Abstract
As a signaling hub, p62/sequestosome plays important roles in cell signaling and degradation of misfolded proteins. p62 has been implicated as an adaptor protein to mediate autophagic clearance of insoluble protein aggregates in age-related diseases, including age-related macular degeneration (AMD), which is characterized by dysfunction of the retinal pigment epithelium (RPE). Our previous studies have shown that cigarette smoke (CS) induces oxidative stress and inhibits the proteasome pathway in cultured human RPE cells, suggesting that p62-mediated autophagy may become the major route to remove impaired proteins under such circumstances. In the present studies, we found that all p62 mRNA variants are abundantly expressed and upregulated by CS induced stress in cultured human RPE cells, yet isoform1 is the major translated form. We also show that p62 silencing exacerbated the CS induced accumulation of damaged proteins, both by suppressing autophagy and by inhibiting the Nrf2 antioxidant response, which in turn, increased protein oxidation. These effects of CS and p62 reduction were further confirmed in mice exposed to CS. We found that over-expression of p62 isoform1, but not its S403A mutant, which lacks affinity for ubiquitinated proteins, reduced misfolded proteins, yet simultaneously promoted an Nrf2-mediated antioxidant response. Thus, p62 provides dual, reciprocal enhancing protection to RPE cells from environmental stress induced protein misfolding and aggregation, by facilitating autophagy and the Nrf2 mediated antioxidant response, which might be a potential therapeutic target against AMD.
Keywords: Autophagy, aging, Nrf2, Oxidative stress, p62
1. Introduction
The scaffolding adaptor protein p62/SQSTM1 (p62) is a stress-inducible intracellular protein. Because it contains several protein-protein interaction domains, p62 influences multiple functions by regulating various signal transduction pathways. In addition, it is involved in protein trafficking, aggregation, and degradation. Efficient removal of damaged or misfolded proteins is crucial for maintaining cellular homeostasis and survival, yet proteasomes, the primary machineries for the degradation of polyubiquitinated proteins, are unable to digest stable protein complexes or aggregates[1]. Thus, p62-mediated selective autophagic clearance of aggregated proteins (aggrephagy) via its ubiquitin-associated (UBA) domain[2], provides an important compensatory mechanism, especially when organisms are under the influence of mutation, aging, and/or environment stress. Its protective role has been previously investigated in a number of age-related neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease, where oxidative stress leads to protein misfolding and their toxic accumulation in the cytosol and within intracellular inclusions[3-6].
Age-related macular degeneration (AMD), the most common cause of blindness among the elderly, shares several clinical and pathological features with age-related neurodegenerative diseases, and likewise, is thought to develop through a complex interaction of genetic and environmental factors. AMD mainly involves the degeneration of neurosensory retinal photoreceptors and the retinal pigment epithelium (RPE). The RPE, a single layered epithelium, is located on Bruch’s membrane between the photoreceptors and the choriocapillaris. The RPE has multiple functions that maintain photoreceptor health, including the daily phagocytic uptake, degradation, and recycling of 30,000 shed apical photoreceptor outer segments (POS)[7, 8]. POS phagocytosis is a metabolically expensive process, requiring high oxygen consumption. Combined with the unique, constant exposure to light that generates photo-oxidative stress, the RPE is under heavy oxidative challenge, making it an ideal model for studying the cell’s response to oxidative stress. One consequence of this high oxidative stress environment is the accumulation of oxidized/damaged protein complexes, which is a recurrent event in many age-related diseases, including AMD and AD[9]. RPE homeostasis is therefore, dependent upon the efficient removal of harmful protein aggregates via the proteasomal and autophagic protein clearance systems, both of which gradually decline with aging[10-13].
Cigarette smoking is the single most important environmental risk factor for developing AMD in part, by adding to the high oxidative burden[14]. Through generating oxidative stress, it is known to damage proteins, lipids, and DNA of RPE cells. In response to cigarette smoke, the antioxidant transcription factor nuclear factor erythroid-2 related factor 2 (Nrf2) is activated in the RPE, which regulates a comprehensive and coordinated transcriptional program that helps to maintain cellular redox homeostasis and mitigates against oxidative injury[15]. Nrf2 activates transcription by specific binding to antioxidant response elements (ARE) in the promoters of target genes. In the absence of stress, Nrf2 is sequestered in the cytosol by Kelch-like ECH-associated protein 1 (Keap1), and is subsequently targeted to the proteasome for degradation. Interestingly, p62 can compete with Nrf2 for Keap1 interaction, resulting in Nrf2 stabilization, and the activation of Nrf2 downstream genes[16]. Meanwhile, p62 itself, contains ARE sites, and can be transcriptionally upregulated by Nrf2[17].
We previously showed that a non-lethal dose of cigarette smoke extract (CSE), inhibited proteasome activity, but as part of a comprehensive cytoprotective response by RPE cells, up-regulated a series of antioxidant and autophagy related genes, including p62[18]. p62 has two protein isoforms that are generated by three mRNA variants due to alternative splicing. Transcript variant 1 encodes p62 isoform1, while variant 2 and 3, having the same coding sequence and differing slightly in their 5′ UTR, encode isoform2. Currently, the expression pattern and functional differences of these isoforms are unknown. While previous reports have identified multiple functions of p62, studies of these functions simultaneously when a cell is under stress, are lacking. Given the high oxidative stress of the RPE, we are particularly interested in determining the extent that p62 influences both the removal of damaged proteins and the antioxidant response. In this manuscript, we address the hypothesis that cigarette smoking leads to an accumulation of misfolded/damaged proteins in the RPE, and activates the differential expression of p62 isoforms, which on the one hand, helps to activate the Nrf2 antioxidant response via its interaction with Keap1, and on the other hand, facilitates aggrephagy via its UBA domain interaction with polyubiquitinated proteins. Understanding the molecular mechanisms for RPE dysfunction and pathology, which are caused by environmental stress, will enhance our ability to design potential therapeutic strategies for AMD.
2. Materials and Methods
2.1 Reagents and antibodies
Cell culture media, Phosphate Buffered Saline pH 7.4, EDTA solution, and DNase/RNase free water were from Life Technologies (Carlsbad, CA). Cigarette smoke extract, containing 40mg/ml condensate and 6% nicotine, was purchased from Murty Pharmaceuticals (Lexington, KY), and was prepared by smoking University of Kentucky’s 3R4F Standard Research Cigarettes on an FTC Smoke Machine[15]. The smoke on the filter is calculated by the weight gain of the filter after smoking. The amount of DMSO is calculated that will dissolve a 4% (40mg/mL) solution. DMSO and Bafilomycin A1 were purchased from Sigma-Aldrich, Inc. (St. Louis, MO).
For Western blot and immunohistochemistry analysis, the following primary antibodies were used: anti-human polyubiquitin mouse monoclonal IgG1 antibody (1:1000; Enzo Life Sciences, Farmingdale, NY), anti-mouse polyubiquitin antibody (1:1000; Millipore Inc., Billerica, MA); anti-p62 mouse monoclonal IgG1 antibody (1:2000; MBL Technologies, Arlington VA); guinea pig polyclonal anti-p62 C-terminus antibody (1:2000; American Research Products, Waltham, MA), rabbit polyclonal anti-LC3B antibody (1:1000; Sigma-Aldrich, St. Louis, MO); anti-V5 mouse monoclonal IgG2a antibody (1:1000; Life Technologies); rabbit polyclonal anti-αB crystallin (1:200; Abcam, Inc., Cambridge, MA), anti-beta actin mouse monoclonal [AC-15] (1:2000; Abcam, Inc), and anti-guinea pig IgG (Alexa Fluor 594), HRP-conjugated anti-rabbit IgG and anti-mouse IgG antibodies, were also purchased from Abcam.
2.2 Cell Culture and transfection
The established human ARPE-19 cell line[19] was maintained in Dulbecco’s Modified Eagle Medium:F12 50/50 mix, supplemented with 10% inactivated fetal bovine serum and 2 mM L-glutamine, at 37°C in a humidified atmosphere containing 5% CO2. Cells were seeded at 50,000 cells/cm2 in 12-well plates for 16hrs, and transfected using Lipofectamine 2000 transfection reagent (Life Technologies) in OPTI-MEM media, according to the manufacturer’s instructions. Briefly, for gene knockdown studies[20], 5nM non-targeting control siRNA or siRNA targeting human Nrf2 or human p62 were used to transiently transfect ARPE-19 cells, by incubating for 8hrs, using our published protocol[21]. For gene over-expression studies[15], 0.5-1.0μg of plasmid DNA was used for transfection, and incubating for 24 hrs. Cells were collected 48hrs after transfection, and RNA was isolated for RT-qPCR, or protein was extracted for immunoblotting or ELISA.
