Summary
Fragile X syndrome (FXS) is the most common form of inherited mental retardation and it is caused by loss of function of the Fragile X Mental Retardation Protein (FMRP). FMRP is an RNA-binding protein that is involved in the translational regulation of several neuronal mRNAs. However, the precise mechanism of translational inhibition by FMRP is unknown. Here, we show that FMRP inhibits translation by binding directly to the L5 protein on the 80S ribosome. Furthermore, cryo-electron microscopic reconstruction of the 80S ribosome•FMRP complex shows that FMRP binds within the inter-subunit space of the ribosome such that it would preclude the binding of tRNA and translation elongation factors on the ribosome. These findings suggest that FMRP inhibits translation by blocking the essential components of the translational machinery from binding to the ribosome.
Introduction
Fragile X syndrome (FXS) is a neurodevelopmental disorder caused by the loss of function of a single gene, the fragile X mental retardation 1 gene (FMR1) (Pieretti et al., 1991; Siomi et al., 1993; Verkerk et al., 1991). FXS is typically caused by a triplet repeat expansion in the 5′ untranslated region of the FMR1 gene leading to abnormal methylation of the gene and the repression of transcription (Penagarikano et al., 2007; Pieretti et al., 1991; Sutcliffe et al., 1992; Verkerk et al., 1991). The absence of FMR1 gene expression results in intellectual disability and behavioral problems and is the leading cause for inherited mental retardation, with an average prevalence of ~1:2,500 males and ~1:5,000 females (Hagerman, 2008; Penagarikano et al., 2007). The altered expression of the FMR1 gene has also been linked to autism spectrum disorders, fragile X-associated tremor/ataxia syndrome and fragile X-associated primary ovarian insufficiency (Kenneson and Warren, 2001; Penagarikano et al., 2007).
FMR1 encodes an RNA binding protein, fragile X mental retardation protein (FMRP) that is highly expressed in the brain (Ashley et al., 1993; Devys et al., 1993; Siomi et al., 1994; Siomi et al., 1993; Hinds et al., 1993) and FMRP appears to regulate the expression of many proteins throughout the brain (Ashley et al., 1993; Brown et al., 2001; Miyashiro et al., 2003; O’Donnell and Warren, 2002). FMRP has three RNA-binding domains: one RGG domain that is rich in arginines and glycines and two hnRNP K homology domains (KH domains) (Figure 1A) (Ashley et al., 1993; Siomi et al., 1993). Consistent with its proposed role in regulating protein synthesis, the majority of FMRP in the cell is associated with polyribosomes (Corbin et al., 1997; Darnell et al., 2011; Feng et al., 1997a; Feng et al., 1997b; Li et al., 2001; Mazroui et al., 2002; Stefani et al., 2004; Tamanini et al., 1996). Interestingly, a missense mutation in the KH2 domain (Ile304Asn of human FMRP) abolishes the binding of FMRP to polyribosomes and causes an aggravated form of FXS in humans (Brown et al., 1998; De Boulle et al., 1993; Feng et al., 1997a; Laggerbauer et al., 2001; Siomi et al., 1994). This suggests that RNA binding by FMRP plays a key functional role in the brain. In vitro selection experiments identified a G-quadruplex structure (Brown et al., 2001; Darnell et al., 2001; Schaeffer et al., 2001) and a pseudoknot structure (Darnell et al., 2005) as the potential RNA ligands for the RGG and KH2 domains, respectively. Based on these results it was proposed that FMRP may bind to mRNAs that possess G-quadruplex- or pseudoknot-forming sequences and repress their translation (Brown et al., 2001; Darnell et al., 2005; Darnell et al., 2001; Phan et al., 2011). Additionally, many proteins, microRNAs and noncoding RNAs have been proposed to be important for translational repression by FMRP (Jin et al., 2004a; Jin et al., 2004b; Zalfa et al., 2003).
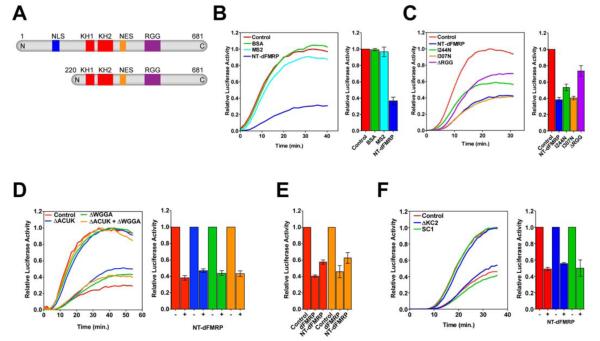
Figure 1. Inhibition of translation by FMRP.
(A) Domain organization of dFMRP (1-681 amino acids) and NT-dFMRP (220-681 amino acids). NLS, nuclear localization signal; KH, K-homology domain; NES, nuclear export signal; RGG, motif rich in arginine and glycine. (B) Time course of luciferase mRNA translation. Red trace, control translation without NT-dFMRP; light blue trace, translation with 1.2 μM bovine serum albumin (BSA); cyan trace, translation with 1.2 μM RNA-binding MS2 coat protein; blue trace, translation with 0.6 μM NT-dFMRP. The addition of NT-dFMRP to the IVTS inhibited luciferase mRNA translation. In contrast, the addition of BSA or MS2 coat protein to the IVTS did not inhibit the synthesis of luciferase. (C) Inhibition of translation by NT-dFMRP mutants. Time course of luciferase mRNA translation in the presence of the indicated mutant NT-dFMRP proteins. (D) Inhibition of translation by FMRP is independent of WGGA and ACUK sequences. Red trace, control mRNA; green trace, mRNA without WGGA sequence; blue trace, mRNA without ACUK sequence; orange trace, mRNA without both WGGA and ACUK sequences. In all cases, the data were normalized with respect to the control translation without NT-dFMRP. The bar graphs next to each time course show the mean ± SD from three independent experiments. (E) Inhibition of translation by FMRP in cells. Translation of control luciferase mRNA (red bars) and luciferase mRNA without WGGA and ACUK sequences (orange bars) are inhibited to a similar extent by full-length dFMRP and NT-dFMRP, as indicated. Data were normalized with respect to control cells, which were co-transfected with an empty plasmid and the appropriate luciferase plasmid. The transfection experiments were done in duplicates and the mean ± SD from three independent transfection experiments are shown. (F) Inhibition of translation by FMRP is independent of G-quadruplex and pseudoknot forming sequences in the mRNA. Red trace, control mRNA; blue trace, mRNA with ΔKC2 pseudoknot forming sequence; green trace, mRNA with SC1 G-quadruplex sequence. The bar graph shows the mean ± SD from three independent experiments. See also Figures S1 to S4.
