without the involvement of complex IV (6). As shown in Fig. 3, the ΔmotAB, ΔflgD, ΔflhB, and ΔflgH strains all displayed increased resistance to menadione when 20 mM fumarate was supplemented into the agar. Indeed, in the presence of fumarate, the ΔmotAB, ΔflgD, ΔflhB, and ΔflgH mutants were no longer significantly sensitive to menadione compared to the parental C. jejuni strain. Importantly, the reduced susceptibility of the flagellar mutants to menadione is specific to the motility defects observed in these strains and not due to any general antioxidant properties that fumarate supplementation may provide upon oxidant exposure. This observation is supported by the phenotypes obtained for the wild-type, ΔsodB, and Δmaf7 strains, which did not show a statistical difference in oxidant sensitivity in the presence of fumarate (Fig. 3). Furthermore, the restoration of the phenotype was specific to menadione exposure. No significant decrease in sensitivity to either H2O2 or cumene hydroperoxide was observed for the ΔmotAB, ΔflgD, ΔflhB, and ΔflgH mutants (see Table S5 in the supplemental material).
Complex IV of the ETC is the primary site of menadione-induced oxidative stress in the nonmotile flagellum mutants.
The use of fumarate as an alternative terminal electron acceptor to oxygen alleviated the menadione-sensitive phenotype of the ΔmotAB, ΔflgD, ΔflhB, and ΔflgH mutants. This result suggested that complex IV of the ETC (which interacts directly with molecular oxygen) may be a major contributor in generating endogenous superoxide stress when oxygen is present in these mutants. This increased endogenous superoxide production may in turn make these mutants more susceptible to menadione-induced stress. Given that we had successfully constructed a deletion mutant of the CcoQ subunit of cytochrome c oxidase (23), we subsequently constructed ΔmotAB ΔccoQ, ΔflgD ΔccoQ, ΔflhB ΔccoQ, and ΔflgH ΔccoQ double-deletion mutants to determine if complex IV plays an important role in generating oxidative stress. Deletion of ccoQ in tandem with the flagellum mutations significantly reduced the hypersensitivity phenotype of the individual flagellum mutants toward menadione (Fig. 3). Thus, complex IV of the ETC contributes to the increased sensitivity to menadione in the flagellum mutants. Similar to the results in the presence of fumarate, no significant restoration of the phenotype toward either H2O2 or cumene hydroperoxide was observed for the double-deletion mutants relative to their respective single-deletion mutants (see Table S5 in the supplemental material), showing that complex IV specifically contributes toward menadione sensitivity. Overall, these findings suggest that in nonmotile flagellum mutants, superoxide stress originates specifically at complex IV of the ETC, leading to the hypersensitivity observed in these strains.
DISCUSSION
Bacterial pathogens that colonize the intestine are threatened by the host innate and adaptive immune responses and face hostile conditions, including the production of ROS. Under the auspice of the gut, oxygen is reduced to superoxide anions (O2·−) which can freely diffuse across cell membranes, damaging DNA, proteins, and lipids. Moreover, the presence of pathogenic bacteria triggers activation of the NOX family of enzymes and yields potent O2·− (27). Pathogens have evolved mechanisms to detoxify and protect themselves from ROS in order to survive and colonize the gastrointestinal tract. To date and despite the physiological importance of oxidative stress defenses, only eight major detoxification enzymes/proteins have been identified and characterized within C. jejuni: AhpC (alkyl hydroxyperoxide reductase), SodB (superoxide dismutase), KatA (catalase), Tpx (thiol peroxidase), Bcp (thiol peroxidase), Dps (bacterioferritin), MsrA/B, and Cj1386 (an ankyrin-containing protein involved in heme trafficking to catalase) (11, 13, 14, 28–31). To deepen our knowledge of genes and/or mechanisms involved in oxidative stress defense, we constructed a collection of 109 mutants with targeted mutations in genes that were previously shown to be induced by oxidant exposure and/or to be regulated by the peroxide stress regulator PerR. This collection constitutes a unique resource to comprehensively identify the measures employed by C. jejuni to cope with oxidative stress. All 109 mutants were screened for sensitivity or resistance to H2O2, cumene hydroperoxide, and/or menadione sodium bisulfite using disc inhibition assays. Strikingly, only 20% of the constructed mutants had measurable phenotypes under the conditions tested. There are several reasons why phenotypes were not observed for all 109 mutants screened. First, it is important to note that the microarray analysis of the oxidant stimulons and PerR regulon were conducted in iron-restricted MEMα medium, whereas the oxidant inhibition assays were performed under iron-replete conditions in Mueller-Hinton (MH) agar. Changes in the growth medium can significantly alter gene expression