Abstract
Background
Multidrug resistant Klebsiella pneumoniae have caused major therapeutic problems worldwide due to the emergence of the extended-spectrum β-lactamase producing strains. Although there are >10 major facilitator super family (MFS) efflux pumps annotated in the genome sequence of the K. pneumoniae bacillus, apparently less is known about their physiological relevance.
Principal Findings
Insertional inactivation of kpnGH resulting in increased susceptibility to antibiotics such as azithromycin, ceftazidime, ciprofloxacin, ertapenem, erythromycin, gentamicin, imipenem, ticarcillin, norfloxacin, polymyxin-B, piperacillin, spectinomycin, tobramycin and streptomycin, including dyes and detergents such as ethidium bromide, acriflavine, deoxycholate, sodium dodecyl sulphate, and disinfectants benzalkonium chloride, chlorhexidine and triclosan signifies the wide substrate specificity of the transporter in K. pneumoniae. Growth inactivation and direct fluorimetric efflux assays provide evidence that kpnGH mediates antimicrobial resistance by active extrusion in K. pneumoniae. The kpnGH isogenic mutant displayed decreased tolerance to cell envelope stressors emphasizing its added role in K. pneumoniae physiology.
Conclusions and Significance
The MFS efflux pump KpnGH involves in crucial physiological functions besides being an intrinsic resistance determinant in K. pneumoniae.
Introduction
Drug efflux, in particular multidrug efflux, is a serious problem to drug therapy as they efflux broad range of substrates including clinically relevant antimicrobial agents [1], [2], [3], [4], [5], [6]. Nosocomial pathogen Klebsiella pneumoniae is a Gram-negative bacillus associated with healthcare infections, known to cause variety of diseases in humans with significant morbidity and mortality [7]. K. pneumoniae causes various serious illnesses and the problem is aggravated due to its propensity to acquire multidrug resistance determinants [8], [9]. Recently hypervirulent, hyperviscous K. pneumoniae NTUH-K2044, that belongs to the K1 serotype, known to cause pyogenic liver abscess, sometimes complicated by endophthalmitis or meningitis, has emerged in Taiwan, Singapore, Korea and other Asian countries [10]. Analysis of the available genome sequence of different K. pneumoniae isolates from NCBI database www.ncbi.nlm.nih.gov revealed that more than 10% of total genes were annotated as transporter proteins or efflux pumps, of which, to date only few have been studied and demonstrated to have a role in drug resistance. The 5.2-Mb genome of K. pneumoniae strain, NTUH-K2044 (encoding 4,992 proteins, GC content: 57.7%) is reported to harbor >15 open reading frames encoding for putative efflux pumps from different families (accession number NC_012731) [10]. The efflux systems functionally characterized in K. pneumoniae so far include acrAB and kexD from resistance/nodulation/cell division (RND), family [11], [12], [13]; the kdeA efflux gene from multi drug and toxic compound extrusion (MATE) family [14]; the kmrA gene from major facilitator super family (MFS) family [15]; and kpnEF from small multidrug resistance (SMR) family [16]. In Gram-negative bacteria, a subset of inner membrane proteins in the MFS act as efflux pumps to decrease the intracellular concentrations of multiple toxic substrates and confer multidrug resistance [17], [18]. The MFS type of transporters perhaps the most largest and diverse among all the efflux super families are found in all kingdoms of life. Well-studied examples such as QacA and NorA of Staphylococcus aureus and SmvA of Salmonella enterica serovar Typhimurium belong to the latter family [19], [20], [21]. Analysis of the K. pneumoniae NTUH-K2044 genome reveals the presence of a novel two component efflux pump operon, an emrAB homolog that belongs to MFS super family; whose functions have remained completely unexplored so far [10]. The objective of the present study was to investigate the role of putative MFS efflux system, an emrAB homolog (denoted kpnGH) in K. pneumoniae with respect to cellular physiology and broad spectrum antimicrobial resistance.
Materials and Methods
Bacterial Strains, Plasmids and Media
K. pneumoniae NTUH-K2044 (a strain that resulted in pyogenic liver abscess in a 66 year old patient) was kindly provided by Dr. Jin Town Wang of the National Taiwan University Hospital, Taipei, Taiwan [10]. E. coli SM10 lambda pir and pUT-Km was used to create isogenic mutants. Genomic DNA, plasmid, restriction digestion, DNA elution (Qiagen), ligation, transformation, conjugation, DNA sequencing (Applied Biosystems) were performed as previously described [22], [23], [24]. Primers used in the present study were custom-synthesized (Eurofins MWG operons, Germany).
Cloning of kpnGH in Hyper Susceptible E. coli Strain KAM32
The putative efflux genes kpnG and kpnH were amplified by standard PCR protocol using primers kpnG-F; kpnG-R and kpnH-F; kpnH-R (Table 1) respectively and cloned into EcoRI and PstI (New England Biolabs, MA, USA) site of pUC18. The resulting recombinant plasmid pkpnG, pkpnH and pkpnGH were transformed into E. coli KAM32 (ΔacrAB and ΔydhE) for functional characterization.
Table 1. Primers used in this study.
| Primer name | Primer sequences (5′–3′) |
| ΔkpnGH-F | GATATCTATGGCGATGACGTGAAATACACCGGTAA |
| ΔkpnGH-R | GATCAGCGGTCCGGCGACGATCCCCTGAAT |
| kpnG-F | CATAGGATCCATGAGTGCAAATGCGGAGAGCCAAACC |
| kpnG-R | ATTACTCGAGTTATCCGGCGTTGGCCTGAATGATCTC |
| kpnH-F | AGATCATATGCAACAGCAAAAACCCCTGGAAGGCGCGCAGCTGGTC |
| kpnH-R | ATTAGGATCCAGACCCGGAGGTCCCTTTATGTGAGGCTTAGTG |
| primer R-1 | TATAGGATCCATTAACGAGACATTATTTATGGCGCTGATT |
| primer R-2 | ATTAGAATTCGGTACTTTTCAGCAGGATGGCTCAGCATAGT |
Construction of the kpnGH Mutant in K. pneumoniae
The putative emrAB homolog, KP1_4279/KP1_4280 (designated kpnGH) is located starting from nucleotides 4108112 to 4109284 bps (kpnG: 1173 bp, 390 aa, 42.59 Kda and kpnH: 1536 bp, 511 aa, 55.589 Kda) in K. pneumoniae [10]. A 720 bp internal fragment was amplified by PCR using ΔkpnGH-F and ΔkpnGH-R primers and cloned in EcoRI digested pUT-Km and transformed into E. coli SM10 lambda pir strain (thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir). The recombinant plasmid pUT-kpnGH was mobilized into recipient K. pneumoniae from donor E. coli SM10 lambda pir as described previously to create inactivation (insertional inactivation means the operon is present on the chromosome but is disrupted, therefore the kpnGH efflux pump is non-functional) in kpnGH [22], [23]. The intact kpnGH operon along with its promoter was amplified with primer R-1 and primer R-2 and cloned into pCRIITOPO-CAT. The resulting construct was electroporated into ΔkpnGH and selected on LB agar plates supplemented with 50 µg/mL kanamycin and 100 µg/mL chloramphenicol to get the transcomplemented strain ΔkpnGHΩkpnGH.
Bacterial Growth Curves and Growth Inactivation Assay
The growth kinetics of WT (control strain: K. pneumoniae), ΔkpnGH and Δ kpnGHΩkpnGH were monitored in LB at different pH (5.0, 6.0, 7.0, 8.0, 10.0 and 12.0). Optical densities were measured for 18 h at 37°C shaking using Bioscreen C automated growth analysis system (Labsystems, Helsinki, Finland) at 600 nm and automatically recorded for each well after every 30 mins. The WT, ΔkpnGH and ΔkpnGHΩkpnGH cultures at mid log phase (OD600 nm = 0.01) were inoculated into LB broth containing antibiotic such as cefepime (0.1 µg/ml), tetracycline (0.01 µg/ml), ethidium bromide, EtBr (2 µg/ml), rhodamine (2 µg/ml), safranine (2 µg/ml) in independent experiments either alone or with efflux pump inhibitors, protonophores carbonyl cyanide 3-chlorophenylhydrazone; CCCP 2.5 µg/ml; 2, 4-Dintrophenol (2, 4 DNP); 2.5 µg/ml calcium channel blocker verampamil; 2.5 µg/ml and plant alkaloid reserpine; 2.5 µg/ml, (Sigma, St. Louis, MO) [23]. The growth at 37°C was analysed by measuring the absorbance at OD600 nm periodically for 18 h in Bioscreen C automated growth analysis system (Labsystems, Helsinki, Finland). The experiment was performed with freshly autoclaved medium in triplicates at least three independent times.
