Abstract
Brassinosteroids (BRs) are essential phytohormones that play crucial roles in plant growth and development. Perception of BRs requires an active complex of brassinosteroid-insensitive 1 (BRI1) and BRI1-associated kinase 1 (BAK1). Recognized by the extracellular leucine-rich repeat (LRR) domain of BRI1, BRs induce a phosphorylation-mediated cascade to regulate gene expression. Here we present the crystal structures of BRI1-LRR in free and brassinolide (BL)-bound forms. BRI1-LRR exists as a monomer in crystals and solution independent of BL. It comprises a helical solenoid structure that accommodates a separate insertion domain at its concave surface. Sandwiched between them, BL binds to a hydrophobicity-dominating surface groove on BRI1-LRR. BL recognition by BRI1-LRR is through an induced-fit mechanism involving stabilization of two inter-domain loops that creates a pronounced non-polar surface groove for the hormone binding. Together, our results define the molecular mechanisms by which BRI1 recognizes BRs and provide insight into BR-induced BRI1 activation.
Ubiquitously distributed throughout the plant kingdom, BRs are a class of low-abundance phytohormones that play crucial roles in many aspects of growth and development1, 2. Studies in Arabidopsis have led to the identification of a number of genes involved in BR perception and signaling3–9. One of them, BRI1 when mutated, abolishes BR-mediated responses of plants3. BRI1 belongs to a large family of plant LRR receptor like kinases (RLKs) with more than 200 members in Arabidopsis and over 300 in rice10. LRR-RLKs are characterized by an extracellular LRR domain, a single-pass transmembrane segment and a cytoplasmic kinase domain. BRI1 has been established as a bona fide receptor of BRs by genetic and biochemical investigations3, 11, 12. The most convincing evidence for this comes from the biochemical study showing that BRI1-LRR is essential and sufficient for recognition of BRs12. A 70-residue island domain (ID, residues 587-656) is indispensable for BR recognition. Binding of BRs to BRI1 initiates a phosphorylation-mediated cascade, transducing the extracellular steroid signal to transcriptional programs13, 14.
A second gene named BAK1 involved in BR signaling also encodes an LRR-RLK, albeit with only five extracellular LRRs4, 5. While not involved in BR binding, BAK1 promotes BR-induced signaling by physically associating with BRI14, 5, 15. Several models2, 4, 5, 8, 16–19 of BR-induced BRI1 activation converge at transphosphorylation within the BRI1-BAK1 complex, which involves BR-enhanced BRI1 homodimerization16, release of BRI1 kinase inhibitor 1 (BKI1)19 from BRI1 and BRI1-BAK1 heterodimerization17–19. It is still not fully understood how BRs are perceived by BRI1 to give rise to these events.
In the present study, we report the crystal structures of free BRI1-LRR and its complex with brassinolide (BL, the most active form of BR) (Supplementary Table 1). The structures not only reveal the molecular mechanisms underlying BR recognition by BRI1 and provide insight into BR-induced BRI1 activation, but also explain the structure-activity relationship of BRs and serve as a foundation for the rational design of nonsteroidal mimetics.