patterns. This is especially true for the three main oxidative defense genes in C. jejuni, katA, ahpC, and sodB. The expression of these three genes is iron responsive, and both katA and ahpC are regulated by iron-dependent transcriptional regulators (13, 17). In addition, both katA and sodB require iron or iron cofactors for their catalytic function (14). These changes in C. jejuni's antioxidant potential could influence the phenotypes observed. Second, many antioxidant proteins have overlapping and compensatory roles in detoxifying oxidants. This has already been reported for C. jejuni with the redundancy observed between the Tpx and Bcp proteins (11). While mutants with single mutations in these genes were not affected in oxidant sensitivity, the double mutant displayed significant sensitivity to oxidants. Thus, several mutants with mutations in genes identified using transcriptomic approaches may not exhibit a phenotype unless these are deleted in tandem with other genes. Indeed, we have demonstrated that tandem deletion of the flagellum mutant genes with ccoQ restored the oxidant sensitivity of these mutants. Third, oxidant exposure likely induces genes involved not only in oxidative stress resistance but also in protection against other stresses. This last hypothesis would explain the unchanged phenotype of several mutants, including those with mutations in drug efflux pumps (cmeA and cj0309c), the stringent response (spoT), sulfonamide resistance (folP), and potential osmotic and/or temperature resistance (truB), with regard to oxidative stress defense. Finally, we also identified genes involved in C. jejuni's growth, energy metabolism, and biosynthesis that were not directly related to ROS detoxification and protection, which are likely a response of the bacteria to oxidant exposure and thus do not display a phenotype.
Despite these limitations, however, a total of 22 mutants displayed a phenotype toward oxidant exposure. Importantly, to confirm the observed phenotypes and rule out possible polar effects or secondary mutations, the mutants were complemented, which resulted in full or partial restoration of the wild-type phenotype, in agreement with the role of these genes in oxidative stress defense. These genes encode two major groups of proteins, those directly involved in protective mechanisms and those contributing to metabolic rearrangements which indirectly affect the endogenous production of ROS. Work from our lab and others has previously identified several genes involved in the direct detoxification of oxidants, including the well-characterized katA, ahpC, and sodB genes (13, 14, 16, 28, 29). Despite our expectations, results from the mutant screening identified relatively few novel genes directly involved in detoxification. Indeed, with the exception of both rrc (cj0012c) and cj0358, we did not identify any genes with phenotypes toward oxidants that are directly involved in detoxification. Rrc is a unique C. jejuni protein and appears to be a chimera of rubredoxin oxidoreductases (Rbo) and rubreythrins (Rbr) found in other bacteria (32). From our study, although just below our very stringent cutoff for statistical significance (P < 0.001), inactivation of rrc led to increased sensitivity to menadione sodium bisulfite and H2O2 (P = 0.00522 and 0.0052, respectively). Furthermore, cj0358, encoding a putative cytochrome c peroxidase (23), displayed increased resistance to H2O2. Although the in vitro results suggest important roles for Rrc and Cj0358 in cellular defense against oxidants, the Δrrc mutant and the Δcj0358 mutant (CJJ0382 in C. jejuni 81-176) were not significantly affected in their ability to colonize the ceca of chicks (our work and reference 33). Thus, it appears that C. jejuni relies primarily on KatA and SodB to detoxify H2O2 and superoxide, respectively, during colonization despite deletion of Rrc or Cj0358.
In addition to rrc and cj0358, our screen identified 20 genes indirectly contributing to protection against oxidant exposure. These genes encode proteins involved in flagellum biogenesis, energy metabolism, cation transport, and general bacterial physiology. The larger number of genes involved in secondary or indirect protection mechanisms reflects the paucity of genuine oxidative defense pathways in C. jejuni. Indeed, it is now clear from our work that this organism primarily relies on KatA, AhpC, and SodB to prevent cellular damage from ROS. As a result and compared to other bacteria, C. jejuni processes a relatively small number of genes involved in direct ROS detoxification, suggesting a rudimentary oxidative stress defense system. This somewhat simple defense system is surprising given the continuous exposure of C. jejuni to ROS in the gastrointestinal tract and during inflammation. In particular, the lack of enzyme isoforms present in other bacteria (e.g., multiple superoxide dismutase [SOD] enzymes) suggests that these multiple defense pathways might not be essential for C. jejuni gut colonization.
