Abstract
Bacterial cytokinesis is commonly initiated by the Z-ring, a cytoskeletal structure assembling at the site of division. Its primary component is FtsZ, a tubulin superfamily GTPase, which is recruited to the membrane by the actin-related protein FtsA. Both proteins are required for the formation of the Z-ring, but if and how they influence each other’s assembly dynamics is not known. Here, we reconstituted FtsA-dependent recruitment of FtsZ polymers to supported membranes, where both proteins self-organize into complex patterns, such as fast-moving filament bundles and chirally rotating rings. Using fluorescence microscopy and biochemical perturbations, we found that these large-scale rearrangements of FtsZ emerge from its polymerization dynamics and a dual, antagonistic role of FtsA: recruitment of FtsZ filaments to the membrane and a negative regulation on FtsZ organization. Our findings provide a model for the initial steps of bacterial cell division and illustrate how dynamic polymers can self-organize into large-scale structures.
As in eukaryotic cells, proteins related to actin and tubulin provide the key structural components coordinating cellular functions in bacteria1,2. For example, cell division in most bacteria depends on the tubulin-related GTPase FtsZ and the widely conserved actin-related protein FtsA, which form an annular structure at the middle of the cell3,4. Purified FtsZ assembles into polar, straight or gently curved protofilaments in the presence of GTP5–8 and lateral interactions between FtsZ protofilaments can lead to higher-ordered structures, like tubules, bundles, circles and sheets5,6. In most bacteria, FtsZ is recruited to the membrane by FtsA, which binds to the membrane via a C-terminal amphipathic helix9,10. Although binding of ATP is required for FtsA to interact with FtsZ, no ATPase activity of FtsA was found9–12. In Escherichia coli and other Gammaproteobacteria, FtsZ is also recruited to the membrane by the trans-membrane protein ZipA13–15 and both membrane anchors are required for successful cell division. Although structurally not related, FtsA and ZipA bind to the same C-terminal peptide of FtsZ, which is connected to the rest of the protein via a flexible linker16,17. Most previous models for Z-ring formation mainly assumed both proteins to be passive membrane anchors for FtsZ18–20, an idea also followed in reconstitution studies, where the requirement for physiological membrane anchors was circumvented by supplying FtsZ with its own membrane targeting peptide21–23. However, in vitro experiments using an FtsA mutant suggested that the membrane anchor can change the properties of FtsZ assemblies24, raising the question if and how ZipA or FtsA might influence the organization of FtsZ filaments into large-scale, dynamic cytoskeletal structures. To probe the mechanism underlying Z-ring assembly, we set out to reconstitute FtsZ polymerization mediated by wild-type FtsA or ZipA on supported lipid bilayers in vitro.
RESULTS
FtsZ and FtsA self-organize into a rapidly reorganizing filament network
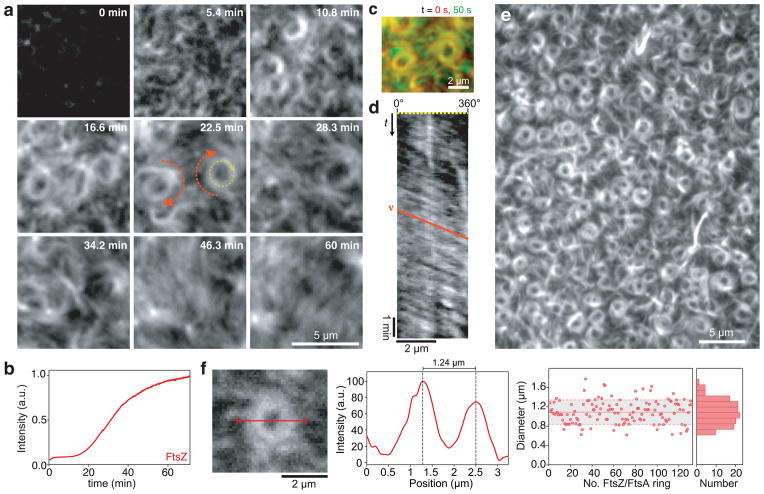
Earlier attempts to reconstitute FtsA-FtsZ interaction were frustrated by wildtype FtsA being notoriously difficult to purify. Here, we circumvented this problem by using a SUMO-fusion system for overexpression of FtsA followed by removal of the SUMO-tag25. We then used total-internal-reflection-fluorescence (TIRF) microscopy to reveal the behavior of fluorescently labeled proteins on a supported membrane (Supplementary Fig. 1). First, we added FtsZ labeled with Alexa488 together with unlabeled FtsA and ATP to the buffer above the bilayer to a concentration ratio of [FtsZ]:[FtsA]=3:1 to 5:1, corresponding to the one found in vivo26. Under these conditions, we saw no FtsZ bound to the membrane. Next we added GTP to promote FtsZ polymerization. After a lag time of about 5 min, short FtsZ filaments started to attach to the membrane. These filaments increased in density and then formed dynamic bundles, which self-organized into coherent, motile assemblies (Fig. 1a and b, Supplementary Video 1 and 2). Dominant features of this large-scale organization were streams of traveling filament bundles, apparently moving in one direction. These streams often formed rotating vortices that persisted for tens of minutes during which their diameter did not change (Fig. 1 c). To analyze the rotation of the FtsZ swirls, we generated kymographs along their circumference and found that these rings rotated with a velocity of 6.56 ± 0.69 μm/min (s.e.m, n = 40), corresponding to a radial velocity of 11.47 ± 1.21 degrees/s, which means that one full rotation required 31.4 ± 3.3s (Fig. 1d). In some cases a field of vortices covered large areas of the membrane (Fig. 1e, Supplementary Video 3). Although our in vitro system did not provide any spatial cues like membrane curvature or geometric confinement, the FtsZ filaments organized into rings with a similar diameter as the E. coli cell (1.09 ± 0.24 μm (s.d., n = 132) compared to 0.7–1.4 μm, ref. 27) (Fig. 1f). Furthermore, these vortices were chiral, since all rings rotated clockwise when viewed from the membrane. This dynamic reorganization of FtsZ filaments in vitro was reminiscent of the rapidly moving helical pattern during Z-ring assembly seen in E. coli cells28 and B. subtilis29.
Figure 1. FtsZ and FtsA self-organize into a rapidly reorganizing filament network.
(a) Snapshots showing typical cytoskeletal patterns of FtsZ emerging from its interaction with FtsA on a supported membrane. Similar results were obtained in more than 100 experiments. After adding GTP (to 3 mM at t = 0 min), short filaments of FtsZ start attaching to the membrane. While their density increases, they self-organize into a rapidly reorganizing filament network, forming traveling streams and rotating swirls (orange arrows) (FtsZ, 1.5 μM with 10 % FtsZ-Alexa488; FtsA, 0.5 μM). (b) Representative time-intensity curve corresponding to the amount of FtsZ polymerizing on the membrane after adding GTP (c) Overlay of two individual frames from Supplementary Video 2 separated by 50s. While FtsZ bundles outside of the ring are constantly rearranging, the bundles inside of the vortex are more persistent. (d) Representative kymograph along the circumference of a vortex shown in (a), see yellow dashed arrow at 22.5 min. The slope of the orange line corresponds to the velocity (x/t) of the vortex. Kymographs were obtained and analyzed for 40 different vortices. (e) Array of dynamic rings of FtsZ. (f) Ring diameters were determined by measuring the peak-to-peak distance in the intensity profile (red line shown in micrograph). Right, scatterplot of ring diameters (red dots), average value (solid red line, n = 132), standard deviation (dashed line and grey background) and corresponding histogram. Source data is given in Supplementary Table 1.