A helical solenoid structure of BRI1-LRR
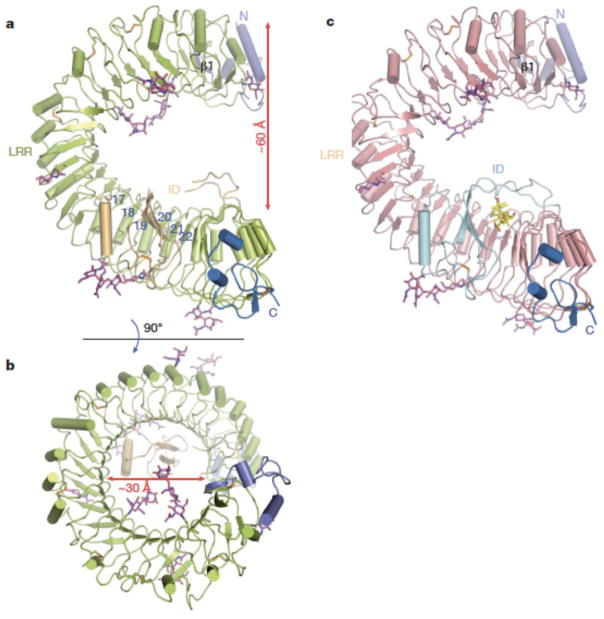
The three-dimensional structure of BL-free BRI1-LRR (residues 30-589, 596-643 and 647-772) contains 25 LRRs as predicted3, including 24 regular ones and an irregular one abutting the N-terminal side. The LRRs packed in tandem assemble into the anticipated but a highly curved solenoid structure (Fig. 1a), with the overall rotation angle about the central axis approximately 360 degrees (Fig. 1b). Compared to the regular horseshoe-shaped structures of other LRR proteins, one strikingly distinct feature of BRI1-LRR structure is that it is exceptionally twisted, resulting in formation of a whole turn of right-handed helix with an inner diameter of about 30 Å. Relative to LRR1, LRR25 shifts about 60 Å along the central axis of the helix (Fig. 1a). An unprecedented structural feature of BRI1-LRR is that it has an insertion domain (the ID) anchoring to the inner surface of the solenoid and running across six LRRs (Fig. 1a). The N-terminal capping domain (residues 30–70), consisting of one β-strand (β1) and two α-helices, is integrated into the LRR structure by forming an anti-parallel β-sheet with the β-strand from the irregular repeat, whereas the C-terminal capping domain (residues 752-772) employs two short helices to tightly pack against the last repeat (Fig. 1a). Eight potential glycosylation sites (Supplementary Fig. 2) as defined by sufficient electron density were found in BRI1-LRR. An assignment of functions to them awaits further investigations.
Fig. 1. BRI1-LRR has a helical solenoid structure.
a. and (b) are overall structures of BL-free BRI1-LRR shown in two different orientations. The N-linked sugars (N-acetylglucosamines) are shown in magenta stick. Colored in orange are disulfide bonds. The N- and C-terminal cap is shown in slate and marine respectively. The blue numbers in (a) indicate the positions of LRRs. The β-strand (β1) from the N-terminal cap is labeled.
c. Overall structure of BL-bound BRI1-LRR with the same orientation as (a). BL molecule is shown in stick and colored in yellow.
Like canonical LRR proteins, the concave surface of BRI1-LRR solenoid is of parallel β-sheet comprising 25 continuously running parallel β-strands. Compared to many other LRR proteins, however, BRI1-LRR also possesses parallel but distorted β-sheets following the inner β-structure in most of the repeats (Fig. 1a and 1b). This is likely due to the plant-specific consensus sequence L/fXGxI/vP (X and x stand for polar and any amino acids respectively)20 of BRI1-LRRs (Supplementary Fig. 2) found in many LRR-RLKs, as a similar structural feature also exists in the plant LRR protein PGIP2 (ref. 21). In contrast with the concave side, the convex outer surface consists of varied second structure elements, including 310 helices, α-helices and different length of loops (Fig. 1a and 1b). Some of the loops are stabilized through disulfide bonds formed between two consecutive repeats (LRR2-LRR3, LRR5-LRR6, LRR7-LRR8, LRR10-LRR11 and LRR14-LRR15) (Fig. 1a and Supplementary Fig. 1). The overall structure of BRI1-LRR (residues 30-775) remains nearly unchanged upon BL binding, with an r.m.s. (root mean square) deviation 0.70 Å over 734 Cα atoms. The BL molecule is sandwiched between the ID and the concave surface of the solenoid structure (Fig. 1c).
Interruption of BRI1-LRRs by a folded domain
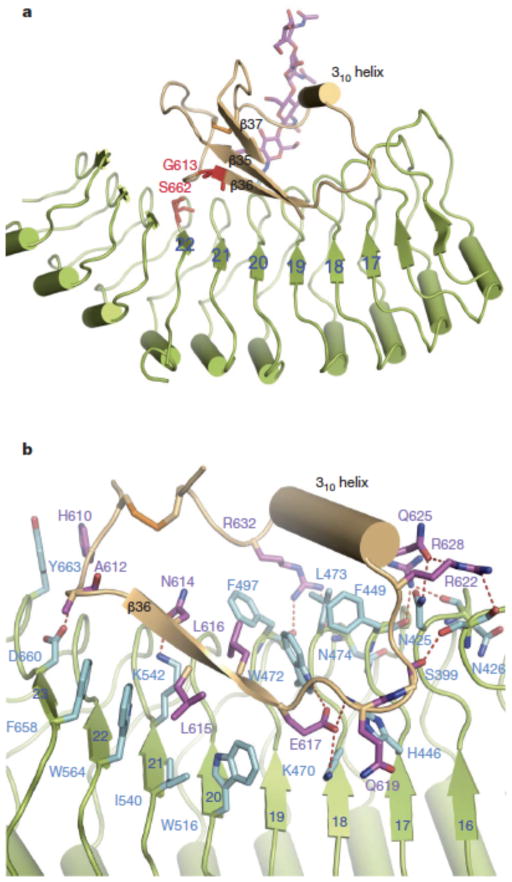
Two short segments (residues 590-595 and 644-646) from the ID do not have interpretable electron density in the structure of BL-free BRI1-LRR but become well defined following BL binding (Supplementary Fig. 3). Residues 596-643 form a separate folded domain that consists of one three-stranded antiparallel β-sheet and one 310 helix (Fig. 2a). In addition, this domain contains one disulfide bridge (Cys609-Cys635) that can further contribute to its structural integrity. Data base search using DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server) did not reveal known structures that share significant homology with the ID, indicating that it represents a novel fold.