Several independent studies have identified hundreds of mRNAs as potential targets for FMRP (Brown et al., 2001; Chen et al., 2003; Darnell et al., 2001; Miyashiro et al., 2003). However, very little overlap was found among various studies that identified the putative mRNA targets of FMRP. Recently, Darnell and co-workers used high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) to identify neuronal mRNAs regulated by FMRP in the mouse brain (Darnell et al., 2011). They identified 842 unique mRNA targets of FMRP. About 24% of the newly identified FMRP target transcripts showed overlap with targets identified in a previous study (Brown et al., 2001). Although previous studies suggested that FMRP inhibits translation by binding to either a G-quadruplex (Brown et al., 2001; Darnell et al., 2001; Schaeffer et al., 2001) or a pseudoknot (Darnell et al., 2005) forming sequence in the mRNA, none of the FMRP binding sites identified in the new study can be folded into a G-quadruplex or pseudoknot structure (Darnell et al., 2011). A more recent study indicated that the KH1 and KH2 domains of FMRP bind to frequently occurring ACUK and WGGA sequences (in which K = G or U and W = A or U), respectively, in the target mRNAs (Ascano et al., 2012). However, it is unclear how these sequences can provide the binding specificity to FMRP for regulating the translation of select mRNAs in the cell because they are present in all mRNAs (the frequency of WGGA or ACUK occurring in a random sequence is 1 in 128 nucleotides). Thus, the precise mechanism of translational repression by FMRP remains unknown.
In mammals there are two autosomal paralogs of FMRP, FXR1P and FXR2P, which are also expressed in the brain (Agulhon et al., 1999; Zhang et al., 1995). The presence of three related proteins with some degree of functional redundancy makes it more difficult to genetically study their function in mammals (Agulhon et al., 1999; Wan et al., 2000). In contrast, Drosophila contains only one Fmr1 gene whose expression product shares 56% overall amino acid similarity with mammalian FMRP (Wan et al., 2000) (Figure S1A) (Lee et al., 2003; Wan et al., 2000; Zhang et al., 2001). Importantly, Fmr1 gene knockout flies exhibit phenotypes that are consistent with the synaptic defects found in human FXS and Fmr1 gene knockout mice (Comery et al., 1997; Greenough et al., 2001; Hinton et al., 1991; Lu et al., 2004; Nimchinsky et al., 2001; Rudelli et al., 1985; Zang et al., 2009; Zhang et al., 2001; Zhang et al., 2004). Specifically, Fmr1 gene knockout flies display increased growth and branching of dendritic processes as observed in patients with FXS (Zhang et al., 2001). Furthermore, the neurological and behavioral defects of Fmr1 gene knockout flies can be fully rescued by the human FMR1 gene demonstrating the relevance of using Drosophila to study FXS (Coffee et al., 2010; Coffee et al., 2012). The availability of sophisticated genetic, cellular and molecular tools make Drosophila an attractive model system to study the function of FMRP. Here, we analyzed the mechanism of translational inhibition by the Drosophila FMRP (dFMRP). Our biochemical experiments and structural studies show that dFMRP binds to the 80S ribosome near the binding site for tRNA and translation factors. These results suggest that FMRP would inhibit translation by interfering with the binding of essential translation factors to the ribosome.
Results and Discussion
Inhibition of translation by FMRP
To understand the mechanism of translational inhibition by FMRP we used the Drosophila FMRP (dFMRP) homolog, in which the RNA-binding domains are nearly 75% identical to human FMRP (Wan et al., 2000) (Figure S1A). We purified both the full-length and a N-terminally truncated dFMRP (NT-dFMRP) and used an in vitro translation system (IVTS) made from Drosophila embryo extract to test the activity of dFMRP (Gebauer and Hentze, 2007) (Figure S2). We used Renilla luciferase mRNA as the reporter for protein synthesis because it has three G-rich sequences that potentially form G-quadruplex structures (Kikin et al., 2006), and additionally has 7 ACUK and 6 WGGA sequences (Figures S1B and S1C). The time course of protein synthesis was monitored by bioluminescence. The addition of dFMRP or NT-dFMRP to the IVTS inhibited the synthesis of luciferase (Figure 1B and S2). We used NT-dFMRP in our further studies because it was equally active in inhibiting translation as the full-length protein and easier to purify than the full-length dFMRP (see Experimental Procedures).
Titration experiments showed that the inhibition of translation depends on the concentration of NT-dFMRP added to the IVTS (Figure S3A). NT-dFMRP also inhibited the translation of luciferase mRNAs that do not have a N7-methyl guanosine cap at the 5′ end or a 3′ poly(A) tail, indicating that translation inhibition is 5′ cap and poly(A) tail independent (Figures S3B and S3C). To confirm that the inhibition of translation by NT-dFMRP is 5′ cap-independent, we synthesized uncapped luciferase mRNA with an internal ribosome entry site (IRES) from the Reaper mRNA at the 5′ end (Hernandez et al., 2004). IRES-dependent translation of luciferase mRNA was as efficient as the translation with the 5′ capped mRNA (Figure S3D). NT-dFMRP inhibited the translation of luciferase mRNA having the IRES element, confirming that the 5′ cap is not essential for inhibition (Figure S3E). These results also suggest that FMRP does not affect the initiation step of protein synthesis.
We next investigated the importance of the different RNA binding domains of dFMRP in suppressing translation. We made constructs with a missense mutation in the KH1 (Ile244Asn) or KH2 (Ile307Asn) domains or with the RGG domain deleted (residues 414 to 681) (Siomi et al., 1994) (Figure S4). Circular dichroism spectroscopy indicated that the mutations or the deletion did not change the overall structure of the mutant proteins (Figures S4C and S4D). In agreement with an earlier study (Wan et al., 2000), the KH1 mutant was less active than the KH2 mutant and wild type NT-dFMRP in inhibiting translation (Figure 1C). Interestingly, the ΔRGG mutant inhibited translation poorly suggesting that this region is also important for the function of NT-dFMRP (Figure 1C).