The emergence of complex dynamic patterns of FtsZ depended on the presence of both ATP and GTP. When GTP, but no ATP was added, we observed only transient recruitment of FtsZ filaments to the membrane, presumably because some nucleotide from the purification was bound to FtsA12. As soon as fresh, exogenous ATP was added, FtsZ reassembled on the membrane (Supplementary Fig. 2a and Video 4). We did not find any ATPase activity of FtsA, and similar dynamic patterns were formed when we substituted ATP with ADP or ATPγS, a non-hydrolyzable analog of ATP, consistent with previous suggestions that FtsA requires nucleotide to interact with FtsZ, but functions without hydrolyzing ATP11,12,30. When we added GMPCPP, a GTP analog that cannot be hydrolyzed by FtsZ, FtsZ formed static bundles of filaments, which did not reorganize over time (Supplementary Video 5). Furthermore, to test the role of FtsA self-interaction, we repeated our experiment with mutants of FtsA, which were found to fail to polymerize in vivo10,12,31,32. With these FtsA mutants, FtsZ assembled into the same dynamic patterns as with wild-type FtsA, indicating that FtsA polymerization does not play a role for the formation of cytoskeletal patterns (Supplementary Fig. 2b).
To conclude, although GTP and ATP were required for FtsZ-FtsA self-organization, the energy that drives the dynamics of their filament pattern originates solely from GTP hydrolysis coupled to FtsZ polymerization.
The FtsZ filament network reorganizes via FtsZ polymerization dynamics
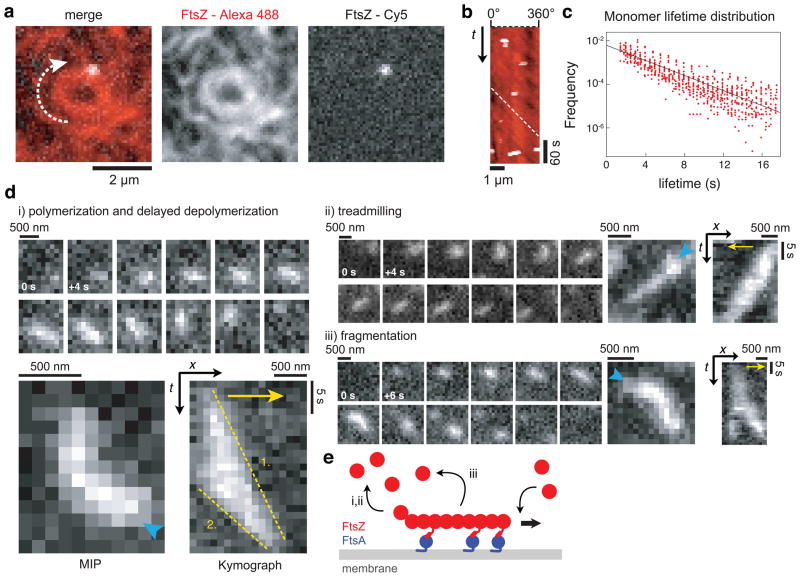
Next, we were interested in how the dynamics of these cytoskeletal patterns of FtsZ and the directionality of their movement arise. Two models for the remodeling of the Z-ring during cell septum constriction have been proposed33–35: First, FtsZ filaments could slide along each other. In this case, single FtsZ subunits would move together with the filament network. Second, FtsZ filaments could reorganize by polymerization dynamics, in which case single FtsZ subunits would remain static. To visualize individual FtsZ molecule dynamics in our in vitro experiment, we added small amounts of FtsZ labeled with a Cy5 to a background of FtsZ labeled with Alexa488. We found that while the filament network was continuously moving and rearranging, single FtsZ subunits appeared and remained at the same position (Fig. 2a and b, Supplementary Videos 6–8). This observation rules out a sliding mechanism and supports network reorganization by polymerization dynamics, most likely treadmilling. When we analyzed the lifetime of FtsZ monomers using single particle tracking, we found an exponential lifetime distribution corresponding to a first-order reaction and an average lifetime of 7.16 ± 1.23 sec (s.d., n = 24 analyzed videos from 5 independent experiments with more than 1500 particles in total) (Fig. 2c). This exponential distribution is consistent with the observation that monomer exchange is not limited to the filament ends36, but can occur along the whole filament37–39.
Figure 2. Reorganization of the FtsZ filament network emerges from FtsZ polymerization dynamics and not from filament sliding.
(a) Typical micrograph of a single FtsZ molecule (white, FtsZ-Cy5) in an FtsZ vortex (red, FtsZ-Alexa488) (FtsZ, 1.25 μM with 30 % FtsZ-Alexa488 and 0.4 % FtsZ-Cy5; FtsA, 0.4 μM). (b) Kymograph along the circumference of the vortex shown in (a). The slope of the white dashed line shows corresponds to the velocity of the vortex. While the FtsZ ring is rotating, individual FtsZ proteins appear for one to three frames (3 to 9 sec) without moving (Supplementary Videos 6–8). Similar results were obtained in more than 20 experiments. (c) Distribution of individual FtsZ lifetimes (red circles) in a linear-log plot and linear fit (black line). See text for details. (d) FtsZ filaments can show different kinds of polymerization dynamics when recruited to the membrane by FtsA. Representative snapshots, maximum intensity projections (MIP) and kymographs of (i) filament polymerization and delayed depolymerization, (ii) treadmilling and (iii) fragmentation. Cyan arrowhead in MIPs indicate start of FtsZ polymerization on the membrane, yellow arrows indicate the direction of polymerization. The slopes of the yellow dashed lines correspond to the polymerization (1.) and depolymerization rate (2.) of the FtsZ filament. All scale bars correspond to 500 nm or 5 s. (38 filaments from 5 independent experiments were analyzed, see also Supplementary Fig. 3). (FtsZ, 0.4 μM with 10 % FtsZ-Alexa488; FtsA, 0.2 μM) (e) Illustration of a FtsZ filament recruited to the membrane by FtsA. After binding and unipolar polymerization, the filament detaches either after polymerization from the opposite end (i,ii) or after fragmentation (iii).
When we lowered the concentrations of FtsA and FtsZ, we could observe short individual filaments on the membrane, which often appeared as diffraction-limited spots. Those filaments that were dynamic did not simply diffuse on the membrane; instead we found them to show different kinds of polymerization dynamics. We analyzed the behavior of individual, isolated filaments and found that most of them polymerized from one end and then started to depolymerize from the opposite end at a faster rate (1.89 ± 0.48 μm/min and 3.97 ±1.07 μm/min, s.d, n=18 from 5 independent experiments, p<0.0001) (Fig. 2d and e, Supplementary Fig. 3 and Video 9). In other cases, both rates were similar and as a result, the polymer traveled across the membrane at a constant length, i.e. showing treadmilling dynamics (2.58 ± 0.64 μm/min and 2.61 ± 0.67 μm/min, s.d, n=16, p=0.8978). Alternatively, filaments appeared to break into shorter fragments before detaching from the membrane (4 out of 38 analyzed filaments). Fragmentation of FtsZ filaments supports the idea that FtsZ monomers can exchange along the filament, while monopolar elongation and treadmilling of FtsZ filaments explains the directionality of the reorganizing filament network. Importantly, the polymerization rate of single filaments was about three times slower than the rotation velocity of FtsZ rings, suggesting that at higher protein concentrations FtsZ assembles into oligomers before it is incorporated into the cytoskeletal patterns on the membrane. In summary, we propose that the motion of these structures emerges from polymerization dynamics and not from a sliding mechanism.