Fig. 2. Interaction of the ID with LRRs.
a. Cartoon representation of BRI1-LRR highlighting interaction of the ID with the concave surface. The positions of two genetic mutants are shown in red and stick.
b. Detailed interactions of the ID with LRRs. The side chains from the ID are shown and labeled in purple, and those from LRRs in cyan. Red dashed lines represent hydrogen bonds.
Extensive non-covalent including van der Waals and polar interactions exist between the ID and the solenoid structure. While making few contacts with the remaining parts of the ID, the 310 helix packs tightly against Phe449 and Leu473 from underneath and forms a large network of hydrogen bonding interactions with those from one flanking side (Fig. 2b). In contrast with the 310 helix, β36 primarily binds the concave surface (Fig. 2a). Around this binding site, Trp516, Ile540, Trp564 and Phe658 establish close contacts with one side of β36, whereas Trp472 and Phe497 wedge between β36 and the 310 helix (Fig. 2b). Hydrogen bonds dominate the interactions of the loop linking the 310 helix and β36 with the concave surface. Additionally, packing of the carbonhydrate moiety of the glycosylated Asn545 against the loop connecting the 310 helix and β37 seems to play a role in positioning the ID in the concave surface (Fig. 2a). Two mutations, G613S and S662F, generated weak hormone-insensitive phenotypes2. Our structure suggests that they can perturb local conformations and consequently generate a deleterious effect on BRI1 recognition of BRs. Substitution of Gly613 with serine would produce steric clash with the carbonyl oxygen of Ile600 or the benzene ring of Tyr599 and consequently generate a damaging effect on the β-sheet of the ID (Supplementary Fig. 4). Ser662 is limited within a small pocket and its mutation to the bulky residue phenylalanine is expected to generate serious clash with its neighboring residues, in particular Gly611 that is N-terminal to β36.
BL binds to a surface groove on BRI1-LRR
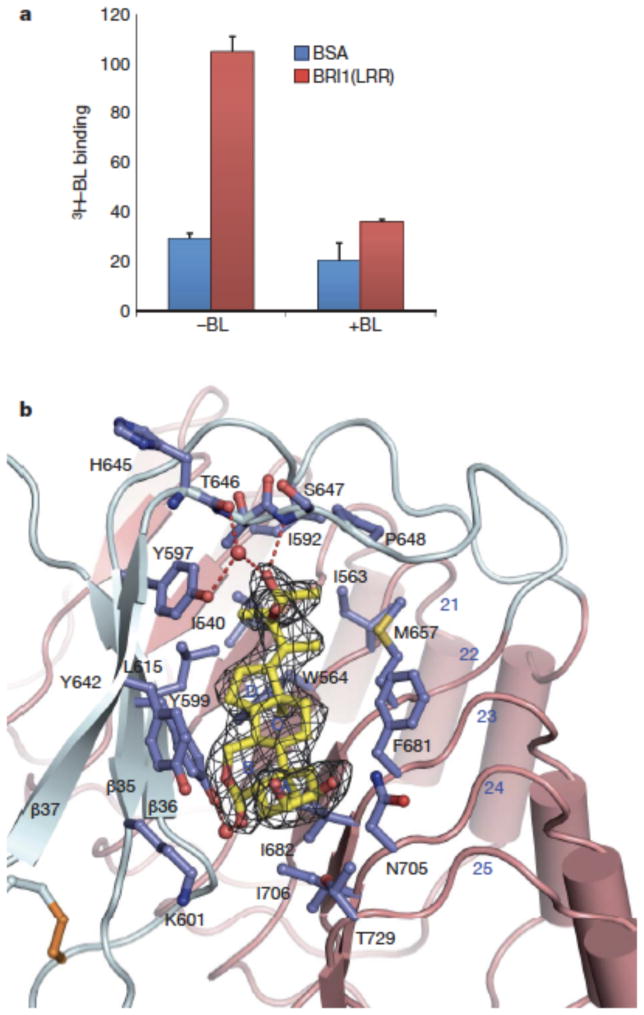
We used 3H-labelled BL to test if the purified recombinant BRI1-LRR was active in binding BL. As shown in Fig. 3a, the protein exhibited a comparable BL-binding activity to that previously reported12. In BL-bound BRI1-LRR structure, the electron density unambiguously defines the BL molecule that binds to a pronounced hydrophobic groove between the ID and the concave side of the solenoid (Fig. 3b). Most of the residues lining the surface groove are hydrophobic and highly conserved (Supplementary Fig. 1). The fused ring moiety of BL (Supplementary Fig. 5) occupies most of the surface groove (Fig 3b). A-ring makes marginal contacts with the concave surface, whereas B ring tightly stacks Tyr642 from one lateral side. The other two rings are sandwiched between