This study measured changes in global mRNA translation in response to ER stress. The analysis suggests that translation of a majority of gene transcripts is either repressed or resistant, whereas select key regulators are subject to preferential translation. From this last group, IBTKα is identified as a novel target of the UPR central to cell fate.
Abstract
Disruption of protein folding in the endoplasmic reticulum (ER) triggers the unfolded protein response (UPR), a transcriptional and translational control network designed to restore protein homeostasis. Central to the UPR is PKR-like ER kinase (PERK/EIF2AK3) phosphorylation of the α subunit of eIF2 (eIF2α∼P), which represses global translation coincident with preferential translation of mRNAs, such as activating transcription factor 4 (ATF4) and C/EBP-homologous protein (CHOP), that serve to implement UPR transcriptional regulation. In this study, we used sucrose gradient ultracentrifugation and a genome-wide microarray approach to measure changes in mRNA translation during ER stress. Our analysis suggests that translational efficiencies vary over a broad range during ER stress, with the majority of transcripts being either repressed or resistant to eIF2α∼P, whereas a notable cohort of key regulators are subject to preferential translation. From the latter group, we identified the α isoform of inhibitor of Bruton's tyrosine kinase (IBTKα) as being subject to both translational and transcriptional induction during eIF2α∼P in both cell lines and a mouse model of ER stress. Translational regulation of IBTKα mRNA involves stress-induced relief of two inhibitory upstream open reading frames in the 5′-leader of the transcript. Depletion of IBTKα by short hairpin RNA reduced viability of cultured cells coincident with increased caspase 3/7 cleavage, suggesting that IBTKα is a key regulator in determining cell fate during the UPR.
INTRODUCTION
Translational control allows for a rapid and efficient means of reprogramming gene expression. A key regulatory mechanism for translational control involves the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α∼P; Wek et al., 2006; Baird and Wek, 2012). In response to environmental stresses, eIF2α∼P causes a general repression of translation resulting from the reduced ability of eIF2 to couple with GTP and deliver the initiator Met-tRNAiMet to ribosomes for start codon recognition and the commencement of mRNA translation. Mammals express four different eIF2 kinases, each of which is activated during distinct sets of endogenous and external stress stimuli (Wek et al., 2006). For example, disruption of protein homeostasis in the endoplasmic reticulum (ER) activates the eIF2 kinase PKR-like endoplasmic reticulum kinase (PERK/EIF2AK3), which reduces protein synthesis and influx of nascent polypeptides into the secretory pathway (Walter and Ron, 2011). Translational control by PERK is central to the unfolded protein response (UPR), which also features signaling from other sensory proteins for ER stress that collectively lead to changes in transcription and mRNA decay genome wide (Walter and Ron, 2011). Loss of PERK abrogates translational control during ER stress, leading to Wolcott–Rallison syndrome, a genetic disorder characterized by insulin-dependent neonatal diabetes, osteoporosis, digestive dysfunctions, and hepatic complications (Wolcott and Rallison, 1972; Delepine et al., 2000; Julier and Nicolino, 2010).
Coincident with repression of global translation initiation, eIF2α∼P by PERK selectively enhances the translation of activating transcription factor 4 (ATF4) mRNA, encoding a basic zipper (bZIP) transcriptional activator of genes involved in the UPR (Harding et al., 2000; Vattem and Wek, 2004). Induction of ATF4 mRNA translation in response to eIF2α∼P involves a “delayed reinitiation” mechanism featuring two upstream open reading frames (uORFs) preceding the ATF4 coding sequence (CDS; Vattem and Wek, 2004). In this model, the 5′-proximal uORF1 is readily translated by scanning ribosomes, and ribosomes are then proposed to be retained on the ATF4 mRNA and resume scanning for subsequent reinitiation at a downstream coding sequence. During nonstressed conditions, when eIF2α∼P is low and there is abundant eIF2/GTP/Met-tRNAiMet complex, ribosomes readily reinitiate at the next coding sequence—uORF2, which is situated out-of-frame with the ATF4 CDS, leading to sharply reduced ATF4 CDS translation. However, during eIF2α∼P, it is suggested that lowered eIF2 ternary complex delays ribosome reinitiation, allowing for the scanning ribosome to proceed through the inhibitory uORF2 and instead initiate translation at the ATF4 CDS. This translational control mechanism, first described for the bZIP regulator GCN4 in yeast by Hinnebusch and colleagues (Hinnebusch, 2005), relies on the coordinated interplay between a positive-acting uORF1 and one or more downstream repressing uORFs to sense the levels of eIF2α∼P and active eIF2.
Increased expression of ATF4 leads to transcription of downstream target genes involved in protein folding and assembly, metabolism, cellular redox status, and feedback regulation of eIF2α∼P signaling (Harding et al., 2003; Walter and Ron, 2011). Moreover, during episodes of unresolvable ER stress, ATF4 facilitates increased expression of the bZIP transcription factor C/EBP-homologous protein (CHOP; DDIT3/GADD153), which directs cells toward an apoptotic fate (Harding et al., 2000; Tabas and Ron, 2011). CHOP mRNA is also subject to preferential translation during eIF2α∼P by a “bypass” mechanism involving ribosome scanning through a single inhibitory uORF (Palam et al., 2011). In this way, CHOP transcriptional and translational expression is tightly linked with eIF2α∼P levels, and as a consequence, the amount of CHOP protein is believed to serve as a measure of sustained ER stress. Enhanced levels of CHOP during stress would lead to increased expression of a collection of proapoptotic genes, combined with repression of those serving prosurvival functions (Rutkowski et al., 2006; Tabas and Ron, 2011). Included among the CHOP-dependent genes that contribute to increased apoptosis during ER stress is ATF5 (Teske et al., 2013). ATF5 is yet another bZIP transcriptional activator whose expression is both transcriptionally and translationally induced in response to eIF2α∼P during ER stress. Like ATF4, mRNA encoding ATF5 is preferentially translated by a mechanism of delayed translation reinitiation involving two uORFs (Watatani et al., 2008; Zhou et al., 2008).
Prevailing views of translational control in the UPR suggest that there are a few select gene transcripts that are preferentially translated coincident with repression predominant among the remaining mRNAs. We propose that the UPR translational control network may be more expansive than the models described so far, including many additional key regulatory genes subject to preferential translation. To address this hypothesis, we carried out a genome-wide study using sucrose gradient ultracentrifugation to separate translating mRNAs on large polysomes from those associated with fewer ribosomes. Gene transcripts in these gradient fractions were then measured globally to address how translational efficiencies for mRNAs changed in response to ER stress. Our study suggests a gradient model of translational control in which many mRNAs are repressed or resistant to eIF2α∼P, whereas a significant subset are preferentially translated. From this latter group, we identified α isoform of inhibitor of Bruton's tyrosine kinase (IBTKα) as being subject to both preferential translation and increased transcriptional expression in response to PERK-induced eIF2α∼P. IBTKα is a multidomain protein suggested to associate with the ubiquitin ligase CUL3 and serve as a substrate adaptor for protein ubiquitylation (Hadjebi et al., 2008; Jin et al., 2012). We show that knockdown of IBTKα increases caspase activation and lowers cell survival, suggesting that IBTKα is central for the efficacy of the UPR.
RESULTS
eIF2α∼P during ER stress induces a gradient of mRNA translational efficiencies
Thapsigargin (TG) is a potent inducer of ER stress by inhibiting the SERCA family of Ca2+ ATPases, effectively reducing levels of calcium in the lumen of the ER and activating the UPR. As a result, eIF2α∼P by PERK and the accompanying reduction in eIF2-GTP lead to an overall reduction in translation initiation (Figure 1A). This decrease of global translation initiation during ER stress in mouse embryonic fibroblast (MEF) cells is visualized by a reduction in large polysomes, accompanied by increased monosomes. By comparison, MEF cells expressing an eIF2α mutant in which the phosphorylated serine 51 was replaced with an alanine residue (eIF2α−S51A) displayed wild-type levels of polysomes independent of treatment with TG (Figure 1B). In addition to reducing genome-wide translation, eIF2α∼P also enhanced expression of ATF4 and CHOP proteins, key downstream targets of PERK (Figure 1C).