Large-scale dynamics emerge from the interaction of FtsZ with FtsA
The observed dynamic patterns could either be an intrinsic property of FtsZ polymerization that simply require the filaments to be attached to a membrane, or they could emerge from a more complex interaction with FtsA. To discriminate between these two possibilities, we first tested a hybrid version of FtsZ that binds autonomously to the membrane through an amphipathic helix at its C-terminus21. We found that this membrane-targeted FtsZ formed a network of filament bundles on the membrane, however these bundles were stationary, and did not show large scale reorganization or dynamic vortices (Supplementary Video 10).
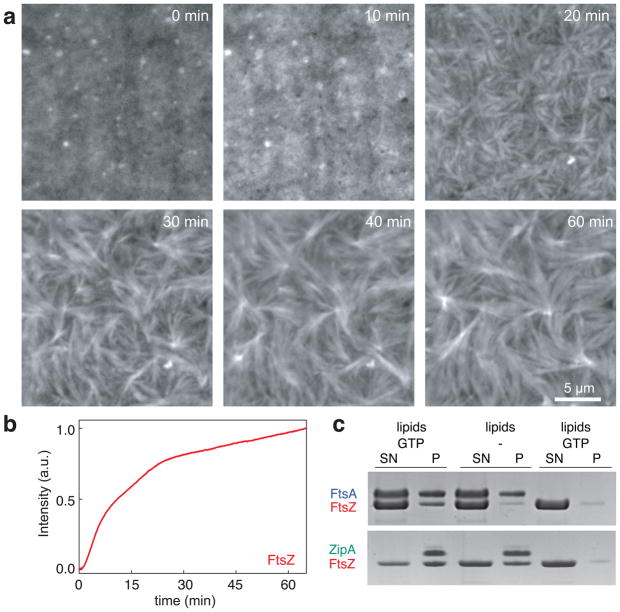
Next, we replaced FtsA with ZipA, the alternative membrane anchor of FtsZ. For these experiments, we substituted the trans-membrane domain of ZipA with a His-tag, which was shown to have no role in FtsZ binding40,41. This allowed us to attach the rest of the protein to a bilayer containing Ni-chelating lipids, thereby mimicking the permanent membrane attachment of full-length ZipA (ref. 42). Like in our experiments with FtsA, we first incubated the membrane with both proteins and then initiated FtsZ polymerization by adding GTP. We found the fluorescent signal of FtsZ to increase homogenously on these membranes, i.e. we did not observe individual filaments starting to bind to the membrane, and a much shorter lag time than in the case of FtsA (Fig. 3a and b, Supplementary Fig. 4). After ~10 min, FtsZ started to condense into long bundles of filaments, in agreement with the observation that ZipA can bundle FtsZ filaments in solution40. Although these FtsZ bundles did not show large-scale reorganizations, single-molecule experiments confirmed that these filaments were still dynamic, with a slightly longer average monomer lifetime (Supplementary Fig. 5a). In agreement with this observation, the GTPase rate of FtsZ did not change in the presence of either membrane anchor (Supplementary Fig. 5b).
Figure 3. FtsZ organizes into static bundles of dynamic filaments with ZipA as membrane anchor.
(a) Representative snapshots showing formation of FtsZ bundles with ZipA as membrane anchor. Similar results were obtained in more than 70 experiments. First, FtsZ intensity increases homogeneously, followed by condensation of FtsZ into long thick filament bundles (FtsZ, 1.5 μM with 30% FtsZ- Alexa488; His-Δ22-ZipA, 0.5 μM; supported membrane contained 2% Ni-chelating lipids). Different concentrations of Ni-chelating lipids (1% to 8%) gave similar results (Supplementary Figure 4). (b) Representative time-intensity curve corresponding to the amount of FtsZ bound to the membrane. (c) SDS-PAGE gel from co-pelleting assay with either FtsA and ATP (top row), ZipA (bottom row) or without membrane anchor (far right lanes). In contrast to ZipA, FtsA only recruits FtsZ to the membrane in the presence of GTP (left lanes versus middle lanes); S: supernatant, P: pellet. Result shown represents eight independent experiments.
Together, these results show that the rapidly reorganizing patterns we observed with FtsZ and FtsA emerged from the co-assembly of these two proteins on the membrane and were not the result of intrinsic FtsZ polymerization dynamics.
To investigate the difference between the two membrane anchors, we tested under which conditions ZipA or FtsA recruit FtsZ to the membrane. First, we incubated FtsZ and FtsA with multilamellar vesicles, which we could sediment by centrifugation without pelleting FtsZ polymers. With GTP, but in the absence of FtsA, no FtsZ was found in the pellet (Fig. 3c). In the presence of both proteins, GTP and ATP, we found a fraction of FtsA and FtsZ to co-sediment with the vesicles, but most of the two proteins were still in the supernatant, consistent with the formation of highly dynamic filaments that constantly exchange monomers. Without GTP but with FtsA, no FtsZ was recruited to the membrane, suggesting that FtsA preferentially interacts with polymerized FtsZ. When we performed the same experiment with ZipA, we found that FtsZ co-sedimented with ZipA-decorated vesicles in the presence of GTP, but also when no GTP was added. Consistent with previous publications41,43, this result shows that in contrast to FtsA, ZipA can also recruit FtsZ monomers to the membrane.
These data can explain why the filament network assembled starting with small FtsZ filaments attaching to the membrane when we used FtsA as a membrane anchor. In the case of ZipA, however, the fluorescence increased homogenously because FtsZ monomers already on the membrane initiate FtsZ polymerization.
