Abstract
To date, the scientific process for generating, interpreting, and applying knowledge has received less informatics attention than operational processes for conducting clinical studies. The activities of these scientific processes — the science of clinical research — are centered on the study protocol, which is the abstract representation of the scientific design of a clinical study. The Ontology of Clinical Research (OCRe) is an OWL 2 model of the entities and relationships of study design protocols for the purpose of computationally supporting the design and analysis of human studies. OCRe’s modeling is independent of any specific study design or clinical domain. It includes a study design typology and a specialized module called ERGO Annotation for capturing the meaning of eligibility criteria. In this paper, we describe the key informatics use cases of each phase of a study’s scientific lifecycle, present OCRe and the principles behind its modeling, and describe applications of OCRe and associated technologies to a range of clinical research use cases. OCRe captures the central semantics that underlies the scientific processes of clinical research and can serve as an informatics foundation for supporting the entire range of knowledge activities that constitute the science of clinical research.
Keywords: ontology, clinical research, eligibility criteria, clinical research informatics, evidence-based medicine, trial registration, epistemology
1. Introduction
Interventional and observational human studies are crucial for advancing our understanding of health, disease, and therapy. Clinical research informatics (CRI) is “the use of informatics in the discovery and management of new knowledge relating to health and disease” [1]. To date, CRI has focused on facilitating conduct of clinical trials and management of clinical data for secondary research use. Yet clinical research is fundamentally a scientific pursuit, and foundational methods for CRI should support the science of clinical research: asking the right question, designing rigorous protocols, conducting protocol-adherent studies, fully reporting all results, and finally, making inferences and applying research results to care decisions and policy.
The underpinning of this broad range of knowledge tasks is the study protocol as the study’s conceptual scientific structure. The planned study protocol drives all key scientific and biomedical activities during study execution and analysis, while the executed study protocol represents the study activities that actually took place. Early CRI work relegated support of protocols to the electronic sharing of text-based study protocol documents. More recently, study protocols have been reified into data models (e.g., BRIDG [2,3]) geared towards supporting the execution of clinical trials intended for submission to the U.S. Food and Drug Administration (FDA) for regulatory approval of therapeutic products, or supporting data management of clinical trial results (e.g., OBX [4], CDISC [5]).
To provide knowledge-based support for the scientific tasks of clinical research, the study protocol should be modeled in a knowledge representation formalism with clear, consistent and declarative semantics that support drawing clinical conclusions from study observations. The Ontology of Clinical Research (OCRe) is such a model. OCRe is an OWL 2 ontology of human studies, defined as any study collecting or analyzing data about humans that explore questions of causation or association [6,7]. OCRe models the entities and relationships of study designs to serve as a common semantics for computational approaches to the design and analysis of human studies.
In this paper, we describe the motivation methods behind OCRe, present highlights of the OCRe model, and review examples of how OCRe supports the science of clinical research. Our use cases illustrate why clinical and research informatics need to be more deeply integrated [8], to create a “learning health system” [9] that generates best evidence and also “drive[s] the process of discovery as a natural outgrowth of patient care” [10]. We posit that the study protocol, representing the essence of clinical research, is the epistemological foundation for a learning health system and that OCRe, representing study protocol elements, is a core informatics foundation for clinical research science.
2. Motivating Use Cases and Background
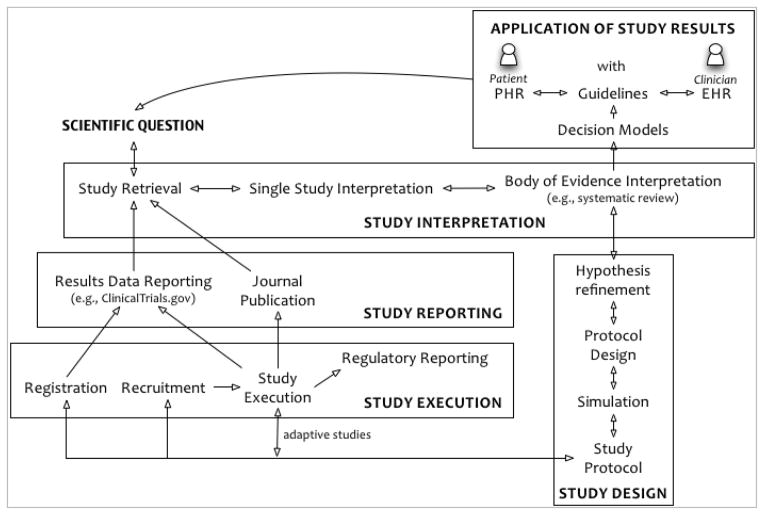
To show the value of OCRe across the breadth of clinical research science, we describe its role in the five phases of a human study’s idealized scientific lifecycle: 1) review and interpretation of results of previous studies to refine a scientific question; 2) design of a new study; 3) study execution; 4) results reporting; and 5) interpretation and application of the results to clinical care or policy (Figure 1). In a learning health system, clinical practice completes the cycle as a source of new scientific questions. The five phases of a study’s lifecycle are closely related and iterative (Figure 1).