Motility Assay
The motility assays were done as reported before [23], where LB grown K. pneumoniae cultures (OD600nm = 1.0) were inoculated with a toothpick on LB plates with 0.25%, 0.45% and 0.7% agar and incubated for 10 hrs at 37°C. In this growth medium bacteria can swim through the soft agar and produce a halo. The diameter of the halo is a measure of the ability to swim.
Crystal Violet Binding Assay
The crystal violet binding assay was done as described before [23].
Various Stress Challenge Assays
The WT, ΔkpnGH, ΔkpnGHΩkpnGH were plated onto LB agar plates containing different substrates at varied concentrations such as bile (0.2, 0.5, 0.75, 1.0, 2.0%), sodium chloride (NaCl) (0.075, 0.15, 0.25, 0.5, 0.75, 1 and 2M), acriflavine (0.5, 4, 16, 64, 256, 512 µg/ml), acridine orange (0.5, 4, 16, 64, 256, 512 µg/ml), safranine (0.5, 4, 16, 64, 256, 512 µg/ml), rhodamine (0.5, 4, 16, 64, 256, 512 µg/ml), EtBr (0.5, 4, 16, 64, 256, 512 µg/ml), sodium dodecyl sulphate, SDS (1024, 2048, 4096, 8192, 16834 µg/ml), chloramphenicol (0.5, 4, 16, 64, 256, 512 µg/ml), ciprofloxacin (0.5, 4, 16, 64, 256, 512 µg/ml), neomycin (0.5, 4, 16, 64, 256, 512 µg/ml), tetracycline (0.5, 4, 16, 64, 256, 512 µg/ml), ticarcillin (0.5, 4, 16, 64, 256, 512 µg/ml), benzalkonium chloride (3.2, 6.4, 12.8, 25.6, 51.2, 102.4 µg/ml), chlorhexidine (3.2, 6.4, 12.8, 25.6, 51.2, 102.4 µg/ml), and triclosan (0.003, 0.005, 0.01, 0.05, 0.1, 0.5 µg/ml) and number of colonies were scored after incubation at 37°C for 16 h. The percentage of survival for each culture WT, ΔkpnGH, ΔkpnGHΩkpnGH was monitored by plating approximately 105 cells onto LB agar containing different concentrations of substrates and counting the number of colonies, then calculated i.e. number of colony forming units (CFU) on substrate containing plate/number of CFU on LB alone (control)×100. Survival on control plate was 100%. These experiments were performed at least three times.
Oxidative and Nitrosative Stress Tolerance Assays
The impact of oxidative stress inducing agent (H2O2) on WT, ΔkpnGH, ΔkpnGHΩkpnGH was performed using disc assay as reported before [23]. The survivability of cells to oxidative stress was tested by exposing stationary-phase bacteria diluted in LB medium (OD600 nm 0.01) at 37°C to 0.07894 mM, 0.7894 mM, 1.5788 mM, 2.3682 mM and 3.1576 mM for 1 h. The growth kinetics in presence of different nitrosative stress inducing agents [sodium nitroprusside (SNP; concentrations 5 mM, 10 mM, 15 mM, 20 mM, 25 mM) and acidified nitrite; (NaNO2; concentrations 5 mM, 10 mM, 15 mM, 20 mM, 25 mM)] was performed as reported before [23], [24].
Antibiotic Susceptibility and Determination of Minimum Inhibitory Concentration (MIC)
In this study the strains WT, ΔkpnGH, ΔkpnGHΩkpnGH were examined for resistance to ampicillin: AMP (10 µg/ml), azithromycin: AZM (15 µg/ml), chloramphenicol: CHL (30 µg/ml), carbencillin: CAR (100 µg/ml), ciprofloxacin: CIP (5 µg/ml), colistin: CST (10 µg/ml), cefepime: CPM (30 µg/ml), ceftriaxone: CTR (30 µg/ml), cephalothin: CTX (10 µg/ml), ceftazidime: CAZ (30 µg/ml), doxycycline: DOX (10 µg/ml), doripenem: DOR (10 µg/ml), ertapenem: ETP (10 µg/ml), linezolid: LZD (10 µg/ml), levofloxacin: LVX (10 µg/ml), oxacillin: OXA (10 µg/ml), polymyxin B: PMB (300 µg/ml), streptomycin: STR (10 µg/ml), tetracycline: TET (30 µg/ml), tigecycline: TGC (10 µg/ml), ticarcillin: TIC (75 µg/ml), tobramycin: TOB (10 µg/ml) and trimethoprim: TMP (5 µg/ml) by using commercial discs (Hi Media, Bombay, India) as described previously according to the interpretation criteria recommended by the Clinical and Laboratory Standards Institute CLSI. E. coli ATCC 25922 was used as a reference strain (control) as recommended [23], [24]. MIC of antibiotics was tested using E-strips (Hi Media, Bombay, India). Interpretation was done as per the criteria approved by the CLSI [25].
Fluorimetric Efflux Studies
Accumulation of EtBr was monitored as described previously [26]. Briefly, bacterial cells were grown to mid-log phase, harvested, and suspended in 1× phosphate buffered saline (0.136 M NaCl, 0.0026 M KCl, 0.01 M Na2HPO4, 0.00176 M KH2PO4; pH 7.0) to an optical density at OD600 nm = 0.2. EtBr was added at a concentration of 10 µg/ml. Aliquots were taken at different time intervals, harvested, suspended in 1 ml of 0.1 M glycine HCl buffer (pH 2.3), and incubated for 6 h at 37°C, and the fluorescence of the supernatant was measured with excitation 530 nm and emission 600 nm. Where indicated, the proton motive force uncoupler CCCP was added to the assay mixture at a final concentration; 25 µg/ml.
OMP Preparation
OMPs were purified by the method as described previously [23].
Bioinformatic Analysis and Statistical Analysis
All data are presented as means ± the standard error of the mean. Plotting and calculation of the standard deviation was performed in Microsoft Excel. Statistical analysis was performed on crude data by using a paired Student t test. P values of <0.05 were considered significant.
Results
Insilico Analysis of kpnGH MFS-type Efflux Pump
The deduced KpnG and KpnH proteins consist of 390 and 511 amino acid residues, respectively, similar to the size of EmrA and EmrB in E. coli. The nucleotide sequence deduced from the 2709 bp DNA fragment obtained from K. pneumoniae shares >90% identity with EmrAB efflux system in E. coli, S. dysenteriae and S. Typhimurium. Thus, KpnG and KpnH are both members of the MFS family of drug transporters. In addition, significant sequence similarities were detected between KpnGH and EmrAB of E. coli, Proteus vulgaris, and Citrobacter freundii. The multiple alignment of KpnGH with EmrAB of E. coli has been shown in Figure S1. The KpnG has two hydrophobic, presumably transmembrane regions spanning from amino acids 23–45; (23 aa in length) and 55–77; (23 aa in length). The KpnF also has thirteen transmembrane regions spanning from amino acids 16–38; (23 aa in length), 52–73; (22 aa in length), 76–98; (23 aa in length), 106–128; (23 aa in length), 138–160; (23 aa in length), 168–188; (21 aa in length), 202–222; (21 aa in length), 230–252; (23 aa in length), 271–2293; (23 aa in length), 307–329; (23 aa in length), 336–358; (23 aa in length), 370–392; (23 aa in length), and 474–496; (23 aa in length). As per SOSUI software average of hydrophobicity was 0.185128 and 0.639921 for kpnG and kpnH respectively.
Characterization of kpnGH in Hyper Susceptible E. coli strain KAM32
The kpnGH harbouring cells (E. coli KAM32/pkpnGH) displayed a elevated resistance to streptomycin (1.5-fold), kanamycin (1.5-fold), erythromycin (4-fold), trimethoprim (2-fold), nalidixic acid (4-fold), SDS (4-fold), bile (4-fold), chlorhexidine (3-fold), benzalkonium chloride (3-fold), and methyl viologen (2-fold) compared to other constructs including E. coli KAM32/pkpnG, E. coli KAM32/pkpnH, and E. coli KAM32/pUC18 (Table 2).