Among the genes indirectly contributing to oxidative defense pathways, the largest single functional category is those that encode proteins involved in flagellum biogenesis. More specifically, we found that defects in bacterial motility were indirectly responsible for the increased sensitivity to the superoxide generator menadione. Mutants that displayed a reduced or nonmotile phenotype likely experience a disrupted proton gradient and consequently electron leakage along the ETC, contributing to increased endogenous O2·− production and thus increased oxidative stress in these strains. In support of this, fumarate supplementation or tandem deletion of ccoQ in nonmotile flagellum mutants significantly reduced menadione-induced cell death. From these results, complex IV of the ETC appears to be a particularly susceptible site for generating oxidative stress. Supplementation of the growth medium with an alternative electron acceptor such as fumarate likely reduces the additional oxidative damage that occurs at complex IV by promoting fumarate respiration, which does not use an oxygen-dependent oxidase. Furthermore, deletion of ccoQ in the nonmotile flagellum mutants restored the oxidant sensitivity to menadione, suggesting that this complex plays an important role in the generation of oxidative stress. Although the precise mechanism by which complex IV generates stress in these strains is unknown, it is clear that bacterial motility and a functional ETC are required to minimize superoxide-induced stress in the presence of menadione.
Mutants with the cation transport and/or binding proteins ChaN, ExbB1, and TonB2 inactivated also displayed significant differences in their resistance to oxidants. The ΔchaN mutant exhibited increased resistance to H2O2 (P = 0.002) and cumene hydroperoxide, whereas ΔexbB1 was more resistant specifically to H2O2. The ΔtonB2 mutant was more sensitive to H2O2 than the wild-type strain. ChaN is an iron-regulated lipoprotein that binds two heme groups per dimer and is thought to be involved in heme trafficking (34). Based on the phenotype of the ΔchaN mutant, it is tempting to speculate that the heme-sequestering capacity of ChaN might limit the amount of heme trafficking to the major H2O2 detoxifier in C. jejuni, KatA, explaining the observed resistance of the mutant to H2O2. Likewise, the exbB1 energy transduction protein was also found to have increased resistance to H2O2. The potential role of ExbB1 in oxidative stress defense is unclear; however, it is interesting to note that exbB1 is located downstream from chaN and also displays a similar phenotype. Moreover, and in contrast to the ΔexbB1 mutant, the ΔtonB2 mutant displayed significant sensitivity to H2O2, indicating that TonB2 plays an important role in hydrogen peroxide defense. Although the precise role that tonB2 plays in oxidative stress defense is unknown, it is possible that deletion of this gene causes disruption in energy transduction for processes important to oxidant defense. While none of the other mutants with deletions in iron acquisition genes were affected in oxidative stress resistance, phenotypic characterization under iron-limited growth conditions would be required to refute or validate their potential role in oxidant defense.
Our previous transcriptomic work pointed to a role for genes involved in energy metabolism in protection against oxidants. Among the 7 mutants constructed with mutations in energy metabolism genes, ald, cj1377c, cj1207c, hypC, ccoQ, cj0073c, and acnB, only the ΔacnB (aconitase) mutant exhibited a phenotype. This mutant was hypersensitive to H2O2 and menadione sodium bisulfite. In E. coli, the AcnA and AcnB proteins have been implicated in posttranscriptional regulation. Following oxidative damage to the iron-sulfur clusters of AcnA or AcnB, the apoproteins specifically bind and stabilize the 3′ untranslated regions (UTRs) of their respective mRNA transcripts, resulting in increased synthesis of their respective proteins (35). Moreover, the apo-aconitase proteins also posttranscriptionally regulate the synthesis of the SodA oxidative stress defense enzyme as well as many other proteins involved in oxidative stress defense (35). While our study shows a critical role for acnB in oxidative stress defense, whether AcnB has a posttranscriptional regulatory role for its own transcript or other oxidative stress defense genes such as sodB remains to be experimentally investigated.