WGGA and ACUK sequences in the mRNA are not important for translational inhibition by FMRP
A recent study showed that the KH1 domain of FMRP binds to mRNA having the sequence WGGA and the KH2 domain of FMRP binds to mRNA having the sequence ACUK (in which W = A or U and K = G or U) (Ascano et al., 2012). To determine whether the 6 WGGA and 7 ACUK sequences in the luciferase mRNA are essential for translational inhibition by FMRP, we systematically replaced them with other sequences but without changing the primary amino acid sequence of the luciferase protein. We made three different mutant luciferase mRNAs: (1) without any WGGA sequence (ΔWGGA mRNA), (2) without any ACUK sequence (ΔACUK mRNA), and (3) without both WGGA and ACUK sequences (ΔWGGA/ΔACUK). Surprisingly, NT-dFMRP inhibited the translation of the ΔWGGA mRNA, ΔACUK mRNA, and ΔWGGA/ΔACUK mRNA to a similar extent as the control luciferase mRNA (Figure 1D). These results suggest that the WGGA and ACUK sequences in the mRNA are not critical for translational inhibition by FMRP. We verified these results by carrying out in vivo studies. HEK293T cells were co-transfected with plasmids expressing dFMRP or NT-dFMRP and either the control luciferase mRNA or the ΔWGGA/ΔACUK mRNA. The amount of dFMRP and NT-dFMRP expressed in the cells was determined by Western blotting and the amount of luciferase expressed in the cells was determined by bioluminescence. Consistent with the in vitro data, full-length dFMRP and NT-dFMRP inhibited the translation of control luciferase mRNA and ΔWGGA/ΔACUK mRNA to a similar extent indicating that these sequences in the mRNA are not essential for translational inhibition by FMRP (Figure 1E). However, we cannot rule out the possibility that mRNAs with WGGA and ACUK sequences are preferentially inhibited by endogenous concentrations of FMRP.
Functional role for G-quadruplex sequences in the mRNA
Previous studies showed that FMRP inhibits the translation of specific mRNAs such as futsch (MAP1B), Rac1, Fmr1, and Chickadee/Profilin by binding to G-rich sequences, which may fold into a G-quadruplex (Coffee et al., 2010; Lee et al., 2003; Schaeffer et al., 2001; Zhang et al., 2001). Therefore, it is likely that FMRP inhibits the translation of the Renilla luciferase mRNA by binding to the three putative G-quadruplex-forming sequences. However, it is impossible to remove the G-quadruplex-forming sequences from the Renilla luciferase mRNA without changing the primary amino acid sequence of the protein. Therefore, we tested whether inserting the well characterized SC1 G-quadruplex- (Darnell et al., 2001) or the ΔKC2 pseudoknot- (Darnell et al., 2005) forming sequences at the 3′ untranslated region of the luciferase mRNA will enhance the inhibition of translation by FMRP. Interestingly, NT-dFMRP inhibited the translation of the control mRNA and mRNAs with the SC1 or the ΔKC2 sequences to a similar extent (Figure 1F), suggesting that the insertion of an additional G-quadruplex-forming sequence or the pseudoknot-forming sequence in the mRNA does not increase the translation inhibition by NT-dFMRP. Finally, adding excess amounts of the ΔKC2 pseudoknot-forming sequence (Darnell et al., 2005) in trans to the IVTS partially relieved the translation inhibition by NT-dFMRP (Figure S3F). These results demonstrate that the inhibition of translation by NT-dFMRP is reversible and suggests that the ΔKC2 sequence competes with the Renilla luciferase mRNA for binding to NT-dFMRP.
FMRP binds directly to the ribosome
Previous studies have suggested that FMRP associates directly with the ribosome (Ishizuka et al., 2002; Khandjian et al., 1996; Mazroui et al., 2003; Siomi et al., 1996). However, other reports showed that FMRP binds to the ribosome via the mRNA or as an mRNP complex (Corbin et al., 1997; Darnell et al., 2011; Feng et al., 1997b; Li et al., 2001; Mazroui et al., 2002; Tamanini et al., 1996). It is not clear whether mRNA or other components are required for the association of FMRP with the ribosome. We used gel filtration chromatography and SDS-PAGE to show that NT-dFMRP could indeed bind directly to the ribosome in the absence of mRNA (Figure 2A, lanes 3 and 4). Furthermore, the binding of NT-dFMRP to the ribosome is stoichiometric even though excess amount of NT-dFMRP was present in the binding reaction. We next tested the binding of NT-dFMRP with functionally relevant mutations in the KH1 (I244N) or KH2 (I307N) domains. The KH1 mutant showed a 2-fold reduced binding to the 80S ribosome, while the KH2 mutant bound to a similar extent as NT-dFMRP (Figures 2B and 2C). The binding results are consistent with our functional data, which showed that the KH1 domain is important for translational inhibition by NT-dFMRP.
Figure 2. FMRP binds directly to the ribosome.
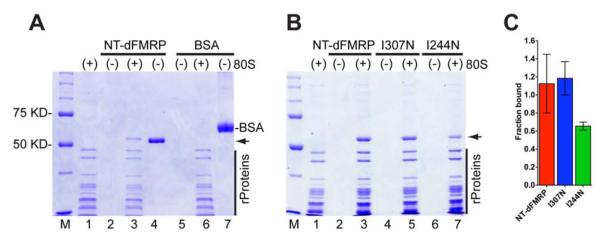
(A) SDS-PAGE gel showing that NT-dFMRP elutes with the ribosome, whereas BSA remains in the gel filtration column. Lanes: M, molecular weight ladder; 1, ribosome only; 2, NT-dFMRP only; 3, ribosome + NT-dFMRP; 4, input NT-dFMRP directly loaded on the gel; 5, BSA only; 6, ribosome + BSA; 7, input BSA directly loaded on the gel. Positions of NT-dFMRP (51 KD, arrow) and BSA (67 KD) are indicated. Vertical black bar indicates ribosomal proteins. (B) KH1 domain of FMRP is important for binding to the ribosome. Lanes: M, molecular weight ladder; 1, ribosome only; 2, NT-dFMRP only; 3, ribosome + NT-dFMRP; 4, KH2 (I307N) mutant only; 5, KH2 ((I307N) mutant + ribosome; 6, KH1 (I244N) mutant only; 7, KH1 (I244N) mutant + ribosome. (C) Bar graph of ribosome binding data showing mean ± SD from three independent experiments.
