Abstract
The field of bioengineering has pioneered the application of new precision fabrication technologies to model the different geometric, physical or molecular components of tissue microenvironments on solid-state substrata. Tissue engineering approaches building on these advances are used to assemble multicellular mimetic-tissues where cells reside within defined spatial contexts. The functional responses of cells in fabricated microenvironments has revealed a rich interplay between the genome and extracellular effectors in determining cellular phenotypes, and in a number of cases has revealed the dominance of microenvironment over genotype. Precision bioengineered substrata are limited to a few aspects, whereas cell/tissue-derived microenvironments have many undefined components. Thus introducing a computational module may serve to integrate these types of platforms to create reasonable models of drug responses in human tissues. This review discusses how combinatorial microenvironment microarrays and other biomimetic microenvironments have revealed emergent properties of cells in particular microenvironmental contexts, the platforms that can measure phenotypic changes within those contexts, and the computational tools that can unify the microenvironment-imposed functional phenotypes with underlying constellations of proteins and genes. Ultimately we propose that a merger of these technologies will enable more accurate pre-clinical drug discovery.
1. Introduction
The road to translating pre-clinical anti-cancer targets into clinically successful drugs is strewn with billions of dollars worth of disappointments. A large number of compounds that cure diseases in rodent model systems failed to provide meaningful clinical benefit in humans. Although there is startling conservation between the expressed genome of in mice and man, significant differences arise at the level of physiology and tissue architecture that can impact drug responses. Modern drug discovery has moved towards use of rationally designed molecules that are screened in high-throughput systems for activity and against off-target effects. In spite of these advances, ~85% of new cancer drugs fail in phase 2 clinical trials because although they meet minimal safety standards, they exhibit no efficacy [1]. Thus a major challenge is to identify preclinical screening strategies using model systems that more faithfully reflect the biologies of human tissues. This review will discuss how biomimetic microenvironments have revealed emergent properties of cells and cellular communities, the platforms that can measure phenotypic changes within those microenvironments, and the computational tools that can unify the microenvironment-imposed functional phenotypes comprising different constellations of proteins and genes. Ultimately we propose that a merger of these technologies will enable more accurate pre-clinical drug discovery.
Our understanding of the molecular basis of cancer has evolved remarkably in the past two decades. Indeed, The Cancer Genome Atlas program has identified a broad range of recurrent gene mutations and structural rearrangements that putatively drive tumor genesis. Improved medicinal and computational chemistry methods have generated unprecedented numbers of experimental therapeutic agents to target pathways affected by recurrent genome modifications. In spite of this progress, molecularly targeted therapies are yet to generate a durable response in the metastatic setting, and cancer remains the leading cause of death world wide, accounting for an estimated 13% of deaths [2]. The confounding reality for anti-cancer drug development is the heterogeneity of tumors [3, 4]. Far from a homogeneous expansion of neoplastic cells, tumors are more appropriately viewed as abnormal organs, comprising multiple cell types and dynamic extracellular matrix (ECM). This wayward “organ” interacts with the body via unique vascular systems and via an immune homeostasis that leads to evasion of immune responses. The complexes of ECM, growth factors, cytokines, inflammatory mediators, immune cells, oxygen tension, and tensile forces that control malignant progression conspire to subvert cancer drug effectiveness.
Understanding the interactions between cells and their natural tissue microenvironment is of fundamental importance for tissue engineering and regenerative medicine. The principles established in this field are relevant for developing tissue model systems for drug development and have revealed contextual drug responses [5–7]. The microenvironment comprises chemical and physical signals that direct cells to organize into functional multicellular architectures. The paradigm of microenvironmental influence is the stem cell niche, a spatially restricted locoregional tissue site that presents specific cell-cell and cell-ECM interactions that control cell proliferation and differentiation. Biofunctionalization of materials endeavors to mimic the nano- and micro-scale interaction mechanisms characteristic of native biological systems. These bioengineering-based approaches also can be applied to model disease. During the development of cancer, the natural tissue architecture breaks down and the microenvironment is distorted. We have proposed that immortal tumor cells respond inappropriately to cues from the surrounding normal tissue, establishing a dynamic interplay between the growing tumor and reciprocal microenvironmental signals that engender malignant characteristics via epigenetic reprograming [8]. The cellular plasticity facilitated by these gene expression changes, exemplified by the epithelial-to-mesenchymal transition (EMT), produces tumor cells with migratory and stem cell-like characteristics (“cancer stem cells”). The unique repertoire of functions gained by these tumor cells enables metastatic spread to distant anatomical sites where new constellations of microenvironmental signals are encountered. The ability of tumor cells to metastasize and survive in foreign microenvironments is strongly correlated with resistance to anti-cancer treatments, and therapeutic response failures are the leading cause of cancer patient deaths.
In vitro modeling of the diverse microenvironments encountered by malignant cells is crucial to reveal contextual drug responsiveness. The majority of pre-clinical investigations are performed in human cell lines or rodent xenograft models that do not always accurately model the human context. In a number of inbred mouse models of different diseases, the gene expression patterns can differ strikingly from the orthologous human disease [9, 10]. Although inbreeding mouse strains was meant to provide tractable genetic backgrounds for experimentation, a confounding side effect has been that each strain has unique properties. Indeed tumor growth in both xenograft and mouse genetic models of cancer can vary dramatically between strains [11, 12]. One solution for this has been to use outbred mouse cohorts, which are genetically diverse and may offer better mimicry of some human diseases and aging at the population level [13].
Established human cell lines and primary cells propagated in 2D culture are amenable to high-throughput experimentation, however, they often lose the tissue-specific gene expression patterns and cell surface proteins that are characteristic of the cells in their cognate tissues [14–16]. A potential solution is to use higher-order multi-lineage human cell systems, to more accurately recapitulate the emergent properties of tissues and organs. Bioengineering technologies are now used to fabricate components of tissue microenvironments on solid-state substrata, while tissue engineering approaches are similarly applied to assemble multicellular mimetic-tissues where cells reside in a defined spatial context. Importantly, the phenotype of cells in these fabricated microenvironments revealed that the microenvironment can be dominant over genotype [17–19]. Hence precisely defining the role of individual and combinations of microenvironmental components is crucial to allow reliable prediction of cellular responses to therapeutics.