FIGURE 1:
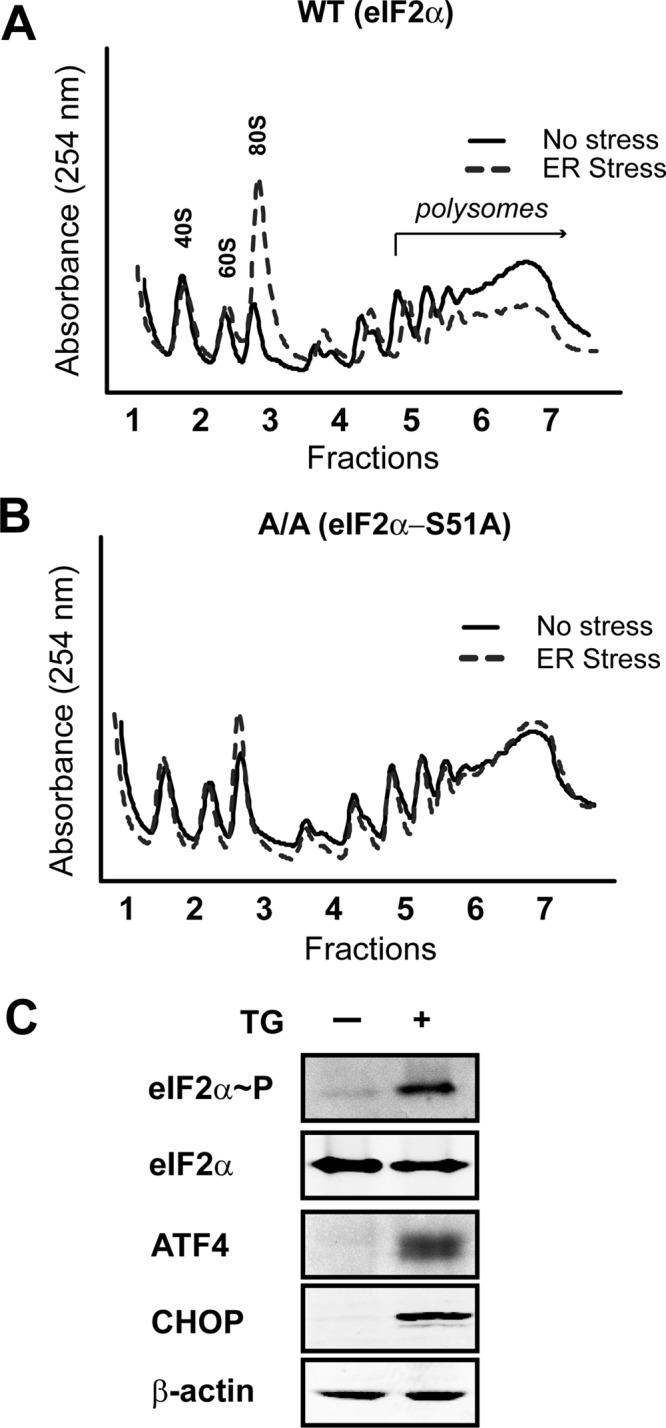
eIF2α∼P represses global translation initiation during ER stress. Polysome profiling was carried out using wild-type (A) or eIF2α−S51A (B) knock-in MEF cells treated with TG (ER stress) for 6 h or no stress treatment. (C) Immunoblot analysis of lysates prepared from wild-type MEF cells treated with TG for 6 h (+) or no stress. The indicated proteins were measured using antibodies specific to each.
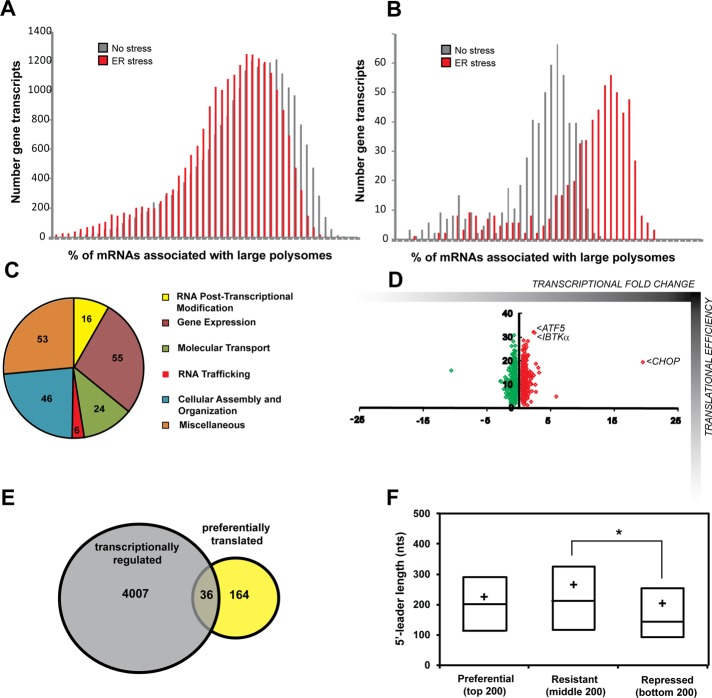
To address gene-specific changes in the translatome after ER stress, wild-type MEF cells were exposed to TG for 6 h—a time that potently increases known PERK targets, such as CHOP—or left untreated. After treatment, cycloheximide was used on both treatment groups to arrest elongating ribosomes on mRNAs. Lysates were then subjected to sucrose gradient ultracentrifugation, and sample fractions were pooled into three groups: free ribosomal subunits (fractions 1 and 2), monosomes and small polysomes (fractions 3 and 4), and large polysomes (fractions 5–7; Teske et al., 2011a). RNA was purified from each pooled group, and expression microarray analyses were conducted for the pools from ER stress and nontreatment groups. To further investigate mRNA changes during ER stress, we also performed microarray analyses on whole-cell lysate RNA. In response to stress, there was a global decrease in the number of mRNAs associated with large polysomes, indicative of an overall decrease in translating ribosomes (Figure 2A). By comparison, a smaller collection of gene transcripts showed significantly greater association (≥15%) with large polysomes upon ER stress, suggesting preferential translation (Figure 2B).
FIGURE 2:
Genome-wide analyses of gene-specific translation during ER stress. (A) Number of gene transcripts as defined by analysis of the 22,862 probe sets associated with the indicated percentage of large polysomes in wild-type MEF cells treated with TG for 6 h or no stress. (B) Percentage association with large polysomes of the top 200 gene transcripts suggested to be preferentially translated in response to ER stress. (C) The categories of molecular and cellular functions as determined by ingenuity pathway analysis of the top 200 genes suggested to be subjected to preferential translation. (D) Scatterplot illustrating percentage shift toward large polysomes during stress suggestive of translational control (y-axis) vs. their relative mRNA fold changes in response to stress (x-axis, transcriptional control). (E) Venn diagram of the overlap between the 4007 genes encoding mRNAs that were significantly increased in response to ER stress (p < 0.05) and the top 200 genes suggested to be preferentially translated during ER stress. (F) Repressed genes encode mRNAs with significantly shorter 5′-UTRs than the stress-resistant cohort (one-way ANOVA with Tukey's HSD post hoc test, p = 0.0042). Lines of the box-plot diagram illustrate 75th, 50th, and 25th percentile values, and the + symbol indicates the arithmetic mean.
We selected the bottom, middle, and top 200 genes as the repressed, resistant, and preferentially translated groups during eIF2α∼P, respectively (Supplemental Table S1). Included among the top 200 genes, which corresponded to ∼15% or greater increased association with ribosomes during stress, are ATF4, ATF5, and CHOP, each of which was previously shown to be preferentially translated (Vattem and Wek, 2004; Zhou et al., 2008; Palam et al., 2011). To investigate categories of gene function, we performed an ingenuity pathway analysis (IPA) on those members suggested to be preferentially translated during ER stress. The results indicate that this group encodes a diverse class of proteins, with the majority involved in gene expression, cellular assembly and organization, and molecular transport (Figure 2C).
By performing expression microarray analyses on both polysome fractions and whole-cell RNA lysates, we were able to address the dynamic nature between transcriptional and translational regulation during ER stress. As mentioned previously, key regulators of the UPR are regulated both transcriptionally and translationally. For example, ATF5 mRNA was shown to increase threefold while increasing 33% toward higher polysomes (Figure 2D). Similarly, the levels of CHOP mRNA were increased 20-fold and shifted 20% toward large polysomes. Of interest, of the top 200 genes suggested to be preferentially translated during ER stress, only 36 (18%) also showed significant increases in mRNA levels (p < 0.05; Figure 2E), which is comparable to the 18% (4007/22,862) of gene transcripts that were significantly increased in response to ER stress in our genome-wide analysis. This finding suggests that the PERK pathway relies largely on translational control for regulation of many of its critical target genes.
The information encoded in the 5′-leaders of gene transcripts is suggested to be critical for their preferential translation in response to eIF2α∼P. The composition of the 5′-leader sequences and the placement of uORFs for the top 200 gene transcripts shows enhanced association with large polysomes in response to ER stress, as illustrated in Supplemental Table S1. The median value for leader length in nucleotides for preferentially translated mRNAs was 226 (±10.6 SEM), for resistant was 260 (±14.8 SEM), and for repressed was 199 (±13.0 SEM), with a significant difference in the length of the 5′-leaders identified between the resistant and repressed groups (one-way analysis of variance [ANOVA], p = 0.004; Figure 2F).
