Abstract
The assembly of amyloid β (Aβ) proteins into nanostructures is currently considered a major pathway of Alzheimer’s disease development, but the molecular mechanisms of this self-assembly process remains unclear. Recently, we showed that single-molecule AFM force spectroscopy (SMFS) is capable of probing the dynamics and interaction between Aβ40 peptides, and these studies allowed us to shed new light on transiently existing Aβ40 misfolding states. In this study, we applied the same SMFS approach to characterize the misfolding of Aβ42 peptide, the most toxic Aβ alloform. The quantitative analysis of SMFS data demonstrated that Aβ interaction leads to the formation of dimers with a lifetime in the range of a second. Interaction via C-terminal segments prevailed at pH 7, but interaction within the peptide center prevailed at acidic pH levels. The difference in the misfolding properties for Aβ40 and Aβ42 peptides and the mechanisms of amyloid nanoassembly are discussed.
Keywords: Amyloid beta, misfolding, single-molecule force spectroscopy, contour length, dimerization pathways
Aberrant folding, misfolding, and aggregation of amyloid proteins are pathological hallmarks of a large class of neurodegenerative diseases.1, 2 The assembly of amyloid β (Aβ) proteins in nanoaggeragtes is a ubiquitous phenomenon for a number of neurodegenerative disorders, including Alzheimer’s disease (AD) (reviewed in 3, 4). Aβ proteins containing 42 and 40 residues (Aβ42 and Aβ40, respectively) are the major species, and Aβ42, which differs from Aβ40 by two extra C terminal residues (I41 and A42), has an elevated aggregation propensity and neurotoxicity compared to Aβ40.5, 6 Traditional structural techniques, such as solid-state NMR, AFM, and electron microscopy, are widely used for their characterization of large aggregates, such as amyloid fibrils. However, these powerful techniques are not amenable to studies of oligomers that exist transiently and convert from one species to another, so novel approaches are needed for oligomers studies. The photocrosslinking method, developed for the stabilization of oligomers in Teplow lab 7 was instrumental in the spectroscopic characterization of Aβ42 oligomers 8. It was shown that the transition from the monomer to the dimer is rapid and then proceeds gradually for higher oligomers. The application of ionization mass spectrometry led to the conclusion that tetramers and hexamers of Aβ40 and Aβ42 were potential nuclei for peptide oligomerization, but they follow different assembly pathways. 9
We have developed a single-molecule AFM force spectroscopy (SMFS) approach to characterize the interactions of amyloidogenic proteins.10–13 In this approach, the protein is end-immobilized covalently on the AFM tip and the substrate, and the interaction between the proteins is the measure of their propensity to form dimers, the smallest nanoaggregate. The correlation of the interaction forces with the protein aggregation propensity validated the application of the SMFs methodology to the probing of protein transient misfolded states.13 The combination of this methodology with the dynamics force spectroscopy (DFS) approach of a number of amyloid proteins and peptides revealed that misfolded dimers are extremely stable compared to highly dynamic monomeric states of the same proteins.14 Specifically, the recent application of the developed approach to Aβ40 interaction showed that misfolded Aβ40 dimers have a lifetime as long as 0.1 s.10 This value is almost 106 times higher than the monomer dynamics leading us to hypothesize that dimerization Aβ40 is the means by which the misfolded state is stabilized.
Here, we have applied a similar approach to Aβ42 peptide, attempting to identify features of this protein upon its misfolding and dimerization processes. We found that Aβ42, similarly to Aβ40, forms stable dimers, but its lifetime is higher than that of Aβ40. The contour length analysis revealed a set of transients conformational of the dimers, but the patterns of their formation for Aβ40 and Aβ42 are different, suggesting that both peptides can form oligomers with different structures.
Materials and Methods
Cysteine modified Aβ42
The Aβ42 terminated with cysteine at N-terminus (AnaSpec, Inc., Fremont, CA) was dissolved in TFA (2 mg/ml) with sonication for 5 min to destroy the peptide oligomeric forms and then instantly dried under a vacuum (Vacufuge, Eppendorf). The white film of Aβ42 was dissolved in DMSO (2 mg/ml) as a stock solution and then diluted to 1 μM in DMSO before being used. The monomer state of Aβ42 diluted in different pH buffers from the stock solution was confirmed by SDS-page gel experiment (Fig. S1).
