We have witnessed great success in the identification of new type 2 diabetes susceptibility loci through genome‐wide association (GWA) analysis in the past 5 years. The number of loci robustly implicated in type 2 diabetes risk; that is, those that have attained a genome‐wide significance level (P < 5 × 10−8) and also have been repeatedly validated in independent samples, has climbed from just three in 2006 to >50 today. As these GWA studies were carried out almost exclusively in European‐descent populations, studies in non‐European populations will allow us to assess the relevance of the findings to other ethnic groups. To address this point, a consortium‐based GWA meta‐analysis of type 2 diabetes was recently carried out in East Asians with a multistage study design (involving >25,000 cases and >29,000 controls in total)1; eight new loci were confirmed to significantly associate with type 2 diabetes. This large‐scale GWA meta‐analysis was principally carried out in the Asian Genetic Epidemiology Network (AGEN), in which our group participated together with investigators from Japan, Korea, China, Taiwan, Singapore and the USA.
Before the AGEN GWA meta‐analysis, GWA studies in East Asian populations already reported several type 2 diabetes loci (e.g. KCNQ1, UBE2E2 and C2CD4A‐C2CD4B), which had not been identified in European‐decent populations until then. However, in the course of such efforts made in East Asians, it has been widely recognized in European‐decent populations that for common complex diseases, such as type 2 diabetes, a major part of susceptibility loci can individually exert modest genetic impacts; that is, ∼10–20% increased risk of developing the disease, and hence might not attain a genome‐wide significance level unless a large number of individuals are analyzed in both the discovery and replication (or follow up) stages of GWA scans.
In this line, it is reported that the number of discovered variants (or loci) is strongly correlated with experimental sample size in GWA studies of type 2 diabetes2. This might particularly hold true in the case that effect sizes (measured as an odds ratio [OR] in case–control studies) of undiscovered variants are almost equivalent to those of discovered ones. In addition to effect sizes, effect allele frequencies (of target variants) have a substantial influence on our chance of identifying significant associations in a given size of samples. When inconsistent results are observed between populations, we should carefully claim the presence of ‘population specific’ genetic association with type 2 diabetes, as discussed later.
Because GWA studies interrogate a huge number of single nucleotide polymorphism (SNP) markers simultaneously, a stringent statistical threshold (P < 5 × 10−8) should be set in order to avoid false positives from multiple testing. Based on the simulations, such a high significance level of association could be detected in the discovery stage (stage 1) of AGEN GWA meta‐analysis (involving 6952 cases and 11,865 controls) for loci with OR ≥ 1.20 assuming 80% power. Indeed, three previously‐identified loci –CDKAL1, CDKN2A/2B and KCNQ1– did satisfy the threshold in the discovery stage (OR = 1.17–1.21). This indicates that effect sizes for undiscovered (or new) type 2 diabetes loci are likely to be rather modest (OR < 1.2) and that a larger number of samples (for cases and controls) are required to attain P < 5 × 10−8, and also to confirm the associations independently. Here, it has to be kept in mind that in the GWA scan, the actual genetic effect is typically smaller than its estimate based on the discovery stage data, whose results are affected by an ascertainment bias known as the ‘winner’s curse’3.
Another issue to be considered in the overall GWA meta‐analysis design is the cost and time of SNP genotyping in the replication stage(s). Although it costs a great deal for the initial GWA genotyping, look‐up of target SNPs in the existing GWA‐scanned datasets does not require additional cost. In contrast, de novo genotyping in independent samples requires additional cost and a great deal of time. Thus, a combination of the two approaches was taken in the replication/follow‐up stages of AGEN GWA meta‐analysis1. That is, modest association signals (3756 SNPs from 297 independent loci, showing P < 5 × 10−4 in the discovery panel) were followed up with a stage 2 in silico replication analysis (involving 5843 cases and 4574 controls) and then 19 SNPs showing the most compelling evidence for association (P < 10−5 in stages 1 + 2) were subjected to stage 3 de novo genotyping (involving up to 12,284 cases and 13,172 controls). A total of eight new loci were finally confirmed to show significant evidence of association in East Asians.
The replicated associations for a limited number of candidate gene loci have broadly shown the tendency of interethnic similarity. Even though the common (or cosmopolitan) effect of type 2 diabetes susceptibility variants is known for several loci, the extent to which the causation of the disease differs or overlaps between populations remains unknown. With in silico replication analysis, just two of eight loci (25%) discovered in East Asian samples also showed nominally (P < 0.05) significant associations in European‐descent populations. For the remaining six loci without reproducible evidence of association, minor allele frequencies were relatively high (0.14–0.46) in European‐descent populations. This suggests that failure of replication is not simply a result of cross‐population differences in risk allele frequency; that is, it is estimated that low allele‐frequency leads to reduced power, even though effect sizes are similar4.