1.1. The tumor microenvironment is a potent determinant of drug responses
The microenvironment is defined as the sum total of cell-cell, -ECM, and -soluble factor interactions surrounding each cell in a tissue. These components exchange information with cells via a combination of physical, chemical, and electrical signals, frequently activating or suppressing the same pathways triggered by oncogenes [20–25]. Exposure of a cell to a particular microenvironment elicits dramatic changes to normal and malignant cell behaviors, such as stem cell-like activity [26–28]. The influence of the microenvironment can be so profound as to correct the otherwise malignant behavior of mutant cells within an intact normal tissue structure [25]. Cytotoxic drugs are subject to microenvironmental effects, usually through cell cycle modulation. For instance, addition of therapeutic antibodies to VLA-4 was shown to prevent minimal residual disease following treatment of acute myeloid leukemia (AML) with the nucleoside analog AraC, VLA-4 antibody prevented the tumor cells from binding fibronectin, which would produce AML cell quiescent and evasion of the DNA replication dependent cytotoxic effect [29]. It has been known for 30 years that the microenvironment can exert dominance over certain oncogenic mutations [30, 31]. Microenvironmental-induced phenotypic changes in tumors such as EMT are associated with broad resistance to anti-cancer agents [32]. The activities of the new generation of anti-cancer drugs developed to target specific oncogenic ‘drivers’ are affected different microenvironments. In a comprehensive study of prostate cancer cell lines on 2-dimensional (D) tissue culture plastic or in 3-D Matrigel, it was reported that PI3-kinase inhibitors were most effective in preventing invasive cell growth in 3-D [33]. Culturing HER2/neu amplified breast cancer cell lines in 3-D versus 2-D revealed distinctive therapeutic activities of the HER2-targeted agents Lapatinib, Trastuzumab, and Pertuzumab [34]. β1 integrin-blocking antibodies could modulate these contextual drug responses in 3-D cultures, implicating the role of the ECM and demonstrating the opportunity for potential microenvironmental intervention as a therapeutic approach.
As the importance of microenvironment in therapeutic response has become more widely accepted the urgency to identify tractable organotypic culture systems for studying human tissues in vitro has manifested. Matrigel, HuBiogel, HuMatrix, and a number of other commercially available laminin-rich ECM are widely used to provide 3-D cell growth environments, and these gels are used increasingly to study the impact of drugs on cells grown in 3-D. Matrigel, which is harvested from a rodent sarcoma cell line, is comprised of hundreds of proteins that can vary significantly in the exact composition between production lots [35]. In fact, laboratories that use these commercial gels in large quantity routinely screen multiple lots for their ability to reproduce data from previous studies (M.Bissell personal communication). Recent adaptions of 3D culture systems to high-throughput screening (HTS) systems is an important advance and use of 3-D gels in HTS studies is now a less daunting prospect [36]. However, placing human cells in an undefined rodent sarcoma 3-D context may not mimic the intended in vivo microenvironment, and variability in the molecular components may confound interpretations and reproducibility of the results.
1.2. Deconstructing cell-microenvironment interactions
Tissues are collections of cells and ECM organized into unique spatial configurations that collectively carry out a specialized function in the body. Remarkably, tissues with an intact architecture can maintain many basic functions in spite of the presence of gene mutations that cause dysfunctions when introduced into cells on tissue culture plastic. Why are tissues so robust? Seminal studies showing that wound-healing microenvironments unleash malignant potential demonstrated the principle that tissue architecture is a crucial component of cellular function [21]. Organized asymmetry is therefore an important basic feature of tissues; there must be distinctive topologies on which receptors assemble in order to correctly integrate the signaling patterns associated with tissue-specific functions. Tumor microenvironments should as well possess combinatorial signaling asymmetries, though the microenvironments may be less obviously organized. One hypothesis is that the normal and tumor microenvironments integrate the signaling apparatuses differently, and thus therapeutic targets could be identified to selectively harm the tumor cells, and microenvironment composition will be a determinant of drug efficacy. Those potential differences in signal integration can be revealed by technologies that recapitulate in vivo microenvironments, using defined physical, geometric, and molecular elements, and allowing one to assess the contribution of each property to the emergent functional outcomes.
2. Combinatorial Microenvironment Microarrays
Cell-based functional screening of interactions with combinatorial microenvironment microarrays (MEAarrays) enables molecular dissection of more complicated 3D microenvironments (Fig 1) [26–28, 37]. These platforms are amenable to high-throughput scale-up using a number of imaging modalities for quantification. Because the ECM, growth factors and other microenvironmental components are adsorbed to a substrate surface, the cells experience the microenvironments asymmetrically. The challenges of these approaches are: access to purified extracellular proteins, managing the combinatorial complexity to minimize cost and maximize the combinatorial space that is evaluated, data visualization, and statistical analysis to identify microenvironment components that contribute a given outcome.
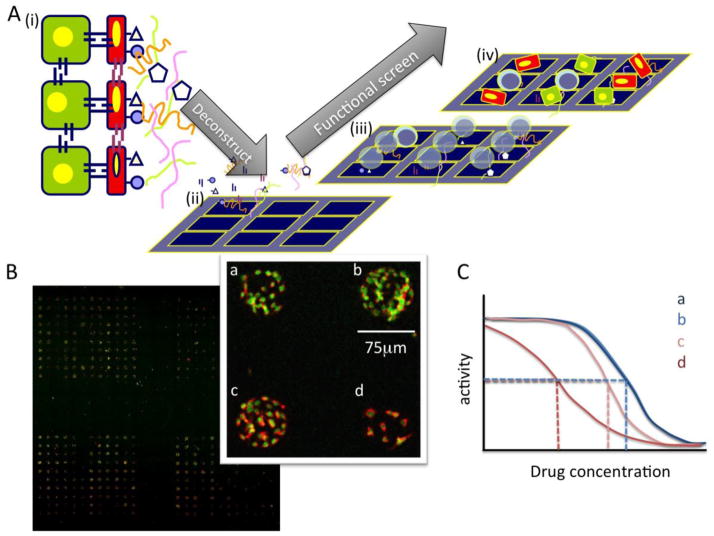
Figure 1.