FtsZ and FtsA co-assemble on the membrane
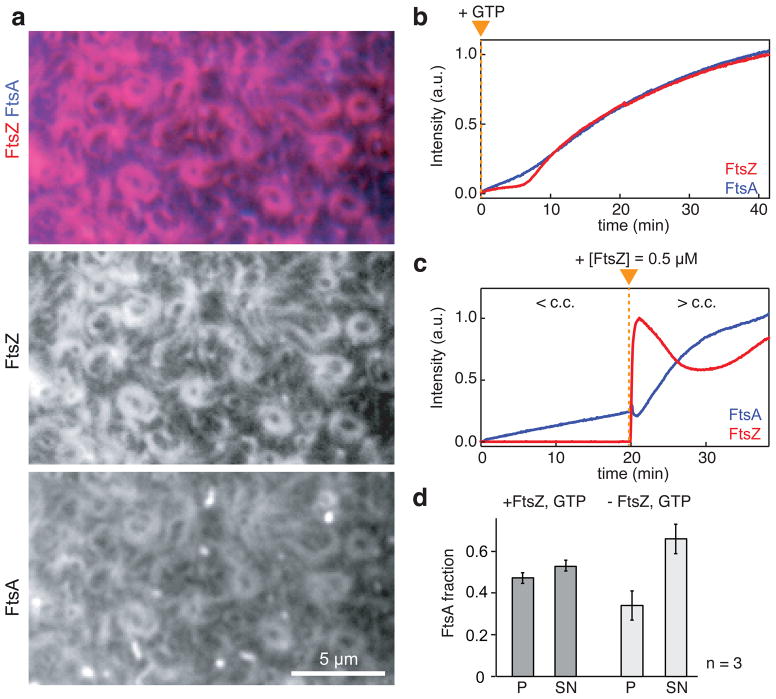
In contrast to ZipA, FtsA can reversibly bind to the membrane. Therefore, we were interested in how FtsA membrane binding relates to FtsZ filament recruitment. To probe this relationship, we repeated our self-organization experiment with both FtsZ and FtsA fluorescently labeled. Before adding GTP to initiate FtsZ polymerization, the intensity corresponding to both proteins on the membrane was low. After addition of GTP, the fluorescence intensity of FtsA increased diffusely just before FtsZ filaments started to assemble simultaneously with FtsA on the membrane (Fig. 4 a, b). Subsequently, the intensity of both proteins increased with similar rates while a dynamic network containing both FtsA and FtsZ assembled on the membrane. FtsZ and FtsA showed synchronized behavior in motile bundles and rotating vortices (Supplementary Video 12) and on the single filament level (Supplementary Video 13).
Figure 4. FtsZ and FtsA co-assemble on the membrane.
(a) Typical micrograph of reorganizing filament patterns with fluorescently labeled FtsZ and FtsA (FtsZ, 1.1 μM with 30% FtsZ-Alexa488; FtsA, 0.4 μM with 10 % Cy5-GG-FtsA). (b and c) Representative intensity traces corresponding to the amount of FtsZ (red) and FtsA (blue). (b) After adding GTP (orange arrowhead), FtsZ and FtsA assemble simultaneously on the membrane. Similar micrographs and intensity curves were obtained in more than 50 experiments. (c) Binding of FtsA is slow at a low concentration of FtsZ (< c.c. = critical concentration of FtsZ allowing for filament network formation), but facilitated at higher FtsZ concentration (> c.c.). Orange triangle indicates time of FtsZ addition. Similar intensity traces were obtained in five independent experiments. (d) Bar plot representing the amount of FtsA co-pelleting with vesicles (P) or remaining in the supernatant (SN) after 30 min of incubation (see also Supplementary Fig. 7b). The presence of FtsZ filaments (+FtsZ, GTP), increases the amount of FtsA bound to the membrane (p=0.0391). Error bars represent s.d. from n=3 independent experiments. Source data is given in Supplementary Table 1.
The synchronous assembly of FtsA and FtsZ after we initiated FtsZ polymerization suggested a mutually enhancing interaction between the two proteins. To probe this hypothesis, we initiated the experiment with a low FtsZ concentration, retaining a high concentration of FtsA (Fig. 4c, Supplementary Video 14). Under these conditions, we saw a slow, linear increase of FtsA intensity and short FtsZ filaments transiently binding to the membrane. We then rapidly added FtsZ to a concentration, which supports FtsZ filament network formation. Right after the increase in FtsZ, its intensity peaked as short FtsZ filaments covered the membrane. These filaments partially dissociated and then reattached to organize into a dynamic filament network. Remarkably, at this high FtsZ concentration, the intensity of FtsA increased faster and nonlinearly, showing that binding to polymerized FtsZ facilitates membrane-attachment of FtsA during initiation of the cytoskeletal pattern and in equilibrium (Fig. 4d and Supplementary Fig. 7b).
Next, we asked if the different effects of FtsA and ZipA on FtsZ polymerization could be due to FtsA being able to bind to the membrane reversibly. Therefore, we replaced the C-terminal amphiphatic helix of FtsA with a His-tag and attached FtsA to a membrane containing Ni-chelating lipids as for ZipA. With this permanently attached FtsA, we found FtsZ to form the same dynamic cytoskeletal patterns as with wildtype FtsA (Supplementary Video 15).
These findings show that the formation of a dynamic FtsZ filament network does not depend on the reversible attachment of its membrane anchor. However, the mutual dependence between FtsZ and FtsA for membrane targeting could be important to prevent FtsA from binding to the membrane and interacting with downstream targets anywhere outside of the Z-ring.
FtsA destabilizes the FtsZ filaments network
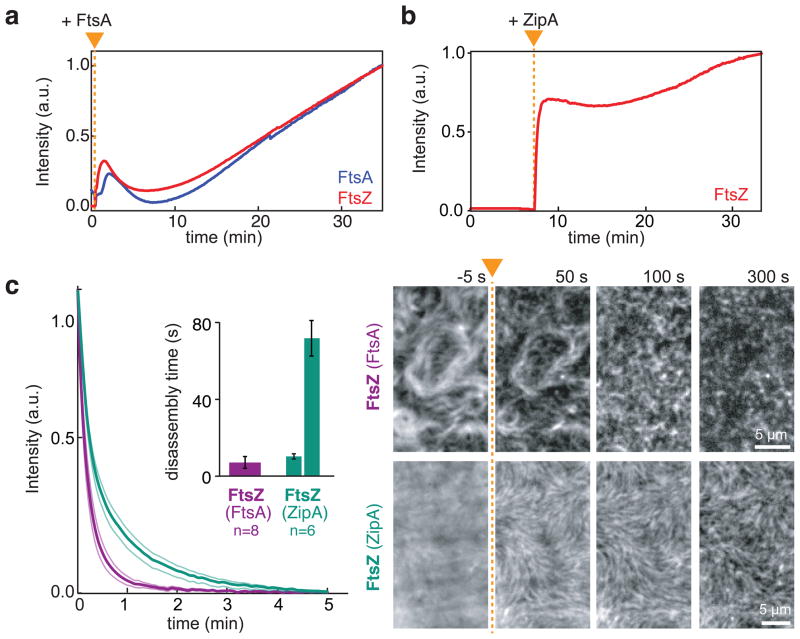
The initial peak of FtsZ fluorescence and subsequent decrease (Fig. 4c) was reminiscent of damped oscillations observed in biochemical regulation networks, which are typically a consequence of a delayed negative feedback44–46. Overshoots and damped oscillations have been seen for tubulin polymerization under certain conditions47, but we hypothesized that a negative feedback from FtsA to FtsZ might be involved here, since a hypermorphic mutant of FtsA has been found to disassemble FtsZ filaments in solution without increasing GTP hydrolysis rate by FtsZ11. To probe the influence of FtsA on FtsZ organization we asked whether FtsA could promote disassembly of pre-polymerized FtsZ filaments. Therefore, we first allowed FtsZ to polymerize in solution and then added FtsA to promote their recruitment to the membrane. Consistent with a delayed negative feedback, we found that after adding FtsA, short FtsZ filaments first appeared on the membrane, but then rapidly shortened and detached. Subsequently, both proteins reattached to self-organize into a dynamic polymer network (Fig. 5a, Supplementary Video 16). When we performed the same experiment with ZipA, we did not observe a disassembly of pre-polymerized FtsZ filaments (Fig. 5b, Supplementary Video 16).