As previously noted, uORFs have been shown to be critical for translational regulation, and we hypothesized there may be an enrichment of uORFs in the preferentially translated cohort. However, an analysis of uORF frequency among groups failed to show a significant difference in the presence, number, or length of uORFs among the groups (Table 1). Of the 600 total transcripts analyzed, 271 (45%) possessed uORFs. This percentage is representative of previous studies reporting that 35–50% of mouse and human transcripts contain uORFs (Iacono et al., 2005; Matsui et al., 2007). Furthermore, there was no significant difference in the GC composition among the predicted 5′-leaders of the mRNAs in the repressed, resistant, and preferentially translated groups (Table 1). The initiation codon context can be important for ribosome selection of a CDS (Kozak, 1987; Hinnebusch, 2011), with the optimized sequence gcc(A/G)ccATG(G), with the initiation codon (underlined) and purine residues at −3 and +4 (bold) being most critical. It is suggested that alteration at either purine reduces a strong initiation codon to an adequate one and that loss of both purines renders an initiation codon to a weak context. Among mRNAs suggested to be preferentially translated that contain a single uORF, 58% of the transcripts showed a stronger start codon context for the CDS than for the uORF. By comparison, those in the repressed group were significantly different, with only 31% of transcripts adopting this configuration (chi-squared, p = 0.01). This suggests that the initiation context of uORFs can be important for translational control during ER stress.
TABLE 1:
The 5′-leader characteristics for gene transcripts suggested to be preferentially translated, resistant, or repressed during ER stress.
| Characteristic | Preferential (top 200) | Resistant (middle 200) | Repressed (bottom 200) |
|---|---|---|---|
| 5′-Leader length (mean nts) | 225.7 | 260.2 | 199.8 |
| 5′-Leader length (median nts) | 201.5 | 212.5 | 143.0 |
| uORF frequency (%) | 38.5 | 48.5 | 48.5 |
| uORF length (mean nts) | 120.5 | 112.8 | 93.9 |
| GC content of 5′-leader (%) | 61 | 62 | 64 |
| Transcripts with optimal Kozak context for CDS (%) | 52 | 53 | 40 |
| Transcripts with uORF in poor Kozak context (%) | 58 | 61 | 31 |
For 5′-leader length, a one-way ANOVA reported a significant difference between groups (p = 0.004), with a Tukey HSD post hoc test indicating a difference in leader length between the repressed and resistant groups. For uORF length, when multiple uORFs were present, the longest uORF was used in the analysis. A one-way ANOVA indicated no significant difference between groups (p = 0.155). nts, nucleotides.
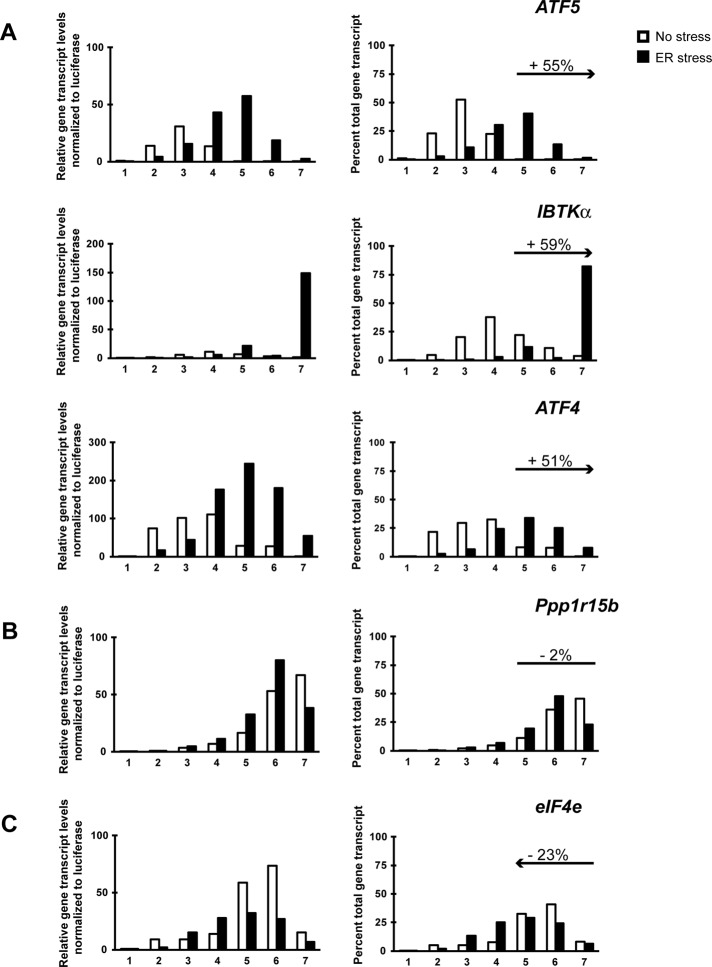
To validate the results of the translational microarray analysis, we performed quantitative PCR (qPCR) analysis using RNA prepared from sucrose gradient fractions to measure specific examples of key regulatory genes that fell into each of the three categories. ATF4 and ATF5 mRNAs, both of which are preferentially translated during ER stress, shifted 55 and 51% toward large polysomes, respectively (Figure 3A). The left side of the figure illustrates the levels of gene transcripts present in each sucrose gradient fraction relative to an exogenous, polyadenylated Luciferase spike-in control mRNA. There were increased levels of ATF4 and ATF5 mRNAs in response to ER stress, along with a shift of the gene transcripts to the large polysome fractions. The right side depicts the percentage of the ATF4 and ATF5 gene transcripts among the gradient fractions for each of the two conditions, highlighting changes in ribosome association with these mRNAs independent of changes in total transcript levels (Figure 3A). After ATF5, the second-largest shift toward polysomes during ER stress in our microarray analysis was IBTKα (Figure 2D and Supplemental Table S1), which was confirmed by qPCR analysis (Figure 3A). IBTKα is a focus of this study.
FIGURE 3:
Changes in polysome association of gene transcripts suggest preferential, resistant, or repressed translation during ER stress. Fractions were collected by sucrose gradient analyses of lysates prepared from wild-type MEF cells treated with TG for 6 h (ER stress) or no stress. Relative levels of the indicated gene transcripts were then determined by qPCR for each fraction (left), with preferentially translated gene transcripts in response to ER stress (A), resistant (B), and repressed (C). For these values, gene expression was normalized to an exogenous polyadenylated Luciferase spike-in mRNA control. Right, percentage of total gene transcripts in each of the seven fractions prepared from the TG-treated and nonstressed cells. Percentage changes in association with large polysomes (fractions 5–7) in response to ER stress are indicated in each gene transcript panel. For example, ATF5 showed a 55% increase in transcripts levels into fractions 5–7 during ER stress compared with no stress.
Among the resistant cohort, we verified minimal changes in large polysome association between the stressed and nonstressed conditions for PPP1R15b (CReP), a gene encoding the protein phosphatase regulatory subunit that constitutively targets PP1c to dephosphorylate eIF2α∼P (Figure 3B). We found the mRNA encoding the 5′-cap-binding protein, eIF4e, to be shifted 23% away from large polysomes during ER stress (Figure 3C). Furthermore, the total levels of eIF4e mRNA were sharply reduced in the stress lysates (Figure 3C, left). This finding illustrates dynamic coordination between transcription and translation that can serve to dampen gene expression during ER stress.