Figure 1.
Idealized scientific lifecycle of a human study within a Learning Health System.
This remainder of this section presents use cases for each of the five phases. Based on these use cases, Section 3 presents the foundational capabilities that would transform informatics support for clinical research and describes the OCRe model. Section 4 then applies OCRe to selected use.
2.1 Pose a scientific question, retrieve and interpret prior studies
The first step of a clinical study’s lifecycle is highly iterative: potential scientific questions are posed and revised many times as prior studies are retrieved and interpreted over time.
2.1.1 Retrieve prior studies
Investigators often frame their research questions using the “PICO” mnemonic (for Patient, Intervention, Comparison, Outcome[11], sometimes with a T added for outcome timing [12]). For example, a broad initial question about vitamin D and cardiovascular risk (e.g., “does vitamin D supplementation reduce LDL cholesterol levels?”) could be phrased as Intervention = vitamin D and Outcome = cardiovascular endpoints (e.g., hypertension, high cholesterol, body weight). Running this PICO query at the PubMed PICO interface [13] returned 265 studies on vitamin D’s effect on high cholesterol (hyperlipidemia) at the writing of this paper.
These results are not directly helpful to an investigator because the PICO structures of these studies are buried within PDFs. A better search interface might be CTSearch with its interactive tag cloud PICO display [14], or interactive visualizations of the scientific structure of human studies like the tools that biomedical researchers have for visual exploration and query of gene sequences, pathways, and protein structures.
Even so, PICO elements alone are insufficient to support the full retrieval task. Different study objectives are best addressed by different study design types [15]. PubMed Clinical Queries [16] allows narrowing a search to appropriate study designs (e.g., prospective cohort studies to explore the association of vitamin D levels with cardiovascular outcomes, randomized controlled trials (RCTs) to explore questions of therapy), but two major problems attend this approach. First, this interface returns 2247 citations for a Narrow search for Therapy studies of vitamin D. An investigator would still need visualizations that reveal PICO and study-design features of large numbers of studies. Second, because there is no established study-design taxonomy, design types are poorly indexed in PubMed entries and searches by design type are correspondingly inaccurate [17]. There have only been a handful of published study-design taxonomies [18–20], including the Cochrane Collaboration’s taxonomy [21] and Hartling’s which showed a reliability of κ = 0.45 and is the basis for an AHRQ taxonomy [17]. In Section 3.2.1, we describe our OCRe-based study-design typology, which showed a moderate inter-rater agreement of Fleiss’ kappa of 0.46 in a preliminary evaluation [22].
2.1.2 Interpret prior studies
Once investigators have retrieved a set of relevant studies whose designs are appropriate for the scientific question, they need to assess their evidentiary strength [23]. In statistical terms, they need to appraise the “internal validity” of each study, including the comparability of comparison groups and the existence and nature of follow-up bias [24], Critical appraisal remains somewhat of an art, as many study quality scales and bias instruments are poorly correlated, imprecise, and irreproducible [25,26]while not predictive of observed effects [27].
Many researchers lack the methodological skills embodied in guides like those from JAMA [28], BMJ [29], the Cochrane Collaboration [30], and others [31] to carry out these appraisal tasks. While projects like THOMAS [32] and others [33] have developed relevant computational methods for automated critical appraisal, no computer-based tools are widely available to provide knowledge-based decision support for this critical task. A crisp, computable formulation of study design and study-design strength like OCRe would underpin this field (see 4.3). In contrast, major clinical research models such as BRIDG and CDISC SDTM [34] serve operational and administrative needs, but do not attempt to model study validity, confounding, and bias needed for assessing study-design strength.
2.2 Design a new study
There are persistent concerns about the quality of the design of human studies [35]. In 1994, Altman called the prevalence of poor medical research a “scandal” [36]. In a 2005 survey, 13% of NIH grantees acknowledged using “inadequate or inappropriate research designs” [37]. Decision support for study design has traditionally revolved around sample size planning, pharmacokinetic simulation, complex drug-disease modeling [38], or person/societal level simulation [39], but has not covered the full breadth of common design shortcomings. Investigators would benefit from methodological decision support at the point of design on:
Recommending appropriate study design types based on the research question
Defining the appropriate study population by querying a library of computable designs and comparing the content and selectivity of the investigator’s initial criteria with those of related studies
Identifying biases that should be taken into account at the design phase by querying for biases most important for the recommended design
Identifying potential clinical confounders to be accounted for in the design by mining prior articles and outcomes databases
Drawing on historical cohort identification and past recruitment patterns for sample size calculations
Broad tools like Design-a-Trial [40] start to get at these issues but have made little impact in part because they require ongoing maintenance of methodological and clinical knowledge bases, and do not incorporate knowledge from prior studies or outcomes data. By modeling the notion of bias and confounding into OCRe, we have begun to provide an informatics foundation for study design decision support, as discussed in Section 4.3 below.