Figure 5. FtsA destabilizes the FtsZ filaments network.
(a and b) Representative intensity traces corresponding to the amount of FtsZ (red) and FtsA (blue) co-assembling on the membrane. Prepolymerized FtsZ filaments are disassembled after addition of FtsA (a), but remain stable after addition of ZipA (b) (Supplementary Video 16) (FtsZ, 1.2 μM; FtsA or His-Δ22-ZipA, 0.3 μM). Intensity traces correspond to four independent experiments respectively. Orange triangle indicates the time of addition of either protein. (c) Left: Mean intensity traces for FtsZ depolymerization upon rapid dilution (FtsZ and FtsA (purple); FtsZ and ZipA (turquoise). Inset: mean disassembly times obtained from single-exponential (FtsZ with FtsA) and double-exponential fits (FtsZ with ZipA). Thin lines and error bars illustrate s.e.m (n=8 (FtsZ with FtsA) and n=6 (FtsZ with ZipA)). Right: Representative snapshots showing dilution-induced FtsZ disassembly. After adding buffer (at time point indicated by orange arrowhead and dashed line), FtsZ filaments rapidly disassembled when recruited by FtsA. In contrast, with ZipA FtsZ bundles can persist for more than 5 min. Intensities of micrographs have been normalized to have constant overall intensity. Raw data is shown in Supplementary Video 17.
To better compare the effect of FtsA and ZipA on the stability of FtsZ filaments, we performed rapid dilution experiments: at steady state, we diluted the sample five-fold to induce disassembly of FtsZ filaments and then determined the rate of decrease in FtsZ fluorescence corresponding to an exponential decay. In the case of FtsA, we could fit the decay curve to a single-exponential with a rate of 0.144 ± 0.06 s−1 (s.e.m., n=8). In contrast, with ZipA, we obtained a better fit assuming two separate processes, with a similar and a much slower rate than with FtsA (0.097 ± 0.013 s−1, p=0.5198, and 0.014 ± 0.002 s−1 s.e.m., n = 6, p=0.0881) (Fig. 5c). With ZipA, we saw that thick FtsZ bundles could persist on the membrane for more than 5 min, explaining the two rates we obtained from fitting (Fig. 5c, Supplementary Video 17) and a longer residence time for FtsZ on a single molecule level (Supplementary Fig. 5a). Interestingly, with both membrane anchors present, we found similar, intermediate values for two decay rates and no persisting FtsZ bundles on the membrane (Supplementary Fig. 6).
Finally, in agreement with a negative influence by FtsA, the length and dynamics of FtsZ filaments were strongly influenced by the concentration of FtsA. Using the same concentration for FtsZ (1 μM), we found long, static bundles of filaments at a protein ratio much lower (FtsA, 0.15 μM), but short, highly dynamic filaments at ratio 6 times higher than in vivo (FtsA, 2 μM) (Supplementary Fig. 4b and c).
Together, these data illustrate the ability of FtsA to destabilize FtsZ filaments on the membrane, especially compared to ZipA, allowing for a rapid reorganization of the filament network.
DISCUSSION
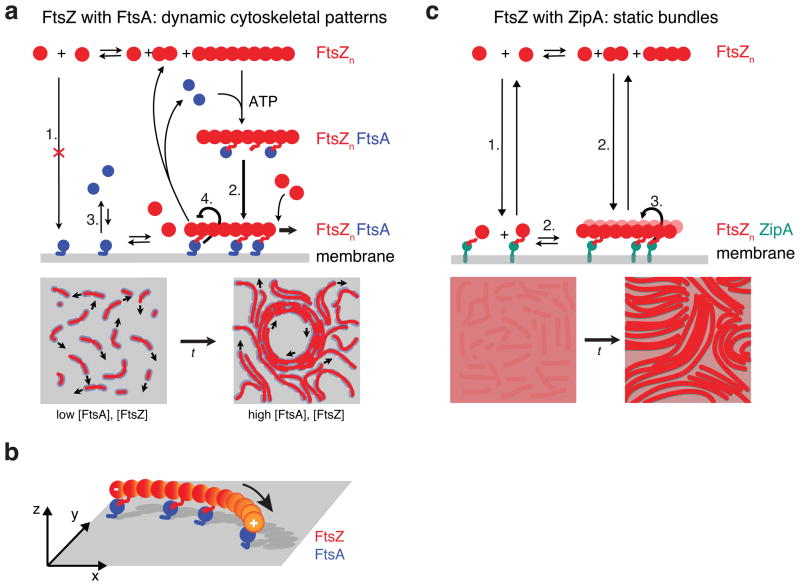
In summary, we found that FtsZ and FtsA can self-organize into highly rapidly reorganizing cytoskeletal patterns. We believe that this behavior emerges from a dual, contradictory influence of FtsA on FtsZ: On the one hand, it enables FtsZ to assemble on the membrane; on the other it provides a negative regulation of FtsZ filament network organization. Accordingly, the dynamics of this filament network reflects the combination of two interactions superimposed on the intrinsic polymerization dynamics of FtsZ: a fast, mutually positive, bidirectional interaction between FtsA and FtsZ during membrane-binding and a delayed, negative unidirectional feedback from FtsA to FtsZ, which destabilizes the filament network. This negative interaction can explain why with FtsA, FtsZ forms a rapidly reorganizing cytoskeletal pattern. Based on our results, we suggest a model of how FtsA and FtsZ self-organize on the membrane (Fig. 6a): FtsA does not recruit FtsZ monomers to the membrane, probably because the binding constant of the C-terminal peptide of FtsZ to FtsA is too weak12. An FtsZ-filament-FtsA complex would exhibit multiple bond interactions allowing for attachment to the membrane, where FtsZ filaments can further polymerize. These filaments are highly dynamic, partially because FtsA can cause fragmentation of FtsZ polymers independent of GTP hydrolysis by a yet unknown mechanism. Fragmented filaments might be too short for a strong enough interaction with FtsA, which leads to their detachment from the membrane. With increasing density of short treadmilling filaments on the membrane, lateral interactions, but no filament sliding, give rise to dynamic higher-order structures, such as streams and vortices. The observed chirality of the swirls could be explained by FtsZ filaments being curved6. Attachment of curved, polar polymers to a surface through one face automatically creates chiral asymmetries48 (Fig. 6b). Combined with bundling and treadmilling, these curved dynamic filaments give rise to the rotating rings with a preferred directionality.
Figure 6. Model for membrane-based FtsZ polymerization.