Table 2. Determination of MIC for E. coli KAM32/pUC18 and KAM32/pkpnGH.
| Compounds | KAM32/pUC18 | KAM32/pkpnGH | Fold changea |
| Erythromycin | 4 | 16 | 4 |
| Kanamycin | 2 | 4 | 2 |
| Nalidixic acid | 1 | 4 | 4 |
| Streptomycin | 2 | 4 | 2 |
| Trimethoprim | 0.125 | 0.250 | 2 |
| Benzalkonium chloride | 1 | 3 | 3 |
| Chlorhexidine | 2 | 6 | 3 |
| Deoxycholate | 1024 | 8096 | 4 |
| Methyl viologen | 64 | 128 | 2 |
| Pyronin Y | 4 | 8 | 2 |
| Rhodamine 123 | 8 | 16 | 2 |
| SDS | 50 | 200 | 4 |
The MIC of various antimicrobial agents such as erythromycin, kanamycin, nalidixic acid, streptomycin, trimethoprim, benzalkonium chloride, chlorhexidine, deoxycholate, methyl viologen, pyronin Y, rhodamine 123, SDS, for E. coli (KAM32/pUC18 and KAM32/pkpnGH) strains used in this study.
Fold change is the ratio of MICs for pkpnGH to pUC18.
Contributions of MFS-type Efflux kpnGH in K. pneumoniae Growth
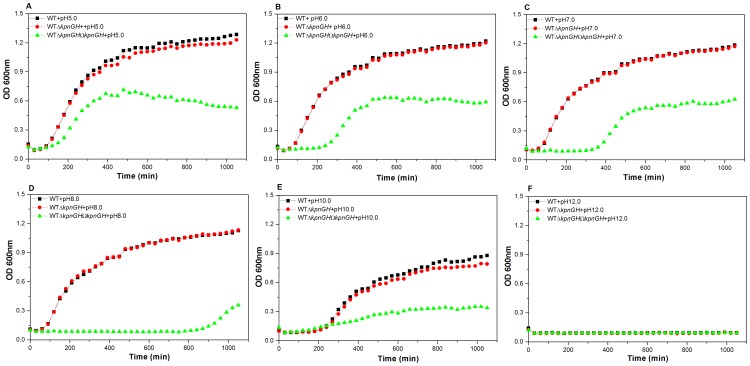
Experimentally the growth characteristics of WT, ΔkpnGH (inactivation confirmed by southern and RT-PCR) and ΔkpnGHΩkpnGH (confirmed by RT-PCR) were determined over a period of ∼18 h in LB medium with different pH and analysis revealed unique patterns. We tested the growth kinetics at pH 5.0, 6.0, 7.0, 8.0, 10.0 and 12.0 respectively. The mutant exhibited 1.782 fold (±0.0002) reduced growth compared to WT in LB at pH 5.0 [P = 0.0003404] (Figure 1-A). The mutant exhibited 2.466 fold (±0.00047) reduced growth compared to WT in LB at pH 6.0 [P = 0.000337] (Figure 1-B). The mutant exhibited 3.22 fold (±0.000011) reduced growth compared to WT in LB at pH 7.0 [P = 0.000208] (Figure 1-C). The mutant exhibited 8.46 fold (±0.00058) reduced growth compared to WT in LB at pH 8.0 [P = 0.000134] (Figure 1-D). The mutant exhibited 1.909 fold (±0.00032) reduced growth compared to WT in LB at pH 10.0 [P = 0.000164] (Figure 1-E). The other tested pH was 12.0 (Figure 1-F). The motility and biofilm forming ability of kpnGH mutant was found to be diminished (Data not shown). Results demonstrate that kpnGH influences growth and biofilm forming ability in K. pneumoniae.
Figure 1. Growth kinetics.
Effect on bacterial growth was monitored in WT, ΔkpnGH and ΔkpnGHΩkpnGH in LB medium at pH 5.0, 6.0, 7.0, 8.0, 10.0 and 12.0. The patterns of representative pH (5.0 (A), 6.0 (B), 7.0 (C), 8.0 (D), 10.0 (E), 12.0 (F)) are shown here. The mutant exhibited 1.782 fold (±0.0002), 2.466 fold (±0.00047), 3.22 fold (±0.000011), 8.46 fold (±0.00058), 1.909 fold (±0.00032), reduced growth compared to WT strain in LB at pH 5.0, 6.0, 7.0, 8.0, and 10.0 respectively. The data presented is the means of triplicate measurements performed three times.
Role of Membrane Transporter in Gastrointestinal (GI) Stress Challenges
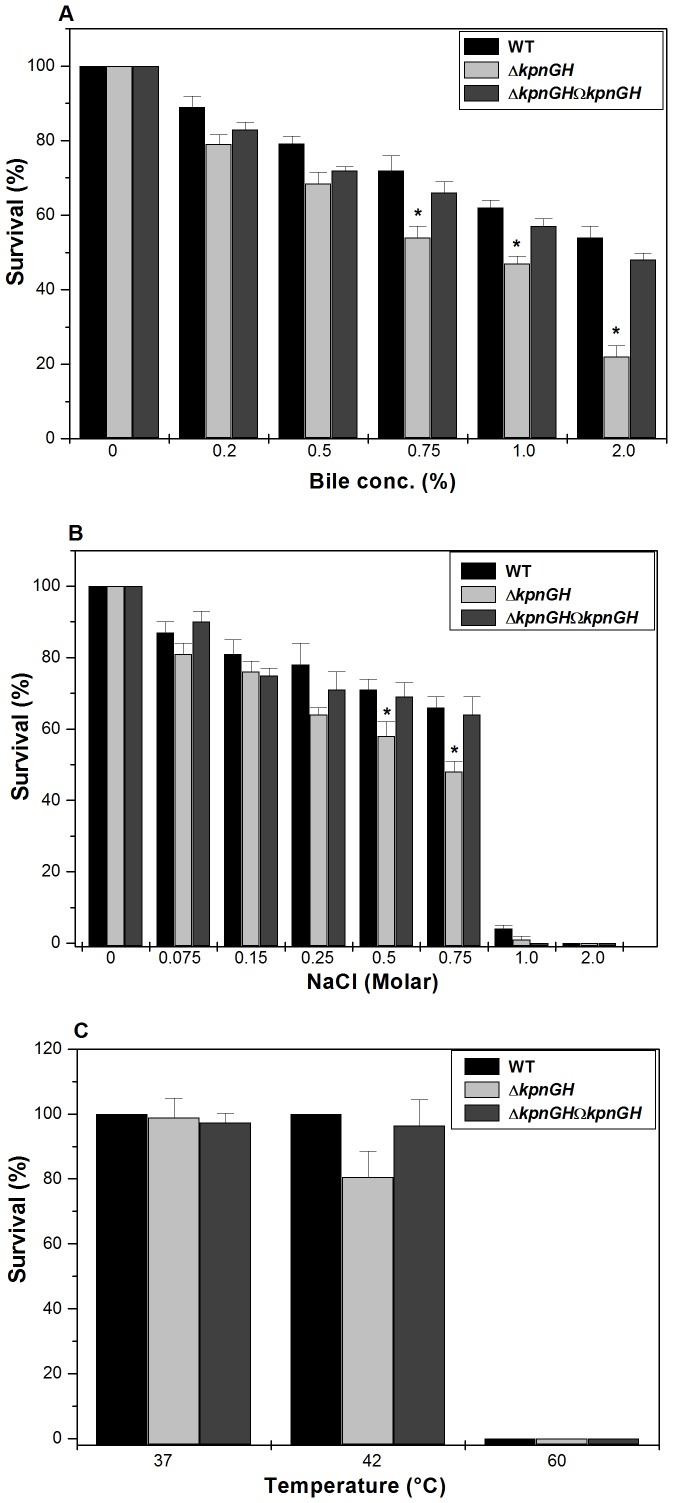
To determine the role of kpnGH in intestinal colonization, WT, ΔkpnGH and ΔkpnGHΩkpnGH underwent specific GI stress associated with bile and osmotic challenges. In the bile resistance assay, different strains were exposed to different concentrations (physiological concentration is 0.2% to 2%, [27]) of bile. When mid-log phase cultures were exposed to different concentrations of bile it was observed that the total CFU count of control (measure of surviving capacity) was higher as compared to ΔkpnGH. The ability of control to grow in the presence of 0.5% bile was 1.15 fold (±0.042), 0.75% bile was 1.3 fold (±0.024), 1% bile was 1.3 fold (±0.002) and 2% was 2.45 fold (±0.045) higher when compared to ΔkpnGH, while transcomplemented ΔkpnGHΩkpnGH strain restored the ability to tolerate stress (Figure 2-A) [P = 0.02157191].