2.3 Execute the study
Study execution steps include IRB application, study set-up, study registration, recruitment and enrollment of participants, protocol execution, and adverse events monitoring and reporting. The CRI field has developed many tools and methods for these tasks (reviewed in [41]). A computable study protocol model could serve to integrate these tools across the study lifecycle phases (Figure 1). In particular, a computable representation of eligibility criteria is useful for cohort identification and eligibility determination for study execution and for determining the applicability of studies to a target patient or population.
Cohort identification refers to computer-based matching of a study’s eligibility criteria against clinical data (e.g., an EHR) to identify a cohort of potentially eligible patients. Eligibility determination is the converse problem: matching one patient’s EHR data against a database of studies and their eligibility criteria to identify studies that the patient may be eligible for. In both cases, a prose criterion needs to be transformed into a computable representation, and then matched against EHR data. Complicating this task is the complexity of eligibility criteria. In an analysis of 1000 randomly drawn eligibility criteria from ClinicalTrials.gov, Ross et al. found significant semantic complexity, with 20% of criteria having negation, 30% having two or more complex semantic patterns, and 40% having temporal features [42]. Given these complexities, the state of the art in formalizing eligibility criteria is still rudimentary [43], as is eligibility determination performance. Cuggia found that among 28 papers reporting on clinical trial recruitment support systems, most were small scale tests that were not formally evaluated, none matched criteria against unstructured patient notes, and very few systems used interoperability standards [44]. OCRe includes a model of eligibility criteria capable of capturing much of their observed semantic complexity as discussed in 3.3 below.
2.4 Register and report the study
For clinical research to advance care and science, study results and the study designs that gave rise to those results must be fully reported. Indeed, transparent, unbiased study registration and reporting of interventional studies is a fundamental ethical duty as codified in the Declaration of Helsinki [45], and is arguably an ethical duty for observational studies as well [46]. OCRe provides a common ontology for describing study designs that can complement both registration and study reporting. Study reporting is going “beyond PDF” as medical journals move towards publishing full datasets (e.g., BMJ [47], Trials [48], Annals of Internal Medicine [49]), and as regulatory agencies like the European Medicines Agency begin to require disclosure of individual-patient level data [50]. To ensure retrieval of all relevant studies (see 2.5.1), federated data querying will be needed across these data sources as well as trial registers and other trial databases (e.g., AHRQ’s Systematic Review Data Repository, Cochrane’s Central Register of Studies). OCRe can serve as a well-formed semantics for federated data querying of study designs, as we discuss below for the Human Studies Database project (4.1.3),
2.5 Synthesize bodies of evidence and apply to care and policy
While study reporting is the last interaction of an investigator with a study, the study itself lives on to be retrieved, interpreted, synthesized with other studies, and applied to clinical care or policy. In the Application phase of a study’s lifecycle, we become interested in the entire body of evidence for a clinical or policy question, and less interested in single studies in isolation [51]. The evidence application subtasks include retrieving evidence, synthesizing it, and applying it in a learning health system.
2.5.1 Identifying and retrieving whole bodies of evidence
To ensure that clinical or policy decisions are made on best available evidence, all relevant studies and their full results need to be identified and retrieved. As study results become available through textual and database sources such as trial registers, journal publications, and regulatory databases, a federated search approach becomes necessary. Because study design greatly affects the validity of clinical research results, retrieval of bodies of evidence must also include retrieval by study design features. In our Human Studies Database (HSDB) project, we used OCRe to demonstrate federated query access to study protocol information from multiple academic medical institutions [7] (4.1.3). In 4.2, we also describe how OCRe can be used to conceptualize study retrieval as a phenotype-matching problem. The PICO-based phenotype of a study comprises its enrolled population, the intervention(s) tested, the outcomes assessed, and key clinical and non-clinical covariates (e.g., care setting). This study phenotype can be matched to a search query, to another study to determine study similarity, or to a patient’s clinical phenotype (e.g., via the EHR) to determine study applicability.
2.5.2 Systematic review
A high quality systematic review of high quality randomized trials is now the gold standard clinical evidence for questions of therapy [52]. Systematic reviews of other study designs are also critical for evidence-based medicine. Systematic reviews are, however, very time and labor intensive and current practices are not sufficient to keep up with the flood of new studies [53]. The holy grail of automated systematic reviews [54] remains elusive for both technical and methodological reasons. Current systems are mainly workflow support tools like the Cochrane Collaboration’s RevMan [55] and AHRQ’s SRDR system [56] that do not provide knowledge-based support for comparing, critiquing, and meta-analyzing studies. Meta-Analyst, now available as OpenMetaAnalyst [57], was one of the earliest decision support systems for meta-analysis with a Bayesian model of studies at its core. A fuller model of study designs and their attendant biases could offer a foundation for knowledge-based systematic review systems and for their interoperation with other phases of the study lifecycle. This is a wide-open area for substantial advances in clinical research informatics.