(a) Model for FtsZ-FtsA co-assembly on the membrane starting from top left: FtsA does not recruit monomeric FtsZ to the membrane (1.). After FtsZ polymerization, an FtsA-FtsZ filament complex forms and attaches to the membrane, which facilitates binding of FtsA to the membrane (2. versus 3.). Here, FtsZ filaments can further polymerize or disassemble into short filaments or monomers, which are not longer recruited to the membrane by FtsA leading to their detachment. The dual, antagonistic role of FtsA is highlighted by two thick arrows: FtsA co-assembles with FtsZ (2.), but allows for the rapid disassembly of FtsZ filaments (4.). Bottom: With increasing density, lateral interactions between short, dynamic filaments give rise to a rapidly reorganizing filament network, i.e. streams and vortices. (b) Illustration of a polar, curved FtsZ filament recruited to the membrane by FtsA. Anchoring a polar filament with a curved rigid conformation to a surface creates chiral asymmetries of the system, i.e. the membrane-bound filament is not identical to its mirror image. (c) Model for the formation of static bundles of dynamic FtsZ filaments mediated by ZipA. In contrast to FtsA, ZipA can recruit polymerized and non-polymerized FtsZ to the membrane (1.). Membrane-recruited FtsZ initiates FtsZ polymerization (2). Lateral interactions allow FtsZ filaments to organize into thick bundles, which is further enhanced by ZipA (3.). Short filaments and monomeric FtsZ can remain on the membrane after depolymerization of fragmentation. Illustration of GTP hydrolysis by FtsZ has been omitted for clarity.
In contrast to FtsA, ZipA can also recruit FtsZ monomers to the membrane. One possible explanation for this observation could be that the binding affinity for FtsZ to ZipA is stronger than to FtsA. FtsZ monomers recruited to the membrane can then nucleate the formation of longer filaments, which further organize into thick bundles (Fig. 6c). Filaments in these bundles are dynamic (Supplementary Fig. 5a), but do not show the same large-scale reorganization as with FtsA, since short filaments and even FtsZ monomers remain recruited to the membrane. As a result, FtsZ organizes into static bundles of dynamic filaments with ZipA instead of a rapidly reorganizing filament network.
Self-organized filament patterns have been described for eukaryotic cytoskeletal filaments and molecular motors49–52, however pure solutions of actin filaments or microtubules usually show only nematic ordering and no large-scale rearrangement53,54. Dynamic filament patterns in these systems either depend on motor proteins generating shear stress or flow, or multiple other cofactors. Importantly, the FtsA/FtsZ system does not contain any molecular motor, which could transform chemical energy into mechanical force to actively move filaments. Instead, based on our finding we favor a model where the large-scale rearrangement of the FtsZ filament network is based on collective polymerization dynamics of FtsZ, which emerge from its interaction with FtsA on the membrane. The chiral organization of FtsZ filaments is analogous to chiral filaments of MreB in vivo55, which were suggested to give rise to chiral ordering of the cell wall during cell elongation. Our findings support the view of the Z-ring continuously adapting to the decreasing diameter of the constricting division septum. A highly dynamic Z-ring could act as a scaffold to spatially organize the enzymes required for the inward growth of the cell wall. In this case, the mechanical force would primarily arise from bond formation during cell wall biosynthesis, as described for MreB56,57, before the Z-ring itself performs the final scission event24.
Methods
Protein Biochemistry
Untagged FtsZ was purified as His-SUMO fusion from pTB567. Since FtsZ does not contain a native Cystein, we modified FtsZ with an N-terminal Cystein residue (Cys-FtsZ) by introducing seven additional aminoacids (AEGCGEL) to the N-terminus of the protein to obtain pT567-GCG-FtsZ (both plasmids were gifts from Thomas Bernhardt, Harvard Medical School). Both versions of FtsZ were expressed in C41 cells grown in 2XYT medium at 37°C and incubated to an optical density of OD600 of 0.8. Expression was induced by adding IPTG to 1 mM and growing the cells at 37°C for 5 hours. The harvested cells were pelleted and resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 50 mM KCl, 20 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol and Protease Inhibitor (Complete EDTA free, Roche Molecular Biochemicals)) and stored at −80°C. For purification, cells were thawed, incubated with Lysozyme (10 mg/mL, L-6876, Sigma-Aldrich) and DNase (1 mg/ml, DN25, Sigma-Aldrich) and lysed by sonification. The resulting extract was centrifuged at 45 000 g for 20 min at 4°C to pellet cell debris and membranes. The clarified lysate was then loaded onto a Ni2+ agarose column (HisPur Ni-NTA Resin, Thermo Scientific). The column was washed with lysis and wash buffer (50 mM Tris-HCl pH 8.0, 50 mM KCl, 30mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol and protease inhibitors (Complete, Roche)) and eluted with elution buffer (50 mM Tris-HCl pH 8.0, 50mM KCl, 200 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol). Peak His-Sumo-FtsZ fractions were pooled. The H-SUMO tag was cleaved with 6xHis-tagged SUMO protease during dialysis into 50 mM Tris, pH 8.0, 300 mM KCl, 10% glycerol. The protease and released tag were removed from the preparation by passing it over Ni-NTA Resin. After removal of the tag, Cys-FtsZ was labeled by thiol-reactive dyes (Alexa488-maleimide, Molecular Probes; Cy5-maleimide, GE Healthcare) according to the protocols of the manufacturer. Polymerization-competent proteins were enriched by CaCl2-mediated polymerization. The FtsZ preparation was dialyzed against FtsZ polymerization buffer (50 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.7, 10 mM MgCl2) and warmed to room temperature. CaCl2 and GTP were added to the final concentrations of 10 mM and 5 mM, respectively, and the reaction was centrifuged at 20,000 x g for 2 min at room temperature to pellet polymeric FtsZ. FtsZ pellets were resuspended in storage buffer (50 mM Tris, pH 7.4, 50 mM KCl, 1 mM EDTA, 10% glycerol) and dialyzed extensively to remove CaCl2 and free GTP.
FtsA was PCR amplified from MG1655 cells and cloned into pTB146 (a gift from Thomas Bernhardt (Harvard Medical School) restriction digested with by SapI and XhoI to obtain pML29. For labeling of FtsA we introduced a pentaglycine tag (Gly5-FtsA) at the N-terminus of FtsA to obtain pML60. Both versions of FtsA were expressed as His-SUMO fusion in C41 cells grown in 2XYT medium at 37°C and incubated to an optical density of OD600 of 1 to 1.5. The SUMO-construct allowed for increased solubility of the protein and decreased toxicity for the cells. Expression was induced by adding IPTG to 1 mM and growing the cells at 18°C overnight. Harvested cells were resuspended in lysis buffer (50 mM Tris -HCl pH 8, 20 mM Imidazol, 500 mM NaCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 1 mM ADP and protease inhibitors). After lysis, the lysate was loaded onto a Ni2+ agarose column, which was washed with wash buffer (50 mM Tris-HCl pH 8, 500 mM KCl, 30 mM Imidazol, 10 mM MgCl2, 10 mM β-mercaptoethanol and 1 mM ADP). His-SUMO-FtsA was eluted at 200 mM Imidazol and peak protein fractions were pooled. The SUMO tag was removed by adding Ulp1 to 5 mM and incubation on ice for 3 1/2 hours. The protease and released tag were removed from the untagged FtsA by gel filtration using a Superdex 200 column and FtsA storage buffer (50 mM HEPES pH 7.55, 500 mM KCl, 10 mM MgCl2, 10 % Glycerol and 1 mM ADP). Peak fractions of FtsA were pooled and concentrated to 50 μM, flash frozen in liquid nitrogen and stored at −80°C. FtsZ was fluorescently labeld by “sortagging”(ref. 58): His-SUMO-Gly5-FtsA was incubated with 5 mM Ulp1 and additionally 50 μM His-SrtΔ59 in the presence of 1 mM Cy5-labeled pentapeptide LPTEG and 5 mM CaCl2. The protease, sortase, tag and excess Cy5-pentapeptide were removed by gel filtration to obtain Cy5-GG-FtsA. Mutants of FtsA were obtained by site directed mutagenesis (Quikchange, Agilent).