Tip and mica surface modification
The process for the cleaning and modification of the tip and mica surfaces was similar to a previously reported protocol.10, 12, 15 Briefly, the tips (MLCT, Veeco, Santa Barbara, CA) were cleaned by ethylene alcohol followed by UV irradiation for 30 min and were modified by maleimide polyethylene glycol silatrane (MAS). The freshly cleaved mica surface was modified by 1-(3-aminopropyl) silatrane (APS), after which N-hydroxysuccinimide-polyethylene glycol-maleimide (NHS-PEG-MAL) (MW = 3400 g/mol, Laysan Bio Incorporation, Arab, AL) was coupled to the amine group of APS; the reaction was performed in DMSO. The functionalized tip and mica were incubated with ~20 nM of Aβ42 diluted in HEPES buffer (pH 7) with TCEP (~100 nM) for 30 min and then mildly rinsed with HEPES buffer. The surface density of the Aβ42 peptide was estimated using α-Syn 10, 11 and the 42 base-pair of dsDNA (See SM, Fig. S2–4). The prepared tips and mica were kept in HEPES buffer within 24 hours.
Single-molecule AFM force spectroscopy
The single-molecule interactions of Aβ42 terminated with cysteine at N-terminus were measured using the AFM force spectroscopy method. The force–distance (F-D) curve of Aβ was measured at two different pH values (pH 2.7; 20 mM glycine-HCl, and pH 7; 20 mM HEPES) at room temperature using MFP 3D AFM (Asylum Research, Santa Barbara, CA). In both buffers, ionic strength was adjusted to 150 mM by addition of NaCl. The spring constant of the tips (MSNL, Veeco) was 70–120 pN/nm, which was calibrated by the thermal noise analysis method. The triggering contact force was in the range 100~150 pN. At high retracting velocities (1 μm/s and above), the tip remained at the surface for 0.3 s (dwell time) after making contact. The retracting velocity varied from 100 nm/s to 4 μm/s with 7~8 discrete steps, corresponding to an apparent loading rate (ALR) of 1,000~200,000 pN/s. The F-D curves were collected more than 2,000 times for each retracting velocity. The selected F-D curves were analyzed using the worm-like chain (WLC) model with Igor Pro 6.01 software. The persistence length was an adjustable parameter with mean values (± SD) of 0.14 (±0.07) nm and 0.2 (±0.1) nm at pH 7 and 2.7, respectively (Fig. S5). The probability density function (p(F)) was used for fitting the histogram to find the most probable rupture force 10, 11:
where koff(0) is the off-rate constant of the complex at zero force, xβ is the distance of the transition state to be bound state, kB is Boltzman constant, and T is the absolute temperature. The r is the loading rate (dF/dt) given by the formula:
Where, , kc is the spring constant (pN/nm), v is the tip velocity (nm/s), Lc and Lp are the parameters from the worm-like chain model (WLC) fitting, Lc is the contour length (nm) and Lp is the persistence length (nm). F is rupture force (pN) and r is the apparent loading rate (pN/s).
A contour length analysis was also performed with a protocol described in previous publications.10 The distribution shape of the contour length in the histograms was approximated by four Gaussian peaks. All Gaussian peaks had similar full-width at half-maximum (4.5–5 nm) values, and each Gaussian curve was integrated to calculate the area under the curve for the estimation of the each conformer population. The correlation between rupture force and retraction velocity was revealed by the DFS methods.14 Based on the parameters of the DFS result, the energy landscape along the reaction coordinate was constructed.16
RESULTS
1. Interactions between Aβ42 peptides
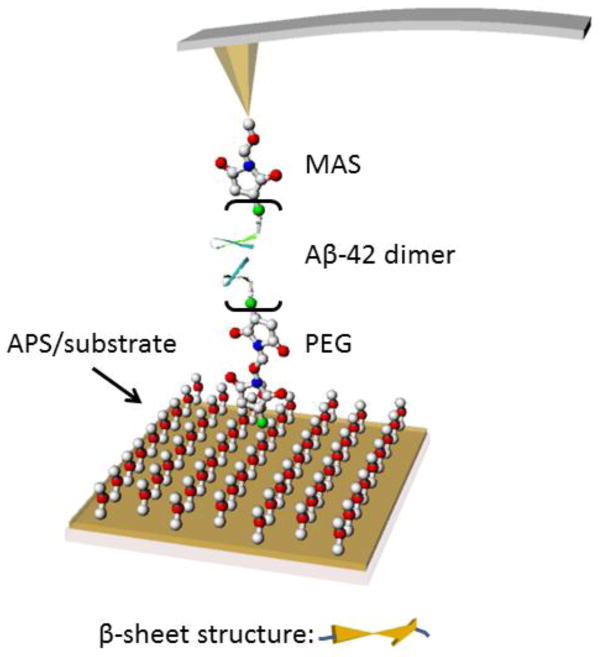
Figure 1 schematically illustrates the experimental approach. The peptide with N-terminated Cys was immobilized on the surface and the tip via tethers (MAS and PEG respectively) through coupling of thiol groups of Cys to maleimide termini of the tethers.17 The use of N-termini for immobilization is justified by the NMR data of Aβ42 oligomers demonstrating that the N-terminal segment as opposed to the C-terminus is not involved in the assembly of peptides into oligomers.5 The interactions between the immobilized Aβ42 peptides were measured by multiple approach-retraction cycles of the AFM tip. The buffer condition of measurement was pH 7 and 2.7. Acidic pH was used, as this condition is widely used for in vitro Aβ aggregation.18–20
Figure 1.