2.5.3 Evidence application and the Learning Health System
Rather than a one-way pipeline of research evidence to practice, translational medicine embraces the concept of a learning health system in which evidence is both applied and generated at the point of care. In this vision, the study lifecycle intertwines with the care system, and the informatics requirements discussed above become pertinent to frontline health information systems as well. Although clinical research and clinical informatics are still more separate than they should be [8], research innovations such as “point of care research” [58] embedded n-of-1 studies [59,60] and digital epidemiology[61] are knocking down silos between clinical research and health care informatics. By modeling study-design features and eventually study results, OCRe places human studies in a formally characterized relationship to patient data, and provides an epistemologically sound informatics foundation for clinical research and the learning health system.
3. The OCRe Ontological Infrastructure
The above use cases describe clinical research as a scientific enterprise with a wide range of informatics requirements. We identify the following requirements as representing the foundational capabilities that would transform informatics support of clinical research.
Ability to describe a study’s PICOT structure and the design of the study that facilitates identification and classification of studies in term of their scientific and design features (use case 2.1.1)
Ability to ensure the completeness and internal coherence of study instances (use cases 2.2 and 2.4)
Ability to ingest and instantiate study metadata from multiple manual and electronic sources (use case 2.4 and also a pre-requisite of requirement 4).
Ability to query for ontology-conformant study instances within or between institutions, using concepts from the ontology as well as from controlled vocabularies (use cases 2.1.1 and 2.5.1)
Ability to assist or enable selected tasks in the execution of a study (use case 2.3)
Ability for investigators to visualize and assess the strengths and weaknesses of study designs either singly or in the aggregate (use cases 2.1.2 and 2.5.1)
Ability to incorporate assessments of evidentiary strength into results interpretation, synthesis, and application (use cases 2.1.2 and 2.5)
At the core of these capabilities is a unifying representation of the study protocol. We wish to classify and query studies by their PICOT characteristics and by their design characteristics. We wish to constrain relationships among components of study protocols by placing various types of restrictions. We decided that description logic, in the modern form of Web Ontology Language (OWL), provides the most expressive and tractable knowledge representation formalism for capturing the formal semantics of study protocols.
The first three requirements above call directly for an ontological infrastructure upon which specific methods can be developed to address the use cases described in Section 2. At the center of our framework is an Ontology of Clinical Research (OCRe) and, as part of it, a study design typology that serves as the reference semantics for reasoning and data sharing. We developed OCRe to satisfy the first two requirements directly, and also developed a suite of OCRe-based approaches to address the other requirements: a data representation format and data acquisition methods for requirement #3; data federation technologies for requirement #4; an eligibility criteria annotation method for requirement #4 and #5; visualization methods for requirement #6; and representational and analytic methods for requirement #7.
In this section, we first review the ground methods and principles behind the development of OCRe. Then we describe OCRe’s major components, including a study-design typology and ERGO Annotation for capturing eligibility criteria. Finally, we describe how XML schemas can be generated from OCRe and used for data acquisition and use-case–specific querying.
3.1 Methods and principles of modeling
Rector[62] makes a distinction between representations of the world (models of meaning) versus data structures (information models). Models of meaning make formal statements about entities and relationship in the world. Information models (e.g., Health Level 7 Reference Information Model [63]) specify which data structures are valid for the purpose of exchanging and reusing them in different information systems. Usually models of meanings are formulated as ontologies using logic-based representations such as OWL. Information models may be represented in OWL or object-oriented languages such as Unified Modeling Language (UML). While most other CRI models, e.g., CDISC’s Study Design Model [34], are information models, OCRe encapsulates both models of meaning and information models. On one hand, we conceptualize a study as a real-world entity and we use the OWL formalism to make assertions about the structural components and features of human studies as well as assertions about particular studies. The core structures of a study protocol, including the design typology, design features, subgroups in the study, and interventions and exposures, are readily captured in the OWL formalism. On the other hand, some aspects of study design – eligibility criteria in particular – are difficult to formulate ontologically and are better modeled using an information model approach.
To illustrate, the eligibility criteria of a study define the target population of the study in terms of descriptions that may involve prior assessments, treatments, and test results and their temporal relationships. An ontological modeling of such descriptions is extremely complex and non-scalable. Instead of attempting to formalize such complex descriptions, we defined an information model called ERGO Annotation that allows us to annotate the criteria more simply but still capture the essential meaning of the criteria. These annotations, while not direct assertions about the world like others in OCRe, are data structures that allow the application of natural language processing (NLP) methods to automate the recognition of coded concepts and relations in free-text eligibility criteria and that can be transformed into computable formats to satisfy use cases from the lifecycle of studies that require the characterization of subject populations. Recently, we re-formulated ERGO Annotation, previously represented in Protégé’s frame language, as a component of the OWL-based OCRe, and extended the syntax to capture temporal and non-temporal constraints on entities and events referenced in eligibility criteria. Representing such informational model entities in OWL serves two purposes: 1) it enables, within a single representation, the transformations of the annotations into OWL expressions so that we can reason about a study’s target population; and 2) it completes the representation of a study in a single formalism so that applications can reason about a study’s target population together with other components of the study protocol. Section 3.3 presents more details about this hybrid modeling approach.