eIF2α∼P leads to translational expression of IBTKα mRNA
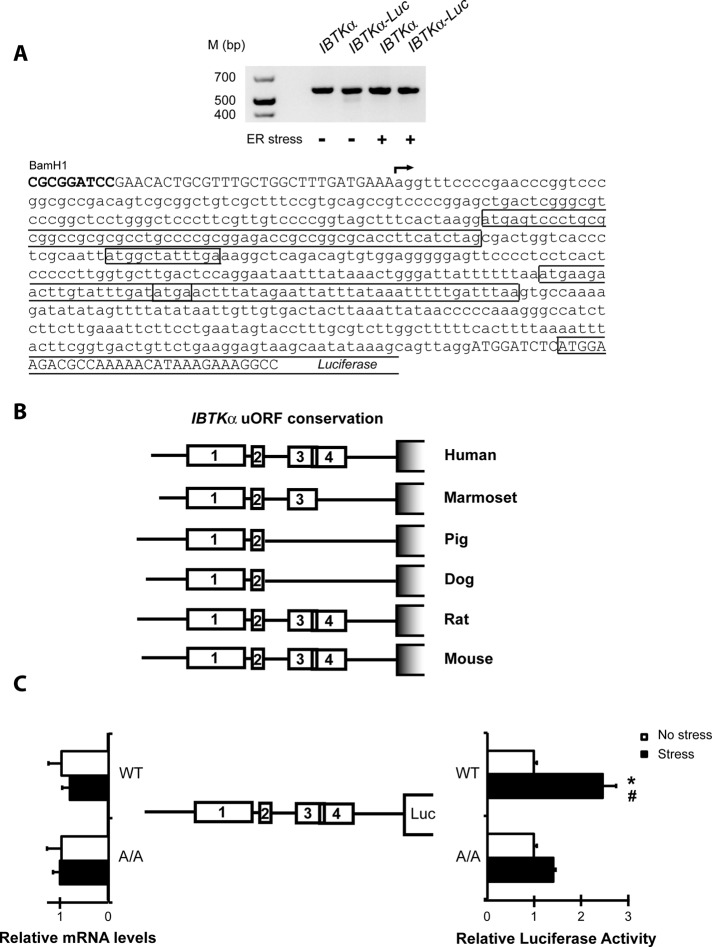
Two transcripts can be generated from the mouse IBTK gene, designated α and γ, by a mechanism suggested to involve transcription from different promoters (Spatuzza et al., 2008). The transcript that we measured in the microarray and qPCR analyses is the longer version, IBTKα, which is 5679 nucleotides in length, with an encoded protein of 150 kDa suggested to be widely expressed among tissues (Spatuzza et al., 2008). To define the 5′-leader of the IBTKα transcript, we performed 5′-rapid amplification of cDNA ends (RACE) to determine the transcription start site using RNA that was isolated from wild-type MEF cells in the absence and presence of ER stress. The 5′-leader of IBTKα is 588 nucleotides in length and encodes four uORFs (Figure 4A). A phylogenetic analysis among mammals illustrates a high level of conservation for both leader length and placement of the uORFs, with uORFs 1 and 2 being conserved between each of the orthologues (Figure 4B).
FIGURE 4:
The 5′-leader of the IBTKα gene transcript confers preferential translation in response to eIF2α∼P. (A) 5′-RACE was performed to establish the transcriptional start site for IBTKα in the presence and absence of stress. The arrow indicates the transcriptional start site, and each of the four uORFs present in the IBTKα 5′-leader is boxed. 5′-RACE assays were performed using RNA-containing endogenous transcript and the luciferase translational reporters prepared from cells treated with TG or no stress. The image of the cDNA products that were analyzed by agarose gel electrophoresis is presented above the IBTKα sequence. (B) Schematic of phylogenetic conservation of uORFs in the 5′-leader of the IBTKα mRNA among different mammalian species. (C) IBTKα translational control was measured by a dual luciferase assay. The PTK-IBTKα-Luc reporter, which contains the IBTKα leader sequence, and a control Renilla luciferase plasmid, were introduced into wild-type or eIF2α-S51A MEF cells and treated with TG or no stress. Three independent experiments were conducted for each measurement, and relative values are represented, with the SD indicated. In parallel, levels of IBTKα-Luc mRNA were measured by qPCR, and relative values are presented, with error bars representing SD. The asterisk indicates a significant difference in wild-type MEF cells in response to ER stress, and the number sign (#) between wild-type and eIF2α-S51A cells during TG treatment (p < 0.001).
To determine whether the 5′-leader of IBTKα confers preferential translation in response to eIF2α∼P, we inserted the cDNA segment encoding the 5′-leader of the mouse IBTKα mRNA between a constitutive TK promoter and a firefly luciferase reporter gene. The resulting PTK-IBTKα-Luc plasmid was transfected into wild-type MEF cells, and these cells were exposed to TG or left untreated. In response to ER stress, there was ∼2.5-fold induction of luciferase activity despite minimal changes in mRNA levels (Figure 4C). Of importance, when this same reporter construct was analyzed in MEF cells expressing eIF2α-S51A, which cannot be phosphorylated by PERK, this induction of luciferase expression was abolished (Figure 4C). As expected, there were no significant changes in levels of reporter transcripts in these MEF cells and treatment conditions. Moreover, we performed 5′-RACE assays on the IBTKα-Luc construct to rule out the possibility of truncation or alternative splicing events in the 5′-leader (Figure 4A). These results suggest that IBTKα mRNA is preferentially translated in response to eIF2α∼P and ER stress.
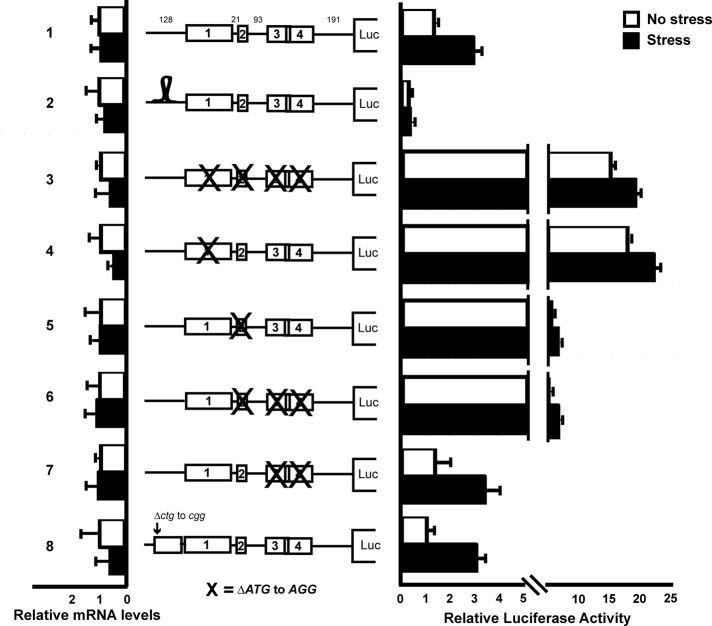
To address the mechanism underlying preferential translation of IBTKα in response to eIF2α∼P, we generated multiple mutant constructs of the wild-type PTK-IBTKα-Luc reporter. An illustration of these constructs, along with their luciferase activity and mRNA levels in response to TG or no stress treatment, is depicted in Figure 5. To address whether translation of IBTKα occurs by cap-dependent scanning, we generated a stem-loop construct in which a highly structured palindromic sequence with a high free energy (ΔG = −41 kcal/mol) was inserted 30 nucleotides downstream of the cap structure (construct 2). This highly structured stem-loop sequence, which was shown previously to impede ribosome scanning (Vattem and Wek, 2004), caused significant reduction in basal luciferase expression and loss of stress induction. There were no significant changes in the mRNA levels among these reporter constructs and those that followed.
FIGURE 5:
Translation of IBTKα mRNA is regulated by a scanning model involving two inhibitory upstream ORFs. Wild-type and the depicted mutant versions of the PTK-IBTKα-Luc reporter were analyzed in MEF cells subject to TG or no stress treatment. The 5′-leader of the IBTKα mRNA is illustrated upstream of the firefly luciferase reporter. Boxes indicate uORFs 1–4, and the numbers in the wild-type leader depiction represent the number of nucleotides separating the ORFs. Relative luciferase activities are shown after ER stress or no stress treatment, with error bars indicating SD. Left, relative levels of the reporter mRNAs as measured by qPCR. The stem-loop structure (ΔG = – 41 kcal/mol) adjacent to the 5′-end of the reporter is illustrated, and the X indicates mutations of the start codon for the indicated uORF.
To further dissect the mechanism of regulation, we mutated AUG start codons for the uORFs, both individually and in combination, to noninitiating AGG codons. Mutating the start codons for each of the four uORFs led to a 15-fold increase in luciferase activity independent of stress conditions (Figure 5, construct 3). Of interest, the same level of enhanced reporter activity was observed when only the start codon for uORF1 was altered (construct 4), indicating that uORF1 is a major repressive element for translation of the IBTKα downstream CDS. Conversely, mutating the start codon of uORF2 resulted in a modest fivefold increase in basal levels of luciferase expression (construct 5), whereas uORFs 3 and 4 were wholly dispensable (constructs 6 and 7). We further showed that uORFs 1 and 2 alone were sufficient to facilitate the threefold stress induction observed in the wild-type construct (construct 7).
Previous genome-wide ribosome footprinting studies in human and murine cell lines reported initiating ribosomes in IBTKα at uORFs 1 and 2 and a noncanonical uCUG start codon at position −468 nucleotides upstream of the CDS start codon (Ingolia et al., 2011; Lee et al., 2013). We addressed whether the latter uCUG had any functional role in translation during basal and stress conditions by mutating the upstream CUG to a CGG in the reporter construct. There was no observable difference between wild-type and the mutant ΔuCUG luciferase expression values, indicating that this noncanonical start codon does not have a role in IBTKα translational control (Figure 5, construct 8). Together these studies suggest that uORF1 and uORF2 can serve as repressing elements in IBTKα translation, with uORF1 being predominant. Phosphorylation of eIF2α is suggested to lead to a partial bypass of these repressing uORFs.