Schematic of the experimental setup. The peptide was covalently attached at the Cys terminus at the N-end to the functionalized AFM tip and the substrate surfaces via PEG tethers. As the AFM tip approaches the mica surface, two monomers interact with each other forming a misfolded dimer.
Fig. 2A shows a representative force-distance (F-D) curve measured from the interaction between Aβ42 at pH 7. The main signature of this curve is the appearance of the rupture peak located at a certain distance indicated by the arrow on the F-D curve. The approximated contour length and rupture force with the WLC model are ~58 nm and ~83 pN, respectively. Multiple measurements over several hundred rupture events for pH 7 and 2.7 collected from independent experiments with different batches of tip and substrate produced the histograms shown in Figs. 2B and C. The most probable rupture force (Fr) at a constant retracting velocity corresponding to the apparent loading rate (ALR) ~4000 pN/s was calculated by approximating the histograms with a probability density function. 10, 11 The Fr values measured at pH 7 and 2.7 were 81 (±3) pN and 82 (±0.6) pN, respectively. Typical force curves illustrating the variability of the rupture forces depending on the loading rate as shown in Fig. S6. In the control experiment shown in Fig. S7, non-specific rupture forces were distributed in a broad range (20 pN~1.3 nN), characterized by the rupture lengths less than 30 nm with a yield < 1%. The yield of specific events was ~5%.
Figure 2.
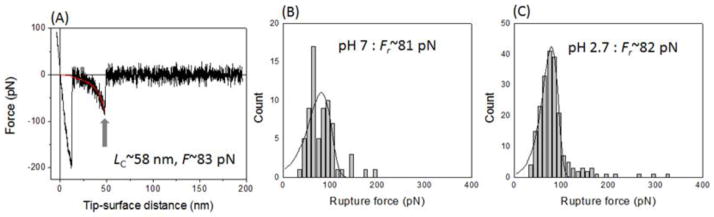
A representative force-distance curve and histograms for the rupture force distribution: (A) A typical force-distance curve measured at pH 7 with ALR~4,000 pN/s. Rupture peak fitted by the WLC model shows a contour length of ~58 nm and a rupture force of ~83 pN. (B and C) Histograms of the multiple measured rupture forces at pH 7 (B) and 2.7 (C) with ALR lower than 20,000 pN/s. Each histogram was fitted by probability density function, and the estimated most probable rupture force was ~81 (±3) pN (C) and ~82 (±0.6) pN (D), respectively.
2. Dynamic force spectroscopy
Dynamic force spectroscopy (DFS) analysis was performed in a pulling rate range of 0.1 μm/s and 4 μm/s much lower speed than ~5×104 μm/s at which hydrodynamic interactions possibly start to affect the rupture forces of proteins.21 Note that the tip was kept at the surface for 0.3 s (dwell time) when the pulling speed was over 1 μm/s. Implementation of dwell time provided the protein with sufficient time for multiple refolding and minimized the fluidic flow effect. 21, 22 Typically, this methodology led to the yield of the rupture events of ~5 % enabling us to obtain sufficient statistics for the data analysis.