Figure 2. Gastrointestinal stress challenge assays.
Survival of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in different concentrations of A) Bile (0.2%, 0.5%, 0.75%, 1.0%, and 2.0%), B) Osmotic stress (0.075M, 0.15M, 0.25M, 0.5M, 0.75M, 1M and 2M) and C) Different temperatures (37°C, 42°C and 60°C). Percentage survival was calculated by comparing the number of viable cells in LB medium alone. *, Significant difference (P<0.05, Student t test).
The ability of control to grow in the presence of NaCl (physiological concentration being 150 mM, [28]) at 0.25M was ∼1.218 fold (±0.033), 0.5M was ∼1.23 fold (±0.015), and 0.75M was ∼1.375 fold (±0.14), higher when compared to ΔkpnGH regardless of the inoculum size (Figure 2-B) [P = 0.018152256].
To deduce the role of kpnGH in temperature tolerance, we performed the heat shock assay. The temperature dependent assay showed that the kpnGH mutant displayed 15% reduced survival than the wild type at 42°C, thereby demonstrating that the response of K. pneumoniae kpnGH mutant in temperature stress (Figure 2-C). Overall these results imply that kpnGH had a contributory role towards varied stress tolerance in K. pneumoniae.
Role of kpnGH in Oxidative Stress Response
To deduce the role of kpnGH in oxidative stress, we performed the hydrogen peroxide challenge assay. Disc diffusion assay showed that the kpnGH mutant exhibited 1.07 fold greater sensitivity to 30% H2O2 (inhibition zone = 42±1.0 mm) than the wild-type (inhibition zone = 39±1.5 mm) (Figure S2-A) [P = 0.225], clearly demonstrating the role of K. pneumoniae kpnGH in oxidative stress.
The sensitivity of stationary-phase cultures to oxidative stress was tested by exposing them to a range of H2O2 concentrations (0.07894 mM, 0.7894 mM, 1.5788 mM, 2.3682 mM to 3.1576 mM) for 1 h. Only 42% and 17% of the ΔkpnGH cells survived upon treating with 0.07894 mM, 0.7894 mM of hydrogen peroxide as compared to the 91% and 77% survival observed in control cells respectively (Figure S2-B).
Involvement of kpnGH in Nitrosative Stress Response
SNP is a nitrosative stress inducer. It was interesting to note that kpnGH mutant exhibited 3.024-fold (±0.032; P = 0.000216), 3.38-fold (±0.072; P = 0.000241), 2.58-fold (±0.066; P = 0.000232), 1.702-fold (±0.012; P = 0.000141), 2.194-fold (±0.054; P = 0.000581), 1.132-fold (±0.076; P = 0.0002814), reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively (Figure S2, C I–IV).
To further evaluate the potentiality of K. pneumoniae kpnGH against other reactive nitrogen species, we tested tolerance of kpnGH inactivated cells towards acidified sodium nitrite. Protonated nitrite quickly degrades to generate numerous species of nitrogen oxides for example nitric oxide [27]. It was interesting to note that kpnGH mutant exhibited 2.88-fold (±0.045; P = 0.000115), 1.838-fold (±0.087; P = 0.000316), 1.629-fold (±0.026; P = 0.000211), 1.643-fold (±0.097; P = 0.000510), 1.454-fold (±0.032; P = 0.000409), 1.602-fold (±0.74; P = 0.000311), reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively (Figure S2, D I–IV). Overall, these results imply that kpnGH had a role towards nitrosative stress tolerance in K. pneumoniae.
Functions of KpnGH in Antibiotic Resistance
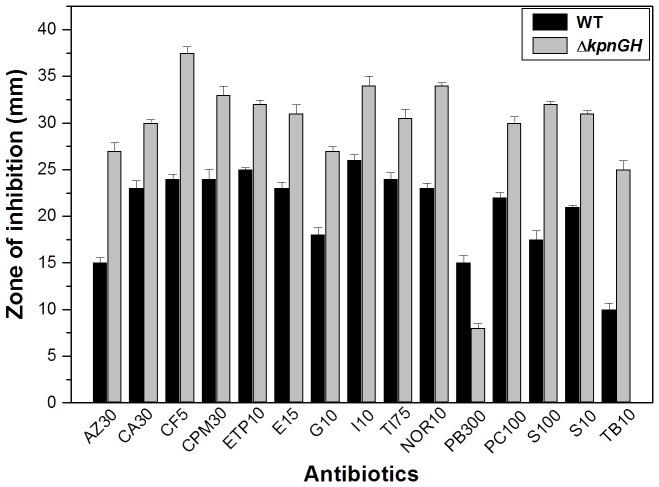
To evaluate the role of kpnGH in drug resistance, antibiotic susceptibilities of WT and ΔkpnGH was monitored. The results of disc diffusion assay displayed that upon inactivation of the efflux pump, the bacterial cells displayed significantly altered susceptibility to azithromycin, ceftazidime, ciprofloxacin, ertapenem, erythromycin, gentamicin, imipenem, ticarcillin, norfloxacin, polymyxin-B, piperacillin, spectinomycin, tobramycin and streptomycin (Figure 3).
Figure 3. Antimicrobial Susceptibility assays.
The Kirby Bauer disc diffusion assay was performed with different antibiotics using commercial discs. Data for representative drugs have been shown here.
The precise minimum inhibitory concentration (MIC) was further evaluated by following the guidelines of CLSI by E-test [25]. The MIC of K. pneumoniae for different antibiotics namely was, ceftazidime 0.256 µg/ml, cefepime 2.56 µg/ml, ceftriaxone 2.56 µg/ml, nalidixic acid 0.1 µg/ml, tobramycin 0.1 µg/ml, streptomycin 0.1 µg/ml, spectinomycin 0.1 µg/ml, tetracycline 5 µg/ml whereas MIC for ΔkpnGH {fold increase in brackets} for the same line of drugs were ceftazidime 0.064 µg/ml {4 fold}, cefepime 0.64 µg/ml {4 fold}, ceftriaxone 0.64 µg/ml {4 fold}, nalidixic acid 0.05 µg/ml {2 fold}, tobramycin 0.05 µg/ml {2 fold}, streptomycin 0.01 µg/ml {10 fold}, spectinomycin 0.01 µg/ml {10 fold}, tetracycline 1 µg/ml {5 fold} (Table 3). Complementation restored the defective phenotype. Overall the data indicates that MFS-efflux pump has multiple substrates.
Table 3. Determination of MIC for WT (K. pneumoniae), ΔkpnGH and ΔkpnGHΩkpnGH.
| Antibiotics | WT (µg/ml) | ΔkpnGH (µg/ml) | Fold changea | ΔkpnGHΩkpnGH |
| Cefepime | 2.56 | 0.64 | 4 | 2.56 |
| Ceftazidime | 0.256 | 0.064 | 4 | 0.256 |
| Ceftriaxone | 2.56 | 0.64 | 4 | 2.56 |
| Ciprofloxacin | <0.01 | <0.005 | 2 | <0.01 |
| Erythromycin | >4 | 2 | 2 | 4 |
| Spectinomycin | 0.1 | 0.01 | 10 | 0.1 |
| Streptomycin | 0.1 | 0.01 | 10 | 0.1 |
| Tetracycline | 5 | 1 | 5 | 5 |
| Tobramycin | 0.1 | 0.05 | 2 | 0.1 |
Fold change is the ratio of MICs for WT to ΔkpnGH.