Induction of IBTKα gene expression requires PERK in both cultured cell and animal models of ER stress
Transcriptional induction is also central for expression of PERK targets ATF4, CHOP, and ATF5 in the UPR. To determine whether IBTKα mRNA is increased by PERK signaling, we measured IBTKα transcripts in wild-type and mutant MEF cells treated with TG or not subjected to stress. There was a threefold increase in the amount of IBTKα mRNA (p = 0.05) in the wild-type cells in response to ER stress. Of importance, this stress-responsive increase in IBTKα mRNA was abrogated in eIF2α-S51A, PERK−/−, and ATF4−/− mutant cell lines (Figure 6A). These results suggest that in addition to preferential translation, the PERK/eIF2α∼P/ATF4 pathway facilitates increased IBTKα mRNA levels in response to ER stress.
FIGURE 6:
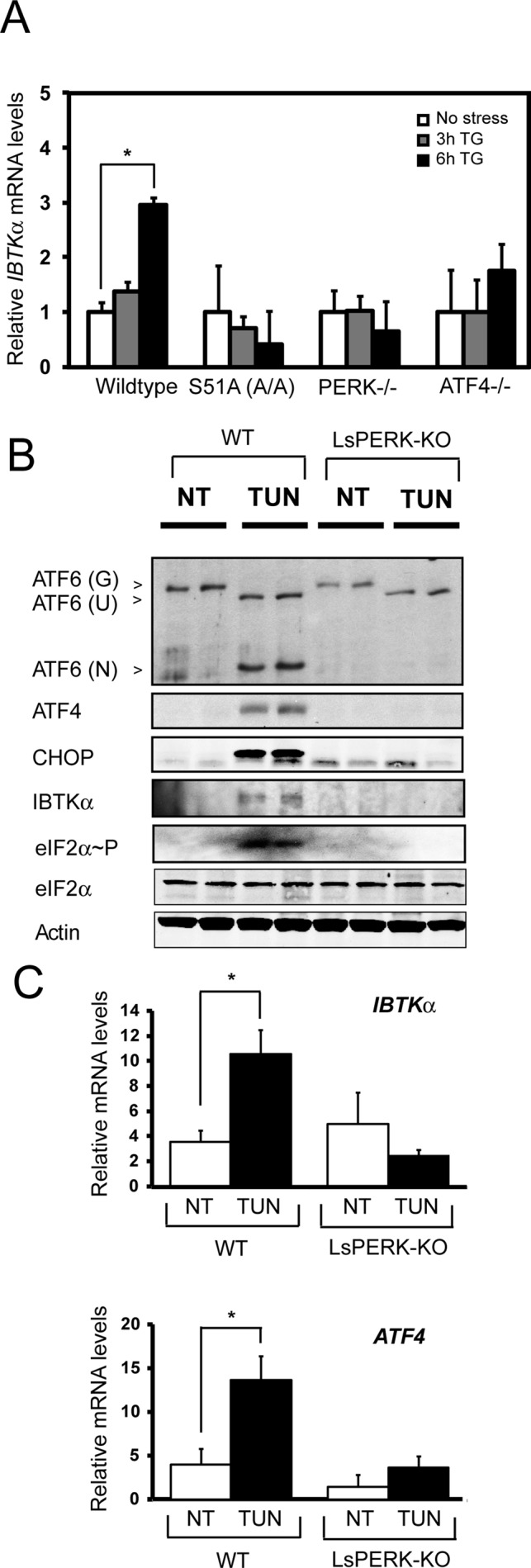
Enhanced IBTKα gene expression requires PERK in both cultured cell and animal models of ER stress. (A) IBTKα mRNA levels were measured by qPCR in wild-type and mutant MEF cells treated with TG for 3 or 6 h or no stress (0). The error bars indicate SD, and the asterisk indicates significant enhancement in transcript levels in response to the ER stress (p < 0.05). (B) Wild-type and liver-specific PERK knockout (LsPERK-KO) mice were subjected to a single intraperitoneal injection of tunicamycin or saline control and killed after 6 h. Lysates were prepared and immunoblot analyses carried out using antibodies against the indicated proteins. (C) Alternatively, RNA was prepared from the LsPERK-KO livers and qPCR analyses were carried out to measure IBTKα or ATF4 mRNA levels. The error bars indicate SD, and the asterisk indicates significant increase in transcript levels in response to ER stress (p < 0.005).
We further addressed the requirement of PERK for IBTKα induction during ER stress in a mouse model system. For this, liver-specific knockout (LsPERK-KO) mice and their wild-type counterparts were subjected to a single intraperitoneal injection of tunicamycin. Tunicamycin inhibits N-linked glycosylation and as a result acts as a potent inducer of ER stress. We previously reported that loss of PERK in livers exposed to tunicamycin disrupted liver homeostasis and increased apoptosis (Teske et al., 2011b). There was significant induction of eIF2α∼P, ATF4, CHOP, and IBTKα proteins 6 h after treatment of the ER stress agent in the wild-type mice (Figure 6B). Furthermore, there was also a substantial increase in cleavage and release of ATF6 N-terminal protein, a hallmark of UPR activation. As expected, the LsPERK-KO mice failed to show increase in expression of ATF4 and CHOP or ATF6 activation in response to the tunicamycin treatment. Of importance, induction of the IBTKα protein after tunicamycin injection was also undetected upon loss of PERK (Figure 6B). We also measured IBTKα mRNA levels in livers of wild-type and LsPERK-KO mice treated with tunicamycin. There was a significant ∼3-fold increase (p = 0.005) of IBTKα mRNA during ER stress that was completely abrogated in the LsPERK-KO samples (Figure 6C). Consistent with prior reports (Lu et al., 2004; Teske et al., 2011b), PERK was also required for increased levels of ATF4 mRNA in response to tunicamycin treatment. These results indicate that PERK signaling facilitates increased IBTKα expression in response to ER stress in both cultured cells and mouse model systems.
Loss of IBTKα expression results in lowered cell viability
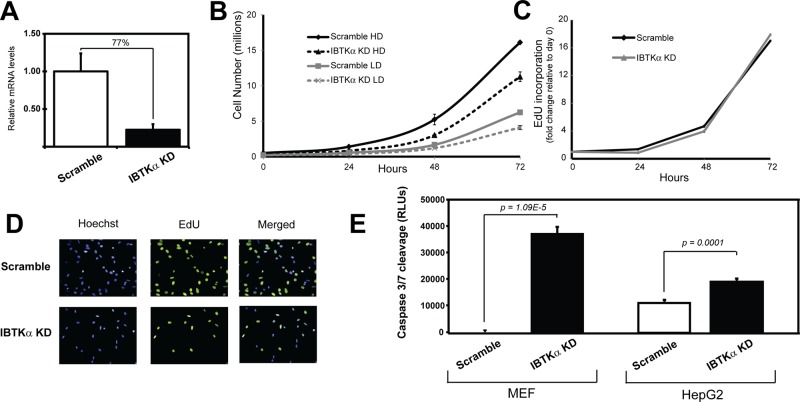
To address the importance of IBTKα in cell viability, we used short hairpin RNA (shRNA) and a lentiviral delivery system to knock down expression of IBTKα in MEF cells. We present an analysis using lentivirus targeted against IBTKα (TRCN0000088505; Sigma-Aldrich), with a second shRNA (TRCN0000088503; Sigma-Aldrich) showing similar results. There was a significant depletion of IBTKα mRNA in MEF cells compared with the control scrambled shRNA (Figure 7A). Knockdown of IBTKα led to a reduced number of cultured MEF cells compared with the control cells when plated at either high density or low density in the medium (Figure 7B). The reduction in cell number was not the result of reduced cell division as measured by 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Figure 7, C and D). Instead, there was sharp enhancement of cleaved caspase 3/7 that occurred in the absence of stress treatment (Figure 7E, left). There was also enhanced caspase 3/7 when IBTKα was reduced using shRNA in other cell types, including HepG2 human hepatocytes (Figure 7E, right), supporting the idea that IBTKα has prosurvival functions in both mouse and human cell types.
FIGURE 7:
Knockdown of IBTKα reduces cell viability and increases caspase 3/7 cleavage. shRNAs against IBTKα and a lentiviral delivery system were used to knock down IBTKα expression in wild-type MEF cells. (A) qPCR measurements of IBTKα mRNA in knockdown cells (KD) and control shRNA–expressing cells (scrambled). (B) IBTKα-KD and control MEF cells expressing scrambled shRNA were plated in culture dishes at low or high density. Cell numbers were determined upon culturing for up to 72 h. (C) Measurements of cell proliferation by EdU incorporation during cell culture normalized to day 0. (D) Hoescht-stained, EdU-stained, and merged IBTKα-KD and scrambled control cells. (E) Measurements of caspase 3/7 cleavage in MEF cells and HepG2 cells expressing IBTKα shRNA (IBTKα-KD) or scramble shRNA.