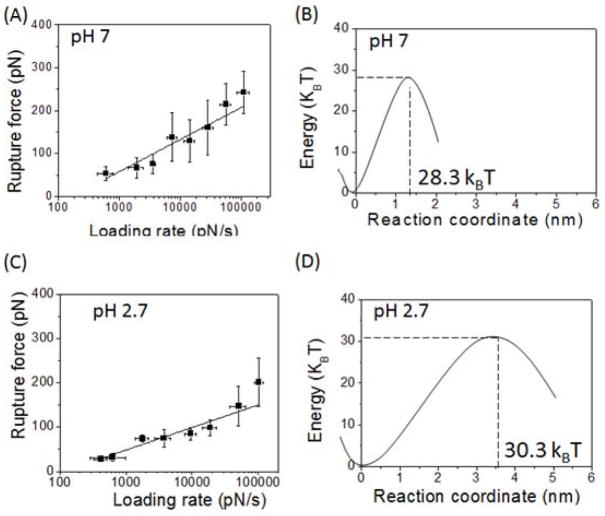
The Fr values obtained for various pulling rates were plotted against the logarithm of the apparent loading rate (ALR), and the Bell-Evans model was used for an analysis. 14 Fig. 3A shows a plot of Fr vs. ALR measured at pH 7. The data set was approximated by a single linear line indicating to the one-barrier landscape for the Aβ42 dissociation. Two major parameters, the intercept and the slope, were used to obtain the position of the barrier (xβ) relative to the stable state and off-rate constant for the dimer dissociation (koff), respectively. The barrier height (ΔG) in the energy landscape and the dimer lifetime were calculated by using the koff values.14 The ΔG and lifetime for the dissociation of the Aβ42 dimer at pH 7 were ~28.3 kBT and 0.46 (±0.25) s, respectively. Using the values of ΔG and xβ, the energy landscape profile for the dimer dissociation was reconstructed as shown in Fig. 3B. A similar DFS plot for the data obtained at pH 2.7 is also represented by a single line (Fig. 3C). The corresponding energy landscape profiles are shown in Fig. 3D. The lifetime and xβ are 2.5 (±0.7) s and 1.3 (±0.5) Å, respectively. The major difference between the data for pH 2.7 and that for pH 7 is about a 5-fold increase of the lifetime, which can be responsible for the aggregation acceleration at acidic pH levels.18–20
Figure 3.
Semi-logarithmic plots of the rupture force (Fr) vs. apparent loading rate (ALR; A and C) and the corresponding energy landscape profiles (B and D). The data sets of Fr vs. ALR are fitted with a linear plot according to the Bell-Evans model.35 The parameters extracted from these plots for the generation of the energy landscapes (B and D) are shown in Table 1.
3. Patterns of interpeptide interactions in Aβ42 dimers
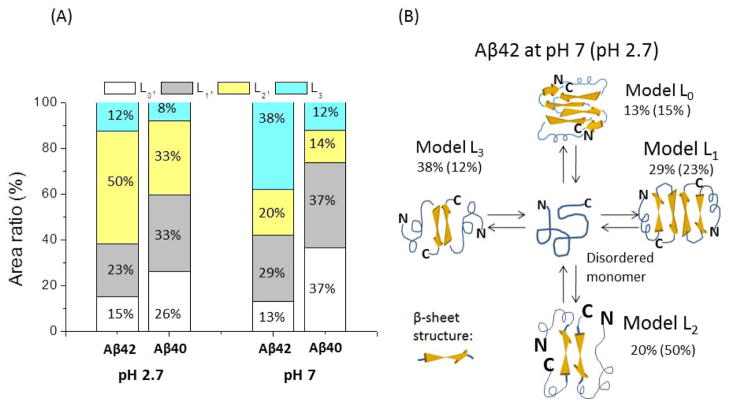
The force curves in the AFM force spectroscopy experiments provide another important characteristic for peptide structure within the dimer: the contour length (Lc). According to Fig. 1, the contour length primarily comprises the length of the flexible linker and the N-terminal segments of the peptide not involved in dimer formation. Therefore, subtracting the lengths of the linkers from the experimentally measured contour length yields the length of the N-terminal segments preceding the structured segments of Aβ42 involved in the interpeptide interaction. This approach was verified in our recent work on misfolded Aβ40 and α-synuclein.10, 12, 23 However, the rupture position in different force curves varies. This is illustrated by Fig. 4A, in which four different force curves obtained with similar ALR are shown. They have very close rupture forces (150±20 pN) but different rupture positions between ~30 nm (L0) and ~50 nm (L3) (Fig. 4A). Figures 4B and C show histograms of hundreds of contour lengths with relatively lower ALR between 100 pN/s and 20,000 pN/s measured at pH 7 and 2.7. The contour lengths varied between 28 nm and 62 nm. The histograms at both pH levels showed broadly distributed contour lengths with different overall profiles. At pH 7 (Fig. 4B), the most probable lengths (L3) are located at the right of the distribution, but these events localized around the center of the distribution for pH 2.7 (Fig. 4C). The variability of the contour lengths could be attributed to the polydispersity of the PEG linkers and the dependence of the contour lengths on the rupture force. However, the analysis shown in supplemental Fig. S8 demonstrates that these contributions are considerably less than contour length variability observed in the experiments (see also ref. 10).