Deconstructing complex microenvironments into tractable pieces using combinatorial microenvironment microarrays (MEArrays). A) (i) A cartoon a tissue microenvironment in which different cell types interact with each other and with ECM and soluble factors to generate a functional tissue. (ii) Purified ECM, growth factors, and recombinant cell surface receptors are mixed into combinations and printed on substrata that will support cell adhesion. (iii) Live cells are then added and cultured until an endpoint, (iv) when the relevant phenotypic responses are measured. (B) A low resolution scan of a breast cancer cell line on an MEArray that were treated with an anti-cancer agent. Red fluorescence shows the staining of a receptor tyrosine kinase, and green shows nuclei. Inset, shows a higher magnification image in four cells on four distinct microenvironments (a,b,c,d). (image credit: Dr. Tiina Jokela, LBNL) (C) Hypothetically, drug activities (e.g. IC50) (dashed lines) should be shifted in some combinatorial microenvironments.
MEArrays have been used to profile cell-ECM adhesion biases [38], to optimize growth of cultured cells [39], and to better understand the interactions of human stem cells with putative niche proteins and other tissue-specific proteins that were relevant to embryonic [26], neural [28], mammary [27], and hepatic stem cells [37]. Taking a combinatorial approach, relative to a candidate-based approach, allows screening combinations of multiple tissue-specific microenvironment proteins to identify extracellular cues that are the basis for emergent cell behavior. Functional roles for a number of molecules known to be expressed in human mammary gland and brain, but hitherto had not been ascribed respective roles for mammary or neural stem and progenitor cell regulation, were discovered using this type of approach.
2.1. Fabrication substrata
Combinations of ECM and other extracellular proteins are usually printed on modified glass using standard quill-pin or piezoelectric microarray printers, allowing functional screening on hundreds or thousands of defined combinatorial microenvironments. Printing substrata range from aldehyde-, nitrocellulose-, polydimethylsiloxane (PDMS) or poly acrylamide (PA)-coated glass slides, and polystyrene plates. Nickel-modified gold-coated glass can adsorb histidine-tagged proteins. Aldehyde-derivatized glass facilitates covalent protein attachment, but the covalent bond may destroy the activity of the printed molecules in some cases. PDMS is cost-effective, readily adsorbs proteins in a nearly irreversible electrostatic interaction, and is capable tuning the elastic modulus to mimic that of tissues like cartilage, skin, and tendon (<1MPa to 10MPa) [40]. PA coated slides are not as good at adsorbing proteins, having weak electrostatic interactions, but proteins seem to get stuck in the pores that are created, persist during washing steps, and ultimately support cell attachment. PA gels can be tuned to mimic the elastic modulus of soft tissues, e.g., brain, lung, and breast (0.1kPa to 100kPa) [40]. MEArrays printed on PA gels are reported to remain stable stored for up to three months [41]. It is possible that treatment of PA gels with cross-linking reagents such as sulpho-SANPAH may enhance protein stability. Nickel-modified gold-coated surfaces will enable quantification of protein on the arrays by surface plasmon resonance and a high level of specificity to binding only his-tagged proteins. Aldehyde- and PA-coated glass are hydrophilic and most contact-printed features are thus circle-shaped, whereas features printed on hydrophobic PDMS will be the exact shape of the pin-head and can thereby take advantage of defined geometric shapes. Thus molecular composition, stiffness, and geometry are all potentially tunable features in MEArray platforms.
2.2. Analysis and visualization
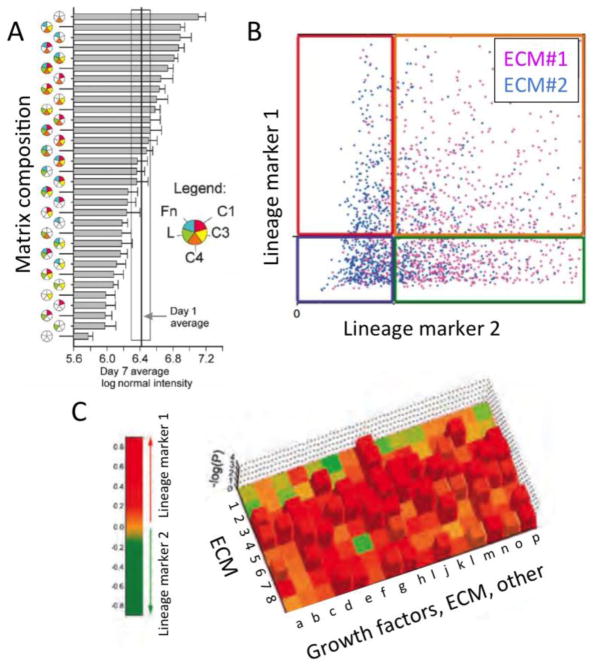
The combinatorial complexity of a MEArray experiment increases rapidly when taking into account stiffness, geometry, and molecular composition. The statistical analysis of MEArray experiments is a rate-limiting step for this technology and there have been multiple solutions to this problem. Data are first normalized either to the ensemble average of the total signal from all array features, or to the signal emanating from cells on a control microenvironment that is known a priori to reproducibly bias towards a given phenotype. Of the published reports, each microenvironment is replicated from 4 to 12 times per array to enable calculation of means and standard deviations, different modes of visualization have been used that emphasize different aspects of the data. Flaim et al. used a combination of pie charts, which showed matrix compositions, with bar graphs that represented a measured functional response (Fig 2A) [26]. Soen et al. measured GFAP and TUJ1 signals, markers of neuronal differentiation, in every neuronal progenitor on a given feature and visualized the data with X-Y intensity plots (Fig 2B). The authors assigned a response magnitude in progenitor cells upon adhesion to a given microenvironment by normalizing to the response on laminin only. They used hierarchical clustering and Pearson correlations as the similarity metric to generate heatmaps of the data and identify trends in responses [28]. In Konagaya etal a relatively small number of growth factor combinations were tested to optimize neural progenitor microenvironments, and they used hierarchical cluster analysis to reveal three major clusters of microenvironment combinations that caused growth, astrocyte or neuron differentiation[39]. The combinatorial activity of microenvironment proteins was revealed using this cluster method in that EGF combined with either BDNF or IGF-1 grouped into the astrocyte-inducing cluster, but EGF alone was in the growth cluster and IGF-1 or BDNF alone were in the neuron-inducing cluster. Brafman et al. utilized Z-score standardization to identify microenvironments that imposed phenotypes distinct from the global mean [41], and they and others [26] employed factorial analyses to reveal complex interactions between microenvironmental components. In LaBarge et al. we primarily printed pair-wise combinations of microenvironmental components, which allowed for streamlined visualization, using a standard heat-map in which each row represented a mammary ECM and each column represented an ECM or a growth factor [27]. The heat-map colors corresponded to the magnitude of log2 transformed fluorescence intensities of two meaningful markers of mammary stem cell differentiation (keratin 14 and keratin 8), and the height of the Z-axis corresponded to the –log of the p-value (Fig 2C). Dunnette’s tests were used to compare the means in each microenvironment to type 1 collagen-only controls, because when many conditions are compared to one control condition the test has a narrow confidence interval and fewer false positives than other T-tests. In order to distinguish between cellular subsets that selectively adhered to a given environment from microenvironment-imposed differentiation we compared arrays just after all array features were saturated with cells with an cells on arrays after some time had elapsed [42]. In all cases, it must be acknowledged that microarray-based methods have inherent variability necessitating that conclusions be validated with multiple orthogonal assays.