A potential point of confusion is that much of what we are modeling, such as the description of a study in a clinical trial protocol, are informational entities themselves. For example, a study protocol contains specifications of what phenomena the study variables represent or operationalize and at what frequency and how they should be measured. Similarly, instead of actual interventions that are carried out, the protocol specifies how interventions should be performed on study subjects. We model the structural components of these informational entities. For example, a protocol contains specifications of subgroups whose members may be subject to different interventions or exposures. We also model these informational entities in terms of their characteristic descriptors. For example, a protocol may specify that it has a parallel group study design, which is defined as an interventional design that has two design descriptors (written in Courier font): having multiple regimens and making main comparison across experimental units. We represent the relationships between descriptor-based and structural-based descriptions as axioms that must hold. A simple example is that a study with a design descriptor of multiple regimen must contain more than one study arm (sub)population. These axioms can be used as integrity constraints to check the consistency and completeness of a study specification, as described in Section 4.1.2.
Another point of contact between OCRe and informational entities is in OCRe’s references to controlled terminology. OCRe models the design of human studies, as expressed in study protocols, independent of any specific clinical domain. Study protocols, as informational entities, make references to real world phenomena through natural-language text or through structured codes. In OCRe, the semantics of clinical content are expressed through references to external ontologies and terminologies such as SNOMED. OCRe entities (e.g., outcome phenomenon) are related to external clinical concepts (e.g., acute myocardial infarction) through terminologies and their associated codes (e.g., SNOMED code for acute myocardial infarction). Thus, if we strip labels that are necessary to aid human understanding from clinical concepts referenced in OCRe, we have a formal and machine-processable representation of a study description.
Besides being domain independent OCRe is also independent of the requirements of specific applications in the way it models the entities and relationships of human studies. We made the decision that the semantics of application-specific ontological extensions must be expressible in terms of the underlying OCRe concepts. For example, our HSDB project to federate descriptions of human studies among CTSA institutions [7] defined an extended OCRe model called HSDB_OCRe that has additional categories of roles specific to our CTSA use case that organizations can play. HSDB_OCRe also implements “shortcuts” in navigating OCRe entities and relationships to enable the efficient acquisition and federated querying of large volumes of study information. These shortcuts add no additional semantic content, but create alternative categories that are convenient for application purposes. For example, OCRe reifies Funding as a class of administrative relationship associated with studies, where the reified relationship is identified by some funding number and has some person or organization as actors. This reification is necessary because a study, its funder, and the funding number represent an n-ary relationship that cannot be captured directly using OWL’s binary properties. It is not necessary, however, for OCRe to have a Funder class because this category can be derived completely as a defined class in OWL. Nevertheless, in the HSDB project, we found it convenient to have a Funder class and a ‘has funder’ object property fully defined in terms of underlying OCRe concepts. In this way, OCRe can be extended to satisfy arbitrarily different use cases without having to model those use case domains directly.
A different challenge arises in satisfying the requirement of instantiating study design information in OCRe-compliant form. A formal ontology such as OWL consists of a collection axioms and additional assertions that can be inferred using the Open World Assumption (OWA): an assertion is possibly true unless it can be explicitly shown to be false. There is no requirement that a specific collection of facts has to be asserted explicitly. As a consequence, completeness of data entry cannot be checked, unless we add closure axioms to say that only explicitly asserted data elements exist. Furthermore, an OWL class does not “own” a set of properties in the sense of having a set of attribute or participating in a set of relationships. It is not appropriate, therefore, to instantiate descriptions of specific studies into OCRe OWL. Rather, a data model (schema) is needed, consisting of templates of fields and value sets for acquiring entities and relationships in a domain. For these reasons, the OCRe infrastructure includes a method for extracting data models suitable for the acquisition and maintenance of large amounts of instance data for specific use cases, as described in Section 3.4.
3.2 The OCRe OWL Model
OCRe is organized as a set of modular components with the main modules related by import relationships as shown in Figure 2.
Figure 2.
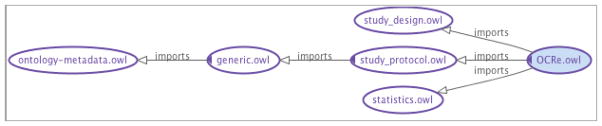
OWL import graph for OCRe.