Figure 4.
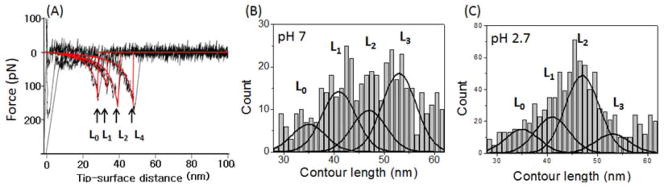
Representative rupture force curves with various contour lengths and histograms of the contour length. (A) The representative rupture force curves with various rupture lengths obtained at pH 7. The force curves were obtained in the same pulling regime (~20,000 pN/s) and have rupture forces of ~150 pN. (B and C) Histograms for the contour length distribution measured at pH 7 (B) and 2.7 (C). The overall profile of each histogram was approximated by four Gaussians with centers located around 35 nm (L0), 41 nm (L1), 47 nm (L2), and 53 nm (L3).
DISCUSSION
The strength of the interpeptide interaction within Aβ42 dimers
The rupture force of Aβ42 dimers was in the range of 80 ~120 pN at a loading rate of 4~20 nN/s (200 nm/s ~ 1 μm/s) at pH 7. This value is comparable to the unfolding force of titin Ig domains (170~180 pN) which mainly consists of β-sheet structures 25, suggesting that β sheets of shorter sizes are formed in Aβ42 monomers and the interactions between them stabilized the dimer. This conclusion is consistent with our very recent MD simulations for Aβ13-23 peptide showing that antiparallel β-sheet structure stabilized the peptide.26 Interestingly, considerably less force (~60 pN) is applied for unfolding of the α-helical structure of T4 lysozyme27 suggesting that Aβ42 dimers are more stable than dimers formed by misfolded α-helical structures of T4 lysozyme. The rupture forces for Aβ40 dimers are quite similar to those for Aβ42, indicating that the forces stabilizing both types of peptides are rather close. However, some difference exists, and other characteristics of SMFS as described below provide the explanation.
Dynamics of misfolded dimers
Although rupture forces characterize directly the interpeptide interactions, DFS provide such an important property to the dimers as the stability defined by the lifetimes. At a neutral pH level, this value is 0.46 (±0.25) s. Compared to the monomer’s time scale (10−6 – 10−9 s) 28, the dimers of Aβ42 show extremely high stability, suggesting that the dimerization of the peptide is the mechanism by which the misfolded state of Aβ42 acquires advantages compared to other conformations that may not be disease-prone. The Aβ40 peptide, similarly to Aβ42 peptide, also forms stable dimmers, but its lifetime is four times shorter.10 The dimers are the smallest oligomers of Aβ peptides that are further involved in aggregation, so the elevated stability of Aβ42 dimers gives the advantage of these species compared to those for Aβ40 peptides in the formation of oligomers and other species. The aggregation process of Aβ peptides accelerates at acidic pH levels 18–20, and the elevated stability of Aβ42 dimers at pH 2.7 measured in this paper (τ=2.5±0.7 s) is fully in line with this observations. Note that a similar high stability of dimers was observed for another amyloid protein, α-synuclein 12 and the peptide of Sup35 prion 29, lending further support to our hypothesis on the critical role of dimers in the amyloid aggregation phenomenon 14.
Structures of misfolded Aβ dimers
Probing interactions of Aβ42 within a dimer by SMFS provides a pattern of peptide interactions within the dimers. According to the immobilization procedure (Fig. 1), the peptide is covalently bound at its N-terminal cysteine. The experimentally measured contour length consists of two major components, the lengths of the flexible tethers used for peptide immobilization and the length of the peptide segment between the N-terminus and the segment of the peptide involved in interpeptide interactions. Therefore, the position of the peak on the force distance curve is defined by the peptide length between the N-terminus and the interacting segment. If Aβ peptide adopts various conformations in these transient states, the positions of interacting segments, and hence the length of the nonstructural segments, will vary. The closer the structured segment is to the N-terminus, the shorter the contour length will be when measured by AFM. The variable contour length measurements for Aβ42 at both pH values are broad (Fig. 4) suggesting that the monomers in the dimer adopt various conformations, but the patterns are pH dependent. We propose the following model explaining the Aβ42 folding in dimers shown schematically in Fig. 5.
Figure 5.