Figure 2.
Three successful approaches to visualizing functional consequences cellular interactions with combinatorial microenvironments in a highly parallel experimental environment. (A) Mixed use of pie charts for detailing microenvironmental composition, together bar graphs depicting functional responses (adapted from [26]). (B) Scatter plots showing single cell functional responses on different ECM combinations (adapted from [28]). (C) Heat maps showing functional consequences of cells interacting with different pair-wise microenvironments (composed of ECM 1–8 with a-p other), statistical significance is on the z-axis (adapted from [27]).
2.3. Managing combinatorial complexity
The potential complexity in a microenvironment microarray such that combinations are not repeated can be determined by the equation n!/(n-x)!(x!); where n is the number of microenvironmental components, and x is the number of components per microenvironment. The number of permutations possible rises sharply in an array design that incorporates three or more components per microenvironment. Pairwise combinations are relatively straightforward for identifying the components that drive the emergence of a particular functional phenotype, and factorial analysis might provide insight into the driving components in more complex microenvironments. However, a hypothetical MEArray design could take advantage of methods that more efficiently sample across combinatorial space to determine the different combinations of microenvironmental proteins to be tested, but that do not test every possible permutation. The Taguchi Method, which is used in engineering for process and manufacturing optimization [43], is an example of orthogonal array optimization that could be employed in the design of microenvironment arrays that have three or more components per microenvironment that differ in concentration. The goal is to sample across the potential spectrum of microenvironments to identify those which elicit the strongest desired functional outcome, then optimize and validate based on those initial findings.
The utility of the MEArray approach for identifying drug targets or determining how different microenvironments might impact drug activity has not been explicitly demonstrated. However, using MEArrays we discovered that the notch ligand Jagged1 was involved in maintaining a stem cell-like phenotype in human mammary progenitors. As a control, gamma secretase inhibitors were added to antagonize notch signaling, and a number of microenvironments were revealed that modulated the effects of the inhibitor with respect to mammary stem cell fate decisions [27]. Those data demonstrated the principle that drug-microenvironment interactions could be revealed using this type of an array approach. MEArray-type approaches could be combined with small interfering RNA (siRNA) technology to aid in target identification or in determining the molecules that underlie a given functional phenotype. “Cell spot microarrays” are essentially siRNA libraries printed in type 1 collagen microenvironments that support cell adhesion and reverse transfection of the siRNAs [44]. The content of the microenvironments could be altered to explore the interactions between given genes, microenvironments, and desirable functional phenotypes. There is tremendous potential for using these HTS methods for identifying drug targets in context, and for identifying the key properties of tissues that will alter a drug responses.
3. Highly parallel fabrication of microtissues with reproducible architectures
Tissues in vivo have exquisite architectures, and most tissue culture models are poor substitutes. Cell-based microtissue models compensate for gaps in knowledge that impact the completeness of MEArray designs. Indeed, recent co-culture models that recapitulate vascularized stromal microenvironments in 96-well plates demonstrated that micron-scale changes in cellular neighborhoods was the difference between enabling and quenching metastatic behavior of breast cancer cells [45]. Typical 3D cultures in which acini or other structures are derived from a single cell provide some elements of tissue-like architecture, but there is poor control over the cellular composition, structural morphology, and cell positioning. To accurately mimic a tissue, control of single cell positioning is required, because in locations such as stem cell niches even a one-cell diameter change in position can have discernible impact. Additionally, an optimal high-throughput experimental environment would incorporate significant reproducibility of composition among tissues so that phenotypic outputs are comparable.
3.1. Self-organizing microtissues
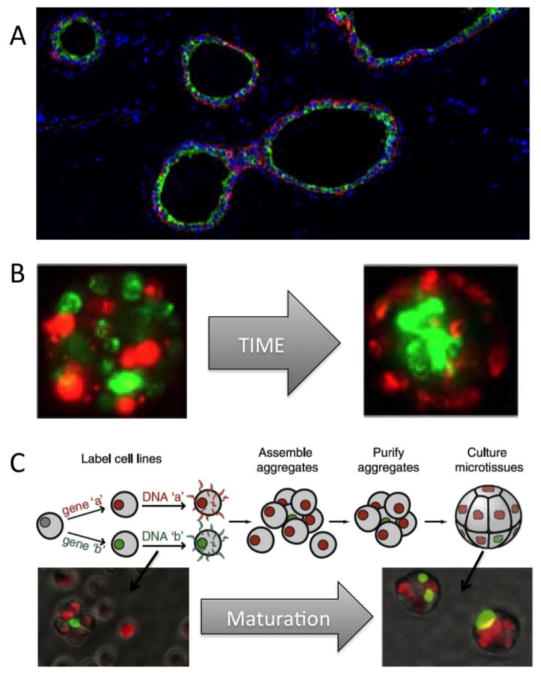
In some cases, mixtures of cell types have the ability to organize into higher-order structures based on differential cell-cell and cell-ECM adhesion [46, 47]. Experiments to understand self-organizing behavior of cells are often performed in hanging droplets or upon agarose plates, which are challenging imaging environments where there is poor control of size or final morphology of the structures, and the microenvironments may be more representative of those experienced by pond dwellers rather than human tissues. Using silicon wafers to micropattern arrays of cylindrical microwells in PDMS, it was possible to control the ultimate geometry of self-organized bilayered structures generated from differentiated lineages of human mammary epithelial cells (Fig 3B)[48]. Whereas this micropatterned approach was amenable to high-throughput imaging, and to quantifying changes in higher-order tissue structure following exposure to different blocking antibodies and small molecule inhibitors, it still lacked precision control at the single cell length-scale.