Mutants of FtsA were obtained by site directed mutagenesis (Quikchange, Stratagene) following the instructions of the manufacturer to obtain pML35, pML52 and pML53.
ZipA without the N-terminal periplasmic and transmembrane domains, i.e. amino acids 23–328, was cloned into pET28a to obtain pML21 and purified as His-Δ22-ZipA fusion. Truncated ZipA was expressed in C41 cells grown in 2XYT medium at 37°C and incubated to on optical density of OD600 of 0.8. Expression was induced by adding IPTG to 1 mM and growing the cells at 37°C for another 3 hours. The protein was purified using a Ni2+ agarose column (HisPur Ni-NTA Resin, Thermo Scientific). Purity of the proteins was confirmed by SDS-PAGE stained with Coomassie Blue (Supplementary Fig. 1). His-SrtΔ59, His-Ulp1 and FtsZ-YFP-mts were purified according to previously published protocols59–61.
All plasmids were verified by DNA sequencing. Oligonucleotide primers used in these study are given in Supplementary Table 2.
Preparation of liposomes
E. coli polar lipids dissolved in chloroform were transferred into a glass vial and the solvent was evaporated under a gentle stream of nitrogen. Any residual solvent was further removed by drying the lipid film in a vacuum for 1 h. The lipids were then rehydrated in SLB buffer (50 mM Tris pH 7.5, 300 mM KCl, 5 mM MgCl2) to a lipid concentration of 4 mg/ml and incubated at 37°C for 30 min. The lipid film was then completely resuspended by vortexing rigorously to obtain multilamellar vesicles of different sizes. This mixture was then placed in a bath sonicator where shear forces help to reduce the size of the vesicles giving rise to small unilamellar vesicles (SUVs). This SUV dispersion was stored at −20°C as 20 μl aliquots.
Vesicle sedimentation assay
Liposomes were prepared as described above in 50 mM PIPES pH 6.7, 50 mM KCl, 10 mM MgCl 2, and sonicated only briefly to obtain multilamellar vesicles of different sizes. Purified proteins (5 μM FtsZ, 3 μM FtsA or 3 μM ZipA) were added to a suspension of 2 mg/ml SUVs in reaction buffer. Reactions were started by adding GTP (and in case of FtsA with additionally ATP) to 1 mM and incubated at room temperature for 10 min. Vesicles were sedimented by centrifugation at 25 000 rpm in a Beckmann TLA-100 rotor in a table top ultracentrifuge at room temperature for 10 min. The supernatant of each mixture was collected and the pellet was resuspended in reaction buffer to the original volume. Amounts of FtsZ, FtsA or ZipA were estimated by SDS-PAGE. Following electrophoresis, gels were stained with Coomassie Blue.
Preparation of supported lipid bilayers
Glass cover slips were cleaned by sonicating in 2 % Hellmanex II solution (Hellma, Müllheim, Germany) for 15 min, followed by extensive washing with milliQ H2O. Finally, glass cover slips were washed, sonicated and stored in 95% Reagent grade ethanol. Before usage, glass cover slips were blown dry with compressed air and cleaned in an air plasma for 15 min. The reaction chamber for the self-organization assay was prepared by attaching a plastic ring on a cleaned glass cover slip using UV glue (Norland optical adhesive 88). For supported lipid bilayer formation, the SUV dispersion was diluted in SLB buffer to 0.5 mg/ml of which 75 μl were added to the reaction chamber. Adding CaCl 2 to a final concentration of 3 mM induced fusion of the vesicles and the formation of a lipid bilayer on the mica surface. After 10 min of incubation at 37°C, the sample was rinsed with 2 ml pre-warmed reaction buffer.
FtsZ self-organization assay
For the self-organization assay of FtsZ and FtsA, FtsZ with 30 % Alexa488-labeled FtsZ, FtsA with optionally 30 % Cy5-GG-FtsA at different concentrations and ATP (1 mM) were added to the reaction buffer (50 mM Tris-HCl pH 7.5, 150 mM KCl, 5 mM MgCl2) above the supported lipid membrane in a self-made reaction chamber (Supplementary Fig. S1). The final volume of the assay was 100 μl. The reaction chamber was closed with a lid of a Eppendorf reaction tube during observation. Polymerization of FtsZ was induced by adding GTP to 2.5 mM to the buffer, which allowed polymerization for about 2 hours. For experiments on FtsZ and ZipA, we first added His-ZipA (1 μM) to the buffer and allowed it to bind to bilayers containing 4% 18:1 DGS-NTA (Ni2+). After 10 min, we washed the system four times with 200 μl reaction buffer to remove unbound protein, before we added FtsZ to the buffer. All experiments were performed with oxygen scavenger system (40 mM D-glucose, 0.040 mg/ml glucose oxidase, 0.016 mg/ml catalase, 20 mM DTT and 1 mM Trolox) added to the buffer to prevent photobleaching.
TIRF microscopy imaging
All experiments were performed on a Nikon Ti Total Internal Reflection Fluorescence equipped with a 100x TIRF NA 1.49 DIC objective lens and the Perfect Focus System for continuous maintenance of focus. Alexa488 was excited with 100mW 491 nm solid state laser of Solamere laser launch, Cy5 from a 30 mW 640nm laser. The laser line was selected via a fiber-optic delivery system and 4-channel AOTF. Images were acquired with a Hamamatsu ImagEM 512×512 back-thinned electron multiplying cooled CCD camera controlled with Nikon Elements software. For time-lapse experiments, images were obtained every 2–5 sec, with 30 ms exposure time, with illumination light shuttered between acquisitions.
Single particle tracking
Tracking of individual FtsZ monomers using performed as described in ref. 62. During each experiment, 5–10 videos were acquired. For each video (between 200 and 500 frames with a total run length of 50 – 160 s), a minimum number of 100 protein tracks were analyzed. Therefore, one experimental value represents the average of at least 500 analyzed particles. The averaged value of 5 experiments represents at least 1500 analyzed tracks. At the imaging conditions we used, we found that the observed number of fluorescent molecules did not decrease with observation time, suggesting that the bleaching time was longer than the lifetime of FtsZ on the membrane. As a further control, we decreased the frame rate during data acquisition, such that the time between two successive frames increased from 0.5 s and 0.8 s and did not see a significant difference in the observed lifetimes indicating that bleaching occurred on a longer time scale than the binding/unbinding events and that it did not contribute significantly to the measured lifetimes.
Image analysis and processing
Image analysis and processing was carried out with Image J (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2007). The frames of time- lapse movies were normalized to have a constant overall intensity, thus compensating for the increasing intensity over time due to protein binding to the membrane. As a consequence of this normalization, non-specific bound fluorescent particles appear disproportionately bright at the beginning of a movie. Kymographs of ring rotation were obtained using an ImageJ kymograph plugin from Jens Rietdorf and Arne Seitz (EMBL, Heidelberg; http://www.embl.de/eamnet/html/kymograph.html).