Schematics for dimer structures. (A) Models for Aβ42 dimers. The monomer in intrinsically disordered conformation is shown in the middle. Four different folding models corresponding to specific contour lengths are shown. The populations of each conformer calculated from histograms in Fig. 4 are indicated. The data for pH 2.7 are shown in parentheses. (B) The stacked histograms for the populations of each conformer for Aβ42 dimers are shown on the left. The right histogram is taken from ref. 10.
The initially structurally disordered Aβ42 peptides (scheme in the middle) at pH 7 assemble into a dimer that is stabilized primarily by C-terminal interaction (38%; model L3) or middle segments (29%; model L1) rupture, which leads to contour lengths L3 and L1, respectively. Based on our force values estimates, we hypothesize that these are interactions between the β sheets indicated in the schemes with arrows. The interactions very close to N-termini and in the middle of the peptide are minor (13% for model L0 and 20% for model L2). Similar folding models can be found at the acidic pH of 2.7, but the distributions of the conformers’ populations are very different. The primary folding is model L2, in which the dimers are stabilized by the interaction of the central regions of the peptide. There is a three-fold drop in the interaction between the C-termini (model L3) and essentially the same populations of models L0 and L1. Given the difference in the fibrillization kinetics of Aβ42 at acidic and neutral pH levels, we speculate that conformers stabilized by the interpeptide interactions within the central and C-terminal segments of Aβ42 (Model L2 and L3 in Fig. 5B) are primarily involved in the formation of fibrils.
It is instructive to compare the folding models of Aβ42 and Aβ40 presented in our previous paper 10, which also showed broad distributions for the contour lengths. The results are shown schematically in Fig. 5A as histograms, in which the populations of each conformer are shown in different colors. The most dramatic differences were observed at pH 7. If the interpeptide interactions for Aβ42 primarily involve C-terminal segments, the major interactions (L0 and L1; 74% in Fig. 5A) for Aβ40 occur within the N-terminal segment corresponding to Model L0 and L1 in Fig. 5B. This is a surprising observation as Aβ42 differs from Aβ40 by two extra peptides at C terminus. Therefore, these findings suggest that misfolding conformations of the peptides involve long-range interactions within the peptide chain, so the extra two residues at the C-terminus of Aβ42 change the peptide folding in such a way that interactions of the N-termini for Aβ40 are replaced by the C-termini interactions for Aβ42. Given the difference in neurotoxic activities of the oligomers formed by these peptides, we speculate that the conformations involving N-terminal interactions for Aβ40 lead to less toxic oligomers compared assembly which involves the C-terminal segments of the Aβ42 peptide.
Structure and aggregation propensity of Aβ dimers
As we described above, the common feature of Aβ peptides is their ability to form different conformers and different conformer occurrence frequency distributions. However, the structures of the amyloid fibrils formed by Aβ40 and Aβ42 peptides have similar features. Solid-state NMR studies of Aβ40, combined with X-ray fiber diffraction analyses, and electron microscopy have produced a model in which the first ~10 residues of the peptide are structurally disordered in the fibril.30 Residues 12–24 and 30–40 adopt β-strand conformations and form parallel β-sheets through intermolecular hydrogen bonding. Similarly, Aβ42 fibrils are also stabilized by the in-register β-strand arrangement of residues 18–42, with residues 1–17 being disordered.31 The common feature of these structures is an unstructured N-terminal segment of the peptide. However, the conformers with interactions between the N-termini are highly represented in Aβ40 dimers. Moreover, the conformations of this peptide that could give rise to fibrils are minor, thus explaining the low aggregation propensity of the Aβ40 peptide.
There are two primary models for amyloid fibril formation.5, 9, 14, 32 In one model, the fibrils are formed from oligomers after their conformational transition to the fibril-prone conformation. In the second model, oligomers of different types are formed, and the fibrils are formed from those that have the necessary conformation. Our data (Fig. 4) show that dimers with different conformations are formed, and those with the fibril-type conformation are present in the pool. Therefore, our combined data supports the second model. Note that the dimers are dynamic. The most stable of these are the conformers at acidic pH values with a lifetime of 2.5 s, suggesting that conformational change at the dimer level can occur. However, compared to dimer dynamics, the conformational dynamics of monomers are significantly faster and take place at the nanosecond time-scale.