2.3 Real-time RT-qPCR
Total RNA from cultured ARPE-19 cells or mouse tissue (retina or RPE/choroid) was isolated using an RNeasy Mini Kit (Qiagen, Valencia, CA), with on-column DNA digestion by RNase-free DNase (Qiagen), following the manufacturer’s instructions. cDNA was synthesized using a High Capacity cDNA Reverse Transcription kit using random hexamers (Applied Biosystems, Foster City, CA). Each real-time PCR reaction consisted of 10μl of cDNA template, 10μl of SYBR green master mix (ABI), and 4pmol of forward and reverse primers on an ABI’s StepOne Plus (Applied Biosystems) for 40 cycles (95°C for 3sec, 60°C for 30sec) after an initial 20sec of incubation at 95°C. The primers used for real-time PCR are listed in Table 1. Amplicon size and reaction specificity were confirmed by melting curve analysis and DNA agarose gel electrophoresis. The percentage change in expression of each gene was calculated using the comparative Ct method, with cyclophilin A (PPIA) as internal control.
Table 1.
The Sequences of primers used in the SYBR based real-time PCR.
| Gene Name | Primer Name and Sequence | |
|---|---|---|
| mouse Nqo1 | mNqo1-523f mNqo1-642r |
5′_GGCATCCTGCGTTTCTGTG_3′ 5′_GGTTTCCAGACGTTTCTTCCAT_3′ |
| mouse Gclm | mGclm-431f mGclm-661r |
5′_AGCCTTACTGGGAGGAATTAGAG_3′ 5′_GCAGTTCTTTCGGGTCATTGTG_3′ |
| mouse p62 | mP62-346f mP62-424r |
5′_GAGGCACCCCGAAACATGG_3′ 5′_ACTTATAGCGAGTTCCCACCA_3′ |
| mouse PPIA | mPPIA-67f mPPIA-191r |
5′_GAGCTGTTTGCAGACAAAGTTC_3′ 5′ CCCTGGCACATGAATCCTGG 3′ |
| human Nqo1 | Nqo1-11f Nqo1-206r |
5′_GAAGAGCACTGATCGTACTGGC_3′ 5′_GGATACTGAAAGTTCGCAGGG_3′ |
| human Gclm | Gclm-204f Gclm-421r |
5′_TGTCTTGGAATGCACTGTATCTC_3′ 5′_CCCAGTAAGGCTGTAAATGCTC_3′ |
| human Nrf2 | Nrf2.1-364f Nrf2.1-446r |
5′_CTTTTGGCGCAGACATTCCC_3′ 5′_GACTGGGCTCTCGATGTGAC_3′ |
| human p62 | p62-528f p62-672r |
5′_TGCCCAGACTACGACTTGTG_3′ 5′_AGTGTCCGTGTTTCACCTTCC_3′ |
| human p62 variant1 |
p62v1-153f p62-328r |
5′_ATTCGCCGCTTCAGCTTCT_3′ 5′_GAAAAGGCAACCAAGTCCCC_3′ |
| human p62 variant2 |
p62v2-291f p62-366r |
5′_ TGCAACATGGGGCTTGAGAA_3′ 5′_GGCCATTGTCAATTCCTCGTC_3′ |
| human PPIA | PPIA-77f PPIA-374r |
5′_CAGACAAGGTCCCAAAGACAG_3′ 5′_TTGCCATCCAACCACTCAGTC_3′ |
2.4 RT-PCR
Total RNA was prepared from cultured ARPE-19 cells, using Qiagen’s RNeasy Mini Kit according to the manufacturer’s instructions, and cDNA was synthesized using the SuperScript III First Strand Synthesis System for RT-PCR kit (Life Technologies). In a total volume of 20μl, 1μg of total RNA was incubated with 200U SuperScript III reverse transcriptase and 0.5μg Oligo(dT)12-18 primer, at 50°C for 50min. Heating to 85°C for 10min stopped the reaction, and 1μl first-strand product was amplified in 25μl Platinum PCR SuperMix High Fidelity (Life Technologies) plus 10pmol forward and backward primers. After 35 cycles (denaturation at 94°C for 30sec, annealing at 55°C for 40sec, extension at 72°C for 2min, and a final extension at 72°C for 5min after the last cycle) of amplification on a PTC-200 Pettier Thermal Cycler (MJ Research, Waltham, MA), 10μl of PCR product was separated in a 1% agarose gel, and stained with ethidium bromide.
2.5 Western blot analysis
ARPE-19 cells were washed with cold PBS, scraped off wells into RIPA buffer containing Sigma FAST Protease Inhibitor, and lysed on ice. Dissected mouse tissues (retina or RPE/choroid) were sonicated in cold RIPA buffer (Sigma) containing Sigma FAST Protease Inhibitor. Protein content from ARPE-19 cells or mouse tissue (retina or RPE/choroid) was quantified using the BioRad DC Protein Assay kit (Hercules, CA). For immunoblotting, 5-20μg protein per lane was separated by 4-12% Bis-Tris SDS-PAGE (Life Technologies) and transferred to 0.2μm pore nitrocellulose membranes. Membranes were blocked with 5% non-fat milk (BioRad) at room temperature for 1hr and then incubated with primary antibodies at 4°C overnight. After washing with 1×TBS-T (20mM Tris, 136.8mM NaCl, pH7.6, with 0.1% v/v Tween 20) three times for 10min, the blots were incubated with secondary antibodies (1:5000) at room temperature for 45min, and blots were visualized with enhanced chemiluminescence (ECL; Thermo Fisher Scientific, Inc., Waltham, MA). Images were captured and analyzed using GE Healthcare’s ImageQuant LAS 4010 Digital Imaging System (Pittsburgh, PA).
2.6 Protein Carbonyl ELISA
ARPE-19 cells were washed with cold PBS once and sonicated in Buffer A (25mM HEPES, 150mM NaCl, 10mM MgCl2, 1mM EDTA, 2% Glycerol, pH7.5), supplemented with Sigma FAST Protease Inhibitor. After quantification using the BioRad DC Protein Assay kit, cell lysates were diluted to 5μg/ml and coated onto the 96-well OxiSelect Protein Carbonyl ELISA plate (Cell Biolabs, Inc., San Diego, CA). Protein carbonyls were derivatized by 2, 4-dinitrophenylhydrazine (DNPH) to DNP hydrazone, and then probed with an anti-DNP antibody, followed by an HRP conjugated secondary antibody. The luminescence was measured at 450nm on a Synergy HT plate reader (BioTek, Winooski, VT).
2.7 Assessment of autophagy by Cyto-ID staining
Cells were seeded at 50,000 cells/cm2 in 6-well glass bottom plates coated with laminin (MatTek Corporation, Ashland, MA), grown, and transfected, as described earlier. After incubation with 125μg/ml CSE for 24hrs, the cells were rinsed and incubated with a 1:2000 dilution of Hoechst 33342 Nuclear Stain, and a 1:500 dilution of Cyto-ID Green Detection Reagent (Endo Life Sciences), which specifically stains autophagic vacuoles so that cells were examined at 48 hours[22, 23]. After 30min incubation at 37°C, cells were rinsed and observed under a confocal microscope (ZEN LSM 710, Carl Zeiss Microscopy, LLC, Thornwood, NY).
2.8 Plasmid constructs and site-directed mutagenesis
Human p62 mRNA sequences were retrieved from the NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and analyzed using the Vector NTI software (Life Technologies). Two sets of PCR primers were designed to clone p62 variant 1 and variant 2 into pcDNA3.1D/V5-HIS-TOPO vector, using the V5-HIS Directional TOPO Cloning Kit (Invitrogen, Grand Island, NY) according to the manufacturer’s instructions. For cloning p62 variant 1, the following primers were used: hP62T1fTO forward, 5′-CCAC CCTCCGCGTTCGCTACAAAA-3′, hP62T1r reverse, 5′-TGGCTTCTTGCACCCTAACC-3′. For cloning p62 variant 2, the primers were: hP62T2fTO forward, 5′-CCAC TGCAACATGGGGCTTGAGAA-3′, hP62T2r reverse, 5′-CCCTGATCCTGGAAGAAGGC-3′. Recombinant plasmids were verified by sequencing (DNA Core Facility, Johns Hopkins Univ., Baltimore, MD). For p62 variant 1, an alanine substitution mutation was introduced using the Quick Change Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA), with the primer sets: P62S403Af, 5′-ctctcccagatgctggccatgggcttctctg-3′, P62S403Ar, 5′-cagagaagcccatggccagcatctgggagag-3′. Mutation on the plasmid was then verified by DNA sequencing.