Our phenotypic screen revealed a link between numerous general biological processes and oxidative stress defenses. Mutation of pstC, encoding a putative phosphate transport system permease protein, revealed increased sensitivity to H2O2 and cumene hydroperoxide relative to that of the wild-type strain. Given the requirement for phosphorous for numerous biological processes, bacteria consequently must be able to acquire phosphorous from the surrounding environment. Phosphorous is taken up into the cell in the form of inorganic orthophosphate (Pi). An important feature of Pi is that it can be linked together by enzymes such as Ppk to form polyphosphates. Polyphosphates have important roles in pathogenesis, as studied in several bacterial pathogens, including E. coli, Pseudomonas aeruginosa, Helicobacter pylori, Vibrio cholerae, and Shigella flexneri (36–42). Polyphosphate has been reported to be involved in processes within these bacteria such as motility, biofilm formation, acid, heat and osmotic stress, stationary-phase survival, and resistance to oxidative stress (36–42). The increased sensitivity to oxidants in the ΔpstC mutant suggests a role for phosphate acquisition and polyphosphate synthesis in the oxidative stress response in C. jejuni, which requires further investigation.
The requirement of antioxidant enzymes for successful colonization of C. jejuni in vivo has been previously demonstrated for the major detoxification enzymes KatA, AhpC, and SodB. Indeed, the ΔkatA, ΔahpC, and ΔsodB deletion mutants were all significantly attenuated in colonization of the chick cecum, revealing the significant role that oxidative stress defenses play during colonization (16). Subsequently, we sought to assay key genes identified from our phenotypic screening to assess their biological relevance using the chick colonization model. Among the strains tested, the ΔacnB and ΔpstC mutants were found to be significantly attenuated for colonization of the chick cecum. Mutants with deletions in the flagellum genes flgR and flgP were also affected in colonization; however, the attenuation observed for these mutants may also be a result of the motility defects associated with these strains. The cj0344, cj0947c, and cj0062c mutants all displayed a significant decrease in colonization relative to the wild-type strain. Given that the precise function that these genes have in oxidant defense is unknown, it cannot be ruled out that mechanisms other than oxidative stress defense contribute toward the observed in vivo phenotypes. Overall, the in vivo colonization experiments highlight the importance of these genes for chick colonization.
In this study, we report the construction of an isogenic deletion mutant library with mutations in potentially relevant oxidative stress defense genes as identified by microarray analysis. This genome-wide screening of a constructed library of oxidant-sensitive mutants is the first to be described for C. jejuni. Phenotypic characterization of the constructed mutants in both in vitro and in vivo assays revealed novel functions for genes important for oxidative stress defense within C. jejuni. We report a major role for genes involved in motility as an indirect contributor to oxidative stress through disruption of the ETC. These results have thus revealed an unexpected pathway used by oxidants to induce cell death in C. jejuni. We also identified important roles for acnB and cation transport and binding protein genes as well as roles for previously uncharacterized genes in oxidative stress defense. Future characterization of these oxidant defense genes will provide insight into their function and the role they play in oxidant detoxification within the cell.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by CIHR grant MOP84224 to A.S., a QEIIGSST scholarship to A.F., and a CIHR-Banting graduate scholarship to J.B.
We thank J. E. Algire and the animal care staff for their professional and technical help during the in vivo chick colonization experiments at OLF, CFIA.
Footnotes
Published ahead of print 18 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01528-13.
REFERENCES
- 1. Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28–35. 10.3201/eid0501.990104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food-foodborne diseases active surveillance network, 10 U.S. sites, 1996-2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 3. Ruiz-Palacios GM. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703. 10.1086/509936 [DOI] [PubMed] [Google Scholar]
- 4. Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117:237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 5. Nachamkin I. 2002. Chronic effects of Campylobacter infection. Microbes Infect. 4:399–403. 10.1016/S1286-4579(02)01553-8 [DOI] [PubMed] [Google Scholar]
- 6. Sellars MJ, Hall SJ, Kelly DJ. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187–4196. 10.1128/JB.184.15.4187-4196.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- 8. Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Segal AW. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197–223. 10.1146/annurev.immunol.23.021704.115653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Annuk H, Shchepetova J, Kullisaar T, Songisepp E, Zilmer M, Mikelsaar M. 2003. Characterization of intestinal lactobacilli as putative probiotic candidates. J. Appl. Microbiol. 94:403–412. 10.1046/j.1365-2672.2003.01847.x [DOI] [PubMed] [Google Scholar]