FMRP binds with high affinity to the ribosome
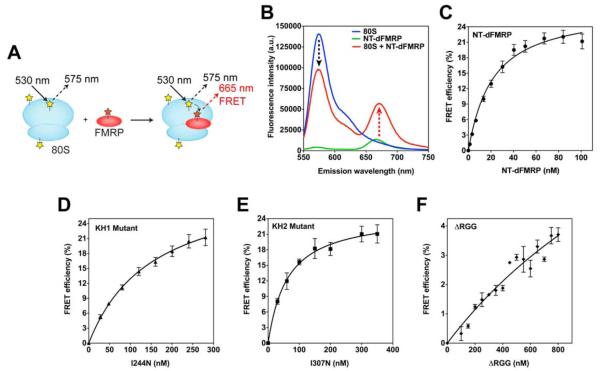
To determine the binding affinity of FMRP for the ribosome we developed an Förster Resonance Energy Transfer (FRET)-based assay using the fluorescent dyes Cy3 (donor) and Cy5 (acceptor) (Figure 3A). Cy3 and Cy5 dyes form an efficient FRET donor-acceptor pair and energy transfer can only occur within distances of 1-100 Å. We attached the fluorescent dye Cy5 to the single cysteine at position 263 in dFMRP. To label the purified Drosophila ribosome, we used an amino-reactive Cy3 dye that reacts with all the exposed amino groups in the ribosomal proteins. The Cy5-labeled NT-dFMRP was incubated with the Cy3-labeled ribosome to form the FMRP•ribosome complex. The sample containing both Cy3-labeled ribosome and Cy5-labeled NT-dFMRP clearly showed a decrease in the Cy3 fluorescence emission (near 575 nm) and an increase in the Cy5 fluorescence emission (near 665 nm) compared to the two controls (Figure 3B). This result also suggests that the NT-dFMRP binds to the 80S ribosome. The increase in FRET efficiency due to NT-dFMRP binding to the ribosome was used to measure the equilibrium dissociation constant (KD). The KD for NT-dFMRP is 20 ± 3 nM showing that it binds with high affinity to the ribosome (Figure 3C). We also analyzed the KH1, KH2, and the ΔRGG mutants using this quantitative FRET-based binding assay. Our results show that the KD for the KH1 and KH2 mutants are 130 ± 5 nM and 61 ± 2 nM, respectively (Figure 3D and 3E). In contrast, the ΔRGG mutant bound poorly to the ribosome with a KD that is > 2 μM (Figure 3F). These results also correlate well with our in vitro translation assays, suggesting that the ΔRGG mutant was defective and the KH1 mutant was slightly defective in inhibiting translation compared to the KH2 mutant and the wild type NT-dFMRP (Figure 1C). Previous studies with the individual KH domains have indicated that the I244N mutation in the KH1 domain of the human FMRP would unfold the protein (Musco et al., 1997), whereas I307N mutation in the KH2 domain of the Drosophila FMRP would adopt a native fold (Pozdnyakova and Regan, 2005). These findings may explain why the KH1 (I244N) mutant binds with lower affinity to the ribosome compared to the KH2 (I307N) mutant in our experiments, which in turn confirm that NT-dFMRP can bind directly to the ribosome without mRNA.
Figure 3. FRET-based assay for determining the binding affinity of FMRP for the ribosome.
(A) Schematic representation of the FRET assay. Ribosome (cyan) is labeled with Cy3 (yellow stars). NT-dFMRP (purple) is labeled with Cy5 (red star). The binding of NT-dFMRP to the ribosome results in FRET (emission at 665 nm). (B) Emission spectrum showing the changes in fluorescence intensity because of FMRP binding to the ribosome. Blue trace, 80S ribosome only control reaction; green trace, NT-dFMRP only control reaction; red trace, 80S ribosome + NT-dFMRP complex. Similar concentration of 80S ribosome and NT-dFMRP were used in the different reactions. (C) Binding curve for wild type NT-dFMRP. (D) Binding curve for KH1 (I244N) mutant. (E) Binding curve for KH2 (I307N) mutant. (F) Binding curve for the ΔRGG mutant. The y-axis shows the %FRET efficiency and the error bars show mean ± SD from three independent experiments.
FMRP binds close to ribosomal protein L5 on the ribosome
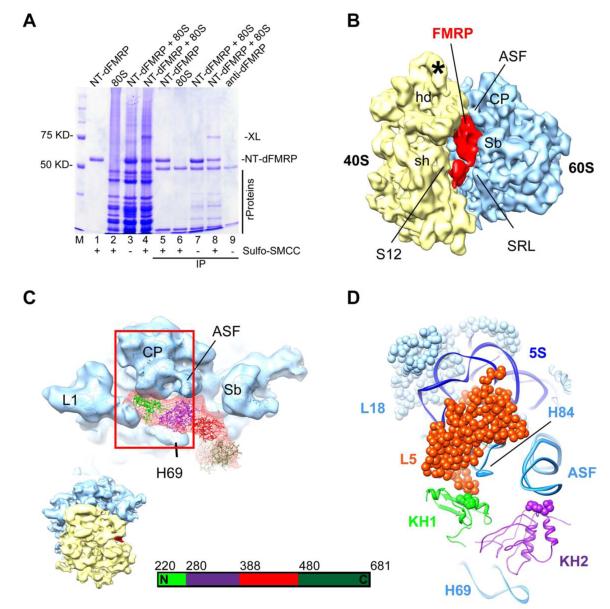
To map the binding site of NT-dFMRP on the ribosome, we used both the chemical crosslinking approach and the structural approach. We used sulfosuccinimidyl-4- (N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC) to crosslink the ribosome-bound NT-dFMRP and observed a new ~80 KD band in the SDS-PAGE analysis, while the amount of NT-dFMRP band was decreased (Figure 4A, lane 4 and labeled XL). Purification of the crosslinked product with the anti-dFMRP 5B6 monoclonal antibody and mass spectrometry analysis indicated that the ~80 KD band consist of FMRP and the large 60S ribosomal subunit protein L5 (previously called L11 in yeast and human ribosomes, but see (Jenner et al., 2012) for more unified nomenclature for the ribosomal proteins). In addition, the mass spectrometry data showed that a peptide fragment from the N-terminal end of NT-dFMRP was crosslinked to L5, which provides important constraint for the arrangement of NT-dFMRP on the ribosome.