2.9 Animals and treatments
All experimental protocols used in this study were in accordance with National Institute Health (NIH) guidelines, and were approved by the Johns Hopkins University Animal Care and Use Committee. Briefly, 2-month old Nrf2 competent (WT) and Nrf2 deficient (Nrf2−/−) mice[24] in a C57BL/6J background were placed in a smoking chamber for 2.5 hours per day, 5 days per week, as described previously[25], or raised in a filtered air environment for 3 weeks. After sacrifice, eyes were enucleated. The RPE/choroid was removed, and either protein or RNA was extracted.
2.10 Statistical analysis
Statistical analysis was carried out using the unpaired t test, with GraphPad software (GraphPad Software, Inc., San Diego, CA). Each experiment was repeated at least three times. Blots are selected as the representative one of specific group of experiments, and graphs represent the mean±SEM of at least three independent experiments.
3 Results
3.1 Expression of alternatively spliced p62 mRNA variants in RPE
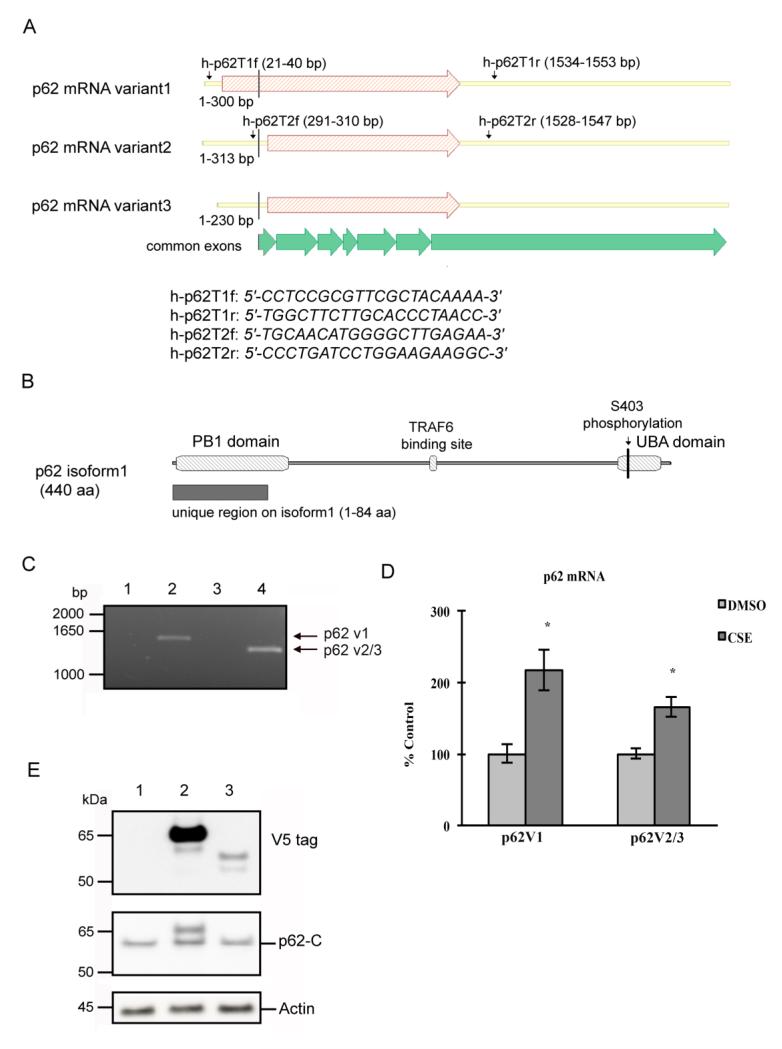
Human p62 pre-mRNA is alternatively spliced and generates three mature mRNA transcripts (Fig. 1A), of which, p62 mRNA variant1 (p62 v1) is the longest and encodes a 440-aa protein. The other two mRNA variants 2 and 3 (p62 v2/3), differ slightly in their 5′UTR regions, and encode p62 isoform2, which is 84 amino acids shorter than isoform1 at the N terminus (Fig.1B). Unlike p62 isoform1, which is abundant in various cell types[26, 27] including RPE cells[11], p62 isoform2’s existence and distribution remain unknown. Previous studies have indicated that the rat expresses three p62 isoforms, and that the isoforms have common interacting partners within the same cell type[28, 29], raising the possibility that human p62 isoforms may be co-expressed in the RPE. Before examining the protective role of p62, we first determined whether human p62 mRNA variant 2/3 is expressed in RPE cells, and whether its expression is coordinately regulated with the p62 mRNA variant1. Total RNA was extracted from cultured ARPE-19 cells and reverse transcribed to amplify the full length coding sequences of p62 variants. As shown in Fig. 1A, primer h-p62T1f is located in the unique 5′UTR of p62 mRNA variant1, while primer h-p62T2f is complementary to a 5′UTR region common in p62 mRNA variant2 and variant3. Primers h-p62T1r and h-p62-T2r are located in the 3′UTR that is common for all three mRNA transcripts. Using primers h-p62T1f and h-p62T1r, a 1533 bp DNA fragment was obtained, and amplification of same cDNA sample with primer h-p62T2f and h-p62T2r generated a product of 1257 bp (Fig. 1C, lanes 2, 4). In both cases, fragments were not obtained using the negative controls, for which the reverse transcriptase was omitted during cDNA synthesis. The PCR products were purified and sequence analysis confirmed that they were identical to the published p62 cDNA sequences. We then conducted SYBR-based qPCR to examine the extent that p62 mRNA variants are differentially expressed by oxidative stress. ARPE-19 cells were treated with DMSO or 125μg/ml CSE, a sublethal dose, for 24hrs. Using primers that specifically amplify p62 v1 or both p62 v2 and p62 v3, we found that the mRNA levels of both p62 v1 and p62 v2/3 variants increased in response to the CSE treatment, suggesting that they are coordinately regulated (Fig. 1D).
Figure 1.
p62 mRNA variants are expressed in the RPE and up-regulated by CSE. (A) Schematic representation showing the structure of human p62 mRNA variants, and the positions of the primers used. The C-terminal regions of all three mRNA variants are transcribed from 7 common exons, as shown in the figure. (B) Schematic representation showing the structure of human p62 isoform 1. S403 phosphorylation site, positions of PB1 domain, TRAF6 binding site and UBA domain are indicated in the figure. (C) Ethidium bromide-stained gel showing the PCR products for p62. Total RNA from ARPE-19 cells was reverse transcribed and amplified using primers h-p62T1f and h-p62T1r (lane 1 and 2), or primers h-p62T2f and h-p62T2r (lane 3 and 4). Negative controls, with reverse transcriptase omitted during cDNA preparation, are shown in land 1 and 3. (D) ARPE-19 cells were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. Total RNA was extracted and RT-qPCR was conducted to quantify the mRNA levels of p62 variant 1, and p62 variants 2/3. Expression was normalized to PPIA and graphed as percent of ctrl (DMSO treated). The graph represents the mean values ± SEM (n=3). *p<0.05. (E) ARPE-19 cells were transfected with empty vector (lane 1), p62 v1 (lane 2), or p62 v2/3 (lane 3) plasmids. The whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibody against V5, p62 C-terminus, or beta actin.
Although the calculated M.W. of p62 isoform1 and isoform2/3 are 47 kDa and 38 kDa respectively, due to complex covalent modifications, the p62 protein band observed in immunoblot analysis is larger than 60 kDa[26]. It is unclear whether this band represents p62 isoform1 or both isoforms. We therefore, cloned p62 v1 and p62 v2/v3 coding sequences into the pcDNA3.1D/V5-HIS-TOPO vector, and over-expressed V5-tagged p62 isoforms in ARPE-19 cells. Whole cell lysates were subjected to immunoblot analysis. Fig. 1E shows that the V5-tagged isoform2 is minimally expressed, with M.W. 10 kDa less than that of V5-tagged isoform1. Endogenous p62 was examined using an antibody against the C-terminus of p62, thus recognizing both isoforms, and a predominant band was present at around 60 kDa, suggesting that isoform2, either tagged or untagged, is barely detectable in ARPE-19 cells. These findings indicate that p62 mRNA variants are present and coordinately stimulated by CSE in RPE cells, yet the p62 v2/3 variants do not generate a significant amount of translational product.