The major effect of the contour length measurements is that the profile is not smooth and characterized by a set peak on the histograms that can be seen without any additional analysis. Moreover, the contour length profile changes with the pH (Fig. 4 B, C). This effect reflects the pattern of interactions within the dimer and we used fitting of the overall contour lengths distribution profiles with a set of peaks to facilitate the data interpretation. The positions of Gaussians were selected by the positions of peaks on the histograms. The correlation of this position with available structural properties of amyloid fibrils led us to the conclusion that observed interactions within dimers are due to the formation of the beta-sheet structures. This assumption is confirmed by our recent MD simulation results performed for 14–23 segments of Aβ42 peptide.33
It is instructive to evaluate the effect of the position of the molecule on the tip on the contour length value as farther the molecule from the apex, the smaller the contour length value. The effect is stronger for sharp tips compared to blunt ones. Note however, that the molecule position factor lead to a smooth distribution of the contour lengths and did not produce nonuniform profiles observed in the experiment. The estimates of potential effects of non-apical peptide positions on the contour length distributions can be obtained from the data in Fig. 4. The distribution is asymmetric with the maximum shifted to large contour lengths ~ 55 nm for histogram (B). If the sample coats the tip uniformly with the same density, the contribution of non-apical location should be larger than for the apical one as the number of molecules along the perimeter increases with the height, so the overall distribution should be shifted to short values rather then large ones as observed in the experiment. Additionally, the population of the short-range events (Lo) is very low compared to longer values with almost no events with L values below 30 nm. This value corresponds to the overall length of tethers, so one should be able to expect events below 30 nm if they appear from non-apical locations of the peptides. Thus this analysis suggests that an apical location is preferential in these probing experiments. A plausible explanation for this finding is that the density of the peptide coverage is very low, so non-apically located peptides are not able to interact efficiently with the counterpart on the substrate. This conclusion is confirmed by our very recent results obtained with DNA pulling experiments in which fully homogeneous PA tether was used 24. The experiments produced the contour length distribution as narrow as 3 nm. The contribution from non-apical location of the DNA molecules would broaden the contour length distribution well beyond 3 nm. Therefore, we attribute the variability of contour lengths to the ability of Aβ42 to adopt different conformations in the dimers.
Given the long lifetimes of dimers, we hypothesize that the dimers can be building blocks for amyloid aggregation. Indeed the analysis of the kinetics of Aβ42 aggregation revealed the pathway by which oligomers with even numbers are formed.9 We also cannot exclude that trimers and the higher-order oligomers can be even more stable than dimers, so the association of small oligomers into larger ones can be another pathway for the aggregation process. However, the high stability even dimers is the most important property, suggesting that careful attention is needed in the development of new approaches to the prevention and treatment of AD, PD and other neurodegenerative protein misfolding diseases. This assumption is also supported by the findings that Aβ42 dimers isolated from AD brains are stable and neurotoxic.34
Conclusions
A number of novel features of the misfolding and self-assembly process of Aβ42 peptides were determined using single-molecule force spectroscopy. The dynamic force spectroscopy analysis revealed strong interpeptide interactions for Aβ42 with elevated stability at acidic pH levels. A complementary single-molecule force spectroscopy approach, in which contour lengths were analyzed, revealed an unexpected long-range interaction effect in Aβ42 misfolding. The comparison of dissociation patterns of Aβ42 and Aβ40 peptides revealed differences in the misfolding pathways of these two amyloid species, suggesting that treatment research can be focused on preventing the disease-prone misfolding pathways. This is a novel direction for the development of treatments for AD with the prospect for novel diagnostic and prevention means. Progress in these areas requires techniques capable of analyzing the transient species present during the processes of Aβ peptide misfolding, interaction and self-assembly, as exemplified by the use of AFM force spectroscopy.
Supplementary Material
Table 1.
The DFS parameters for the force spectroscopy data calculated from the DFS plot in Fig. 3A. Here, xβ and koff are the potential barrier location and dissociation ratio, respectively. The lifetime was calculated by k−1off, and the standard deviation (SD) was calculated from five independent experiments for pH 7 and pH 2.7.
| pH | xβ (Å) (±SD) | Koff (S−1) (±SD) | Lifetime (s) |
|---|---|---|---|
| 7 | 1.3 (±0.2) | 3.1 (±1.7) | 0.46 (±0.25) |
| 2.7 | 3.6 (±0.5) | 0.43 (±0.12) | 2.5 (±0.7) |
Acknowledgments
The work was supported by grants to Y.L.L from National Institutes of Health (5 R01 GM096039-2), the U.S. Department of Energy Grant DE-FG02-08ER64579, National Science Foundation (EPS–1004094) and partially by the grant to B. H. K. from the KUSTAR-KAIST Institute, Korea, under the R&D program supervised by the KAIST.