The ontology is modularized into a main OCRe.owl ontology and sub-ontologies for study design (study_design.owl), the plan components of a study protocol (study_protocol.owl), and statistical concepts related to the specification of study analysis (statistics.owl). Earlier descriptions of OCRe, the genesis of the study-design typology and the application to the HSDB project are presented in [6,7,22].
As mentioned earlier, the HSDB Project extended OCRe with HSDB-specific concepts that are fully defined in terms of OCRe concepts. The main HSDB_OCRe ontology imports the extended OCRe and also uses a set of annotation features (defined in export_annotations_def.owl) to specify ontology elements that should be exported as part of a HSDB_XSD (Section 3.4).
3.2.1 Study design typology
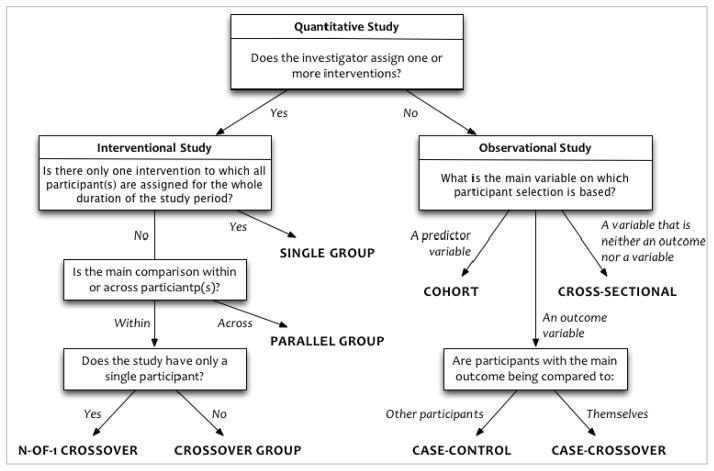
Study data should be interpreted within the context of how those data were collected (e.g., interventional versus observational study) and the purpose for which they were intended (e.g., public health versus regulatory filing). Since each study type is subject to a distinct set of biases and interpretive pitfalls, a study’s design type strongly informs the interpretation and reuse of its data. Through iterative consultation with statisticians and epidemiologists, we defined a typology of study designs based on discriminating factors that define mutually exclusive and exhaustive study types [22]. For example, whether the investigator assigns one or more interventions discriminates between interventional and observational studies (Figure 3).
Figure 3.
The Study Design Typology
The typology classifies studies hierarchically into four interventional and four observational high-level design types. Additional descriptors elaborate on secondary design features that introduce or mitigate additional interpretive features: eight additional descriptors for interventional studies (e.g., comparative intent (superiority, non-inferiority, equivalence); sequence generation (random, non-random)) and six additional descriptors for observational studies (e.g., sampling method (probability, non-probability); whether samples matched on covariates). Each descriptor applies to one or more of the main study types: for example, sequence generation applies to all of the interventional study types except single group; sampling method applies to all of the observational study types. This typology is available at http://hsdbwiki.org/index.php/Study_design_typology.
We incorporated this study design typology into OCRe in the form of an OWL hierarchy in the study-design module (Figure 2). Formally, the study-design descriptors are represented as a hierarchy of “study-design characteristics” (Figure 4). A particular study design is then defined in terms of its parent design and the descriptors that differentiate it from its siblings. Figure 5 shows the definition of a parallel-group design in terms of design characteristics, to illustrate the distinctions that are made in OCRe.
Figure 4.
The hierarchy of study-design characteristics
Figure 5.

The OWL definition of a parallel group study design
3.2.2 Study Arms and Interventions
OCRe models both interventional and observational studies. The Code of Federal Regulation 45 CFR 46 defines a study intervention as “manipulations of the subject or the subject’s environment that are performed for research purposes” [64]. In OCRe interventions or combinations of interventions (i.e., regimens) are attached to arms and study participants are assigned to these arms. Depending on the objective of the analysis, subjects can then be grouped (independent variable) by the intervention they were originally assigned to (intention-to-treat analysis) or the intervention they received (as-treated analysis). In observational studies, the independent variables for study analyses are called exposures, because they are not manipulated (i.e., assigned) by the study investigator.
3.2.3 Study Outcomes and Analyses
As we described in a previous paper [6], the study protocol specifies study activities (e.g., intervention assignment, data collection) that relate to the generation of observations, which are then analyzed to support or refute study hypotheses. To model these relationships, OCRe first defines a study phenomenon as “a fact or event of interest susceptible to description and explanation.” Study phenomena are represented by one or more specific study variables that may be derived from other variables. For example, the study phenomenon of cardiovascular morbidity may be represented as a composite variable derived from cardiovascular death, myocardial infarction (MI), and stroke variables. Each variable can be further described by its type (e.g., dichotomous), coding (e.g., death or not), time points of assessment (e.g., 6 months after index MI), and assessment method (e.g., death certificate). All variables are associated with participant-level and group-level observations. A study protocol may specify several analyses, each having dependent and independent variables that represent various study phenomena. A variable may play the role of dependent and independent variable in different analyses. If the study protocol designates a primary analysis, the dependent variable of that analysis represents what is conventionally known as the primary outcome of the study. To our knowledge, OCRe is the first model to disambiguate study phenomena of interest from the variables that code observations of those phenomena, and from the use of those variables in study analyses. This clarity of modeling should provide a strong ontological foundation for scientific query and analysis for clinical research.