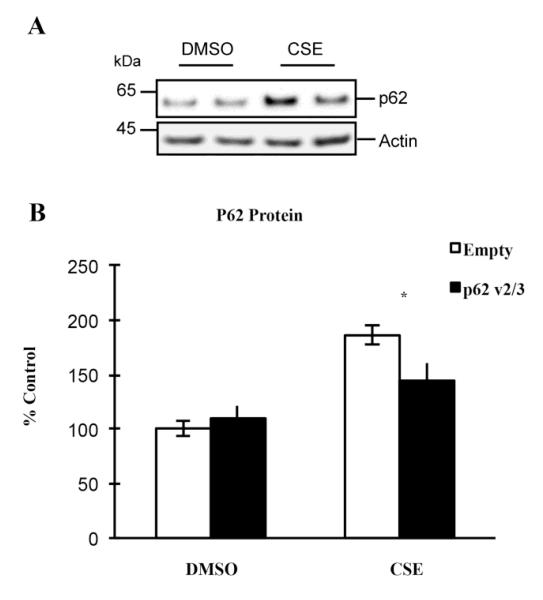
We speculate that translationally inactive p62 v2/3 mRNA transcripts in RPE cells may influence the expression of p62 isoform1 under certain physiological or pathological conditions. To investigate this possibility, we transfected ARPE-19 cells to overexpress p62 v2/3 transcripts, and then treated cells with either DMSO or CSE. By RT-qPCR, we observed that p62 v2/3 variants were highly abundant at the mRNA level (data not shown), yet by western blot, we observed a single p62 band that corresponded to the size of p62 isoform1 (Fig. 2A), which is in accordance with our finding in earlier experiments (Fig. 1E). Our data also show that overexpression of p62 v2/3 transcripts suppressed the expression of endogenous p62 under stress, such as after CSE treatment, but not under basal conditions (Fig. 2A-B), which suggests that these translationally inactive mRNA transcripts have a regulatory role in RPE cells. In all subsequent text, p62 refers to p62 isoform1.
Figure 2.
Over-expression of p62 variant 2/3 suppresses the endogenous p62 expression in RPE, under stressed conditions. (A) ARPE-19 cells were transfected with empty vector, or p62 v2/3 plasmid, and treated with 125ug/ml CSE or an equal volume of DMSO for 24hrs. The whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibody against p62. (B) Band intensity was quantified using GE Healthcare ImageQuant TL software. Data were normalized for the amount of beta actin in each sample and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). Each point represents the average ± SEM of three independent experiments. Asterisk, p < 0.05.
3.2 p62 promotes autophagic clearance of protein aggregates and reciprocally regulates Nrf2 signaling in RPE cells
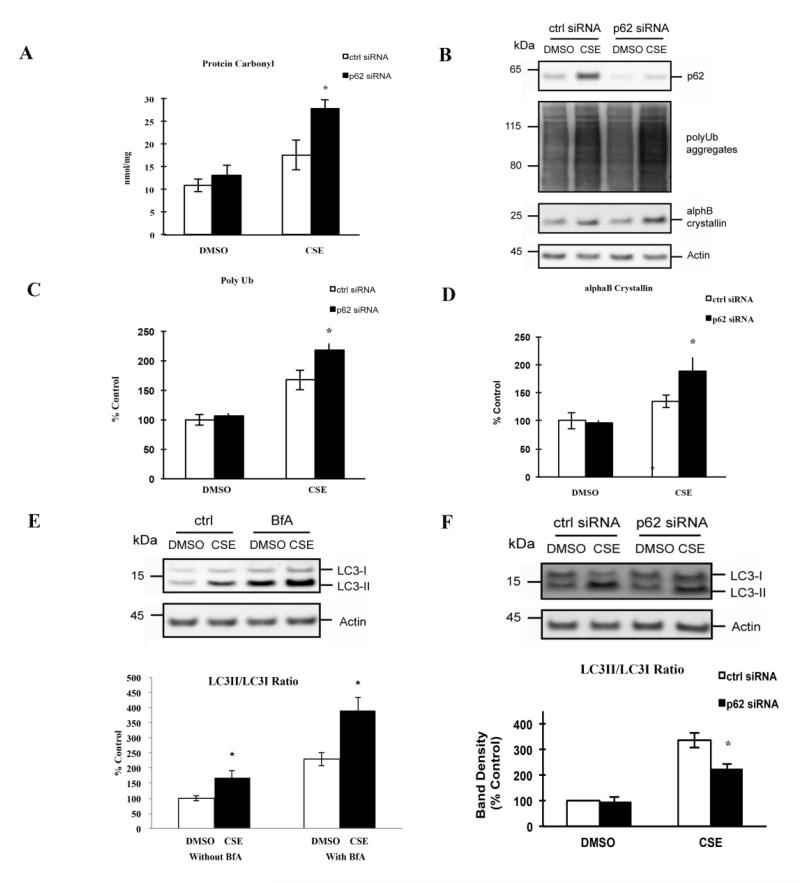
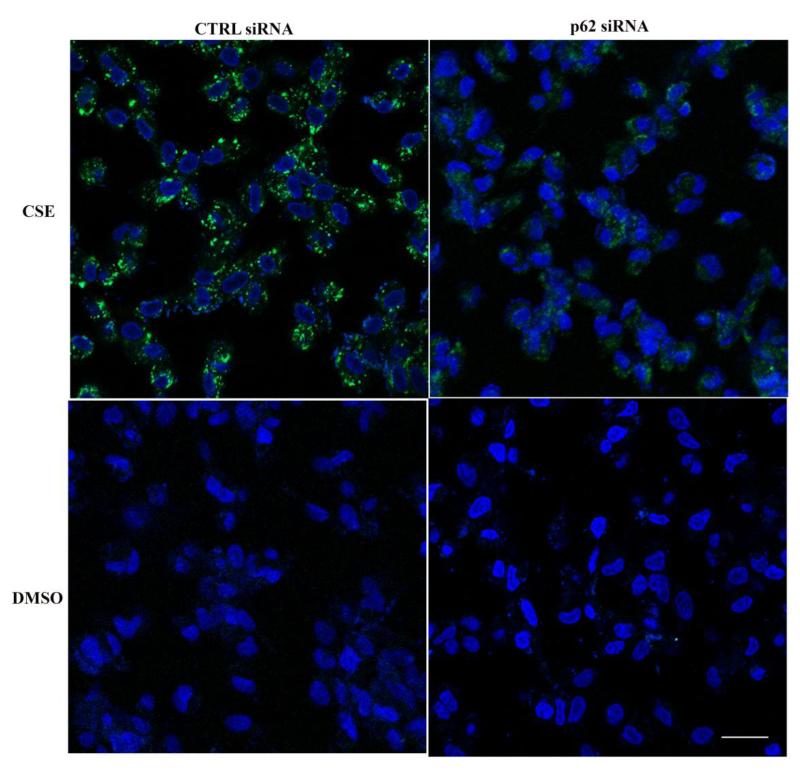
The removal of modified/misfolded proteins relies on several mechanisms, including heat-shock proteins, proteasomes, and autophagy, which are interdependent with one another. An earlier study demonstrated that inhibiting the ubiquitin-proteasome pathway increased p62, which co-localized with polyubiquitinated protein aggregates in ARPE-19 cells[11]. These results suggest that p62-mediated autophagy assumes a central role in processing polyubiquitinated proteins when another clearance mechanism has become dysfunctional. To extend the concepts from this study further, we examined, without completely blocking the proteasome pathway, the extent that p62, and specifically isoform1, actively protects ARPE-19 cells by facilitating autophagy and reciprocal activation of Nrf2 mediated antioxidant responses. After treatment with a sublethal dose of CSE (125μg/ml), ARPE-19 cells had increased protein oxidation, as measured by carbonyl ELISA (Fig. 3A), which was accompanied by an accumulation of polyubiquitinated proteins and αB crystallin (Fig. 3B-D). We chose αB crystallin because it is a chaperone that is induced and becomes aggregated during stress[30, 31], and accumulates in drusen, a hallmark sign of AMD[32], thus, serving as a relevant biomarker for RPE stress. Cells exhibited a protective, activated autophagic response to CSE, as determined by LC3 conversion, rather than accumulation of LC3 by blocking autophagy (Fig. 3E), with the appearance of autophagic vesicles by Cyto-ID labeling (Fig. 4).
Figure 3.
Effect of p62 silencing on the CS-induced protein damages and autophagic protection in RPE. (A) ARPE-19 cells, after transfection with ctrl or p62 siRNA, were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. Whole cell lysates, prepared in Buffer A, were diluted to 5μg/ml and coated onto the protein carbonyl ELISA plate. Data are presented as the mean±SEM of three independent experiments. *p<0.05. (B) ARPE-19 cells, after transfection with ctrl or p62 siRNA, were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62, polyUb, αB crystallin, or beta actin. (C, D) Quantification of polyubiquitinated proteins and αB crystallin. Band intensities were quantified using GE Healthcare ImageQuant TL software. Data were normalized using beta actin and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). The graphs represent the mean±SEM of three independent experiments. *p<0.05. (E) ARPE-19 cells were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs, in the absence or presence of 50nM BfA, an autophagy inhibitor. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against LC3 or beta actin. (F) ARPE-19 cells, after transfection with ctrl or p62 siRNA, were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibody against LC3. Band intensities were quantified using GE Healthcare ImageQuant TL software. The LC3-II/C3-I ratio was calculated and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). The graph below represents the mean±SEM of three independent experiments. *p<0.05.