Figure 3.
Using microtissues to evaluate emergent properties of higher order tissue organization. Tissues have distinctive architecture, which we usually lose in tissue culture dishes. (A) A cross section of a normal human mammary gland shows many ducts with green keratin 19 expressing luminal epithelial cells surrounded by red keratin 14 myoepithelial cells. (B) Primary human mammary epithelial cells possess the ability to self-organize in rudimentary structures, with luminal cells surrounded by myoepithelial cells, over time with confined in a polymer cylinder (adapted from [48]). (C) Defined microtissues formed through a process of ssDNA-guided assembly. In this case cellular stoichiometry was controlled so that one green cell would be surrounded by about 6 red cells (adapted from [53]).
3.2. Assembled microtissues
DNA-programmed assembly is a recently developed technology to direct organization of multicellular “microtissues” with single cell resolution [49]. In this approach, the cohesive properties of cells are controlled through modification of the cell surface with single-stranded DNA (ssDNA) (Fig 3C). There are multiple methods by which cell surfaces have been modified with ssDNA. Cells can be fed azide-modified monosaccharides that get incorporated into cell surface glycans, after which ssDNA is covalently attached to azides by Staudinger reactions or [1,3]-dipolar cycloaddition [49, 50]. N-hydroxysuccinimide-modified ssDNA can be reacted directly with free lysines [51], with lipid-ssDNA conjugates anchoring spontaneously in cell plasma membranes [52]. A number of cell types ranging from lymphocytes to epithelial cells were shown to stay viable through these labeling procedures. Specific multicellular arrangements are achieved by mixing together cells coated with complementary ssDNA, an approach has worked with up to three different cell types. In the case of epithelial cell lines, after the initial ssDNA-directed organization specified the initial location of every cell, the ssDNA was removed through as-yet unknown means, and cadherin, tight, and hemidesmosomal junctions formed [53]. Therefore, once specifying the microtissues’ initial composition and architecture, the novel group of cells will then dynamically maintain and change organization, based upon the first principles of differential cell-cell and cell-ECM interactions.
Using DNA-programmed assembly to generate tissue-level asymmetry in RAS oncoprotein activity demonstrated that emergent properties of tissues only arise in the presence of cell-cell variability [53]. It was only when a single cell of the microtissue, composed of MCF10A, expressed active RAS that they would exhibit extensive extrusion into Matrigel. Thus crucial functional phenotypes may well be obscured in uniform fields of cells cultured on tissue culture plastic. By microprinting the complementary ssDNA in regular arrays on microscope slides, this method combined with quantitative imaging was used to show that membrane dynamics in a non-adherent leukocyte cell line were altered in the presence of different drugs [52]. DNA-programmed microtissues and cell arrays present a new opportunity to precisely control cellular interconnectivity to better understand the role of the cellular niche in drug responses.
4. Measuring drug responses in heterogeneous cancer cell systems
Measuring the effect of a drug on tumor cells within a heterogeneous in vitro microenvironment generally entails high-resolution measurement technologies that capture the complexity of the system. Numerous aspects of cell physiology related to the desired phenotype can be monitored, varying from inhibition of specific target protein activity to induction of cell death. In all cases, the selected parameters of interest must be quantifiable, to enable calculation of meaningful drug IC50 values and to allow for higher-order computation. Early in vitro cellular assays developed to screen for new anti-cancer agents measured tumor cell death in monoculture. This simple binary readout could be monitored using a variety of biochemical or microscopy-based techniques, and data analysis was straightforward. However, due to the necessity to move beyond such simple “average cell” measurements, multiparametric measurement of spatiotemporal events in heterologous in vitro cell systems has gained favor, facilitated by a rapidly expanding spectrum of biological probes in concert with advances in microscopy and spectrometry. These high-content imaging systems (HCS) generate an unprecedented depth of information at the single cell level, creating challenges for data handling and interpretation. For example, simultaneous measurement of signal transduction events via immunofluorescent detection of post-translational modifications of proteins combined with time-dependent morphological changes reflective of cellular function can be used to inform a compound’s mechanism of action [54]. Contemporary organotypic tumor culture systems comprising multiple cell types necessitate multiparametric measurement to capture the complexity inherent in these systems.
4.1. High content screening in context
The mainstay technology for measuring drug phenotypes in complex in vitro cell systems is fluorescence microscopy. Modern fluorescence microscope systems provide a high degree of flexibility and can be readily integrated into high-throughput screening systems to provide single cell functional and morphometric information [36]. This unique feature is particularly relevant when a contextual variance of the phenotype is expected, for example, when only a subset of cells in the system exhibit the phenotype, such as heterogeneous cell cultures that model a metastatic niche [45]. In a recent study we used automated live-cell imaging analysis of temporally regulated microenvironments, to quantify the contextual activity of small compound inhibitors and conduct structure-activity relationship analysis [6]. Numerous algorithms have been developed that automatically quantify information from microscopy images. Acquisition of quantitative information, such as number, intensity, size, morphology, texture, and spatial distribution of objects is used for computational analysis of drug effects. Indeed, IC50 values for specific small-molecule inhibitors from biochemical assays correspond with IC50 values obtained using cell-based HCS [55]. Computational approaches allow in-depth drug profiling by multiparametric imaging, which can be used to derive mechanism-of-action information [54].