Mutation in kpnGH Increases Sensitivity to Structurally Unrelated Compounds
The WT, ΔkpnGH and ΔkpnGHΩkpnGH cultures in this study were tested for their ability to withstand high concentration of different substrates that are structurally unrelated to antibiotics. The ability of ΔkpnGH to withstand different concentrations of EtBr (figure S3-A) and acriflavine (figure S4-B) was reduced by 1.3 and 2.0 fold when compared to WT respectively. Results were similar when done with other dyes such as rhodamine, safranine and acridine orange (data not shown).
Upon exposing the cells to different concentrations of SDS it was observed that the total CFU count of WT at 16384 µg/ml was 1.5 fold higher than ΔkpnGH [P = /0.009962331] (figure S3-C). Analysis indicates the wide substrate specificity for kpnGH in K. pneumoniae.
The growth rate of ΔkpnGH in the presence of 2.0 µg/ml EtBr, was >12 fold lesser when compared to that of WT (P = 0.000477). To distinguish whether this is due to the loss of efflux pump activity in kpnGH mutant, the growth profile was monitored with CCCP, an uncoupler known to collapse the membrane energy and block the energy-dependent efflux pump. As expected a substantial decrease in growth was observed in kpnGH mutant (15.18-fold; P = 0.000697), (1.39-fold; P = 0.000775), (6.32-fold; P = 0.000681) and (15.06-fold; P = 0.000174) in the presence of CCCP, reserpine, verampamil and 2, 4 DNP respectively (Figure S3, D I–IV).
The growth rate of ΔkpnGH in the presence of 2.0 µg/ml rhodamine, was >17.5 fold lesser when compared to that of WT (P = 0.000153). Akin a substantial decrease in growth was observed in kpnGH mutant (10.476-fold; P = 0.000234), (1.39102-fold; P = 0.000796), (8.13-fold; P = 0.000101) and (23.1-fold; P = 0.000196) in the presence of CCCP, reserpine, verampamil and 2, 4 DNP respectively (Figure S3, E I–IV).
The growth rate of ΔkpnGH in the presence of 2.0 µg/ml safranine, was >15.94 fold lesser when compared to that of WT (P = 0.000221). A decrease in growth was observed in kpnGH mutant (5.16-fold; P = 0.000251), (0.988-fold; P = 0.000182), (16.23-fold; P = 0.000215) and (12.5-fold; P = 0.000101) in the presence of CCCP, reserpine, verampamil and 2, 4 DNP respectively (Figure S3, F I–IV). The mutant lacks kpnGH, the efflux pump in its functional form, so possibly leading to decreased efflux activity and thereby we observe a stunted growth profile.
Tolerance to Different Classes of Antibiotics
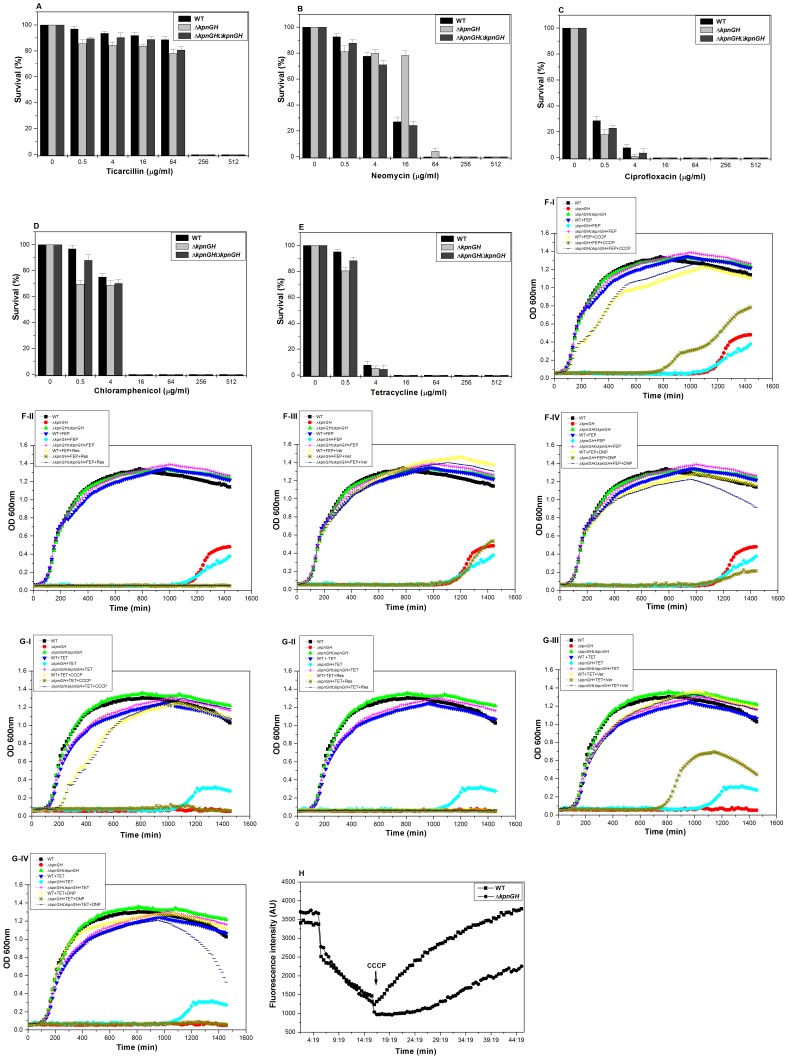
Upon exposing the cells to different concentrations of ticarcillin it was observed that the total CFU count of WT at 0.5 µg/ml was 1 fold, 4 µg/ml was 1.13 fold, 16 µg/ml was 1.11 fold, 64 µg/ml was 1.09 fold, 256 µg/ml was 1.09 fold, 512 µg/ml was 1.13 fold increased than ΔkpnGH [P = 0.027] (Figure 4-A).
Figure 4. Susceptibility, Growth inactivation and accumulation assays towards different antibiotics.
A) Sensitivity of WT, ΔkpnGH and ΔkpnGHΩkpnGH towards ticarcillin. Upon exposing the cells to different concentrations of ticarcillin it was observed that the total CFU count of WT at 0.5 µg/ml was 1 fold, 4 µg/ml was 1.13 fold, 16 µg/ml was 1.11 fold, 64 µg/ml was 1.09 fold, 256 µg/ml was 1.09 fold, 512 µg/ml was 1.13 fold increased than ΔkpnGH [P = 0.027]. The data is the means of measurements made in triplicate and performed three times. B) Sensitivity of WT, ΔkpnGH and ΔkpnGHΩkpnGH towards neomycin. Upon exposing the cells to different concentrations of neomycin it was observed that the total CFU count of WT at 0.5 µg/ml was 1.14 fold, 4 µg/ml was 0.97 fold, 16 µg/ml was 0.34 fold increased than ΔkpnGH [P = 0.042]. The data is the means of measurements made in triplicate and performed three times. C) Sensitivity of WT, ΔkpnGH and ΔkpnGHΩkpnGH towards ciprofloxacin. Exposing the cells to different concentrations of ciprofloxacin it was observed that the total CFU count of WT at 0.5 µg/ml was 1.5 fold, 4 µg/ml was 7.3 fold increased than ΔkpnGH [P = 0.0188]. The data is the means of measurements made in triplicate and performed three times. D) Sensitivity of WT, ΔkpnGH and ΔkpnGHΩkpnGH towards chloramphenicol. Exposing the cells to different concentrations of chloramphenicol it was observed that the total CFU count of WT at 0.5 µg/ml was 1.39 fold, 4 µg/ml was 1.08 fold increased than ΔkpnGH [P = 0.0266]. The data is the means of measurements made in triplicate and performed three times. E) Sensitivity of WT, ΔkpnGH and ΔkpnGHΩkpnGH towards tetracycline. Exposing the cells to different concentrations of tetracycline it was observed that the total CFU count of WT at 0.5 µg/ml was 1.18 fold, 4 µg/ml was 1.45 fold increased than ΔkpnGH [P = 0.0288]. The data is the means of measurements made in triplicate and performed three times. F) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of cefepime. Growth pattern in absence of any drug or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 0.1 µg/ml cefepime, was 15.373 fold lesser when compared to that of WT (P = 0.000574). A decrease in growth was observed in kpnGH mutant (7.474-fold; P = 0.000123), (1.032-fold; P = 0.000159), (16.23-fold; P = 0.000215) and (12.5-fold; P = 0.000101) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. G) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of tetracycline. Growth pattern in absence of any drug or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 0.01 µg/ml tetracycline, was 10.655 fold lesser when compared to that of WT (P = 0.000234). A decrease in growth was observed in kpnGH mutant (10.476-fold; P = 0.000477), (1.102-fold; P = 0.000796), (8.134-fold; P = 0.000101) (15.28-fold; P = 0.000101) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. H) Accumulation studies using EtBr with control strain and ΔkpnGH. Efflux of EtBr in mutant and wild type cells was monitored continuously by measuring fluorescence emission at 600 nm upon excitation at 530 nm. After 5 mins in flourimeter, cells loaded with EtBr were energized by addition of glucose and efflux of EtBr was monitored. After 10 mins, 100 µM CCCP was added as indicated to abolish active transport and fluorescence emission was monitored further. The fluorescence was measured using spectrofluorometer (Hitachi). Each data point represents the mean plus the standard deviation of three independent experiments.