Figure 4.
p62 silencing reduces autophagosomes. ARPE-19 cells, grown on glass bottom plates, were transfected with ctrl or p62 siRNA and then treated with either 125μg/ml CSE or DMSO for 24hrs. Cells were co-stained with Hoechst 33342 for nuclei (blue) and Cyto-ID (green) for autophagosomes, and viewed with a confocal microscope. Bar=25μm.
To determine the functional impact of p62 after CSE induced oxidative stress, we conducted loss- and gain-of-function studies. Under stressed conditions, p62 silencing increased protein carbonylation (Fig. 3A), the aggregation of polyubiquitinated proteins and αB crystallin production (Fig. 3B-D), and reduced the LC3 conversion (Fig. 3F) and Cyto-ID labeled autophagic vesicles (Fig. 4) over control siRNA treatment. The addition of bafilomycin to cells treated with CSE did not alter the reduced LC3 conversion induced by p62 silencing (Fig. 2S). These results suggest that p62 silencing exacerbates cellular stress by impairing autophagic removal of damaged proteins.
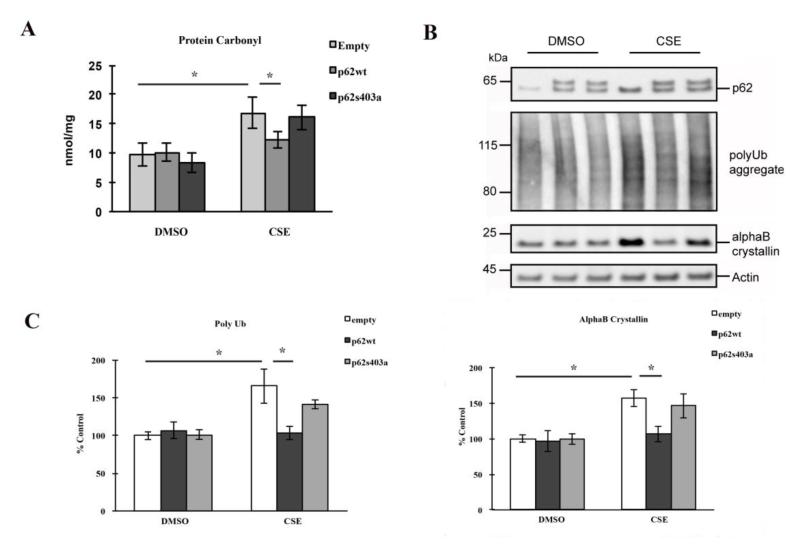
In contrast, over-expression of p62 isoform1, the main isoform produced by RPE cells, reduced protein carbonylation (Fig. 5A), polyubiquitinated proteins, and αB crystallin (Fig. 5B-C), thus rescuing cells from CSE induced stress, compared to empty vector controls treated with CSE. Interestingly, under basal conditions, neither p62 knockdown nor over-expression led to significant changes in the above markers compared to controls, suggesting that p62 mediated autophagy is a proteolytic mechanism that deals with excessive modified/damages proteins exclusively under stressed conditions.
Figure 5.
Over-expression of p62 rescues RPEs by reducing protein damages, and phosphorylation at S403 is crucial for the protection. ARPE-19 cells were transfected with empty vector or wild-type/mutant p62 plasmids, and treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. (A) Whole cell lysates, prepared in Buffer A, were diluted to 5μg/ml and coated onto the protein carbonyl ELISA plate. Data are presented as mean±SEM of 3 independent experiments. *p<0.05. (B) Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62, polyUb, αB crystalline or beta actin. (C) Quantification of polyubiquitinated proteins and αB crystallin. Band intensities were quantified using GE Healthcare ImageQuant TL software. Data were normalized using beta actin and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). The graph represents the mean±SEM of three independent experiments. *p<0.05.
Matsumoto et al. reported that phosphorylation of p62 at S403, which is located in the UBA domain (Fig. 1B), is crucial for its affinity to polyubiquitinated proteins[1]. Lau et al showed that p62 regulates Nrf2 signaling by interacting with Keap1 through a region mapped to 349-DPSTGE-354 (Fig. 1B)[33]. We first wanted to determine the extent that p62 mediated autophagic clearance relies on phosphorylation in its UBA domain under CS induced oxidative stress, and secondly, to test the hypothesis that a nonphosphorylated mutation (S403A) will negatively affect p62’s ability to rescue RPE cells through autophagy without influencing p62’s activation of Nrf2 signaling during CS induced oxidative stress. ARPE-19 cells were treated with CSE or DMSO for 24hrs. While over-expression of wild type p62 isoform1 reduced protein carbonylation, polyubiquitination, and αB crystallin accumulation, in contrast, ARPE-19 cells that over-expressed the p62 S403A non-phosphorylated mutant were unable to reduce the levels of protein carbonylation, polyubiquitination, and αB crystallin (Fig. 5A-C). These results indicate a crucial role for phosphorylation in p62-mediated autophagy in RPE cells.
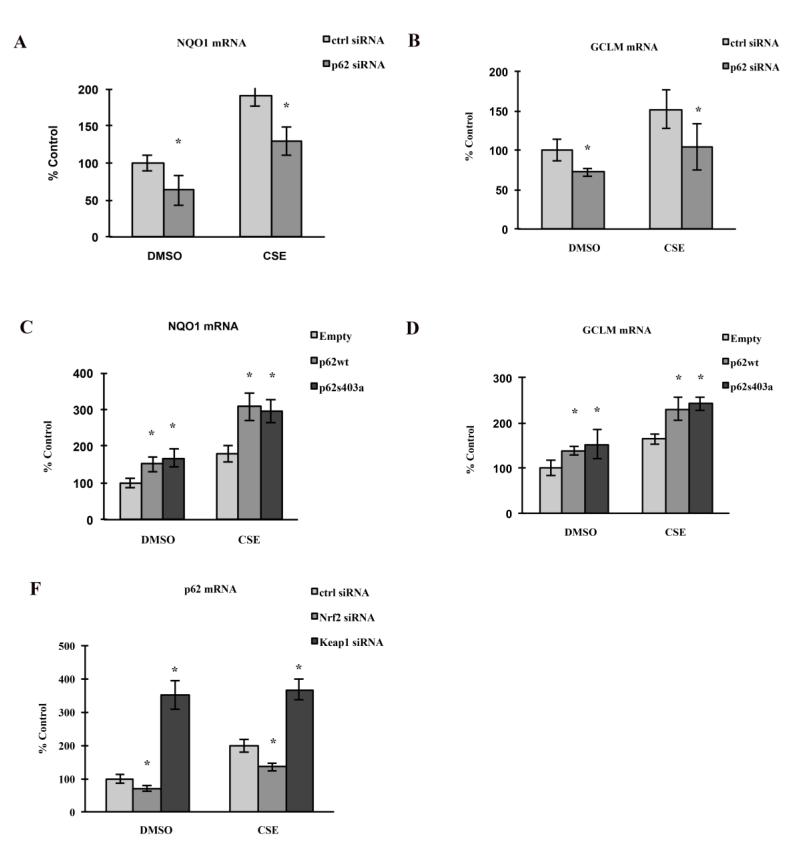
In our previous studies, we showed that CSE transiently induced Nrf2 mediated antioxidant responses in RPE[21]. While p62 has been reported to contain ARE sites in its promoter[34], it also interacts with the Nrf2-binding site on Keap1, a component of Cullin-3-type ubiquitin ligase for Nrf2, thus enhancing the Nrf2 signaling, and possibly generating a positive feedback loop. We predict that p62, in additional to its role in autophagic clearance, also protects the RPE by reciprocally activating Nrf2 signaling. Again, using a loss- and gain-of-function strategy, under both basal and stressed conditions, p62 silencing reduced Nrf2 signaling, as measured by the expression of Nrf2 downstream genes – Nqo1 and Gclm (Fig. 6A-B). In contrast, under both basal and stressed conditions, over-expression of p62 isoform1 significantly enhanced the expression of Nqo1 and Gclm (Fig. 6C-D). Likewise, Nrf2 silencing in ARPE-19 cells decreased p62 expression (Fig. 6F), and Keap1 silencing, which enhances Nrf2 signaling, drastically increased p62 expression (Fig. 6F). These results highlight the interdependent interaction between p62 and Nrf2 that results in an Nrf2 mediated antioxidant response. Based on these data, we conclude that p62 protects against CSE induced oxidative stress by clearing oxidized/misfolded proteins and mediating activation of the Nrf2 antioxidant system.