3.3 ERGO Annotation
Based on the previously described analysis of study eligibility criteria [42], and because of the issues discussed in Section 3.1, we reformulated the problem of representing eligibility criteria from that of formally encoding criteria in some machine-processable expression language to that of classifying and decomposing criteria by identifying noun phrases that capture the basic meaning of the criteria and formalizing the noun phrases as coded terminological expressions[65]. We emphasize that there is closure on the representation of all concepts as controlled terminologies or CUIs from UMLS. An ERGO Annotation statement can be of three types: 1) simple statement that makes a single assertion using a noun phrase that may have recursive modifiers, 2) comparison criterion that is a triplet denoting a measurement value (signified by a noun phrase like “white blood cell count”) a comparison operator (e.g., “greater than”) and a quantitative threshold (e.g., 5000/mm3), and 3) complex statement, which is composed of multiple statements joined by AND, OR, NOT, IMPLIES, or semantic connectors, such as “caused by.” For example, “Tuberculosis of intrathoracic lymph nodes, confirmed histologically” is a simple eligibility criterion with a long noun phrase that is decomposed into a root simple noun phrase (tuberculosis) modified by location (intrathoracic) and by the modifier confirmed_by (histology).
In our current work, we augmented the representation of noun phrases to take into account meaning-altering contexts such as “family history of”. We formulated a constraint framework with different types of constraints. One type is restrictions on values of measurements. Other types are constraints on events (represented by noun phrases, e.g., diagnosis of breast cancer) by qualification of entities, or semantic or temporal relationships among entities and events. The temporal restrictions may be a duration constraint (e.g., event duration < 2 weeks), a time-point constraint that compares the time of the constrained event to that of another event (e.g., 3 weeks after end of chemotherapy), or a time-interval constraint where the time interval associated with the constrained event is compared with another time interval using Allen’s temporal-interval comparison operators [66]. Take for example, “Presence of bone marrow toxicity as defined by WBC < 4,000 3 weeks after the end of chemotherapy.” Here, “Bone marrow toxicity” is the root noun phrase representing the occurrence of some event, and “defined by” is a semantic relation that links it to a complex ERGO Annotation statement consisting of WBC (a noun phrase) constrained by a numeric value constraint (< 4000). “Bone marrow toxicity” is further restricted by the time-point constraint stating that the time of bone marrow toxicity is after a time point defined by “3 weeks after the end of chemotherapy.”
3.4 OCRe XSD Data Models
OCRe’s OWL model is an abstract generic model of the domain of human studies. We previously reported on the rationale for choosing XML Schema as the template formalism [6,7] for acquiring instances of study descriptions. Briefly, we deemed UML too cumbersome for mapping from OWL for our purpose. Without tool support, generating an UML model from an OWL ontology means creating the model in its serialized XMI format. XML Schema, on the other hand, is easy to use, supported by many tools, and follows the closed world assumption (i.e., XML schema types are templates), and has relatively simple serialized XML format. RDF, by contrast, is open world and does not provide a template for targeted export from relational databases, the most common source format for studies. But by keeping our XSD conformant with the logical structure of OCRe, OCRe XML instances can be translated into RDF/OWL format for advanced reasoning.
To ensure that instance data acquired using the schema are consistent with the OCRe ontology, we developed a data-model extractor to automatically derive an XSD file from annotations on OCRe entities. To generate an XSD, annotations are made in OCRe to specify: the root class for the XSD, the properties that should be represented as element tags in an XSD complex type, the subclasses to export as additional complex types, the parent class to use as the extension base in case there are multiple parents, and how value sets for property values should be created from ontology entities (e.g., treating individuals or subclasses of class as enumerations of values). The data-model extractor then acts on these annotations and uses property range axioms and property restrictions on classes to specify the types and constraints on specific XSD elements. For each type and data element in the XSD, its source in the ontology is clearly indexed using Semantic Annotations for SWDL (SASWDL), as shown in Figure 6.
Figure 6.

Example of an XSD element (StudyDesign) indexed to OCRe entity OCRE100056 via a URI. StudyDesign is associated with a 15-element value set partially shown in this figure.
The end product of this process is an XSD schema definition for a hierarchical XML data model. Our automated XSD generation approach allows changes in OCRe OWL to automatically propagate to the XSD and from there to the schemas of local databases, where programmatic approaches can update XML instance data to conform to the revised XSD as necessary. In summary, our OCRe data-model extraction process allows different XSD data models to be generated from different sets of annotations in extensions of OCRe, thus allowing OCRe to support acquisition of instance data for different use cases.