3D cell culture systems comprising primary human cells have been adapted for high-throughput imaging-based compound screening. Common among these approaches is the detection of a fluorescent marker that reports on a feature of cell physiology ranging from a specific signal transduction event to changes in cell morphology. Hence, a prerequisite for using these screening systems is the availability of appropriate fluorescence probes. A plethora of fluorescence-based probes are available that facilitate multiparametric measurement of spatiotemporal phenotypic changes (reviewed in [36]). Limitations to these fluorescence microscopy-based systems are mainly related to the necessity for an invasive staining step or expression of a fluorescent protein; variable fluorophore photostability and phototoxicity; and signal detection at deeper layers of 3D systems. These limitations are at least partially addressed by high-speed multiphoton microscopy [56, 57]. The use of longer-wave excitation greatly reduces photobleaching and allows imaging of live cells hundreds of microns deep within thick, strongly scattering samples.
4.2. Label-free imaging modalities
Label-free noninvasive imaging techniques based on Raman spectrometry are particularly promising alternatives to measure biochemical changes in complex cellular systems. Raman spectroscopy spectra span a broad spectrum of cellular biomolecules and metabolites, providing a ‘biochemical fingerprint’ of the focal field at high spatial resolution. Raman spectra are sensitive to small biochemical changes and acquisition of high-resolution Raman spectra have been used to distinguish between normal and transformed cells, and measure cell cycle, cell death, cell differentiation using computational analysis algorithms (reviewed in [58]. Comparison of confocal Raman spectroscopy with immunofluorescence demonstrated the applicability for comprehensive label-free, functional assessment of live cells [59]. Raman spectroscopy was also used for in vitro monitoring of extracellular matrix (ECM) formation in a 3D culture system [60]. An alternative to Raman is quantitative phase microscopy, which enables real time and label-free quantification of mass transport in living cells, and assessment of 3-D viscoelastic properties of living cells [61, 62]. Moreover, this technique can be combined with fluorescence to maximize information yield. Collectively these advances exemplify the possibility to adopt Raman spectrometry approaches to assess contextual drug responses in mimetic microenvironments.
5. Recapitulating tissue in silico from functional responses in combinatorial microenvironments
Multiparameteric analysis of combinatorial in vitro microenvironments generate large amounts of functional data that must be coupled to specific cell types, microenvironments, and drug responses. There are huge repositories of gene expression data from the cell lines that are commonly used for drug discovery, and those base-line gene expression patterns should serve as a guide to predict how a cell might respond to a given microenvironment. As a result, there are a significant number of new computational opportunities to derive in silico models. Beyond classical bi-clustering strategies that group microenvironmental conditions with the phenotypic responses to infer dependencies, new approaches can be explored for developing improved predictive models through inclusion of (i) advanced regression models, (ii) detailed cellular profiling, and (iii) chemoinformatics analysis. These predictive models can be represented as high-dimensional input/output functions with low sample size requiring careful experimental design that incorporates sufficient genetic diversity for constructing stable computational models.
5.1. Linking genotype with microenvironment-imposed responses to drugs
Although various clustering methods can be used to partition cellular responses into categories, it is also feasible to design experiments that elucidate or hypothesize common regulatory mechanisms. Cellular responses are a function of the microenvironment, cell phenotype, and genotype, and therapeutic targets. Thus, one could hypothesize a common mechanism-of-action for different drugs that elicit similar responses under identical microenvironmental conditions. The availability of transcriptome data allows the construction of elaborate correlative matrices. As an example of how this might be achieved, let transcriptome data be represented as X0 ∈ RC*N, where C is the number of cells, and N is the size of the transcriptome. In particular, the number of cells, C, is selected for their genetic diversity for improved robustness in constructing stable computational predictors. Let D be the number of therapeutic targets, and M be the readout of phenotypic responses. As a result, the problem can be reduced to estimating a regression matrix Td of size N-by-D-by-M, representing the cellular processes affected by a drug treatment. Here, the regression matrix Td can be decomposed into two matrices of TPd, where T is a shared subspace (e.g., a lower dimensional space computed through linear operations) and Pd is determined by specific drugs. This is known as multitask regression, and it can be regularized further for improved stability, by forcing T to be sparse (e.g., most elements of T are zero) [63]. The net result is a subset of genes hypothesizing a common mechanism-of-action. Many variations of the same framework can be envisioned by directly encoding microenvironmental characteristics into the regression matrix.
5.2. Quantifying morphology as a microenvironmental response metric
Cell morphology can be an informative feature in the context of engineered matrices and cell-cell contact [64]. Multiparametric cellular profiling provides a representation of spatial organization and cellular response heterogeneity from single cell information [65, 66]. Response features may include morphometric indices such as cell shape, cell volume, patterns of chromatin organization, and membrane integrity. This cellular profile can be correlated with spatial organization in the context of each microenvironment. Computed indices are then compared between different microenvironments to identify subtypes. Alternatively one can incorporate cellular profile features in the regression matrix for inferring a regulatory network. In each case, the approach can facilitate a unique class of either knowledge organization (e.g., an atlas) or hypothesis generation, at a scale that can be easily disseminated for other investigators.
5.3. Predictive models built on chemoinformatic networks
Chemoinformatics analysis can facilitate the building of predictive models by linking structures to responses. Typically, this is performed by utilizing the Simplified Molecular-Input Line-Entry System (SMILES) code to generate physiochemical properties and/or structural dissimilarities between pairwise molecular graphs of therapeutic targets [67]. Measures for structural dissimilarities can range from simple distances that measures differences between two-dimensional graph structures, corresponding to therapeutic targets, to more elaborate distances that measure differences in three-dimensional graphs. A number of techniques have been proposed for measuring structural and physiochemical distances [68, 69] and some are supported by commercial software such as JChem. However, none of these techniques have yet been applied with the aim of constructing predictive models of microenvironmental drug responses.