Figure 6.
Reciprocal regulation of p62 expression and Nrf2 signaling, under basal and stressed conditions. ARPE-19 cells were transfected for p62 silencing (A, B), over-expression of wild type/mutant p62 (C, D), or silencing of Nrf2 or Keap1 (E). After transfection, cells were treated with 125μg/ml CSE or an equal volume of DMSO for 24hrs. Total RNA was extracted for RT-qPCR to quantify the expression of Nqo1, Gclm or p62. Mean values±SEM (n=3 independent experiments) were normalized to PPIA and graphed as percent of ctrl (transfected with ctrl siRNA/DNA, DMSO treated). *p<0.05.
We then examined the effect of p62 S403A overexpression on Nrf2 signaling, and observed upregulation of both Nqo1 and Gclm (Fig. 6C-D), which indicates that p62 activated Nrf2 signaling in the RPE is independent of the S403 site, and hence, p62 mediated autophagic clearance. Importantly, these results also highlight how p62 augments the Nrf2 mediated antioxidant response to a stressor like CSE, with magnified expression of Nqo1 and Gclm over cells without p62 transfection and treatment with CSE.
3.3 Examination of the protective role of p62 in vivo
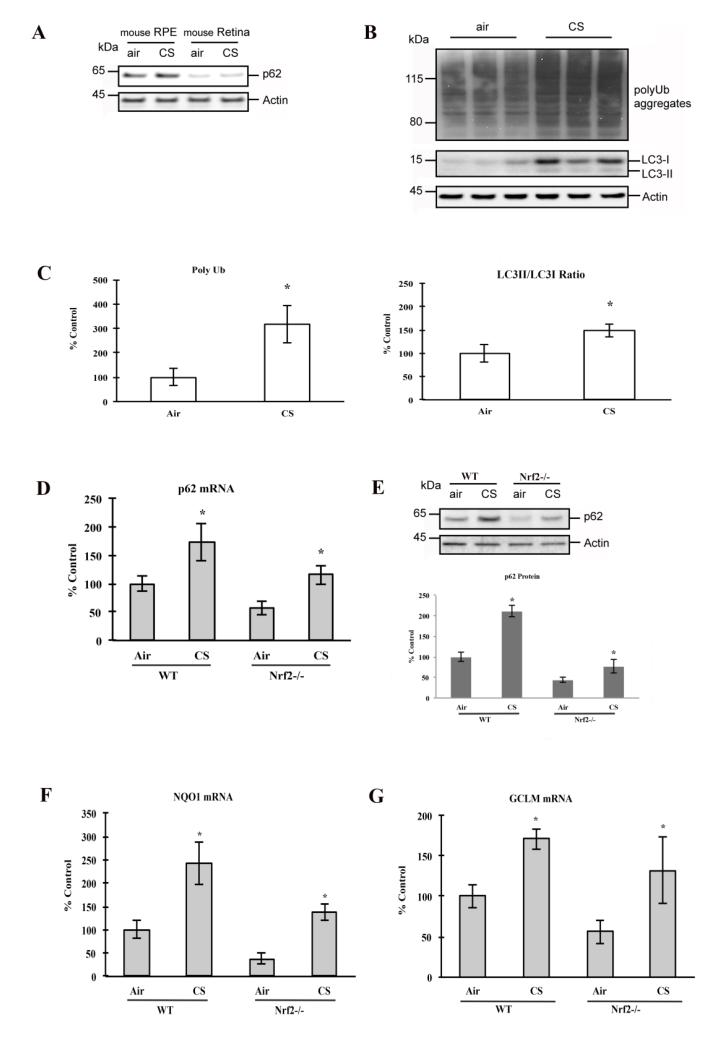
To extend our findings to a complex biological environment as in vivo, we examined the extent that cigarette smoke (CS) exposure induces aggregation of misfolded proteins in mouse RPE, and whether p62 modulates the autophagic clearance of these proteins. Two-month old WT mice were then exposed to CS in a smoking chamber or filtered air environment for 3 weeks. We first examined the expression of p62 in the fundus of 2-month old C57Bl6/J wild type (WT) mice by Western blot analysis, and found that p62 expression was 7.1-fold (p=0.0008; n=4 eyes per group) and 6.9-fold (p=0.0001; n=4) higher in the RPE than in the adjacent neurosensory retina of mice raised in air and CS for 3 weeks, respectively, (Fig. 7A), which is consistent with the RPE’s well known powerful antioxidant system and its role in phagocytosis of photoreceptor outer segments. The RPE of mice exposed to CS had increased protein polyubiquitination, and activated autophagic protection, as measured by LC3 conversion (Fig. 7B-C), compared to mice kept in air. To test the interaction between p62 and Nrf2 signaling, we exposed WT mice and mice deficient in Nrf2 signaling (Nrf2−/−) for 3 weeks in air or CS. With RT-qPCR and western blot, we observed that smoking enhanced p62 expression in both WT and Nrf2−/− mice (Fig. 7D-E). In Nrf2−/− mice, p62 abundance is significantly lower than in WT mice, as are the expression of Nqo1 and Gclm (Fig. 7F-G), which suggests that p62 is regulated in part, through Nrf2 signaling. These in vivo observations correlate with the response seen in ARPE-19 cells, where p62 mediated cytoprotection is in part, through Nrf2 signaling. Our data show that p62 is increased after exposure to CS, and is associated with a cytoprotective response in the RPE against smoking in vivo.
Figure 7.
CS-induced protein damage and protective responses in mouse RPE/choroid. 2-mo old WT and Nrf2−/− mice were raised in a smoking chamber or filtered air environment for 3 weeks and then sacrificed, and eyes were enucleated. (A) Whole cell lysates from RPE/choroid and retina of WT mice were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62 or beta actin. (B) Whole cell lysates from RPE/choroid of WT and Nrf2−/− mice were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against polyUb, LC3, or beta actin. (C) Quantification of polyubiquitinated proteins and LC3. Lane and Band intensity were quantified using GE Healthcare ImageQuant TL software. The amount of polyubiquitinated proteins was normalized using beta actin, and graphed as percent of control (ctrl; WT raised in air). *p<0.05. The LC3-II/LC3-I ratio was calculated, and graphed as percent of control (ctrl; WT raised in air). *p<0.05. (D) Total RNA was extracted from RPE/choroid of WT and Nrf2−/− mice, and RT-qPCR quantified the expression of p62. Mean values±SEM (n=3 independent experiments) were normalized to PPIA and graphed as percent of control (ctrl; WT raised in air). *p<0.05. (E) Whole cell lysates from RPE/choroid of WT and Nrf2−/− mice were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62 or beta actin. Band intensity was quantified using GE Healthcare ImageQuant TL software. Data were normalized using beta actin and graphed as percent of control (ctrl; WT raised in air). Data represent the mean±SEM of three independent experiments. *p<0.05. Total RNA was extracted from RPE/choroid of WT and Nrf2−/− mice, and RT-qPCR quantified the expression of Nqo1 (F) and Gclm (G). Mean values±SEM (n=3 independent experiments) were normalized to PPIA and graphed as percent of control (ctrl; WT raised in air). *p<0.05.
4 Discussion
p62 binds to ubiquitinated proteins and facilitates degradation through autophagic clearance since the proteasome is incapable of removing large protein aggregates[1]. For many age-related diseases including AMD, oxidative stress causes protein misfolding/damage, and upon polyubiquitination, these proteins aggregate and accumulate in cytoplasmic inclusions. As a result, p62 has gained attention as a potential therapeutic target for age-related diseases that would facilitate autophagy, and reduce toxic protein accumulations. p62 however, is a multifunctional protein that mediates different cellular events under different conditions, and therefore, requires more study before it can be considered a therapeutic target. p62 was originally studied because of its tight association with Ser/Thr kinase[26], and has since been found to be involved in diverse signaling events, including phosphorylation of potassium channels in neurons[28] and promoting caspase-8 aggregation during apoptosis[35]. Thus, p62 is a multi-dimensional protein that acts as a signaling hub, interacting with different protein partners to regulate multiple cellular functions, including cell survival, inflammation, apoptosis, and autophagy.