Upon exposing the cells to different concentrations of neomycin it was observed that the total CFU count of WT at 0.5 µg/ml was 1.14 fold, 4 µg/ml was 0.97 fold, 16 µg/ml was 0.34 fold increased than ΔkpnGH [P = 0.042] (Figure 4-B).
Exposing the cells to different concentrations of ciprofloxacin it was observed that the total CFU count of WT at 0.5 µg/ml was 1.5 fold, 4 µg/ml was 7.3 fold increased than ΔkpnGH [P = 0.0188] (Figure 4-C).
Exposing the cells to different concentrations of chloramphenicol it was observed that the total CFU count of WT at 0.5 µg/ml was 1.39 fold, 4 µg/ml was 1.08 fold increased than ΔkpnGH [P = 0.0266] (Figure 4-D).
Exposing the cells to different concentrations of tetracycline it was observed that the total CFU count of WT at 0.5 µg/ml was 1.18 fold, 4 µg/ml was 1.45 fold increased than ΔkpnGH [P = 0.0288] (Figure 4-E). Results indicate the role of KpnGH in drug resistance.
The growth rate of ΔkpnGH in the presence of 0.1 µg/ml cefepime, was 15.373 fold lesser when compared to that of WT (P = 0.000574). A decrease in growth was observed in kpnGH mutant (7.474-fold; P = 0.000123), (1.032-fold; P = 0.000159), (16.23-fold; P = 0.000215) and (13.7-fold; P = 0.000176) in the presence of CCCP, reserpine, verampamil and 2, 4 DNP respectively (Figure 4, F I–IV).
The growth rate of ΔkpnGH in the presence of 0.01 µg/ml tetracycline, was 10.655 fold lesser when compared to that of WT (P = 0.000234). A decrease in growth was observed in kpnGH mutant (10.476-fold; P = 0.000477), (1.102-fold; P = 0.000796), (8.134-fold; P = 0.000101) and (15.28-fold; P = 0.000101) in the presence of CCCP, reserpine, verampamil and 2, 4 DNP respectively (Figure 4, G I–IV).
The EtBr accumulation data indicate that the efflux was most efficient in the control, whereas it was less efficient in ΔkpnGH. The addition of CCCP increased the EtBr levels which eventually reached a plateau in both control and ΔkpnGH (Figure 4-H). Overall, these preliminary results suggest that mutant of kpnGH possibly affects active efflux in K. pneumoniae.
MFS Efflux Pump kpnGH Influences Tolerance to Disinfectants
We tested the susceptibilities of control and ΔkpnGH towards different concentrations of popularly used hospital based disinfectants such as chlorhexidine and benzalkonium chloride [29], [30]. Upon exposing the cells to different concentrations of benzalkonium chloride it was observed that the total CFU count of control was 5 to 12 fold higher than ΔkpnGH [P = 0.047277226] (figure S4-A).
When cells were exposed to different concentrations of chlorhexidine it was observed that the total CFU count of control was 1.3 to 1.7 fold higher than ΔkpnGH [P = /0.080120359] (figure S4-B).
Upon exposing the cells to different concentrations of triclosan it was observed that the total CFU count of control was 1.3 to 1.4 fold higher than ΔkpnGH [P = /0.065323347] (figure S4-C). Decisively, the results imply the broad spectrum antimicrobial resistance property of kpnGH, a MFS-type efflux pump for the first time in K. pneumoniae.
Alterations in Outer Membrane Profile of the kpnGH Inactivated Mutant in K. pneumoniae
A reduction in the permeation of antibiotics is generally related to a decrease in porin expression or an alteration in the porin structure [2], [5], [6]. Thus, we compared the OMP profiles of ΔkpnGH with WT to find out whether a kpnGH inactivated mutant expresses alternative porins/OMPs to maintain normal cellular functions. It was interesting to note that there was a marked difference in the OMP profiles of mutant compared to wild type (Figure S5), and deciphering the identity and function of these proteins is highly warranted.
Discussion
Klebsiella pneumoniae, a member of Enterobacteriaceae family, is an important pathogen in both the community and the clinical setting. K. pneumoniae is largely responsible for causing diseases such as nosocomial pneumonia (7 to 14% of all cases), septicaemia (4 to 15% of all cases), wound infections (2 to 4% of all cases), and neonatal septicaemia (3 to 30% of all cases) [1]. Few reports have assessed the masked role of efflux pumps in multidrug resistant phenotype in K. pneumoniae clinical isolates. Analysis of K. pneumoniae genome sequences namely K. pneumoniae strain 1084 (NC_018522.1); K. pneumoniae strain HS11286 (NC_016845.1); K. pneumoniae strain MGH 78578 (NC_009648.1); as well as K. pneumoniae strain NTUH-K2044 (NC_012731.1) indicates the presence of several putative multidrug efflux pumps [10]. In this study, we report the biological functions of MFS-type efflux pump kpnGH in modulating the cellular physiology and antimicrobial resistance in K. pneumoniae for the first time.
The E. coli KAM32 cells expressing kpnGH mounted significant growth difference than the vector control on a broad range of antimicrobial compounds, including representative antimicrobial agents, antiseptics, cationic dyes and detergents confirming that K. pneumoniae kpnGH, like the emrAB homolog in E. coli, S. Typhimurium, is required for resistance to various antimicrobials [21], [31], [32]. While inactivation of kpnEF resulted in increased susceptibility towards cefepime, ceftriaxone, imipenem, ertapenem, enrofloxacin, norfloxacin and ciprofloxacin, however non functional kpnGH results in increased susceptibility to cefepime, ceftazidime, ceftriaxone, ciprofloxacin, erythromycin, spectinomycin, streptomycin, tetracycline and tobramycin and structurally unrelated compounds such as SDS, deoxycholate, EtBr, and acriflavine. KpnGH displayed sensitivity to benzalkonium chloride, chlorhexidine and triclosan; however KpnEF displayed greater sensitivity to these compounds. Results presented here provide evidence that inactivation of kpnGH diminishes drug efflux capacity in K. pneumoniae.
Where the previously reported efflux pump kpnEF displayed higher sensitivity to high bile (∼4.0 fold) concentration, interestingly, the kpnGH mutant is only 2.5 fold sensitive to varied bile challenges indicating that K. pneumoniae possibly utilizes kpnGH to extrude bile salts from the cytoplasm out of the cell to successfully thrive in the GI tract of human host [23]. Evidence indicating the role of KpnGH in surviving conditions that mimic the upper parts of the GI, where they encounter hyper osmotic condition in a microaerobic environment adds to the multifaceted, unprecedented functions displayed by MFS-type efflux pump in K. pneumoniae. In conclusion, using various molecular approaches, we have demonstrated the functions of kpnGH, a MFS-type efflux pump in cellular physiology and antimicrobial resistance in Klebsiella spp for the first time.
Supporting Information
Sequence alignment of KpnGH with EmrAB homolog. Sequence alignments of Klebsiella pneumoniae EmrA (KP1_4279) with Escherichia coli EmrA EmrA (b2685) were made in CLUSTAL Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). The conserved domain analysis (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) revealed the Biotin_lipoyl_2 domain (pfam 13533) and HlyD family secretion protein domain (pfam 13437) are highlighted in yellow and cyan color respectively. The asterisks indicate fully conserved residues, colons strongly similar residues, dots weakly similar residues.