Appendix A. Supplementary data
Supplementary data to this article can be found online
Footnotes
No conflict of interest was reported by the authors of this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, et al. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17(5):561–7. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–42. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitan G, Teplow DB. Rapid photochemical cross-linking--a new tool for studies of metastable, amyloidogenic protein assemblies. Acc Chem Res. 2004;37(6):357–64. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 8.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci U S A. 2009;106(35):14745–50. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nature Chemistry. 2009;1:326–31. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JF, Lyubchenko YL. Single-Molecule Atomic Force Microscopy Force Spectroscopy Study of Abeta-40 Interactions. Biochemistry. 2011 doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Warnke J, Lyubchenko YL. Nanoprobing of alpha-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine. 2010 doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Malkova S, Lyubchenko YL. alpha-Synuclein misfolding: single molecule AFM force spectroscopy study. J Mol Biol. 2008;384(4):992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 13.McAllister C, Karymov MA, Kawano Y, Lushnikov AY, Mikheikin A, Uversky VN, et al. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354(5):1028–42. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Lyubchenko YL, Kim BH, Krasnoslobodtsev AV, Yu J. Nanoimaging for protein misfolding diseases. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2(5):526–43. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Lyubchenko YL. Early stages for Parkinson’s development: alpha-synuclein misfolding and aggregation. J Neuroimmune Pharmacol. 2009;4(1):10–6. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 16.Tinoco I, Jr, Bustamante C. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys Chem. 2002:101–102. 513–33. doi: 10.1016/s0301-4622(02)00177-1. [DOI] [PubMed] [Google Scholar]
- 17.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zaikova TO, Keana JF, Lyubchenko YL. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1(4):300–5. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valerio M, Porcelli F, Zbilut JP, Giuliani A, Manetti C, Conti F. pH effects on the conformational preferences of amyloid beta-peptide (1-40) in HFIP aqueous solution by NMR spectroscopy. Chem Med Chem. 2008;3(5):833–43. doi: 10.1002/cmdc.200700324. [DOI] [PubMed] [Google Scholar]
- 19.Lin MS, Chen LY, Tsai HT, Wang SS, Chang Y, Higuchi A, et al. Investigation of the mechanism of beta-amyloid fibril formation by kinetic and thermodynamic analyses. Langmuir: the ACS journal of surfaces and colloids. 2008;24(11):5802–8. doi: 10.1021/la703369b. [DOI] [PubMed] [Google Scholar]
- 20.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, et al. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40(12):3525–35. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 21.Szymczak P, Cieplak M. Hydrodynamic effects in proteins. Journal of physics Condensed matter: an Institute of Physics journal. 2011;23(3):033102. doi: 10.1088/0953-8984/23/3/033102. [DOI] [PubMed] [Google Scholar]
- 22.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Current opinion in structural biology. 2004;14(1):76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Krasnoslobodtsev AV, Peng J, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL. Effect of spermidine on misfolding and interactions of alpha-synuclein. PloS one. 2012;7(5):e38099. doi: 10.1371/journal.pone.0038099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong Z, Mikheikin A, Krasnoslobodtsev A, Lv Z, Lyubchenko YL. Novel polymer linkers for single molecule AFM force spectroscopy. Methods. 2013;60(2):161–8. doi: 10.1016/j.ymeth.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–12. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 26.Lovas S, Zhang Y, Yu J, Lyubchenko YL. Molecular mechanism of misfolding and aggregation of Abeta(13-23) J Phys Chem B. 2013;117(20):6175–86. doi: 10.1021/jp402938p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, et al. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc Natl Acad Sci U S A. 2000;97(1):139–44. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein A, Lyubchenko YL, Sherman S. Dynamic properties of pH-dependent structural organization of the amyloidogenic beta-protein (1-40) Prion. 2009;3(1):31–43. doi: 10.4161/pri.3.1.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portillo AM, Krasnoslobodtsev AV, Lyubchenko YL. Effect of electrostatics on aggregation of prion protein Sup35 peptide. Journal of physics Condensed matter: an Institute of Physics journal. 2012;24(16):164205. doi: 10.1088/0953-8984/24/16/164205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, et al. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99(26):16742–7. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102(48):17342–7. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldsbury C, Frey P, Olivieri V, Aebi U, Muller SA. Multiple assembly pathways underlie amyloid-beta fibril polymorphisms. J Mol Biol. 2005;352(2):282–98. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Lovas S, Zhang YL, Yu JP, Lyubchenko YL. Molecular Mechanism of Misfolding and Aggregation of A beta(13-23) Journal of Physical Chemistry B. 2013;117(20):6175–86. doi: 10.1021/jp402938p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci U S A. 2011;108(14):5819–24. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397(6714):50–3. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.