Phe681 and Tyr599. Despite a smaller size, the two methyl groups C18 and C19 form much denser hydrophobic interactions with BRI1 by docking into two cavities at the bottom. In contrast, the other side of the fused ring is nearly solvent-exposed (Fig. 3b). Although extensive shape complementarities exist between BL and the base of the surface groove, a water molecule fills the cavity that is sealed from the solvent region by Lys601, bridging a hydrogen bond between the carbonyl oxygen at C6 and the hydroxyl group of Tyr599. Standing in contrast with the fused ring, the distal carbon atoms C24~C28 of the side chain are completely buried into a hydrophobic pocket (Fig. 3b). Further reinforcing the interactions around this contact interface, the hydroxyl group at C23 establishes a hydrogen bond with the backbone nitrogen of Ser647 and two water-mediated hydrogen bonds with the carbonyl oxygen atoms of His645 and the hydroxyl group of Tyr597.
Fig. 3. BL binds a hydrophobic groove between the ID and the inner surface of LRRs.
a. [3H]-BL binding activity of BRI1-LRR. About 1 mg ml−1 BRI1-LRR-His (red bar) or BSA as control (blue bar) was incubated with 20 nM [3H]-BL in the absence (−BL) or presence (+BL) of 20 μM unlabelled BL. BRI1-bound [3H]-BL was recovered using nickel beads and quantified by scintillation counting. Data represent the average of triplicate assays and error bars are standard deviations.
b. Detailed interactions between BL and BRI1-LRR. Shown in mesh is omit electron density (2.5 δ) around BL. The ID and LRRs are colored in slate and salmon, respectively. The side chains from both the ID and LRRs are shown in slate. Red spheres represent oxygen atoms of water molecules. The three β–strands from the ID are labeled.
Induced-fit BL recognition by monomeric BRI1-LRR
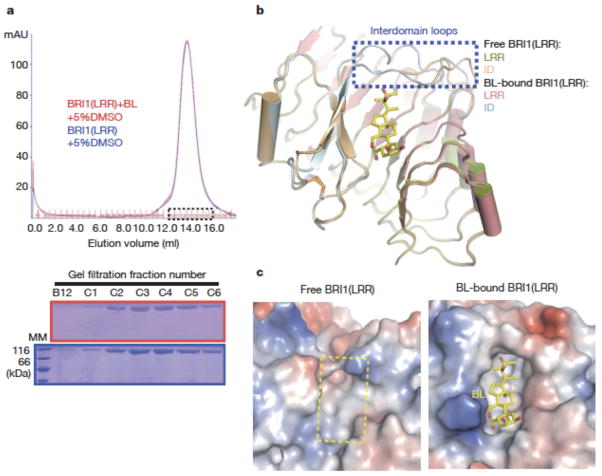
The crystals of both free and BL-bound BRI1-LRRs contain one protein molecule per asymmetric unit and have nearly identical packing. These indicate that homodimers formed by two crystallographic symmetry-related molecules (Supplementary Fig. 6) result from crystal packing rather than BL binding. Consistently, gel filtration assay showed that BRI1-LRR was monomeric in solution and its apparent molecular weight was not affected by the presence of BL (Fig. 4a). The latter is consistent with the observation22 that BL had no effect on BRI1 homo-dimerization in protoplasts. Structural comparison revealed that upon BL binding striking local structural rearrangement occurs to the two loops linking the ID and the LRR structure (referred to inter-domain loops) (Fig. 4b). BL-induced stabilization of the inter-domain loops results in formation of a more striking surface groove where BL binds (Fig 4c), showing that BL recognition by BRI1-LRR is a process of induced fit.