Our investigation of p62 has two purposes. Since the human p62 isoform expression patterns are unknown, we first wanted to examine the expression of these isoforms in RPE cells, and then investigate their individual functional impact in response to oxidative stress. Our results demonstrate that while all three alternatively spliced transcripts of p62 are abundantly expressed in ARPE-19 cells, at the translational level, isoform1, encoded by p62 v1 mRNA, is predominant, and isoform2 protein remained at minimal levels under both basal and stressed conditions. The rat expresses three p62 isoforms[28, 29] in the hippocampus, of which isoform1 differs from isoform2 by 27 amino acids. The ratio of rat p62 isoform1/isoform2 is not only tissue specific, but is also dynamically regulated in response to stimulation within the same cell type[28]. We acknowledge that the distribution of human p62 isoforms in other tissues is unknown, and that these isoforms could have different functions under different physiological and pathological conditions. In our experiments, overexpression of p62 v2/3 suppressed the protein levels of p62 isoform1, but only under the stressed condition of CSE exposure. Human p62 isoform1 and isoform2 are identical, except that isoform2 is missing 84 residues within a major portion of the PB1 domain at the N terminus. Maintenance of a gene that is transcriptionally active, but translationally inactive, may provide a mechanism that facilitates adaptation to environmental stress[36, 37]. The p62 isoforms v1 and v2 have calculated molecular weights of 47 kDa and 38 kDa respectively, yet the predominant p62 isoform1 band was larger than 60 kDa, indicating that it undergoes covalent modification. Since both p62 isoforms undergo covalent modification, we hypothesize that p62 isoform2, due to its missing PB1 domain at N-terminus, might be improperly modified, and thus has a shorter half-life compared to isoform1. Our observation that p62 isoform2 over-expression led to suppression of isoform1 protein could be explained by their competition for common chaperones or enzymes that facilitate protein modifications. This hypothesis will be investigated in future work.
The structure and functional domains of p62 have been identified, and this information can help us unravel the multifunctional nature of p62[38, 39]36,37. Past investigations however, have studied each single p62 function separately, and not in combination. To address this shortcoming, the second purpose of this study was to determine the extent that two of p62’s functions together, provide cytoprotection. We chose p62’s role in aggrephagy and the antioxidant response due to the high oxidative stress burden and phagocytic role of the RPE. Using loss- and gain-of-function studies, we found that p62 isoform1 is critical for promoting autophagic clearance of polyubiquitinated proteins and protein aggregates. Phosphorylation of the UBA domain is crucial for p62 to guide polyubiquitinated protein aggregates to the autophagy machinery[1]. Within the UBA domain, serine 403 is in particular, important for determining the binding affinity of p62 to ubiquitin. By introducing a mutation at S403 that prevents phosphorylation, we showed that p62 mediated autophagic clearance was abrogated. Since the KEAP1 interacting region, which is upstream of the UBA domain35,36, was unaffected by this mutation, Nrf2 signaling was activated after overexpressing this non-phosphorylated p62 mutant, supporting the notion that these two protective mechanisms rely on distinct functional domains of p62. Importantly, we found that p62 loss- and gain-of-function experiments decreased and increased Nrf2 signaling, respectively. Because p62 contains ARE sites in its promoter[17], it can be reciprocally activated by Nrf2 signaling, forming a positive feedback loop by enhancing both p62 production and Nrf2 signaling in RPE cells. These results support the concept that p62 functions as a signaling hub, and through its different functional domains, interacts with different partners mediating separate cellular events. In particular, p62 has a positive impact on autophagy and the Nrf2 mediated antioxidant response under oxidative stress.
The difference between p62 isoform1 and isoform2 is that the majority of the PB1 domain is absent from isoform2 (Fig. 1B). The PB1 domain is involved with p62’s role in activating NF-kB signaling[40, 41]. Since NF-kB activation generates inflammation during age-related diseases[42], including AMD[43-45], it is possible that suppression of isoform1 by isoform2 in RPE cells might be a response to prevent unwanted inflammation without interfering with Nrf2 signaling. Since isoform2 is devoid of the PB1 domain that activates NF-kB, it raises the possibility of facilitating autophagy and Nrf2 mediated antioxidant responses without generating excessive inflammation. When autophagy fails, p62 accumulates in perinuclear inclusions in several neurodegenerative diseases, as well as in macular RPE of AMD samples[4, 10, 46, 47]. If this accumulation is composed predominantly of isoform1, as our results suggest, then this accumulation could foster an Nrf2 cytoprotective response and induce unwanted NF-kB mediated inflammation, including the production of pro-inflammatory cytokines IL-1b and TNF-alpha[48]. On the other hand, if there would be an accumulation of isoform2, then the accumulation could induce Nrf2 signaling without activating NF-kB mediated inflammation. A better understanding of both the isoform distribution and their function will help to provide a greater understanding of the role of p62, whether protecting or contributing to age-related disease.
In conclusion, our study for the first time, shows that in response to a complex oxidant such as CS, p62 protects the RPE by reducing protein aggregates and contributes to activating an Nrf2 antioxidant response. Our in vitro experimental results were strengthened by similar findings in the complex biological environment of mouse experiments, and suggest that both models are suitable for further investigating the protective roles of p62 in response to stress. In our studies, we found that p62 isoform1 was the predominant protective factor, while p62 isoform2 is transcriptionally active but translationally inactive. Our studies were conducted in the ARPE-19 cell line, which is derived from a 19 year old donor. Likewise, our experiments were conducted in young mice. We acknowledge that the responses we report herein could be different in either old cells or old mice. Considering p62’s very diverse roles by interacting with multiple partners, we will further investigate the functional roles of p62 isoforms under different stressed conditions and with aging, aiming to elucidate the precise connection of p62 with the inability of the RPE to manage damaged proteins at different stages of AMD.
Supplementary Material
Supplemental Figure 1: Transfection reagent and ctrl siRNA have negligible influence on Nrf2 signaling or protein polyubiquitination in cultured ARPE-19 cells. ARPE-19 cells were cultured in regular medium, or medium plus transfection reagent and ctrl siRNA, for 8 hrs and continuously cultured in serum free media for 48 hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62, LC3. Band intensity was quantified using GE Healthcare ImageQuant TL software. Data were normalized to beta actin and graphed as percent of control (cultured in media only).
Supplementary Figure 2 Bafilomycin does not influence the decline in LC3II/LC3I ratio by p62 knockdown. ARPE-19 cells, after transfection with ctrl or p62 siRNA, were treated with 125μg/ml CSE or an equal volume of DMSO in the absence or presence of 50nM BfA, an autophagy inhibitor, for 24hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibody against LC3. Band intensities were quantified using GE Healthcare ImageQuant TL software. The LC3-II/C3-I ratio was calculated and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). The graph below represents the mean±SEM of three independent experiments. *p<0.05.
Highlights.