DISCUSSION
A genome-wide analysis was carried out to measure changes in mRNA association with large polysomes in response to ER stress. The majority of genes were either reduced or resistant to changes in polysome association, as exemplified by genes encoding eIF4e and Ppp1r15b, respectively (Figures 2A and 3). However, a significant subset of gene transcripts showed increased association with large polysomes in response to ER stress, suggestive of preferential translation (Figure 2B). These findings suggest that there is a large collection of genes participating in cellular assembly and organization, gene expression, molecular transport, and posttranscriptional modifications whose expression is subject to preferential translation (Figure 2C). We selected one of these genes, IBTKα, for further analysis. IBTKα translation is induced in response to eIF2α∼P by PERK via a mechanism involving relief of two repressing uORFs in the IBTKα mRNA (Figures 4 and 5). In addition, levels of IBTKα mRNA are increased in response to ER stress by the PERK/eIF2α∼P/ATF4 pathway (Figure 6). These findings place IBTKα in the PERK pathway of the UPR (Figure 8A), which features key regulatory proteins each of which is subject to enhanced transcription and translation during ER stress. Each of these key regulators is suggested to be critical for the efficacy of the UPR and, ultimately, cell fate. Indeed, we show that knockdown of IBTKα in cultured cells substantially reduces their viability along with sharply enhancing caspase 3 activity (Figure 7).
FIGURE 8:
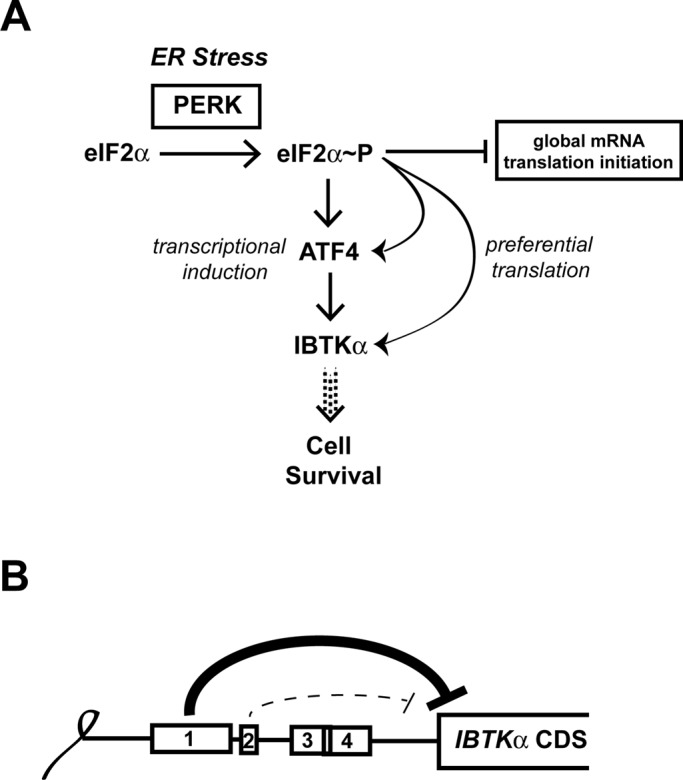
Model depicting IBTKα transcriptional and translational regulation during PERK induced eIF2α∼P. (A) During ER stress, the PERK arm of the UPR directs phosphorylation of eIF2α, resulting in global dampening of translation initiation. During this decrease in global mRNA translation, ATF4 and its downstream target IBTKα are suggested to be both transcriptionally induced and subject to preferential translation, which collectively can determine cell viability during ER stress. (B) The inhibitory uORFs 1 and 2 serve to repress IBTKα translational expression. uORF1 is a major inhibitory element, with uORF2 serving a secondary role. In response to ER stress, eIF2α∼P overcomes these inhibitory elements to translate the downstream IBTKα CDS.
Translational control by eIF2α∼P
Measurement of IBTKα mRNA in sucrose gradient fractions showed a 55% increase in transcript association with large polysomes in response to ER stress (Figure 3). This enhanced association with large polysomes was comparable to that measured for the characterized UPR transcription factors ATF4 and ATF5. However, it is noteworthy that there was a different pattern in the abundance of IBTKα transcripts in the sucrose gradient fractions compared with ATF4 and ATF5. During ER stress, almost 85% of the IBTKα mRNA was present in the largest fraction 7, whereas ATF4 and ATF5 transcripts showed a broader distribution, with the median at fraction 5. The likely explanation for this difference is that IBTKα is a large transcript with a CDS 4056 nucleotides in length, which can accommodate a large number of elongating ribosomes. By comparison, the CDSs for ATF4 and ATF5 are 1047 and 849 nucleotides in length, respectively, and as a consequence can accommodate fewer translating ribosomes. Differences in CDS length are therefore likely to be important when determining changes in ribosome association for each gene transcript that occurs upon ER stress. We selected association with ∼4 or more ribosomes as a measure of efficient translation, which would also account for the ribosomes that are participating in the uORFs of transcripts that are potentially subject to preferential translation.
Enhanced translation of IBTKα in response to eIF2α∼P centers around two uORFs that are well conserved among vertebrates (Figures 4 and 8B). The uORF1 is a major repressing element, whereas uORF2 appears to be less inhibitory. The presence of two repressing uORFs in the 5′-leader of the IBTKα mRNA suggests that the mechanism of translational control governing IBTKα is different from that described for ATF4. ATF4 translational control features a positive-acting 5′-proximal uORF, which allows for ribosomes to scan through an inhibitory uORF due to delayed reinitiation that occurs as a consequence of eIF2α∼P and reduced eIF2/GTP levels required for ribosome acquisition of charged initiator tRNA. However, IBTKα does share features with the translational control mechanism described for CHOP. CHOP contains a single inhibitory uORF that is suggested to be bypassed by scanning ribosomes in response to eIF2α∼P. In the case of IBTKα, uORF1 is suggested to be a major inhibitory element that can be overcome by eIF2α∼P. Although uORF2 is also repressing, uORF2 appears to have an ancillary role to uORF1 in IBTKα translational control. A feature of the CHOP uORF that is believed to contribute to its bypass in response to eIF2α∼P is a weak initiation codon context (Palam et al., 2011), a feature enriched in the preferential list of genes and shared with the major inhibitory uORF1 in the IBTKα transcript. The CHOP uORF is believed to thwart translation elongation, thus reducing reinitiation at the downstream CDS (Palam et al., 2011). It is not known whether translation of uORFs 1 and 2 of IBTKα also serves as elongation barrier. Furthermore, after translation of the IBTKα uORFs, there may be some regulated reinitiation at the downstream CDS in response to eIF2α∼P and ER stress.
Translation of most mRNAs is suggested to be repressed or resistant to eIF2α∼P. Among these, we showed that the gene transcript encoding the 5′-cap-binding protein, eIF4e, displayed reduced levels and was sharply shifted away from polysomes during ER stress. This finding suggests that translation of eIF4e mRNA is repressed during the UPR. Note that during ER stress, ATF4 is also suggested to increase expression of 4E-BP1 (Yamaguchi et al., 2008), a repressor of eIF4E association with eIF4G, suggesting that there can be multiple mechanisms by which PERK/eIF2α∼P/ATF4 can lower cap-dependent translation.
IBTKα facilitates cell survival
In response to acute ER stress, PERK and the UPR are proposed to be critical for survival as cells expand their ER processing capacity to address increased demands on the secretory pathway. Knockdown of IBTKα by shRNA substantially reduces the number of MEF cells in culture (Figure 7). This is not due to lowered cell proliferation, but instead is suggested to occur by enhanced cell death accompanied by increased activation of caspase 3. Increased caspase 3 activation also occurs with knockdown of IBTKα in human hepatoma HepG2 cells, emphasizing that IBTKα also enhances cell survival in cultured human cells. It is noteworthy that reduced cell survival by lowered IBTKα expression occurs even without stress treatments.
In mice there are two isoforms of IBTK—the α isoform that is the focus of this study and a shorter γ form (Spatuzza et al., 2008). IBTKγ, a 26-kDa protein that is highly expressed in hematopoietic tissues, was previously reported to bind to and repress Btk protein kinase, hence the acronym IBTK for “inhibitor of BTK.” IBTKγ suppression of Btk lowers BTK-mediated calcium mobilization and reduces activation of nuclear factor-κB–targeted transcription in B cells (Liu et al., 2001; Janda et al., 2011). Subsequently it was determined the IBTK gene encoded an additional larger product, IBTKα, that is a result of an alternative upstream promoter. IBTKα is a 150-kDa multidomain protein that contains ankyrin repeats, RCC1 repeats, and two BTB/POZ segments (Hadjebi et al., 2008). The BTB is a protein–protein interaction domain, and it was reported that IBTKα can associate with the ubiquitin ligase CUL3, suggesting that IBTKα may serve as a substrate adaptor in protein ubiquitylation (Jin et al., 2012). We do not yet know possible target proteins for the suggested IBTKα ubiquitylation adaptor function, but a related BTB-containing protein, KLHL12, was reported to facilitate monoubiquitylation of SEC31, contributing to the assembly of large COPII vesicle coats (Jin et al., 2012). An important question for the future is whether IBTKα also facilitates ubiquitylation of proteins that facilitate key secretory processes.