- 11. Atack JM, Harvey P, Jones MA, Kelly DJ. 2008. The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J. Bacteriol. 190:5279–5290. 10.1128/JB.00100-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Day WA, Jr, Sajecki JL, Pitts TM, Joens LA. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337–6345. 10.1128/IAI.68.11.6337-6345.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baillon ML, van Vliet AH, Ketley JM, Constantinidou C, Penn CW. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pesci EC, Cottle DL, Pickett CL. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flint A, Sun YQ, Stintzi A. 2012. Cj1386 is an ankyrin-containing protein involved in heme trafficking to catalase in Campylobacter jejuni. J. Bacteriol. 194:334–345. 10.1128/JB.05740-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, Stintzi A. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. 10.1186/1471-2164-10-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Vliet AH, Baillon ML, Penn CW, Ketley JM. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 74:1583–1597. 10.1128/AEM.01507-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714–4729. 10.1128/JB.186.14.4714-4729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367. 10.1038/nature04537 [DOI] [PubMed] [Google Scholar]
- 21. Stahl M, Stintzi A. 2011. Identification of essential genes in C. jejuni genome highlights hyper-variable plasticity regions. Funct. Integ. Genomics 11:241–257. 10.1007/s10142-011-0214-7 [DOI] [PubMed] [Google Scholar]
- 22. Sommerlad SM, Hendrixson DR. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189:179–186. 10.1128/JB.01199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream M-A, Rutherford KM, Vliet AHMv, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- 24. Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39–85. 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- 25. Mertins S, Allan BJ, Townsend HG, Koster W, Potter AA. 2013. Role of motAB in adherence and internalization in polarized Caco-2 cells and in cecal colonization of Campylobacter jejuni. Avian Dis. 57:116–122. 10.1637/10235-050412-ResNote.1 [DOI] [PubMed] [Google Scholar]
- 26. Musatov A, Robinson NC. 2012. Susceptibility of mitochondrial electron-transport complexes to oxidative damage. Focus on cytochrome c oxidase. Free Radical Res. 46:1313–1326. 10.3109/10715762.2012.717273 [DOI] [PubMed] [Google Scholar]
- 27. Lambeth JD. 2007. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radical Biol. Med. 43:332–347. 10.1016/j.freeradbiomed.2007.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grant KA, Park SF. 1995. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 141:1369–1376. 10.1099/13500872-141-6-1369 [DOI] [PubMed] [Google Scholar]
- 29. Purdy D, Park SF. 1994. Cloning, nucleotide sequence and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology 140:1203–1208. 10.1099/13500872-140-5-1203 [DOI] [PubMed] [Google Scholar]
- 30. Atack JM, Kelly DJ. 2008. Contribution of the stereospecific methionine sulphoxide reductases MsrA and MsrB to oxidative and nitrosative stress resistance in the food-borne pathogen Campylobacter jejuni. Microbiology 154:2219–2230. 10.1099/mic.0.2008/019711-0 [DOI] [PubMed] [Google Scholar]
- 31. Ishikawa T, Mizunoe Y, Kawabata S, Takade A, Harada M, Wai SN, Yoshida S. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010–1017. 10.1128/JB.185.3.1010-1017.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamasaki M, Igimi S, Katayama Y, Yamamoto S, Amano F. 2004. Identification of an oxidative stress-sensitive protein from Campylobacter jejuni, homologous to rubredoxin oxidoreductase/rubrerythrin. FEMS Microbiol. Lett. 235:57–63. 10.1111/j.1574-6968.2004.tb09567.x [DOI] [PubMed] [Google Scholar]
- 33. Bingham-Ramos LK, Hendrixson DR. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76:1105–1114. 10.1128/IAI.01430-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan AC, Lelj-Garolla B, Pedersen KA, IR F, Mauk AG, Murphy ME. 2006. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 362:1108–1119. 10.1016/j.jmb.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 35. Tang Y, Quail MA, Artymiuk PJ, Guest JR, Green J. 2002. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology 148:1027–1037 [DOI] [PubMed] [Google Scholar]
- 36. Price-Carter M, Fazzio TG, Vallbona EI, Roth JR. 2005. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J. Bacteriol. 187:3088–3099. 10.1128/JB.187.9.3088-3099.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao NN, Kornberg A. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rashid MH, Rao NN, Kornberg A. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225–227. 10.1128/JB.182.1.225-227.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:9636–9641. 10.1073/pnas.170283397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 187:7687–7695. 10.1128/JB.187.22.7687-7695.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jahid IK, Silva AJ, Benitez JA. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043–7049. 10.1128/AEM.00924-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim KS, Rao NN, Fraley CD, Kornberg A. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U. S. A. 99:7675–7680. 10.1073/pnas.112210499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.