(DOCX)
Oxidative and nitrosative challenge assays. A) The ability of wild type, ΔkpnGH and ΔkpnGHΩkpnGH to combat different levels of hydrogen peroxide was measured by disc diffusion assay. B) Survival of wild type, ΔkpnGH and ΔkpnGHΩkpnGH strains monitored upon exposure to H2O2 at 0.07894 mM, 0.7894 mM, 1.5788 mM, 2.3682 mM and 3.1576 mM. After 1 h of treatment with 0.07894 mM hydrogen peroxide, only 42% of ΔkpnGH cells survived in comparison to 91% of the wild-type cells. C) Growth pattern of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of SNP. The growth kinetics of kpnGH exhibited 3.024-fold (±0.032; P = 0.000216), 3.38-fold (±0.072; P = 0.000241), 2.58-fold (±0.066; P = 0.000232), 1.702-fold (±0.012; P = 0.000141), 2.194-fold (±0.054; P = 0.000581), 1.132-fold (±0.076; P = 0.0002814) reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively. The mean values of three independent experiments have been used for plotting the graph. D) Growth pattern of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of NaNO2. The growth kinetics of kpnGH exhibited 2.88-fold (±0.045; P = 0.000115), 1.838-fold (±0.087; P = 0.000316), 1.629-fold (±0.026; P = 0.000211), 1.643-fold (±0.097; P = 0.000510), 1.454-fold (±0.032; P = 0.000409), 1.602-fold (±0.74; P = 0.000311) reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively. The mean values of three independent experiments have been used for plotting the graph.
(TIF)
Susceptibility and Growth inactivation assays towards different dyes and detergent. A) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards EtBr when cells were exposed to different concentrations of dye (2 µg/ml, 4 µg/ml, 64 µg/ml, 128 µg/ml, 256 µg/ml and 512 µg/ml). The data is the means of measurements made in triplicate and performed three times. B) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards acriflavine when cells were exposed to different concentrations of dye (2 µg/ml, 4 µg/ml, 64 µg/ml, 128 µg/ml, 256 µg/ml and 512 µg/ml). The data is the means of measurements made in triplicate and performed three times. C) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards SDS when cells were exposed to different concentrations of detergent (1024 µg/ml, 2048 µg/ml, 4096 µg/ml, 8192 µg/ml, 16834 µg/ml). The data is the means of measurements made in triplicate and performed three times. D) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of EtBr. Growth pattern in absence of any EtBr or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml EtBr, was >12 fold lesser when compared to that of WT (P = 0.000477). A decrease in growth was observed in kpnGH mutant (15.18-fold; P = 0.000697), (1.39-fold; P = 0.000775), (6.32-fold; P = 0.000681) and (15.06-fold; P = 0.000174) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. E) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of rhodamine. Growth pattern in absence of any rhodamine or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml rhodamine, was >17.5 fold lesser when compared to that of WT (P = 0.000153). A decrease in growth was observed in kpnGH mutant (10.476-fold; P = 0.000234), (1.39102-fold; P = 0.000796), (8.13-fold; P = 0.000101) and (23.1-fold; P = 0.000196) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. F) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of safranine. Growth pattern in absence of any safranine or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml safranine, was >15.94 fold lesser when compared to that of WT (P = 0.000221). A decrease in growth was observed in kpnGH mutant (5.16-fold; P = 0.000251), (0.988-fold; P = 0.000182), (16.23-fold; P = 0.000215) and (12.5-fold; P = 0.000101) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph.
(TIF)
Disinfectant challenge assays. Survival of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of different concentrations (µg/ml) of A) benzalkonium chloride (3.2, 6.4, 12.8, 25.6, 51.2, 102.4), B) chlorhexidine (3.2, 6.4, 12.8, 25.6, 51.2, 102.4), C) triclosan (0.003, 0.005, 0.01, 0.05, 0.1, 0.5). The percent survival was calculated by comparison to the numbers of viable cells obtained in LB medium alone. *, Significant difference (P<0.05, Student t test).
(TIF)
Protein profiling of WT and kpnGH mutants. Membrane protein profiles were compared between the WT strain, and kpnGH mutant. About 20 µg of total protein lysate was loaded in an order as follows; lane 2: WT, lane 3: kpnGH mutant 1, lane 4: kpnGH mutant 2, lane 5: kpnGH mutant3, lane 6: kpnGH mutant 4, lane 7: kpnGH mutant 5 respectively. A similar amount of outer membrane fractions was loaded in an order as follows; lane 8: WT, lane 9: kpnGH mutant 1, lane 10: kpnGH mutant 2, lane 11: kpnGH mutant3, lane 12: kpnGH mutant 4, lane 13: kpnGH mutant 5 respectively. The inner membrane fractions was loaded in an order as follows; lane 14: WT, lane 15: kpnGH mutant 1, lane 16: kpnGH mutant 2, lane 17: kpnGH mutant3, lane 18: kpnGH mutant 4, lane 19: kpnGH mutant 5 respectively. Equal protein concentrations were separated by SDS-PAGE with a 5% stacking gel and a 12% separating gel and stained with coomassie brilliant blue. Lane M has molecular weight standards.
(TIF)
Acknowledgments
We are extremely thankful and grateful to our Director Dr. Girish Sahni, CSIR-Institute of Microbial Technology (IMTECH), Chandigarh, for providing excellent facilities to carry out this work. We are grateful to Dr Jin-Town Wang for providing K. pneumoniae and other basic vectors.
Funding Statement
The research has been supported by intramural funds from IMTECH and DBT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rossolini GM, Mantengoli E, Docquier JD, Musmanno RA, Coratza G (2007) Epidemiology of infections caused by multiresistant Gram-negatives: ESBLs, MBLs, panresistant strains. New Microbiol 30: 332–339. [PubMed] [Google Scholar]
- 2. Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74: 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Piddock LJ (2006) Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol 4: 629–636. [DOI] [PubMed] [Google Scholar]
- 4. Piddock LJ (2006) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19: 382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050. [DOI] [PubMed] [Google Scholar]
- 6. Fernandez L, Hancock RE (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25: 661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daikos GL, Markogiannakis A (2011) Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect 17: 1135–1141. [DOI] [PubMed] [Google Scholar]
- 9. Rapp RP, Urban C (2012) Klebsiella pneumoniae carbapenemases in Enterobacteriaceae: history, evolution, and microbiology concerns. Pharmacotherapy 32: 399–407. [DOI] [PubMed] [Google Scholar]
- 10. Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, et al. (2009) Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191: 4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Padilla E, Llobet E, Domenech-Sanchez A, Martínez-Martínez L, Bengoechea JA, et al. (2010) Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li DW, Onishi M, Kishino T, Matsuo T, Ogawa W, et al. (2008) Properties and expression of a multidrug efflux pump AcrAB-KocC from Klebsiella pneumoniae . Biol Pharm Bull 31: 577–582. [DOI] [PubMed] [Google Scholar]
- 13. Ogawa W, Onishi M, Ni R, Tsuchiya T, Kuroda T (2012) Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae . Gene 498: 177–182. [DOI] [PubMed] [Google Scholar]
- 14. Ping Y, Ogawa W, Kuroda T, Tsuchiya T (2007) Gene cloning and characterization of KdeA, a multidrug efflux pump from Klebsiella pneumoniae . Biol Pharm Bull 30: 1962–1964. [DOI] [PubMed] [Google Scholar]
- 15. Ogawa W, Koterasawa M, Kuroda T, Tsuchiya T (2006) KmrA multidrug efflux pump from Klebsiella pneumoniae . Biol Pharm Bull 29: 550–553. [DOI] [PubMed] [Google Scholar]
- 16. Srinivasan VB, Rajamohan G (2013) KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother 57: 4449–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paulsen IT, Brown MH, Skurray RA (1996) Proton-dependent multidrug efflux systems. Microbiol Rev 60: 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roca I, Marti S, Espinal P, Martínez P, Gibert I, et al. (2009) CraA, a major facilitator super family efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii . Antimicrob Agents Chemother 53: 4013–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown MH, Skurray RA (2001) Staphylococcal multidrug efflux protein QacA. J Mol Microbiol Biotechnol 3: 163–170. [PubMed] [Google Scholar]
- 20. Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M (1990) Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol 172: 6942–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Villagra NA, Hidalgo AA, Santiviago CA, Saavedra CP, Mora GC (2008) SmvA, and not AcrB, is the major efflux pump for acriflavine and related compounds in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 62: 1273–1276. [DOI] [PubMed] [Google Scholar]
- 22. Srinivasan VB, Vaidyanathan V, Mondal A, Rajamohan G (2012) Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae . PLoS ONE 7: e33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srinivasan VB, Venkataramaiah M, Mondal A, Vaidyanathan V, Govil T, et al. (2012) Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 7: e41505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srinivasan VB, Vaidyanathan V, Mondal A, Venkataramaiah M, Rajamohan G (2012) Functional characterization of a novel Mn2+ dependent protein serine/threonine kinase KpnK, produced by Klebsiella pneumoniae strain MGH78578. FEBS Lett 2: 3778–3786. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute (2010) Performance standards for antimicrobial susceptibility testing. CLSI M100-S20U. Clinical and Laboratory Standards Institute. Wayne, PA.