MATERIALS AND METHODS
Cell culture and generation of stable cell lines
Wild-type, eIF2α−S51A, PERK−/−, and ATF4−/− MEF cells were previously described (Jiang et al., 2004). The MEF cells were grown in DMEM (Corning, Tewksbury, MA) supplemented with 1 mM nonessential amino acids, 10% (vol/vol) fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C. The ATF4−/− cells and their wild-type counterpart were supplemented with additional amino acids and 55 μM β-mercaptoethanol due to a predisposed sensitivity of ATF4-depleted cells to oxidative stress (Harding et al., 2003). HepG2 cells were cultured in MEM (Life Technologies, Grand Island, NY) supplemented with 1 mM nonessential amino acids, 1 mM sodium pyruvate, 2 mM glutaMAX, and 10% (vol/vol) fetal bovine serum at 37°C. Cells were cultured to 60–70% confluence and treated with TG for the indicated times.
Stable IBTKα knockdown and scramble control cells were produced by transducing wild-type MEF cells with lentivirus encoding shRNA against IBTKα from validated mission shRNA TRC clones TRCN0000088505 and TRCN0000088503 (Sigma-Aldrich, St. Louis, MO) or control SHC007, or in HepG2 cells, TRC clones TRC0000082575 and TRC0000082577 or control SHC007. Transduced cells were selected for shRNA expression with 5 μg/ml puromycin and maintained in DMEM or MEM. Cell culture maintenance and all assays were performed in the absence of puromycin.
Immunoblot analyses
MEF cells were treated with 1 μM TG for 6 h or without treatment, and protein lysates were prepared, quantitated, and separated by SDS–PAGE, and immunoblot analyses were carried out in three independent experiments using horseradish peroxidase–tagged secondary antibody as previously described (Teske et al., 2013). Antibodies used in the immunoblot analysis are as follows: eIF2α∼P antibody from Cell Signaling Technologies (9721; Danvers, MA), monoclonal antibody for total eIF2α from Scott Kimball (Pennsylvania State University College of Medicine, Hershey, PA), CHOP from Santa Cruz Biotechnology (sc-7351; Dallas, TX), ATF4 prepared against recombinant protein (Zhou et al., 2008), β-actin from Sigma-Aldrich (A5441), ATF6 against recombinant protein (Teske et al., 2011b), and IBTKα from Abnova (H00025998-B01P; Tapei City, Taiwan).
Polysome profiling and sucrose gradient ultracentrifugation
Sucrose gradients ranging from 10 to 50% in a solution containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 50 μg/ml cycloheximide were used for polysome analysis as previously described using a tilted tube rotation method on a gradient station equipped with a Piston Gradient Fractionator and a Gradient Master from BioComp (Fredericton, Canada; Palam et al., 2011; Teske et al., 2011a). MEF cells were cultured in DMEM in the presence or absence of 1 μM TG for 6 h. Before harvesting, cells were incubated in culture media containing 50 μg/ml cycloheximide for 10 min at 37°C. Cells were rinsed twice with chilled phosphate-buffered saline (PBS) containing 50 μg/ml cycloheximide and then lysed with 500 μl of cold lysis buffer consisting of 20 mM Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 0.4% Nonidet P-40, 50 μg/ml cycloheximide, and EDTA-free protease inhibitor cocktail tablet (Roche, Indianapolis, IN). The lysates were sheared with a sterile syringe with a 23-gauge needle, incubated on ice for 10 min, and clarified at 8000 × g for 10 min. A 400-μl amount of supernatant was layered atop the sucrose gradients, which were subjected to centrifugation in a Beckman SW41Ti rotor at 40,000 rpm for 2 h at 4°C. Sucrose fractions and the resulting polysome profiles for each sample were then collected using a Piston Gradient Fractionator and a 254-nm ultraviolet monitor with Data Quest software.
To investigate specific mRNA transcript shifts during stress, 10 ng/ml firefly luciferase control RNA (Promega, Madison, WI) was added to each pooled sample before RNA isolation, allowing for measurements of the relative amounts of the transcript of interest to be normalized to an exogenous RNA control (Palam et al., 2011; Teske et al., 2011a). Samples were then immediately mixed with 750 μl of TRIzol Reagent LS and RNA isolation and cDNA generation performed as described later. To calculate percentage total gene transcript for the seven fractions, 2(−ΔΔCT) was summed for each treatment group, and the 2(−ΔΔCT) value for each fraction was considered as a percentage of the total. This calculation serves to omit changes in the levels of transcript abundance between treatment groups. All polysome profiles and mRNA shifts depicted are representative of three independent biological replicates. The percentage shift was calculated as (percentage total mRNA in fractions 5–7 during ER stress) – (percentage total mRNA in fractions 5–7 during no stress).
Microarray analysis
The MEF cells were cultured in DMEM treated with 5 μM TG for 6 h or no stress. We observed variations in ER stress response between different preparations of TG, and this concentration of this drug lot was optimal for induced eIF2α∼P and downstream translational control. We used 6 h of stress treatment, as this was optimal for expression of downstream targets of PERK, such as CHOP, which require elevated mRNA expression for subsequent preferential translation by eIF2α∼P. Before harvesting, the MEF cells were treated with 50 μg/ml cycloheximide and incubated at 37°C for 10 min. Cell lysates were subjected to sucrose gradient centrifugation and polysome fractionation. Seven fractions were collected from the top of the gradients into cold microfuge tubes and immediately placed on ice. Each fraction was adjusted to 0.5% SDS, and fractions were combined to form three pools as follows: fractions 1 and 2, 3 and 4, and 5–7 were combined as pools 1, 2, and 3, respectively. In parallel, total RNA was isolated from unfractionated lysates for analysis of total gene transcript levels. Synthetic Poly(A) luciferase RNA (10 ng/ml; Promega), along with a bacterial spike-in control RNA (Affymetrix, Santa Clara, CA), was added to each gradient fraction pool. Synthetic luciferase RNA served as a control for the efficiency of RNA isolation. The bacterial spike-in RNA has different concentrations of each of the four exogenous, premixed, polyadenylated prokaryotic RNA controls. These prokaryotic genes have limited cross-hybridization with mammalian sequences but have target sequences on the Affymetrix arrays. These spike-ins normally serve as quality controls for the labeling step and so are added after RNA extraction as the first step of the Affymetrix labeling protocol. Here they are used to normalize the arrays from the different fractions because the amount of RNA in each fraction may not be equivalent. RNA was precipitated at −70°C with 2.5 volumes of 100% ethanol and purified using Qiagen RNeasy midi-columns. For total unfractionated RNA, samples were subjected to ethanol precipitation. Total RNA was isolated, analyzed, and stored in the same way as the RNA from polysomal fractions. The quality of RNA was measured by using an Agilent Bioanalyzer. RNA integrity numbers for the unfractionated total RNA were ≥9.9.
RNA preparations were then labeled using the standard Affymetrix protocol for 3′-IVT arrays (GeneChip Expression Analysis Technical Manual, revision 5, Affymetrix, Santa Clara, CA), starting with 2 μg of total RNA for all samples. Labeled cRNA was hybridized for 17 h to the Affymetrix Mouse Genome 430 2.0 Array. The signal values and detection calls were derived using the MAS5 algorithm in Affymetrix GeneChip Operating Software. Affymetrix arrays were hybridized and scanned at the Center for Medical Genomics at Indiana University School of Medicine (Indianapolis, IN) following standard protocols. Scaling was not used to normalize the arrays. Raw intensity values for the spike-in control RNA probe sets were used to normalize gene expression values across all arrays. Transformations by using the spike-in control probe set values were performed as previously described (Sampath et al., 2008). Probe sets were retained for analysis if a probe set was called present in at least 66.6% of the samples in either the control or treated unfractionated samples. The same probe sets were retained in the fractioned polysomal RNA samples and considered for further analysis.
Differentially translated genes were identified using the data generated from the three pools following a modification of the procedure used for the unfractionated RNA analysis. This analysis is based on the fact that the majority of mRNA bound to multiple ribosomes are in pool 3, whereas pools 1 and 2 contain mRNAs bound to no ribosomes and one to three ribosomes, respectively. Consequently, the percentage of a transcript that resides in pool 3 is a measure of increased mRNA binding to translating ribosomes—a suggested measure of translational efficiency. For each replicate control and treated sample, the fraction of normalized log2- transformed mRNA intensity in pool 3 was divided by the total mRNA intensity (pool 3/[pools 1 + 2 + 3]). Statistical analysis on the biological replicates was performed using Student's t test to derive p values for each probe set. Microarray data are deposited in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under the series number GSE54581. The following link was created to allow review of microarray data: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=inchsgsonnobrcd&acc=GSE54581.
