Dynamic remodeling of the microtubule cytoskeleton is essential for many cell processes including division, migration and differentiation. Microtubules are dynamic polymers of α/β-tubulin dimers and switch stochastically between phases of growth (polymerization) and shortening (depolymerization). Transitions from growth to shortening are referred to as catastrophes, and transitions from shortening to growth are called rescues. Intracellular microtubule organization is controlled by the activity and distribution of nucleation sites, proteins that directly influence polymerization dynamics, proteins that cut or bundle existing microtubules, and proteins that indirectly stabilize microtubules. In cells, microtubule nucleation is mediated by the multi-subunit γ-Tubulin Ring Complex. Associated proteins such as Cep192 recruit the γ-TuRC to the centrosome or spindle poles. However, regulation of microtubule nucleation is incompletely understood. In differentiated cells, microtubule minus ends are often redistributed away from the centrosome. For example, ninein anchors minus ends to cell-cell junctions in polarized epithelial cells and to desmosomes in differentiated keratinocytes. Several different proteins influence microtubule polymerization dynamics by direct interactions with microtubule plus ends. EB-family proteins directly recognize biochemical or structural differences at the growing microtubule end, influence microtubule polymerization dynamics, and may act as adaptors that mediate the association of other proteins with growing microtubule plus ends. Proteins of the XMAP215 family are thought to enhance the microtubule polymerization rate by processively adding tubulin subunits to the growing end. In contrast, neuronal CRMP-2 enhances microtubule assembly by interacting with free tubulin dimers. Microtubule disassembly is promoted by two different mechanisms. Stathmin is a small protein that binds to tubulin dimers and lowers the pool of free tubulin available for polymerization, thus decreasing microtubule growth rate and increasing catastrophe frequency. In contrast, internal motor domain kinesins directly bind to microtubule ends and utilize ATP hydrolysis to catalytically depolymerize microtubules. Microtubule-severing proteins cut existing microtubules generating new minus and plus ends. The generation of short microtubule fragments by these proteins is important for reorganization of the microtubule cytoskeleton without complete microtubule disassembly. Because organization and dynamics of the microtubule cytoskeleton is crucial for many cellular functions, most direct regulators of microtubule nucleation, polymerization and disassembly are involved in multiple cell processes. In addition, the existence of plant-specific families of microtubule regulatory proteins such as SPIRAL emphasizes important differences in microtubule function and organization between plant and animal cells.
Table 1.
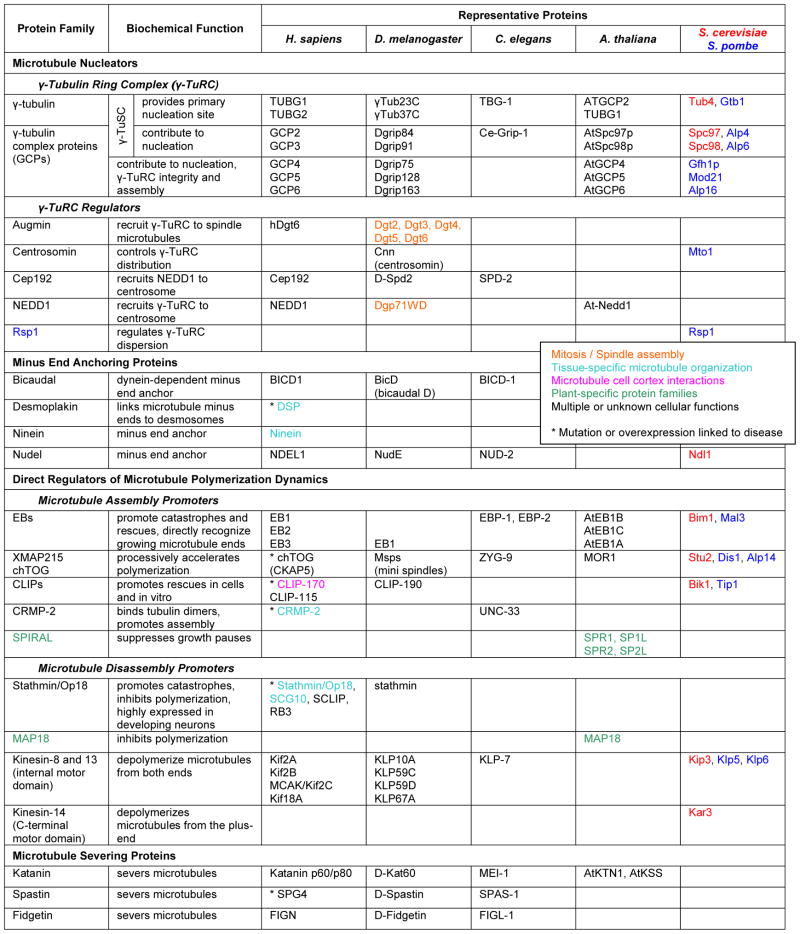
|
Acknowledgments
T.W. is supported by NIH R01GM079139. K.L. is supported by NIH/NIDCR training grant 5T32DE007306.
Abbreviations
- chTOG
colonic, hepatic tumor over-expressed gene
- CLIP
cytoplasmic linker protein
- CRMP
collapsin response mediator protein
- Dgt
dim γ-tubulin
- EB
end-binding protein
- KLP
kinesin-like protein
- MAP
microtubule-associated protein
- NEDD1
neural precursor cell expressed, developmentally down-regulated gene 1
- Spc
spindle pole body component
- γ-TuRC
γ-Tubulin Ring Complex
- γ-TuSC
γ-Tubulin Small Complex
References
- Baas PW, Karabay A, Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15:518–524. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW. Straighten up and fly right: microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr Opin Cell Biol. 2008;20:107–116. doi: 10.1016/j.ceb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Manning J, Kumar S. NEDD1: function in microtubule nucleation, spindle assembly and beyond. Int J Biochem Cell Biol. 2007;39:7–11. doi: 10.1016/j.biocel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Mennella V, Buster DW. KLP10A and KLP59C: the dynamic duo of microtubule depolymerization. Cell Cycle. 2005;4:1482–1485. doi: 10.4161/cc.4.11.2116. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]