Fig. 4. BL induces stabilization of two inter-domain loops but no dimerization of BRI1-LRR.
a. BL has no effect on the oligomeric status of BRI1-LRR in solution. Shown on the top is superposition of the gel filtration chromatograms of BRI1-LRR in the absence (blue) and presence (red) of BL. The vertical and horizontal axes represent UV absorbance (λ=280 nm) and elution volume respectively. Peak fractions are highlighted within the dashed square. The apparent molecular weight of BRI1-LRR was 109.4 kD in the presence or absence of BL, higher than the theoretical BRI1-LRR monomer (83 kD) likely due to existence of multiple glycosylation sites in BRI1-LRR. MM: molecular mass marker. Bottom: Coomassie blue staining of the peak fractions shown on the top following SDS-PAGE.
b. BL binding induces stabilization of two inter-domain loops. Structural superimposition of the free and BL-bound BRI1-LRR around the BL-binding site.
c. BL binding generates a striking hydrophobic surface groove on BRI1-LRR. Shown on the left and right are the electrostatic surfaces of free BRI1-LRR and BL-bound BRI1-LRR (shown in the same orientation) around the BL-binding site respectively. The area highlighted with the yellow dashed square on the left panel is the BL-binding site. White, blue and red indicate neutral, positive and negative surfaces respectively.
Discussion
The hydrophobicity-dominating BL-binding surface groove (Fig 4c, right panel) suggests that BRI1-LRR may be accommodating in ligand binding. This would afford an explanation for why BRI1 is able to respond to a variety of BL derivatives23. BRI1 ligand selectivity, on the other hand, can be conferred by the distal polar groups involved in hydrogen bonding interactions as well as the actual shape of the hormone-binding groove. The structural mechanism of BL recognition by BRI1-LRR rationalizes (Supplementary Fig. 7) the data on structure-activity relationship (SAR) of BRs accumulated during the past decades23, thus opening new perspectives for designing and developing nonsteroidal mimetic of BR with low cost that can be widely applied in agricultural practice.
Our structural analyses reveal that, while the overall structure of BRI1-LRR remains nearly unchanged upon BL binding, significant structural rearrangement occurs to the inter-domain loops around the BL-binding site (collectively referred to BL-created surface), which should be associated with BR-induced signaling. It is not clear how this region contributes to BR-initiated signaling, but it is likely involved in stabilization16 of the kinase domain-mediated BRI1 homodimers19. The absence of BL’s effect on BRI1 homodimerization in current study may be due to the isolated ectodomain used for assays or requirement of another protein or peptide, as suggested by the suppression of bri1 mutants by a secreted peptidase24. If this is the case, BRI1 would be different from toll-like receptors (TLRs) in which ligand-induced homodimerization for activation can be recapitulated through their isolated extracellular domains25, 26. Stabilization of BRI1 homodimers by the BL-created surface is expected to initially activate BRI1 kinase to a basal level, resulting in tyrosine phosphorylation and disassociation of BKI1 (ref. 19); disassociation of BKI1 allows stable association and sequential reciprocal phorphorylation between the kinase domains of BRI1 and BAK1, fully activating the BRI1 kinase as proposed by the sequential phosphorylation model18. Alternatively, BL binding may initiate signaling by altering an interaction between the extracellular domains of BRI1 and BAK1. A role of the extracellular domain of BAK1 in BR signaling has been suggested by the observations that mutations in this domain of BAK1 affect BR sensitivity27 and interaction with BRI128. Although stable BRI1-BAK1 association is a consequence of initial BRI1 activation19, a possible role of BAK1 and its homologs in initial BRI1 activation has not been excluded by genetic studies; for example, it remains unclear whether mutations of BAK1 and its homologs affect BL-induced BKI1 phosphorylation by BRI1. It is interesting to note that BAK1 has five LRRs and the new BL-created surface is also located about five LRRs from the membrane surface. Thus, it is also possible that the BL-created surface is involved in interaction with BAK1-LRR, which triggers BRI1 phosphorylation of BKI1 through unknown mechanisms, allowing formation of more stable BRI1-BAK1 receptor complexes. Although the BL-created surface is proposed to be involved in BRI1-LRR homodimerzation or heterodimerization with BAK1-LRR, further studies are needed to determine if BRs initiate signaling through promoting protein-protein interaction, as many other plant hormones do29, 30.