Measurement of mRNA by qPCR
RNA was isolated from cultured cells using TRIzol reagent (Invitrogen, Life Technologies). Single-strand cDNA synthesis was carried out using the TaqMan reverse transcriptase kit (Applied Biosystems, Life Technologies) following the manufacturer's instructions. Total RNA was extracted from frozen liver preparations as described (Teske et al., 2011b). Levels of mRNA were measured by qPCR using the SYBR Green (Applied Biosystems, Life Technologies) method on a Realplex2 Master Cycler (Eppendorf, Hamburg, Germany). To measure the levels of target mRNAs, transcripts were normalized to either β-actin or luciferase control RNA (Promega) for changes in polysome fraction distribution. The primers used for measuring mRNA levels were as follows: IBTKα, forward primer, 5′-CCACCGTCTGCAGGATTATT-3′, and reverse primer, 5′-CTCGACCTTATCCGAATGGA-3′; ATF5, forward primer, 5′-GGCTGGCTCGTAGACTATGG-3′, and reverse primer, 5′-CCAGAGGAAGGAGAGCTGTG-3′; ATF4, forward primer, 5′-GCCGGTTTAAGTTGTGTGCT-3′, and reverse primer, 5′-CTGGATTCGAGGAATGTGCT-3′; β-actin, forward primer, 5′-TGTTACCAACTGGGACGACA-3′, and reverse primer, 5′-GGGGTGTTGAAGGTCTCAAA-3′; eIF4e, forward primer, 5′-CAGGAGGTTGCTAACCCAGA-3′, and reverse primer, 5′-ATAGGCTCAATCCCGTCCTT-3′; CReP, forward primer, 5′-GGCTACAGTGGCCTTCTCTG-3′, and reverse primer, 5′-CATCCATCCCTTGCAAATTC-3′; and firefly luciferase, forward primer, 5′-CCAGGGATTTCAGTCGATGT-3′, and reverse primer, 5′ -AATCTCACGCAGGCAGTTCT-3′.
Plasmid constructions and luciferase assays
A 5′-RACE (FirstChoice; Ambion, Life Technologies) was performed following the manufacturer's protocol using RNA samples extracted from wild-type MEF cells treated with 1 μM TG for 6 h or no stress agent to determine the transcriptional start site for IBTKα. The cDNA fragment encoding the 5′-leader of IBTKα mRNA was inserted between HindIII and NcoI restriction sites in a derivative of a pGL3 basic luciferase vector (Vattem and Wek, 2004; Palam et al., 2011). The resulting PTK-IBTKα-Luc reporter plasmid contains the 5′-leader of mouse IBTKα mRNA fused to a luciferase reporter downstream of a constitutive TK promoter. ATG start codons were mutated to AGG codons individually for all permutations reported in Figure 5 by site-directed mutagenesis (Stratagene, Agilent Technologies, Santa Clara, CA) following the manufacturer's instructions. For the stem-loop construct impeding cap-dependent scanning, a previously described stem-loop structure (ΔG = –41 kcal/mol) was inserted 30 base pairs downstream of the encoded transcription start site (Vattem and Wek, 2004). All plasmid constructs were sequenced to verify nucleotide substitutions. PTK-IBTKα-Luc constructs were transiently cotransfected with a Renilla reporter plasmid into wild-type or eIF2α-S51A MEF cells for 24 h. Transfected cells were treated with 0.1 μM TG for 12 h, and cells were collected and firefly and Renilla luciferase activities measured as previously described (Vattem and Wek, 2004). Relative values of firefly luciferase activities, normalized for Renilla luciferase control, were determined in triplicate for each of at least three different biological samples.
Animal study
The animal study protocol was approved by the Institutional Care and Use Committee at the Indiana University School of Medicine. LsPERK-KO mice were derived by deletion of floxed PERKfl/fl using cre expression driven by the liver-specific albumin promoter as described previously (Bunpo et al., 2009; Teske et al., 2011b). Mice were genotyped to ensure efficient PERK gene deletion. Mice received intraperitoneal injections of tunicamycin at a dose of 1 mg/kg body weight or an equivolume of excipient (0.3% dimethyl sulfoxide in PBS) as described. Mice were killed by decapitation 6 h after injection, and dissected livers were rinsed in chilled PBS, weighed, and snap-frozen in liquid nitrogen. Preparations of RNA and protein, qPCR measurements of the ATF4 and IBTKα mRNAs, and the indicated protein measurements by immunoblot were prepared as described.
Cell proliferation and viability assays
Scramble and IBTKα KD MEF cells were seeded at either 2 × 105 or 5 × 105 cells in 10-cm dishes and harvested using TrypLE (Life Technologies) for up to 72 h. Harvested cell suspensions were counted for viability by trypan exclusion using a Vi-Cell Cell Viability Analyzer (Beckman Coulter, Indianapolis, IN). For cell proliferation assays, knockdown MEF cells and their scramble counterparts were seeded at 2.5 × 105 cells/well in 96-well plates and allowed to set overnight. Before fixation with 3.7% Formalin, 20 μM EdU was added to cells for 2 h at 37°C. Cells were permeabilized with Triton X-100 and labeled using Click-iT EdU reaction mixture (Invitrogen). Data were normalized to day 0 for each respective cell line. Images for microscopy depict total nuclear staining (10 μg/ml Hoechst) and cells actively in S phase (10 μM EdU). For caspase 3/7 cleavage assays, cells were plated at 10,000 cells/well in a 96-well plate, allowed to grow for 24 h, and then measured using the ApoLive-Glo Multiplex assay (Promega) on a Synergy H1 Microplate reader (BioTek, Winooski, VT).
Supplementary Material
Acknowledgments
We thank current and former members of the Wek lab for useful discussions and acknowledge the Center for Medical Genomics at the Indiana University School of Medicine for the processing of microarray chips. This work was supported by National Institutes of Health Grants GM049164 to R.C.W. and T32DK064466 to T.D.B.
Abbreviations used:
- ATF
activating transcription factor
- bZIP
basic leucine zipper
- CDS
coding sequence
- CHOP
C/EBP-homologous protein
- eIF2
eukaryotic initiation factor 2
- eIF2α∼P
phosphorylation of eIF2α
- IBTKα
α isoform of inhibitor of Bruton's tyrosine kinase
- MEF
mouse embryonic fibroblast
- PERK
PKR-like endoplasmic reticulum kinase
- TG
thapsigargin
- uORF
upstream open reading frame
- UPR
unfolded protein response
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-02-0704) on March 19, 2014.
REFERENCES
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem. 2009;284:32742–32749. doi: 10.1074/jbc.M109.047910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-a kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5’untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda E, et al. Btk regulation in human and mouse B cells via protein kinase C phosphorylation of IBtkgamma. Blood. 2011;117:6520–6531. doi: 10.1182/blood-2010-09-308080. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 (ATF3) is integral to the eIF2 kinase stress response. Mol Cell Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier C, Nicolino M. Wolcott-Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Quinto I, Chen X, Palmieri C, Rabin RL, Schwartz OM, Nelson DL, Scala G. Direct inhibition of Bruton's tyrosine kinase by IBtk, a Btk-binding protein. Nat Immunol. 2001;2:939–946. doi: 10.1038/ni1001-939. [DOI] [PubMed] [Google Scholar]
- Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yachie N, Okada Y, Saito R, Tomita M. Bioinformatic analysis of post-transcriptional regulation by uORF in human and mouse. FEBS Lett. 2007;581:4184–4188. doi: 10.1016/j.febslet.2007.07.057. [DOI] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Spatuzza C, et al. Physical and functional characterization of the genetic locus of IBtk, an inhibitor of Bruton's tyrosine kinase: evidence for three protein isoforms of IBtk. Nucleic Acids Res. 2008;36:4402–4416. doi: 10.1093/nar/gkn413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 2011a;490:333–356. doi: 10.1016/B978-0-12-385114-7.00019-2. [DOI] [PubMed] [Google Scholar]
- Teske BF, Fusakio ME, Zhou D, Shan J, McClintick JN, Kilberg MS, Wek RC. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, Wek RC. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell. 2011b;22:4390–4405. doi: 10.1091/mbc.E11-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y. Stress-induced translation of ATF5 mRNA is regulated by the 5’-untranslated region. J Biol Chem. 2008;283:2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Wolcott CD, Rallison ML. Infancy-onset diabetes mellitus and multiple epiphyseal dysplasia. J Pediatr. 1972;80:292–297. doi: 10.1016/s0022-3476(72)80596-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.