Figure 4. FMRP binds near ribosomal protein L5.
(A) SDS-PAGE gel showing that NT-dFMRP crosslinks to the ribosome. Lanes: M, molecular weight ladder; 1, NT-dFMRP only; 2, ribosome only; 3, ribosome + NT-dFMRP; 4, ribosome + NT-dFMRP. SMCC was added to samples in lanes 1, 2, and 4. Lanes 5 to 8 correspond to samples in lanes 1 to 4 after immunoprecipitation (IP) with anti-dFMRP antibody and purification. Lane 9 shows the heavy and light chains of the anti-dFMRP antibody. Black bar, ribosomal proteins; XL, position of the crosslinked proteins. (B) Isolated density corresponding to NT-dFMRP (red) superimposed onto the 11.2 Å resolution segmented cryo-EM map of the control Drosophila 80S ribosome (40S subunit, yellow; 60S subunit, blue) (see Figure S6 for a stereo representation of the FMRP binding region). (C) A portion of the 60S subunit map is shown from its interface side to reveal the overall binding position of NT-dFMRP. The fitted I-TASSER model of the NT-dFMRP is displayed onto the 60S subunit with fitted X-ray coordinates of the yeast 60S ribosome. Four structural domains of the NT-dFMRP are identified according to the color in the bar-diagram shown at the bottom. Thumbnail at lower left depicts the overall orientation of the ribosome in panels C and D. (D) The boxed area in panel C is enlarged, with cryo-EM densities removed and a cutting plane applied on the far side, to reveal putative interactions of KH1 and KH2 domains of NT-dFMRP with the 60S subunit components. Landmarks of the 40S subunit in panel b: hd, head; sh, shoulder, and S12, protein S12 region; and an asterisk (*) points to a Drosophila specific expansion sequence emerging from helix 39 of the 18S rRNA. Landmarks of the 60S subunit: L1, L1 protein protuberance; CP, central protuberance; Sb, L7/L12 stalk base; 5S, 5S rRNA; H69 and H84, 28S rRNA helices 69 and 84, respectively; ASF, A-site finger, or 28S rRNA helix 38; SRL, α-sarcin-ricin stem-loop; and 60S ribosomal proteins L5 and L18 are shown as space-filled models. See also Figures S5 to S7.
Cryo-EM structure of FMRP•ribosome complex
We obtained a cryo-EM map of the Drosophila 80S ribosome•NT-dFMRP complex to determine the three-dimensional (3D) binding position of NT-dFMRP on the ribosome. Subtraction of the 3D map of the control Drosophila 80S ribosome from that of the 80S ribosome•NT-dFMRP complex shows an elongated mass of density, within the ribosomal inter-subunit space, that spans from central protuberance (CP) to α-sarcin/ricin stem-loop (SRL) region of the 60S subunit (Figure 4B). One end of the elongated difference mass interacts with the CP and A-site finger (ASF) of the 60S subunit, while its other end is situated between the protein S12 region of the small (40S) subunit and SRL region of the 60S subunit. Based on our cross-linking data, suggesting that the N-terminus of the NT-dFMRP construct interacts with the CP protein L5, we assign that portion of the difference map to the N-terminus, and tentatively assign the portion between S12 and SRL to the C-terminus domain of FMRP (Figures 4B-D). Both our crosslinking and cryo-EM results agree with a previous tandem affinity purification analysis of dFMRP from a cytoplasmic lysate, which showed that FMRP could interact with ribosomal proteins L5 and L18 (Ishizuka et al., 2002), both located in the CP of the 60S subunit (Ben-Shem et al., 2011). Indeed, we observe a direct interaction of NT-dFMRP with protein L5. The previous interaction reported with protein L18 could involve the N-terminus of the full-length FMRP that was absent in our construct. Docking of an I-TASSER (Zhang, 2008) homology model of NT-dFMRP into the corresponding cryo-EM map density tentatively places its KH1 and KH2 domains interacting with the CP and ASF, respectively, of the 60S subunit (Figures 4C-D). This region of the 60S subunit would normally be occupied by a tRNA in the peptidyl site (P site) during protein synthesis. Superimposition of the ribosome-bound FMRP and previously known binding position of the tRNA at the ribosomal P site indicated that the KH1 and KH2 domains of FMRP would partially overlap with the anticodon arm of the tRNA (Figure S7). However, future structural studies with a translationally inhibited ribosome•FMRP complex carrying a tRNA in the P site will be essential to understand if and how both the P-site tRNA and FMRP would be accommodated simultaneously on the ribosome.
Model for translational inhibition by FMRP
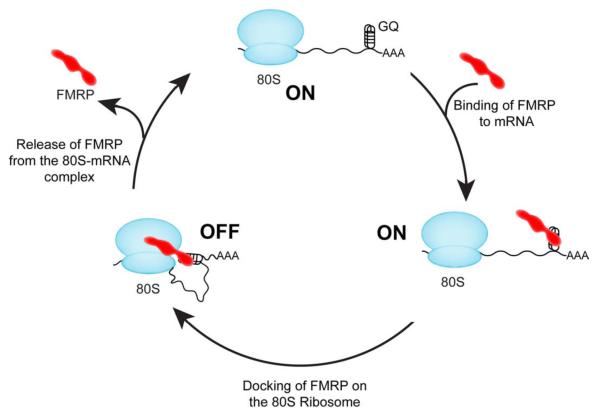
The cryo-EM structure suggests that the N-terminal KH1 and KH2 domains of FMRP bind to the ribosome, whereas the C-terminal RGG domain, which lies closer to the A-site in the small ribosomal subunit may interact with the mRNA. Interaction of the RGG domain of FMRP with G-quadruplex sequences in the mRNA may explain the selective translational inhibition of specific mRNA targets (Coffee et al., 2010; Lee et al., 2003; Schaeffer et al., 2001; Zhang et al., 2001), and is consistent with our results that the ΔRGG mutant inhibits translation poorly. Therefore, as depicted in Figure 5, the interactions of FMRP both with the ribosome and the mRNA may be important for synergistically inhibiting translation. The binding position of NT-dFMRP on the 80S ribosome suggests that FMRP would directly block the binding of several translation factors and tRNA that are known to bind to the overlapping regions on the ribosome (Agrawal et al., 2000), and thereby inhibit protein synthesis. Indeed, a recent study showed that FMRP reversibly stalls 80S ribosomes bound to target mRNAs at the elongation stage of protein synthesis (Darnell et al., 2011). Additionally, the FMRP-stalled 80S ribosomes appeared to form larger complexes consisting of multiple 80S ribosomes bound to the target mRNA (Darnell et al., 2011). These previous results, taken together with our structural data, suggest that FMRP would block the binding of eEF1A•GTP•aminoacyl-tRNA ternary complex and eEF2 to the 80S ribosome to inhibit the transit of the ribosomes on the target mRNAs. Finally, the inhibition of translation by FMRP maybe regulated by the post-translational modification of FMRP such as phosphorylation/dephosphorylation of serine residues (Ceman et al., 2003; Coffee et al., 2012; Muddashetty et al., 2011; Nalavadi et al., 2012; Siomi et al., 2002) and methylation/demethylation of arginine residues (Blackwell et al., 2010; Denman, 2002; Dolzhanskaya et al., 2006). These post-translational modifications may modulate the affinity of FMRP for the ribosome and for the target mRNAs thereby “turning-on” or “turning-off” protein synthesis locally in the neuron in an activity-dependent manner.