6. Conclusions
The integration of new bioengineering approaches with technologies for multiparametric measurement cellular phenotypes to model diverse microenvironments is emerging as a powerful approach to reveal contextual drug responsiveness. Improved 3D cell models that derived from multiple cell types with precise spatial definition, and combinatorial MEArrays comprising hundreds of ECM components are serving to define specific microenvironmental features that determine drug effects. Successful application of bioengineering and cell biology principles has streamlined the processes necessary for MEArray fabrication. Currently the major crux of this technology is inadequate methods of analysis. The ultimate aim is to compare responses in the same cell types across numerous defined microenvironmental conditions, which differ iteratively by one component, in order to develop a complete picture of how molecular microenvironment components and the physical properties of elasticity and shape work together to elicit specific functional phenotypes. However there remains a significant gap between combinatorial array approaches and a comprehensive recapitulation of a tissue microenvironment. Designer microtissues can help fill that gap between because the cellular neighborhood can be precisely controlled without having absolute knowledge of all the microenvironmental components. Both self-organizing and DNA-directed assembly models offer means of fabricating multi-cell type tissue constructs, which can theoretically be constructed to look as we see them in tissue sections without having complete knowledge of the developmental process. These contextual drug development environments will require improvement and adaptation of HCS technologies in order to take full advantage of the emergent properties of cells in tissue contexts. Finally, new computational approaches that build a modular framework for complex queries of genomic data, cellular profiling, and chemical structures will allow exploration of the relative contributions of genomes and microenvironments in drug responses and other emergent phenotypes of high order tissue-level organization.
Acknowledgments
The authors are grateful for support from the National Institutes of Health (R01AG040081, R01CA140663), and from the US Department of Energy (DE-AC02-05CH11231).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mark A LaBarge, Email: MALabarge@lbl.gov.
Bahram Parvin, Email: B_Parvin@lbl.gov.
James B Lorens, Email: Jim.Lorens@biomed.uib.no.
REFERNCES
- 1.Arrowsmith J. Trial watch: Phase II failures: 2008–2010. Nature reviews Drug discovery. 2011;10:328–329. doi: 10.1038/nrd3439. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Cancer Fact Sheet. World Health Organization; 2013. [Google Scholar]
- 3.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 4.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghajar CM, Bissell MJ. Tumor engineering: the other face of tissue engineering. Tissue Eng Part A. 2010;16:2153–2156. doi: 10.1089/ten.tea.2010.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evensen L, Odlo K, Micklem DR, Littlewood-Evans A, Wood J, Kuzniewski C, Altmann K, Hansen T, Lorens JB. Contextual compound screening for improved therapeutic discovery. ChemBioChem. 2013 doi: 10.1002/cbic.201300409. in press. [DOI] [PubMed] [Google Scholar]
- 7.Gjerdrum C, Tiron C, Høiby T, Stefansson I, Haugen H, Sandal T, Collett T, Li S, McCormack E, Gjertsen BT, Micklem DR, Akslen LA, Glackin C, Lorens JB. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. PNAS. 2010 doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mora-Blanco EL, Lorens JB, Labarge MA. The tumor microenvironment as a transient niche: a modulator of epigenetic states and stem cell functions. In: Resende RR, Ulrich H, editors. Trends in Stem Cell Proliferationand Cancer Research. Springer Publishing; 2013. [Google Scholar]
- 9.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauschka TS, Levan A. Cytologic and functional characterization of single cell clones isolated from the Krebs-2 and Ehrlich ascites tumors. J Natl Cancer Inst. 1958;21:77–135. [PubMed] [Google Scholar]
- 12.LaBarge MA. The difficulty of targeting cancer stem cell niches. Clin Cancer Res. 2010;16:3121–3129. doi: 10.1158/1078-0432.CCR-09-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Koning JP, Mao JH, Balmain A. Novel approaches to identify low-penetrance cancer susceptibility genes using mouse models. Recent Results Cancer Res. 2003;163:19–27. doi: 10.1007/978-3-642-55647-0_3. discussion 264–266. [DOI] [PubMed] [Google Scholar]
- 14.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacorre DA, Baekkevold ES, Garrido I, Brandtzaeg P, Haraldsen G, Amalric F, Girard JP. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- 17.Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17:103–110. doi: 10.1007/s10911-012-9251-7. [DOI] [PubMed] [Google Scholar]
- 18.Nelson CM, Tien J. Microstructured extracellular matrices in tissue engineering and development. Current opinion in biotechnology. 2006;17:518–523. doi: 10.1016/j.copbio.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Liu JS, Gartner ZJ. Directing the assembly of spatially organized multicomponent tissues from the bottom up. Trends in cell biology. 2012;22:683–691. doi: 10.1016/j.tcb.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 21.Kenny PA, Bissell MJ. Tumor reversion: correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 23.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bissell MJ, Labarge MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 27.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer M, Petersen OW, Bissell MJ. Human mammary progenitor cell fate decsions are products of interactions with combinatorial microenvironments. Integrative Biology. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Molecular systems biology. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga T, Takemoto N, Sato T, Takimoto R, Tanaka I, Fujimi A, Akiyama T, Kuroda H, Kawano Y, Kobune M, Kato J, Hirayama Y, Sakamaki S, Kohda K, Miyake K, Niitsu Y. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat Med. 2003;9:1158–1165. doi: 10.1038/nm909. [DOI] [PubMed] [Google Scholar]
- 30.Dolberg DS, Bissell MJ. Inability of Rous sarcoma virus to cause sarcomas in the avian embryo. Nature. 1984;309:552–556. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 31.Dolberg DS, Hollingsworth R, Hertle M, Bissell MJ. Wounding and its role in RSV-mediated tumor formation. Science. 1985;230:676–678. doi: 10.1126/science.2996144. [DOI] [PubMed] [Google Scholar]