Online methods
Protein expression and purification
The LRR domain of BRI1 (residues 24-784) from Arabidopsis with an engineered C-terminal 6×His-tag was generated by standard PCR-based cloning strategy and its identity was confirmed by sequencing. The protein was expressed in high five cells using the vector pFastBac 1 (Invitrogen™) with a modified N-terminal Hemolin peptide. 1.0 L of cells (2.0×106 cells ml−1) were infected with 20 ml baculovirus using a multiplicity of infection of 4 at 22°, and protein was harvested from the media after 48 hours. The protein was purified using Ni-NTA (Novagen) and size-exclusion chromatography (Superdex 200, Pharmacia) in buffer (10mM Tris, pH8.0, 100mM NaCl). Samples from relevant fractions were applied to SDS–PAGE and visualized by Coomassie blue staining. Protein purification was performed at 4°. For crystallization of BRI1-LRR, the purified protein was concentrated to about 3.0 mg ml−1 in buffer containing 10mM Tris, pH8.0, 100 mM NaCl.
Crystallization, data collection, structure determination and refinement
Crystals of the BRI1-LRR were generated by mixing the protein with an equal amount of well solution (1 μl) by the hanging-drop vapor-diffusion method. A mixture of BRI1-LRR and BL with a molar ratio 1:10 was used for generating crystals of their complex. The initial buffer producing crystals of both free BRI1-LRR and BL-bound BRI1-LRR contained 0.2 M Na2SO4 and 20% (w/v) polyethylene glycol (PEG) 3,350, which was further optimized by adding 10 mM Trimethylamine-HCl. Crystals grew to their maximum size (0.1×0.1×0.1mm3) within ten days at room temperature.
All the diffraction data sets were collected at the Shanghai Synchrotron Radiation Facility (SSRF) at beam line BL17U1 using a CCD detector. Crystals of both BL-free and BL-bound BRI1-LRR belong to space group C2 with one protein molecule per asymmetric unit. For data collection, the crystals were equilibrated in a cryoprotectant buffer containing reservoir buffer plus 20.0% (v/v) glycerol. The data were processed using HKL2000 (ref. 31). Molecular replacement (MR) with the program PHASER32 was used to solve the crystal structures of both BL-free and BL-bound BRI1-LRR. The atomic coordinates of PGIP2 (ref. 21) (PDB code: 1OGQ) and InlA33 (PDB code: 1O6V) were used as the initial searching model. The model from MR was built with the program COOT34 and subsequently subjected to refinement by the program PHENIX35. The electron density for BL in BRI1-BR/BL complex crystal became apparent after refinement of BRI1-LRR. The final refined model contains residues 30-775 of BRI1-LRR and one BL molecule in BL-bound BRI1-LRR, and residues 30-772 in BL-free BRI1-LRR except the two segments containing residues 590-595 and 644-646 that have no clear electron density and are presumed to be disordered in solution. The structure figures were prepared using PYMOL36.
Gel filtration assay
BRI1-LRR protein purified as described under the section of “Protein expression and purification” was subjected to gel filtration analysis (Superdex200 10/300 GL column; GE Healthcare) in the presence and absence of BL. Buffer containing 50 mM NaH2PO4/Na2HPO4 pH7.4, 100 mM NaCl, 2.0 mM MgCl2 and 5% (v/v) DMSO was used for the assay. A mixture of BRI1-LRR and BL with a molar ratio about 1:10 was used to test the effect of BL on the apparent molecular weight of BRI1-LRR. The assays were performed with a flow rate of 0.5 ml min−1 and an injection volume of 0.5 ml buffer containing BRI1-LRR (0.83 mg ml−1) with or without BL at 20 °C. The column was calibrated using five standard proteins, beta-amylase 200 kDa, alcohol dehydrogenase 150 kDa, albumin 66 kDa, carbonic anhydrase 29 kDa and cytochrome C 12.4 kDa, which had an elution volume (Ve) 12.53 ml, 13.25 ml, 14.25 ml, 16.18 ml and 16.97 ml respectively under the conditions of assay. The void volume (Vo) for the column used was determined to be 8.15 ml using fresh blue dextran solution (1.0 mg ml−1). The calibration curve of the gel-phase distribution coefficient (Kav) vs. log molecular weight (M) was obtained using “Excel”. Kav=(Ve−Vo)/(Vc−Vo), where Ve is the elution volume; Vo is the void volume; Vc is the geometric column volume, 24 ml for the column used. The elution volume of BRI1-LRR in the presence and absence of BL was 13.63 ml under the conditions of assay, corresponding to an apparent molecular weight 109.4 kDa based on the calibration equation , R2 = 0.9814.