Figure 5. Schematic representation of translational inhibition by FMRP.
FMRP (red) uses its RGG domain to bind to mRNAs having G-quadruplex (GQ) forming sequence and then docks on the 80S ribosome (blue) using the KH1 and KH2 domains. Binding of FMRP to the mRNA and the ribosome synergistically inhibit translation (OFF state). Activation of the neuron changes the post-translational modification status of FMRP, which releases FMRP from the 80S•mRNA complex and activates translation (ON state).
Experimental Procedures
Expression and purification of NT-dFMRP and mutant proteins
The Drosophila Fmr1 gene (GenBank accession no. AF305881) was obtained from Prof. Gideon Dreyfuss (University of Pennsylvania). Full-length dFMRP and the N-terminal truncated dFMRP (NT-dFMRP) spanning residues 220 to 681 were subcloned into pTYB1 (New England Biolabs) to produce a fusion protein with a C-terminal intein and chitin binding domain. I244N and I307N NT-dFMRP constructs were then made by QuikChange Site-Directed Mutagenesis (Stratagene). ΔRGG construct spanning residues 220 to 413 was also subcloned into pTYB1. Proteins were expressed in E.coli Rosetta (DE3) strain, and purified using the chitin affinity matrix, as recommended by the manufacturer (New England Biolabs). The protein was stored in 24 mM HEPES (pH 7.5), 250 mM NaCl, 10% glycerol, 2 mM DTT −80 °C. Protein purity was assessed by Coomassie-Blue stained SDS-PAGE and concentrations were estimated by Bradford assay.
Expression of dFMRP in E. coli resulted in about 50% of the protein becoming truncated by ≈20 KD from the N-terminus (Figure S2A). Using a N-terminal hexahistidine tag we were able to purify limited amounts of the full-length dFMRP (Figure S2A). We also separately expressed and purified a N-terminally truncated dFMRP (NT-dFMRP) that is missing the NLS and residues important for FMRP/FXR dimer formation (Figure S1A). The yield of NT-dFMRP was much higher than the full-length dFMRP.
Ribosome purification
Dechlorinated frozen fly embryos were lysed with the Dounce homogenizer and clarified by centrifugation. Supernatant was applied to 30% sucrose cushion in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl, 0.5 mM EDTA, 0.5 mM PMSF, 0.1 mM benzamidine, 5 mM 2-mercaptoethanol and spun at 39,000 RPM for 17 hours at 4 °C. Crude ribosome pellets were resuspended in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 500 mM KCl, 0.5 mM PMSF, 0.1 mM benzamidine, 5 mM 2-mercaptoethanol and spun at 13,000 RPM for 20 min. at 4 °C. Puromycin was added to the supernatant with a ratio of 1 mg puromycin per 100 mg of ribosomes, incubated 30 min. on ice and 15 min. at 37 °C. Solution was clarified by centrifugation for 20 min. at 13,000 RPM. Supernatant was then loaded on 10%–40% sucrose density gradients in a SW-28 rotor and spun at 20,000 rpm for 17 hours and 30 min. Gradients were fractionated using a gradient fractionator and UA-6 detector (ISCO/BRANDEL). 80S ribosome peaks were pooled and diluted 2-fold with 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 25 mM KCl, 5 mM 2-mercaptoethanol and spun at 35,000 RPM 17 hours. Purified ribosome pellets were resuspended in 10 mM HEPES (pH 7), 10 mM MgCl2, 50 mM KCl, 5 mM 2-mercaptoethanol and stored at −80 °C. Ribosome concentration was calculated using 1 A260 unit = 20 pm/ml 80S ribosome.
In vitro translation assay
Renilla luciferase reporter mRNA constructs were capped and/or polyadenylated using the T7 mScriptTM Standard mRNA Production System (CellScript). SC1, and ΔKC2 sequences were subcloned into the 3′-UTR of the luciferase mRNA. Reaper IRES (168 nucleotides) was inserted into the 5′-UTR of luciferase mRNA. In vitro translation assays were conducted with Drosophila embryo extract, as described previously (Gebauer and Hentze, 2007). Coelentarazine (3 μM final conc.) was added to monitor the time course of luciferase synthesis using a 96-well plate reader (Genios, Tecan). Data was normalized with respect to the highest signal for each mRNA in the absence of NT-dFMRP.
In vivo translation assay
Transient transfection of HEK-293 cells were performed with either the control empty vector or plasmids encoding the full length Flag-dFMRP or Flag-NT-dFMRP together with plasmids encoding the wild type Flag-Renilla luciferase or mutant Flag-Renilla luciferase (without any WGGA or ACUK sequences). The amount DNA used for transient transfection was 2.5 μg of control empty vector, full length Flag-dFMRP or Flag-NT-dFMRP and 5 μg of wild type Flag-Renilla luciferase or mutant Flag-Renilla luciferase. Transient transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were collected 48 hr after transfection and lysed using Passive Lysis Buffer (Promega). Coelentarazine (3 μM final conc.) was added to the cell extracts and the amount of bioluminescence produced was measured using a 96-well plate reader (Genios, Tecan). The data were normalized with respect to the signal obtained with the control empty vector for each luciferase mRNA. All experiments were done in duplicates and the mean ± standard deviations from three independent experiments are reported.