By systematically comparing effect sizes (in OR) between East Asians and Europeans for a number of robustly‐confirmed type 2 diabetes loci, we can recognize several points highlighted by the extensive GWA meta‐analysis of East Asian samples (Figure 1). First, for all eight loci newly discovered in East Asians, effect sizes are modest (OR = 1.08–1.13). Second, approximately one‐third (16 of 49 loci included in the plot) of the loci exert a certain level of effect size (OR > 1.1) in both ethnic groups. Here, one of the discovered loci, MAEA, can be categorized to this group, and its minor allele frequency is far less frequent in Europeans than in East Asians (3% vs 42%), similar to the situation for KCNQ14. Third, a number of type 2 diabetes loci that were originally identified in Europeans do not appear to show reproducible associations in East Asians. Roughly, the failure to replicate a nominally significant association between an index SNP and type 2 diabetes in a tested population does not necessarily indicate the absence of an association at the relevant locus. Factors to be considered include sample size, linkage disequilibrium (LD) structure, and the potential impact of gene–gene and gene–environment interactions in the individual population. Despite such caveats, it seems to be helpful to know the extent to which type 2 diabetes associations are reproducible between populations.
Figure 1.
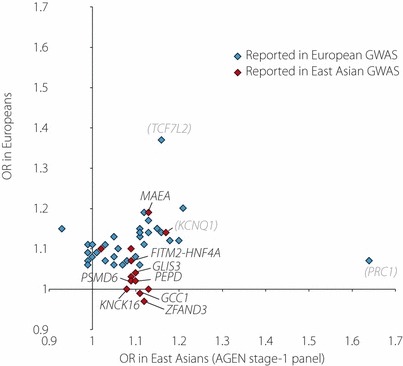
Correlation of effect sizes for type 2 diabetes risk between East Asians (x‐axis) and Europeans (y‐axis) at 49 single nucleotide polymorphism loci, for which the corresponding data are available in the East Asian genome‐wide association (GWA) meta‐analysis1. Only the effect size estimate, odds ratios (OR), but not 95% confidence interval, is shown in the figure for the purpose of readability. Symbols colored in red and blue are loci originally reported in GWA studies/meta‐analysis (GWAS) of East Asians and Europeans, respectively. The gene names are shown for eight loci newly discovered in East Asian samples, as well as three others –TCF7L2, KCNQ1 and PRC1. AGEN, Asian Genetic Epidemiology Network.
Together with the recently reported data on South Asian meta‐analysis, which involves studies in India, Pakistan, the UK, Mauritius, Singapore and Sri Lanka5, an arbitrary picture of cross‐population difference (or overlapping) can be shown in Figure 2, as for a total of 39 loci, which have been originally identified in European GWA studies/meta‐analysis and then subjected to evaluation (i.e. replication) in populations of East and South Asian descent. Almost half of the loci (20 out of 39) are common across three ethnic groups – Europeans, East Asians and South Asians – and 18% of the loci do not show a nominally significant association in non‐European populations. Again, this cannot exclude the possibility that the causal variants are not represented by the index SNPs because of cross‐population differences in LD structure and/or the possibility that sample size is insufficient to attain statistical significance. Nevertheless, it is possible that some of the negative associations reflect true population‐specificity.
Figure 2.
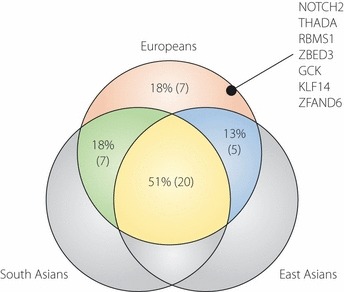
A schematic representation of cross‐population difference (or overlapping) for a total of 39 type 2 diabetes‐associated loci that have been originally identified in European genome‐wide association studies/meta‐analysis, and then subjected to evaluation in populations of East and South Asian descent. Here, an associated locus is assumed to overlap between the ethnic groups when P ≤ 0.05, showing a concordant direction of genetic effect in the population tested for replication.
Among eight loci reaching genome‐wide significance, the GLIS locus alone has previously been reported in the context of metabolic traits or related diseases. This gene encodes a Kruppel‐like zinc finger transcription factor, which has been proposed as a key player in the regulation of pancreatic β‐cell development and insulin gene expression. In accordance with such a biological function, SNPs in high LD with this locus have been implicated in association with type 1 diabetes and fasting plasma glucose levels.
As the number of discovered loci has recently increased, substantial efforts have been made to show that the genes mapping close to type 2 diabetes loci are enriched for particular biological pathways (or processes), although they have thus far met with only limited success. Among the pathways, the most robust finding is related to cell‐cycle regulation; this seems to be consistent with a model in which the regulation of pancreatic islet mass is a principal component of genetic susceptibility to type 2 diabetes, as is shown for GLIS above‐mentioned.
Despite the lack of clear physiological evidence on type 2 diabetes, these GWA findings can provide clues to the precise biological mechanisms underlying appreciable differences in the clinical presentation of type 2 diabetes between populations of European and non‐European origin. Along with the GWA meta‐analyses in individual populations, ‘transethnic’ meta‐analysis is currently being carried out and will allow for a better chance to show novel susceptibility loci and pathophysiological pathways of type 2 diabetes, and might also facilitate the fine mapping of common causal variants by utilizing ethnic differences in LD structure.
References
- 1.Cho YS, Chen CH, Hu C, et al. Meta‐analysis of genome‐wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat Genet 2011; 44: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet 2012; 90: 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case‐control data. Am J Hum Genet 2007; 80: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy MI. Casting a wider net for diabetes susceptibility genes. Nat Genet 2008; 40: 1039–1040 [DOI] [PubMed] [Google Scholar]
- 5.Kooner JS, Saleheen D, Sim X, et al. Genome‐wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 2011; 43: 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]