3H-labelled brassinolide binding assay
His-tagged BRI1-LRR, or BSA as control, was incubated with 20 nM [3H]-BL, or 20 nM [3H]-BL mixed with 20 μM non-radiolabelled BL as competitor, and nickel beads in the BL-binding buffer (0.25 M mannitol, 10 mM Tris-2-[N-morpholino] ethanesulphonic acid (MES), pH 5.7, 5 mM MgCl2, 0.1 mM CaCl2) for 30 min. The beads were washed three times, and His-BRI1-LRR and bound [3H]-BL were eluted by using 1 M immidazole. The eluted radioactivity was measured by scintillation counter.
Supplementary Material
Acknowledgments
We thank S. Huang and J. He at Shanghai Synchrotron Radiation Facility (SSRF) for assistance with data collection; M. Yang from School of Life Sciences, Tsinghua University for structural determination; J. Chory from Salk Institute for generously providing the tritium-labeled brassinolide. This research was funded by the National Outstanding Young Scholar Science Foundation of National Natural Science Foundation of China grant no. 20101331722 to J. C. and NIH R01GM066258 to Z-Y.W.
Footnotes
Author Contributions J.C., Z.H., J.S. and Z.Y.W designed the experiments. The binding assay was performed by T.W.K. and the others by J. S., Z.H., J.J.W, J.W.W, S.S and J.B.C.. Data were analyzed by J.C., Z.H., J.S. and Z.Y.W.. J.C., Z.Y.W. and Z.H. wrote the paper.
The atomic coordinates and structure factors of BRI1-LRR and BRI1-LRR/BL complex have been deposited in the Protein Data Bank under the accession code 3RGX and 3RGZ respectively.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
The authors declare no competing financial interests.
References
- 1.Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 2.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–38. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–22. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 5.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–12. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–77. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–22. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 9.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–60. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–34. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Z, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–3. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–71. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 13.Li J. Brassinosteroid signaling: from receptor kinases to transcription factors. Curr Opin Plant Biol. 2005;8:526–31. doi: 10.1016/j.pbi.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 15.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–29. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–65. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–35. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–7. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–27. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 21.Di Matteo A, et al. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci U S A. 2003;100:10124–8. doi: 10.1073/pnas.1733690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hink MA, Shah K, Russinova E, de Vries SC, Visser AJ. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys J. 2008;94:1052–62. doi: 10.1529/biophysj.107.112003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zullo MTA, Adam G. Brassinosteroid phytohormones - structure bioactivity and applications. Braz J Plant Physiol. 2002;14:143–181. [Google Scholar]
- 24.Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2001;98:5916–21. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 27.Whippo CW, Hangarter RP. A brassinosteroid-hypersensitive mutant of BAK1 indicates that a convergence of photomorphogenic and hormonal signaling modulates phototropism. Plant Physiol. 2005;139:448–57. doi: 10.1104/pp.105.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun HS, et al. Analysis of phosphorylation of the BRI1/BAK1 complex in arabidopsis reveals amino acid residues critical for receptor formation and activation of BR signaling. Mol Cells. 2009;27:183–90. doi: 10.1007/s10059-009-0023-1. [DOI] [PubMed] [Google Scholar]
- 29.Sheard LB, Zheng N. Plant biology: Signal advance for abscisic acid. Nature. 2009;462:575–6. doi: 10.1038/462575a. [DOI] [PubMed] [Google Scholar]
- 30.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–5. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski ZMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert WD, et al. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell. 2002;111:825–36. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. PyMOL Molecular Viewer. 2002 ( http://www.pymol.org)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.