Ribosome binding assay
NT-dFMRP (2.4 μM) or BSA (4.8 μM) were incubated with purified 80S ribosome (0.4 μM) in Binding Buffer (50 mM KOAc, 50 mM TrisOAc at pH 7.7, 10 mM DTT, 5 mM Mg(OAc)2, 30 μg/mL tRNA) for 10 min. at room temperature. Total tRNA was included in the binding reactions to reduce potential nonspecific binding. The reactions were applied to Illustra Sephacryl-300 columns (GE Healthcare) and the eluate was analyzed on 10% SDS-PAGE and stained with Commassie brilliant blue.
Fluorescent labeling
NT-dFMRP, KH1, KH2, and ΔRGG mutants, which each have one natural cysteine at position 263, were labeled with excess of Cy5 mono maleimide, as recommended by the manufacturer (GE Healthcare). Unreacted fluorophores were removed by dialysis and gel filtration columns (Bio-Spin 6, Bio-Rad) and buffer was exchanged to 24 mM HEPES (pH 7.5), 250 mM NaCl, 15% glycerol, 2 mM DTT. The 80S ribosome was diluted to a concentration of 2 mg/ml in 0.1 M sodium carbonate pH 9.1, 10 mM MgCl2, 50 mM KCl, 5 mM 2-mercaptoethanol and then labeled with Cy3 mono-reactive dye (GE Healthcare) for 45 min. at room temperature. Unreacted fluorophores were removed with gel filtration columns (Bio-spin 6, Bio-Rad) and buffer was exchanged to 10 mM HEPES (pH 7), 10 mM MgCl2, 50 mM KCl, 5 mM 2-mercaptoethanol.
FRET assay and KD determination
Cy3-ribosome (2 nM) was titrated with Cy5-NT-dFMRP (0-125 nM) or Cy5-NT-dFMRP mutants (0- 400 nM) in Binding Buffer with excitation at 530 nm. Emission maxima for the donor Cy3-ribosome and the acceptor Cy5-NT-dFMRP or Cy5-mutants were monitored at 575 nm and 665 nm, respectively. Under the same conditions, buffer was also titrated with Cy5-NT-dFMRP or Cy5-NT-dFMRP mutants, and each buffer emission spectrum was subtracted from the corresponding fluorescence emission spectrum. FRET efficiency (%) was then calculated by the equation ID/(ID+IA)*100. ID stands for the intensity of donor at 575 nm and IA stands for the intensity of acceptor at 665 nm. FRET efficiency was then fitted to a quadratic equation using Graphpad Prism to obtain the KD values.
Crosslinking and Immunoprecipitation
NT-dFMRP- ribosome complexes (1 μM) were formed in 1X PBS (100 mM sodium phosphate, 150 mM NaCl, pH 7.2). 30 μM of freshly prepared Sulfo-SMCC (sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate) was added to the complexes and incubated for 40 min. at 4 °C, then analyzed on 10% SDS-PAGE. For immunoprecipitation, monoclonal anti-dFMRP 5B6 (Developmental Studies Hybridoma Bank) was added to the crosslinked reactions and incubated for 1 hour at 4 °C. 25 μl of Protein G Magnetic Beads (New England Biolabs) were then added and reactions were incubated with agitation for 1 hour at 4 °C. Beads were washed extensively and resuspended in 5X SDS Sample Loading Buffer and analyzed on a 10% SDS-PAGE stained with Coomassie brilliant blue. Crosslinked protein bands were then cut out, digested with trypsin, and analyzed by liquid chromatography (LC) in combination with tandem mass spectroscopy (MS/MS) using electrospray ionization. Peptide identifications were made using paragon algorithm executed in Protein Pilot 2.0 (Life Technologies).
Cryo-Electron Microscopy, Image Processing and Three-Dimensional Reconstruction
The 80S•NT-dFMRP complex was prepared under the ribosome binding assay conditions described above, except that the tRNA was excluded from the reaction mixture. The complex was diluted to 35 nM in the same binding buffer, but containing 25-fold molar excess of NT-dFMRP. Cryo-EM grids were prepared according to standard procedures (Grassucci et al., 2007), using Vitrobot (FEI, Hillsboro, OR). Data were collected on a Philips FEI Tecnai F20 field emission gun electron microscope with a magnification of 50,760. 102 micrographs for the control 80S and 98 micrographs for the 80S•NT-dFMRP complex were scanned on a Zeiss flatbed scanner (Z/I Imaging, Huntsville, AL) with a step size of 14 μm, corresponding to 2.78 Å on the object scale and were sorted into 28 and 20 defocus groups for the 80S and the 80S•NT-dFMRP complex, respectively. A total of 78,267 images for the control and 82,481 images for the 80S•NT-dFMRP complex were manually selected. SPIDER (Frank et al., 1996) was used for all image processing, including 2D image classification, reconstruction, and refinement (Shaikh et al., 2008). A previously determined cryo-EM structure of the yeast 80S ribosome (Verschoor et al., 1998) was used as the reference to align images for an initial reconstruction from fewer images, using projection-matching procedure (Penczek et al., 1994). The 3D volume so obtained was low pass filtered and used subsequently for alignment of a larger data set. The original reconstruction and subsequent refinement yielded a weak mass of density corresponding to NT-dFMRP. To enhance the density of the factor in the 80S ribosome complex, the method of supervised classification was applied (Valle et al., 2002), using maps of the empty 80S (control) and the 80S-FMRP complex as two references. 59,313 particle images were used in the supervised classification. 19,520 particle images classified with the reference projection images generated from the original map of the NT-dFMRP-bound 80S ribosome (Figure S5A). The 3D map determined from these images showed an enhanced density for NT-dFMRP. The final resolution determined at 0.5 cutoff of FSC for the maps of control 80S and 80S• NT-dFMRP complex were 11.2 Å and 12.8 Å, respectively (Figure S5B).