- 26. Rajamohan G, Srinivasan VB, Gebreyes WA (2010) Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii . J Antimicrob Chemother 65: 1919–1925. [DOI] [PubMed] [Google Scholar]
- 27. Gunn JS (2000) Mechanisms of bacterial resistance and response to bile. Microb Infect 2: 907–913. [DOI] [PubMed] [Google Scholar]
- 28. Hennequin C, Forestier C (2009) OxyR, a LysR-type regulator involved in Klebsiella pneumoniae mucosal and abiotic colonization. Infect Immun 77: 5449–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gnanadhas DP, Marathe SA, Chakravortty D (2013) Biocides - resistance, cross-resistance mechanisms and assessment. Expert Opin Investig Drugs 22: 191–206. [DOI] [PubMed] [Google Scholar]
- 30. Russell AD (2002) Mechanisms of antimicrobial action of antiseptics and disinfectants: an increasingly important area of investigation. J Antimicrob Chemother 49: 597–599. [DOI] [PubMed] [Google Scholar]
- 31. Furukawa H, Tsay JT, Jackowski S, Takamura Y, Rock CO (1993) Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB . J Bacteriol 175: 3723–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A (2006) Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol 188: 5693–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of KpnGH with EmrAB homolog. Sequence alignments of Klebsiella pneumoniae EmrA (KP1_4279) with Escherichia coli EmrA EmrA (b2685) were made in CLUSTAL Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). The conserved domain analysis (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) revealed the Biotin_lipoyl_2 domain (pfam 13533) and HlyD family secretion protein domain (pfam 13437) are highlighted in yellow and cyan color respectively. The asterisks indicate fully conserved residues, colons strongly similar residues, dots weakly similar residues.
(DOCX)
Oxidative and nitrosative challenge assays. A) The ability of wild type, ΔkpnGH and ΔkpnGHΩkpnGH to combat different levels of hydrogen peroxide was measured by disc diffusion assay. B) Survival of wild type, ΔkpnGH and ΔkpnGHΩkpnGH strains monitored upon exposure to H2O2 at 0.07894 mM, 0.7894 mM, 1.5788 mM, 2.3682 mM and 3.1576 mM. After 1 h of treatment with 0.07894 mM hydrogen peroxide, only 42% of ΔkpnGH cells survived in comparison to 91% of the wild-type cells. C) Growth pattern of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of SNP. The growth kinetics of kpnGH exhibited 3.024-fold (±0.032; P = 0.000216), 3.38-fold (±0.072; P = 0.000241), 2.58-fold (±0.066; P = 0.000232), 1.702-fold (±0.012; P = 0.000141), 2.194-fold (±0.054; P = 0.000581), 1.132-fold (±0.076; P = 0.0002814) reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively. The mean values of three independent experiments have been used for plotting the graph. D) Growth pattern of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of NaNO2. The growth kinetics of kpnGH exhibited 2.88-fold (±0.045; P = 0.000115), 1.838-fold (±0.087; P = 0.000316), 1.629-fold (±0.026; P = 0.000211), 1.643-fold (±0.097; P = 0.000510), 1.454-fold (±0.032; P = 0.000409), 1.602-fold (±0.74; P = 0.000311) reduced growth compared to WT strain in LB at 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 25 mM respectively. The mean values of three independent experiments have been used for plotting the graph.
(TIF)
Susceptibility and Growth inactivation assays towards different dyes and detergent. A) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards EtBr when cells were exposed to different concentrations of dye (2 µg/ml, 4 µg/ml, 64 µg/ml, 128 µg/ml, 256 µg/ml and 512 µg/ml). The data is the means of measurements made in triplicate and performed three times. B) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards acriflavine when cells were exposed to different concentrations of dye (2 µg/ml, 4 µg/ml, 64 µg/ml, 128 µg/ml, 256 µg/ml and 512 µg/ml). The data is the means of measurements made in triplicate and performed three times. C) Sensitivity of wild type, ΔkpnGH and ΔkpnGHΩkpnGH towards SDS when cells were exposed to different concentrations of detergent (1024 µg/ml, 2048 µg/ml, 4096 µg/ml, 8192 µg/ml, 16834 µg/ml). The data is the means of measurements made in triplicate and performed three times. D) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of EtBr. Growth pattern in absence of any EtBr or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml EtBr, was >12 fold lesser when compared to that of WT (P = 0.000477). A decrease in growth was observed in kpnGH mutant (15.18-fold; P = 0.000697), (1.39-fold; P = 0.000775), (6.32-fold; P = 0.000681) and (15.06-fold; P = 0.000174) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. E) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of rhodamine. Growth pattern in absence of any rhodamine or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml rhodamine, was >17.5 fold lesser when compared to that of WT (P = 0.000153). A decrease in growth was observed in kpnGH mutant (10.476-fold; P = 0.000234), (1.39102-fold; P = 0.000796), (8.13-fold; P = 0.000101) and (23.1-fold; P = 0.000196) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph. F) Growth inactivation assay using wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of safranine. Growth pattern in absence of any safranine or inhibitor is included as control. The growth rate of ΔkpnGH in the presence of 2.0 µg/ml safranine, was >15.94 fold lesser when compared to that of WT (P = 0.000221). A decrease in growth was observed in kpnGH mutant (5.16-fold; P = 0.000251), (0.988-fold; P = 0.000182), (16.23-fold; P = 0.000215) and (12.5-fold; P = 0.000101) in the presence of CCCP (I), reserpine (II), verampamil (III) and 2, 4 DNP (IV) respectively. The mean values of three independent experiments have been used for plotting the graph.
(TIF)
Disinfectant challenge assays. Survival of wild type, ΔkpnGH and ΔkpnGHΩkpnGH in the presence of different concentrations (µg/ml) of A) benzalkonium chloride (3.2, 6.4, 12.8, 25.6, 51.2, 102.4), B) chlorhexidine (3.2, 6.4, 12.8, 25.6, 51.2, 102.4), C) triclosan (0.003, 0.005, 0.01, 0.05, 0.1, 0.5). The percent survival was calculated by comparison to the numbers of viable cells obtained in LB medium alone. *, Significant difference (P<0.05, Student t test).
(TIF)
Protein profiling of WT and kpnGH mutants. Membrane protein profiles were compared between the WT strain, and kpnGH mutant. About 20 µg of total protein lysate was loaded in an order as follows; lane 2: WT, lane 3: kpnGH mutant 1, lane 4: kpnGH mutant 2, lane 5: kpnGH mutant3, lane 6: kpnGH mutant 4, lane 7: kpnGH mutant 5 respectively. A similar amount of outer membrane fractions was loaded in an order as follows; lane 8: WT, lane 9: kpnGH mutant 1, lane 10: kpnGH mutant 2, lane 11: kpnGH mutant3, lane 12: kpnGH mutant 4, lane 13: kpnGH mutant 5 respectively. The inner membrane fractions was loaded in an order as follows; lane 14: WT, lane 15: kpnGH mutant 1, lane 16: kpnGH mutant 2, lane 17: kpnGH mutant3, lane 18: kpnGH mutant 4, lane 19: kpnGH mutant 5 respectively. Equal protein concentrations were separated by SDS-PAGE with a 5% stacking gel and a 12% separating gel and stained with coomassie brilliant blue. Lane M has molecular weight standards.
(TIF)