p62 mRNA variants are expressed by RPE cells
p62 isoform2 is not translated, but regulates isoform1 abundance with stress
p62 has dual, reciprocal enhancing protection through its different binding domains
Acknowledgments and Funding
EY019904 (JTH), EY14005 (JTH), Beckman Foundation AMD Grant (JTH), Thome Foundation (JTH), Research to Prevent Blindness Senior Scientist Award (JTH), NIH P30EY001765 core grant, the Robert Bond Welch Professorship (JTH), a gift from the Merlau family, and an Unrestricted grant from RPB to the Wilmer Eye Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: none
References
- [1].Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Molecular cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- [2].Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. International journal of cell biology. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. Journal of neurochemistry. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- [4].Braak H, Thal DR, Del Tredici K. Nerve cells immunoreactive for p62 in select hypothalamic and brainstem nuclei of controls and Parkinson’s disease cases. J Neural Transm. 2011;118:809–819. doi: 10.1007/s00702-010-0508-2. [DOI] [PubMed] [Google Scholar]
- [5].Rue L, Lopez-Soop G, Gelpi E, Martinez-Vicente M, Alberch J, Perez-Navarro E. Brain region- and age-dependent dysregulation of p62 and NBR1 in a mouse model of Huntington’s disease. Neurobiology of disease. 2013;52:219–228. doi: 10.1016/j.nbd.2012.12.008. [DOI] [PubMed] [Google Scholar]
- [6].Salminen A, Kaarniranta K, Haapasalo A, Hiltunen M, Soininen H, Alafuzoff I. Emerging role of p62/sequestosome-1 in the pathogenesis of Alzheimer’s disease. Progress in neurobiology. 2012;96:87–95. doi: 10.1016/j.pneurobio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- [7].Plafker SM, O’Mealey GB, Szweda LI. Mechanisms for countering oxidative stress and damage in retinal pigment epithelium. International review of cell and molecular biology. 2012;298:135–177. doi: 10.1016/B978-0-12-394309-5.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ershov AV, Bazan NG. Photoreceptor phagocytosis selectively activates PPARgamma expression in retinal pigment epithelial cells. J Neurosci Res. 2000;60:328–337. doi: 10.1002/(SICI)1097-4547(20000501)60:3<328::AID-JNR7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [9].Kaarniranta K, Salminen A, Haapasalo A, Soininen H, Hiltunen M. Age-related macular degeneration (AMD): Alzheimer’s disease in the eye? Journal of Alzheimer’s disease : JAD. 2011;24:615–631. doi: 10.3233/JAD-2011-101908. [DOI] [PubMed] [Google Scholar]
- [10].Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D’Agostino VG, Govoni S, Pascale A, Agostini H, Petrovski G, Salminen A, Kaarniranta K. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013;8:e69563. doi: 10.1371/journal.pone.0069563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Viiri J, Hyttinen JM, Ryhanen T, Rilla K, Paimela T, Kuusisto E, Siitonen A, Urtti A, Salminen A, Kaarniranta K. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol Vis. 2010;16:1399–1414. [PMC free article] [PubMed] [Google Scholar]
- [12].Kaarniranta K, Sinha D, Blasiak J, Kauppinen A, Vereb Z, Salminen A, Boulton ME, Petrovski G. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy. 2013;9:973–984. doi: 10.4161/auto.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Uchiki T, Weikel KA, Jiao W, Shang F, Caceres A, Pawlak D, Handa JT, Brownlee M, Nagaraj R, Taylor A. Glycation-altered proteolysis as a pathobiologic mechanism that links dietary glycemic index, aging, and age-related disease (in nondiabetics) Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bertram KM, Baglole CJ, Phipps RP, Libby RT. Molecular regulation of cigarette smoke induced-oxidative stress in human retinal pigment epithelial cells: implications for age-related macular degeneration. Am J Physiol Cell Physiol. 2009;297:C1200–1210. doi: 10.1152/ajpcell.00126.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cano M, Thimmalappula R, Fujihara M, Nagai N, Sporn M, Wang AL, Neufeld AH, Biswal S, Handa JT. Cigarette smoking, oxidative stress, the anti-oxidant response through Nrf2 signaling, and Age-related Macular Degeneration. Vision Res. 2010;50:652–664. doi: 10.1016/j.visres.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- [17].Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cano M, Wang L, Wan J, Barnett BP, Ebrahimi K, Qian J, Handa JT. Oxidative stress induces mitochondrial dysfunction and a protective unfolded protein response in RPE cells. Free Radic Biol Med. 2014;69C:1–14. doi: 10.1016/j.freeradbiomed.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- [20].Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang L, Kondo N, Cano M, Ebrahimi K, Yoshida T, Barnett BP, Biswal S, Handa JT. Nrf2 signaling modulates cigarette smoke-induced complement activation in retinal pigmented epithelial cells. Free Radic Biol Med. 2014 doi: 10.1016/j.freeradbiomed.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Klappan AK, Hones S, Mylonas I, Bruning A. Proteasome inhibition by quercetin triggers macroautophagy and blocks mTOR activity. Histochemistry and cell biology. 2012;137:25–36. doi: 10.1007/s00418-011-0869-0. [DOI] [PubMed] [Google Scholar]
- [23].Lee JS, Lee GM. Monitoring of autophagy in Chinese hamster ovary cells using flow cytometry. Methods. 2012;56:375–382. doi: 10.1016/j.ymeth.2011.11.006. [DOI] [PubMed] [Google Scholar]
- [24].Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fujihara M, Nagai N, Sussan TE, Biswal S, Handa JT. Chronic cigarette smoke causes oxidative damage and apoptosis to retinal pigmented epithelial cells in mice. PLoS ONE. 2008;3:e3119. doi: 10.1371/journal.pone.0003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci U S A. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gong J, Xu J, Bezanilla M, van Huizen R, Derin R, Li M. Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science. 1999;285:1565–1569. doi: 10.1126/science.285.5433.1565. [DOI] [PubMed] [Google Scholar]
- [29].Croci C, Brandstatter JH, Enz R. ZIP3, a new splice variant of the PKC-zeta-interacting protein family, binds to GABAC receptors, PKC-zeta, and Kv beta 2. J Biol Chem. 2003;278:6128–6135. doi: 10.1074/jbc.M205162200. [DOI] [PubMed] [Google Scholar]
- [30].De S, Rabin DM, Salero E, Lederman PL, Temple S, Stern JH. Human retinal pigment epithelium cell changes and expression of alphaB-crystallin: a biomarker for retinal pigment epithelium cell change in age-related macular degeneration. Arch Ophthalmol. 2007;125:641–645. doi: 10.1001/archopht.125.5.641. [DOI] [PubMed] [Google Scholar]
- [31].Sugiyama M, Fujii N, Morimoto Y, Itoh K, Mori K, Fukunaga T, Fujii N. SAXS and SANS observations of abnormal aggregation of human alpha-crystallin. Chemistry & biodiversity. 2010;7:1380–1388. doi: 10.1002/cbdv.200900332. [DOI] [PubMed] [Google Scholar]
- [32].Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Molecular and cellular biology. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci U S A. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stepkowski TM, Kruszewski MK. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic Biol Med. 2011;50:1186–1195. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- [36].Martinez-Campos E, Hernandez-SanMiguel E, Lopez-Sanchez C, De Pablo F, Hernandez-Sanchez C. Alternative splicing variants of proinsulin mRNA and the effects of excess proinsulin on cardiac morphogenesis. FEBS letters. 2013;587:2272–2277. doi: 10.1016/j.febslet.2013.05.060. [DOI] [PubMed] [Google Scholar]
- [37].Yang CK, Yen P. Differential translation of Dazap1 transcripts during spermatogenesis. PLoS One. 2013;8:e60873. doi: 10.1371/journal.pone.0060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moscat J, Diaz-Meco MT, Wooten MW. Of the atypical PKCs, Par-4 and p62: recent understandings of the biology and pathology of a PB1-dominated complex. Cell Death Differ. 2009;16:1426–1437. doi: 10.1038/cdd.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Babu J. Ramesh, Seibenhener M. Lamar, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. Journal of neurochemistry. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- [41].Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang Q, Kang R, Zeh HJ, 3rd, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy. 2013;9:451–458. doi: 10.4161/auto.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tseng WA, Thein T, Kinnunen K, Lashkari K, Gregory MS, D’Amore PA, Ksander BR. NLRP3 inflammasome activation in retinal pigment epithelial cells by lysosomal destabilization: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2013;54:110–120. doi: 10.1167/iovs.12-10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sugano E, Isago H, Murayama N, Tamai M, Tomita H. Different anti-oxidant effects of thioredoxin 1 and thioredoxin 2 in retinal epithelial cells. Cell structure and function. 2013;38:81–88. doi: 10.1247/csf.12025. [DOI] [PubMed] [Google Scholar]
- [45].Ling D, Liu B, Jawad S, Thompson IA, Nagineni CN, Dailey J, Chien J, Sredni B, Nussenblatt RB. The tellurium redox immunomodulating compound AS101 inhibits IL-1beta-activated inflammation in the human retinal pigment epithelium. Br J Ophthalmol. 2013;97:934–938. doi: 10.1136/bjophthalmol-2012-301962. [DOI] [PubMed] [Google Scholar]
- [46].Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- [47].Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathology and applied neurobiology. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- [48].Park S, Ha SD, Coleman M, Meshkibaf S, Kim SO. p62/SQSTM1 enhances NOD2-mediated signaling and cytokine production through stabilizing NOD2 oligomerization. PLoS One. 2013;8:e57138. doi: 10.1371/journal.pone.0057138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Transfection reagent and ctrl siRNA have negligible influence on Nrf2 signaling or protein polyubiquitination in cultured ARPE-19 cells. ARPE-19 cells were cultured in regular medium, or medium plus transfection reagent and ctrl siRNA, for 8 hrs and continuously cultured in serum free media for 48 hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibodies against p62, LC3. Band intensity was quantified using GE Healthcare ImageQuant TL software. Data were normalized to beta actin and graphed as percent of control (cultured in media only).
Supplementary Figure 2 Bafilomycin does not influence the decline in LC3II/LC3I ratio by p62 knockdown. ARPE-19 cells, after transfection with ctrl or p62 siRNA, were treated with 125μg/ml CSE or an equal volume of DMSO in the absence or presence of 50nM BfA, an autophagy inhibitor, for 24hrs. Whole cell lysates were subjected to 4-12% SDS-PAGE and immunoblotted with antibody against LC3. Band intensities were quantified using GE Healthcare ImageQuant TL software. The LC3-II/C3-I ratio was calculated and graphed as percent of ctrl (transfected with ctrl siRNA, DMSO treated). The graph below represents the mean±SEM of three independent experiments. *p<0.05.