- 32.Dave B, Mittal V, Tan NM, Chang JC. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 2012;14:202. doi: 10.1186/bcr2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harma V, Virtanen J, Makela R, Happonen A, Mpindi JP, Knuuttila M, Kohonen P, Lotjonen J, Kallioniemi O, Nees M. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One. 2010;5:e10431. doi: 10.1371/journal.pone.0010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen KC, Kiemele L, Maller O, O’Brien J, Shankar A, Fornetti J, Schedin P. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol Cell Proteomics. 2009;8:1648–1657. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zanella F, Lorens JB, Link W. High content screening: seeing is believing. Trends in biotechnology. 2010;28:237–245. doi: 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Brafman DA, de Minicis S, Seki E, Shah KD, Teng D, Brenner D, Willert K, Chien S. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. Integr Biol (Camb) 2009;1:513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 38.Kuschel C, Steuer H, Maurer AN, Kanzok B, Stoop R, Angres B. Cell adhesion profiling using extracellular matrix protein microarrays. Bio Techniques. 2006;40:523–531. doi: 10.2144/000112134. [DOI] [PubMed] [Google Scholar]
- 39.Konagaya S, Kato K, Nakaji-Hirabayashi T, Arima Y, Iwata H. Array-based functional screening of growth factors toward optimizing neural stem cell microenvironments. Biomaterials. 2011;32:5015–5022. doi: 10.1016/j.biomaterials.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 40.Kim HN, Kang DH, Kim MS, Jiao A, Kim DH, Suh KY. Patterning Methods for Polymers in Cell and Tissue Engineering. Annals of biomedical engineering. 2012 doi: 10.1007/s10439-012-0510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brafman DA, Chien S, Willert K. Arrayed cellular microenvironments for identifying culture and differentiation conditions for stem, primary and rare cell populations. Nat Protoc. 2012;7:703–717. doi: 10.1038/nprot.2012.017. [DOI] [PubMed] [Google Scholar]
- 42.Motulsky H. Intuitive Biostatistics. 2. Oxford University Press; New York, Oxford: 2010. [Google Scholar]
- 43.Phadke MS. Quality engineering using robust design. Prentice Hall; Englewood Cliffs, N.J: 1989. [Google Scholar]
- 44.Rantala JK, Makela R, Aaltola AR, Laasola P, Mpindi JP, Nees M, Saviranta P, Kallioniemi O. A cell spot microarray method for production of high density siRNA transfection microarrays. BMC genomics. 2011;12:162. doi: 10.1186/1471-2164-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY, Chen EI, Lyden D, Bissell MJ. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 47.Steinberg MS, Takeichi M. Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chanson L, Brownfield D, Garbe JC, Kuhn I, Stampfer MR, Bissell MJ, Labarge MA. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1019556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc Natl Acad Sci U S A. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB. Programmable cell adhesion encoded by DNA hybridization. Angew Chem Int Ed Engl. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 51.Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, Bertozzi CR, Francis MB. Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir. 2009;25:6985–6991. doi: 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selden NS, Todhunter ME, Jee NY, Liu JS, Broaders KE, Gartner ZJ. Chemically programmed cell adhesion with membrane-anchored oligonucleotides. J Am Chem Soc. 2012;134:765–768. doi: 10.1021/ja2080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JS, Farlow JT, Paulson AK, Labarge MA, Gartner ZJ. Programmed Cell-to-Cell Variability in Ras Activity Triggers Emergent Behaviors during Mammary Epithelial Morphogenesis. Cell reports. 2012 doi: 10.1016/j.celrep.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8:567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 55.Zanella F, Rosado A, Garcia B, Carnero A, Link W. Chemical genetic analysis of FOXO nuclear-cytoplasmic shuttling by using image-based cell screening. Chembiochem. 2008;9:2229–2237. doi: 10.1002/cbic.200800255. [DOI] [PubMed] [Google Scholar]
- 56.Reddy GD, Saggau P. High-speed two-photon imaging. Cold Spring Harb Protoc. 2013;2013 doi: 10.1101/pdb.top072603. [DOI] [PubMed] [Google Scholar]
- 57.Kim KH, Ragan T, Previte MJ, Bahlmann K, Harley BA, Wiktor-Brown DM, Stitt MS, Hendricks CA, Almeida KH, Engelward BP, So PT. Three-dimensional tissue cytometer based on high-speed multiphoton microscopy. Cytometry A. 2007;71:991–1002. doi: 10.1002/cyto.a.20470. [DOI] [PubMed] [Google Scholar]
- 58.Brauchle E, Schenke-Layland K. Raman spectroscopy in biomedicine - non-invasive in vitro analysis of cells and extracellular matrix components in tissues. Biotechnol J. 2013;8:288–297. doi: 10.1002/biot.201200163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein K, Gigler AM, Aschenbrenner T, Monetti R, Bunk W, Jamitzky F, Morfill G, Stark RW, Schlegel J. Label-free live-cell imaging with confocal Raman microscopy. Biophys J. 2012;102:360–368. doi: 10.1016/j.bpj.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunstar A, Otto C, Karperien M, van Blitterswijk C, van Apeldoorn A. Raman microspectroscopy: a noninvasive analysis tool for monitoring of collagen-containing extracellular matrix formation in a medium-throughput culture system. Tissue Eng Part C Methods. 2011;17:737–744. doi: 10.1089/ten.TEC.2010.0574. [DOI] [PubMed] [Google Scholar]
- 61.Mir M, Wang Z, Shen Z, Bednarz M, Bashir R, Golding I, Prasanth SG, Popescu G. Optical measurement of cycle-dependent cell growth. Proc Natl Acad Sci U S A. 2011;108:13124–13129. doi: 10.1073/pnas.1100506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, Ding H, Mir M, Tangella K, Popescu G. Effective 3D viscoelasticity of red blood cells measured by diffraction phase microscopy. Biomedical optics express. 2011;2:485–490. doi: 10.1364/BOE.2.000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K, Gray J, Parvin B. Sparse multitask regression for identifying common mechanism of response to therapeutic targets. Bioinformatics (ISMB) 2010;26:97–105. doi: 10.1093/bioinformatics/btq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalby MJ, Childs S, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Fibroblast reaction to island topography: changes in cytoskeleton and morphology with time. Biomaterials. 2003;24:927–935. doi: 10.1016/s0142-9612(02)00427-1. [DOI] [PubMed] [Google Scholar]
- 65.Han J, Chang H, Andrarwewa K, Yaswen P, Barcellos-Hoff M, Parvin B. Multidimensional profiling of cell surface proteins and nuclear markers. IEEE Transactions on Computational Biology and Bioinformatics. 2010 doi: 10.1109/TCBB.2008.134. [DOI] [PubMed] [Google Scholar]
- 66.Bilgin C, Kim S, Leung E, Chang H, Parvin B. Integrated profiling of three dimensional cell culture models and 3D microscopy. Bioinformatics. doi: 10.1093/bioinformatics/btt535. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scior T, Benard P, Medina-Franco J, Maggiora G. Large compound databases for structure-activity relationships studies in drug discovery. Mini-Rev Med Chem. 2007;7:851–860. doi: 10.2174/138955707781387858. [DOI] [PubMed] [Google Scholar]
- 68.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genome to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita K, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new development in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]