Repeatability of experiments
Representative micrographs and intensity curves correspond to at least 4 (up to more than 100) successfully repeated experiments. The number of replicated experiments is given in the respective Figure captions. For the analysis of single FtsZ filaments (Fig. 2d and Supplementary Fig. 3), we selected 38 isolated filaments, whose trajectories did not cross with those of neighboring filaments as resolved in our microscopic setup.
Supplementary Material
Acknowledgments
We would like the Mitchison lab and the labs of Tom Bernhardt and Tom Rapoport for discussions and support; Jeffrey Werbin (Harvard Medical School) for the plasmid for Sortase expression, Harold Erickson (Duke University) for the plasmid for FtsZ-YFP-mts expression; Jan Brugues for valuable discussions and advice, Ethan Garner, Karsten Kruse and Stephan Grill for discussions and comments on the manuscript. We would also like to thank the Nikon Imaging Center at Harvard Medical School for their excellent service. M.L.is supported by fellowships from EMBO (ALTF 394-2011) and HFSP (LT000466/2012). Cytoskeleton dynamics research in the T.J.M. group is supported by NIH-GM39565.
Bibliography
- 1.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 2.Ingerson-Mahar M, Gitai Z. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol Rev. 2012;36:256–266. doi: 10.1111/j.1574-6976.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Micro. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 5.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proceedings of the National Academy of Sciences. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, Reedy M, Erickson H. Straight and Curved Conformations of FtsZ Are Regulated by GTP Hydrolysis. Journal of Bacteriology. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliva MA, Trambaiolo D, Löwe J. Structural insights into the conformational variability of FtsZ. J Mol Biol. 2007;373:1229–1242. doi: 10.1016/j.jmb.2007.08.056. [DOI] [PubMed] [Google Scholar]
- 8.Hsin J, Gopinathan A, Huang KC. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109:9432–9437. doi: 10.1073/pnas.1120761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Molecular Microbiology. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 11.Beuria TK, et al. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem. 2009;284:14079–14086. doi: 10.1074/jbc.M808872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale CA, de Boer PAJ. Direct Binding of FtsZ to ZipA, an Essential Component of the Septal Ring Structure That Mediates Cell Division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 15.Mosyak L, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. Journal of Bacteriology. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haney SA, et al. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem. 2001;276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 18.Erickson HP, Osawa M. Cell division without FtsZ - a variety of redundant mechanisms. Molecular Microbiology. 2010;78:267–270. doi: 10.1111/j.1365-2958.2010.07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Curr Opin Microbiol. 2011;14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 2012 doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osawa M, Anderson DE, Erickson HP. Reconstitution of Contractile FtsZ Rings in Liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arumugam S, et al. Surface Topology Engineering of Membranes for the Mechanical Investigation of the Tubulin Homologue FtsZ. Angew Chem Int Ed Engl. 2012;51:11858–11862. doi: 10.1002/anie.201204332. [DOI] [PubMed] [Google Scholar]
- 24.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rueda S, Vicente M, Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. Journal of Bacteriology. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran U, Phillips R, Milo R. SnapShot: key numbers in biology. Cell. 2010;141:1262–1262.e1. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Thanedar S, Margolin W. FtsZ Exhibits Rapid Movement and Oscillation Waves in Helix-like Patterns in Escherichia coli. Current Biology. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 30.Martos A, et al. Isolation, Characterization and Lipid-Binding Properties of the Recalcitrant FtsA Division Protein from Escherichia coli. PLoS ONE. 2012;7:e39829. doi: 10.1371/journal.pone.0039829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pichoff S, Lutkenhaus J. Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol. 2007;64:1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- 32.Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. The EMBO Journal. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu L, Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J. 2008;95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci USA. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Needleman DJ, et al. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol Biol Cell. 2010;21:323–333. doi: 10.1091/mbc.E09-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280:22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mingorance J, et al. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J Biol Chem. 2005;280:20909–20914. doi: 10.1074/jbc.M503059200. [DOI] [PubMed] [Google Scholar]
- 39.Mateos-Gil P, et al. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc Natl Acad Sci USA. 2012;109:8133–8138. doi: 10.1073/pnas.1204844109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. Journal of Bacteriology. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez-Rocamora VM, et al. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J Biol Chem. 2012;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateos-Gil P, et al. FtsZ polymers bound to lipid bilayers through ZipA form dynamic two dimensional networks. Biochim Biophys Acta. 2012;1818:806–813. doi: 10.1016/j.bbamem.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Martos A, et al. Characterization of self-association and heteroassociation of bacterial cell division proteins FtsZ and ZipA in solution by composition gradient-static light scattering. Biochemistry. 2010;49:10780–10787. doi: 10.1021/bi101495x. [DOI] [PubMed] [Google Scholar]
- 44.Tyson JJ, Chen KC, Novák B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current Opinion in Cell Biology. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 45.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 46.Novák B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange G, Mandelkow EM, Jagla A, Mandelkow E. Tubulin oligomers and microtubule oscillations. Antagonistic role of microtubule stabilizers and destabilizers. Eur J Biochem. 1988;178:61–69. doi: 10.1111/j.1432-1033.1988.tb14429.x. [DOI] [PubMed] [Google Scholar]
- 48.Henley CL. Possible Origins of Macroscopic Left-Right Asymmetry in Organisms. J Stat Phys. 2012;148:740–774. [Google Scholar]
- 49.Nédélec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- 50.Schaller V, Weber C, Semmrich C, Frey E, Bausch AR. Polar patterns of driven filaments. Nature. 2010;467:73–77. doi: 10.1038/nature09312. [DOI] [PubMed] [Google Scholar]
- 51.Sumino Y, et al. Large-scale vortex lattice emerging from collectively moving microtubules. Nature. 2012;483:448–452. doi: 10.1038/nature10874. [DOI] [PubMed] [Google Scholar]
- 52.Sanchez T, Chen DTN, DeCamp SJ, Heymann M, Dogic Z. Spontaneous motion in hierarchically assembled active matter. Nature. 2012;491:431–434. doi: 10.1038/nature11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hitt AL, Cross AR, Williams RC. Microtubule solutions display nematic liquid crystalline structure. Journal of Biological Chemistry. 1990;265:1639–1647. [PubMed] [Google Scholar]
- 54.Käs J, et al. F-actin, a model polymer for semiflexible chains in dilute, semidilute, and liquid crystalline solutions. Biophysj. 1996;70:609–625. doi: 10.1016/S0006-3495(96)79630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA. 2012;109:E595–604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garner EC, et al. Coupled, Circumferential Motions of the Cell Wall Synthesis Machinery and MreB Filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 58.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 59.Popp MW-L, Antos JM, Ploegh HL. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci. 2009;Chapter 15(Unit 15.3–56):15.3.1–15.3.9. doi: 10.1002/0471140864.ps1503s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 61.Osawa M, Anderson DE, Erickson HP. Reconstitution of Contractile FtsZ Rings in Liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loose M, Fischer-Friedrich E, Herold C, Kruse K, Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nature Structural &; Molecular Biology. 2011;18:577–584. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.