Atomic structures of small fragments of FMRP and its homologs are known from NMR and X-ray crystallographic (Adams-Cioaba et al., 2010; Ramos et al., 2006; Valverde et al., 2007) studies. The homology model for the full NT-dFMRP construct was obtained using the I-TASSER server (Zhang, 2008), which predicted an elongated structure with four distinct structural domains that were connected to each other through long linkers. Each domain was fitted independently as a rigid body into the cryo-EM density corresponding to NT-dFMRP. Based on the crosslinking data, we first assigned the N-terminus domain (amino acid residues 1-60 of the construct, containing the KH1 motif) to the portion of density closest to the CP of the 60S ribosome. We next fitted the immediately next structural domain (amino acid residues 61-168, containing the KH2 motif), followed by the third structural domain (amino acid residues 169-260, containing first three RGG motifs), and then finally the fourth structural domain (amino acid residues 261-466, containing the remaining two RGG motifs). These independent fittings tentatively place the KH1 motif close to protein L5 and the 28S rRNA helix 84 within the CP of the ribosome, and KH2 motif close to the 28S rRNA helix 38, also known as the A-site finger. The overall features and placements of all four domains match and explain most of the cryo-EM density corresponding to NT-dFMRP. However, the cryo-EM density corresponding to structural domains 3 and 4 are relatively week, suggesting that the RGG motif containing domains and the C-terminus of FMRP are relatively flexible on the ribosome (Figure S6). All modeling, fitting and visualization were done using Chimera software (Pettersen et al., 2004).
Supplementary Material
Highlights.
FMRP binds to the ribosome and to the mRNA to synergistically inhibit translation.
Inhibition of translation by FMRP correlates with its affinity for the ribosome.
FMRP would block the binding of essential translational factors to the ribosome.
Acknowledgements
We are grateful to Prof. Gideon Dreyfuss for providing the plasmid encoding Drosophila FMRP and Prof. James Kadonaga for providing Drosophila embryos. We thank Timothy Booth for some of the cryo-EM data collection and help with image processing, Prem Kaushal for help with image processing and Yidan Li for help with the transfection experiments. We also thank Jing Wang, Uli Muller, Jamie Cate, and Gourisankar Ghosh for useful comments on the manuscript. This work was supported by an NIH Molecular Biophysics Training Grant GM08326 (to E.C.) and NIH grants R01 GM065265 (to S.J.) and R01 GM61576 (to R.K.A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers The cryo-EM maps of the control 80S and the 80S•NT-dFMRP complex were deposited in the EM database (http://emdep.rutgers.edu) with the accession numbers EMD-5805 and EMD-5806, respectively.
Supplemental Information Supplemental Information includes six figures and can be found with this article.
References
- Adams-Cioaba MA, Guo Y, Bian C, Amaya MF, Lam R, Wasney GA, Vedadi M, Xu C, Min J. Structural studies of the tandem Tudor domains of fragile X mental retardation related proteins FXR1 and FXR2. PLoS One. 2010;5:e13559. doi: 10.1371/journal.pone.0013559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulhon C, Blanchet P, Kobetz A, Marchant D, Faucon N, Sarda P, Moraine C, Sittler A, Biancalana V, Malafosse A, et al. Expression of FMR1, FXR1, and FXR2 genes in human prenatal tissues. Journal of neuropathology and experimental neurology. 1999;58:867–880. doi: 10.1097/00005072-199908000-00009. [DOI] [PubMed] [Google Scholar]
- Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT, Jr., Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Coffee RL, Jr., Tessier CR, Woodruff EA, 3rd, Broadie K. Fragile X mental retardation protein has a unique, evolutionarily conserved neuronal function not shared with FXR1P or FXR2P. Dis Model Mech. 2010;3:471–485. doi: 10.1242/dmm.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee RL, Jr., Williamson AJ, Adkins CM, Gray MC, Page TL, Broadie K. In vivo neuronal function of the fragile X mental retardation protein is regulated by phosphorylation. Hum Mol Genet. 2012;21:900–915. doi: 10.1093/hmg/ddr527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Denman RB. Methylation of the arginine-glycine-rich region in the fragile X mental retardation protein FMRP differentially affects RNA binding. Cell Mol Biol Lett. 2002;7:877–883. [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dolzhanskaya N, Merz G, Aletta JM, Denman RB. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J Cell Sci. 2006;119:1933–1946. doi: 10.1242/jcs.02882. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related field. J. Struct. Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Studying translational control in Drosophila cell-free systems. Methods Enzymol. 2007;429:23–33. doi: 10.1016/S0076-6879(07)29002-0. [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci U S A. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ. The fragile X prevalence paradox. J Med Genet. 2008;45:498–499. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Vazquez-Pianzola P, Sierra JM, Rivera-Pomar R. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 2004;10:1783–1797. doi: 10.1261/rna.7154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds HL, Ashley CT, Sutcliffe JS, Nelson DL, Warren ST, Housman DE, Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Current opinion in structural biology. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004a;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004b;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kenneson A, Warren ST. The female and the fragile X reviewed. Semin Reprod Med. 2001;19:159–165. doi: 10.1055/s-2001-15401. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell W T, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Boilard N, Labelle Y, Khandjian EW. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum Mol Genet. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco G, Kharrat A, Stier G, Fraternali F, Gibson TJ, Nilges M, Pastore A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat Struct Biol. 1997;4:712–716. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32:2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pozdnyakova I, Regan L. New insights into Fragile X syndrome. Relating genotype to phenotype at the molecular level. FEBS J. 2005;272:872–878. doi: 10.1111/j.1742-4658.2004.04527.x. [DOI] [PubMed] [Google Scholar]
- Ramos A, Hollingworth D, Adinolfi S, Castets M, Kelly G, Frenkiel TA, Bardoni B, Pastore A. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rudelli RD, Brown WT, Wisniewski K, Jenkins EC, Laure-Kamionowska M, Connell F, Wisniewski HM. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. 1985;67:289–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Higashijima K, Ishizuka A, Siomi H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol Cell Biol. 2002;22:8438–8447. doi: 10.1128/MCB.22.24.8438-8447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tamanini F, Meijer N, Verheij C, Willems PJ, Galjaard H, Oostra BA, Hoogeveen AT. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Verschoor A, Warner JR, Srivastava S, Grassucci RA, Frank J. Three-dimensional structure of the yeast ribosome. Nucl. Acids Res. 1998;26(2):655–661. doi: 10.1093/nar/26.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Matthies HJ, Mancuso J, Andrews HK, Woodruff E, 3rd, Friedman D, Broadie K. The Drosophila fragile X-related gene regulates axoneme differentiation during spermatogenesis. Dev Biol. 2004;270:290–307. doi: 10.1016/j.ydbio.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.