4. ‘Application’ Methods Enabled by the Ontological Infrastructure
This section illustrates the types of methods and solutions made possible with OCRe. First, we briefly review prior work on how study instances can be captured in XSD and exposed for federated querying with OCRe as the reference semantics. Next, we describe how a study’s PICO characteristics constitute a ‘study phenotype’, and propose methods that use study phenotypes to: 1) match studies to a search query; 2) match studies with each other to identify “similar” studies; and 3) match studies to patient data to determine study eligibility. Finally, we introduce the idea of OCRe-indexed bias models and methods for identifying clinical confounders as the basis for computational reasoning about the evidentiary strength of studies, Table 1 shows how these methods address the study lifecycle use cases described in Section 2.
Table 1. OCRe-based Methods and Study Life Cycle Phases.
| Instantiation and Query | Study Phenotype Matching/Comparison | Evidentiary Reasoning | ||||
|---|---|---|---|---|---|---|
| Life Cycle Phase | Constraint Checking | Federated Query | PICO | Design Typology | Bias Models | Clinical Confounders |
| Study Retrieval | ✠ | ✠ | ✠ | ✠ | ||
| Study Interpretation | ✠ | ✠ | ✠ | ✠ | ✠ | ✠ |
| Study Design | ✠ | ✠ | ✠ | ✠ | ✠ | ✠ |
| Study Execution | ✠ | |||||
| Study Reporting | ✠ | ✠ | ||||
| Results Application | ✠ | ✠ | ✠ | ✠ | ✠ | |
The ✠’s denote applications of an OCRe-based method to use cases in that lifecycle phase.
4.1 OCRe-based protocol instantiation and query
Instantiation of study descriptions in OCRe poses both technical and socio-technical challenges. Extraction of study descriptions from clinical trial management systems and electronic IRB systems is limited by incomplete and mostly-text-based information as well as vendor access issues [6]. Protocol documents and journal articles are likewise in free text and highly variable in content. Here we summarize work we have described elsewhere [7] on semi-automated and NLP approaches to data acquisition into OCRe semantics, and on federated data querying using OCRe semantics.
4.1.1 Instance Acquisition
Our HSDB use case was data federation of descriptions of study designs among CTSA institutions. Using the HSDB_XSD data model described in 3.4, we can acquire instances in several ways. With Rockefeller University (RU), we piloted ETL of study design information from their Oracle-based iMedRIS® e-IRB system [7]. Briefly, RU used HSDB_XSD to define SQL queries that mapped constructs of the schema to the iMedRIS relational tables. They then used Oracle’s DBMS_XMLGEN PL/SQL supplied package to issue the SQL-based ETL queries and convert the results “on the fly” into XML instances. An XSL Transform (XSLT) stylesheet was used to transform canonical XML into the XSD-compliant format, and manual post-processing data cleanup and XML validation was performed with the Oxygen editor. With ClinicalTrials.gov, we performed bulk ETL of their data. First, we mapped elements from ClinicalTrials.gov’s XSD to corresponding elements in HSDB_XSD using XSLT [7]. ClinicalTrials.gov entries were then downloaded from their API as separate files and each file was transformed into HSDB_XML files using the Saxon XML processor and XSLT.
NLP approaches are also indicated given that the bulk of study design information is in protocols and journal publications. Examples abound of methods to extract key trial elements such as population description, interventions, and outcomes [67–71] or PICO design features[72]. We ourselves developed the ExaCT semiautomated system for acquiring study-design data from full-text papers [73], as well as NLP techniques to analyze and instantiate eligibility criteria into ERGO Annotation [65]. While none of these systems yet use OCRe-XSD, they can now use OCRe-XSD templates as a common target data schema.
4.1.2 Constraint checking
OCRe’s declared semantics allows for constraint checking at the logical level (using extensions of OWL to enable constraint checking [74,75]), at the data model level (using XSD constraints), and programmatically, based on distinctions in OCRe. These constraint-checking methods can assist with data curation. For example, in reviewing study records from ClinicalTrials.gov, we noticed several types of recurring errors [7,76]. Guided by our OCRe model, we elucidated a number of constraints that could be used to curate these errors, including (using ClinicalTrials.gov names):
All data elements under Study Design should be completed, and N/A used where a specific item is not applicable (e.g., Phase should be “N/A” for behavioral studies)
-
If Allocation = Randomized, then
Study type must be Interventional
Intervention Model must be Parallel or Crossover Assignment
Number of arms must be > 1
-
If Intervention Model = Single Group Assignment, then
Number of arms must be = 1
Allocation must not be Randomized
If Intervention Model not = Single Group Assignment, then Number of arms must be > 1
If Study type = Interventional and Title includes “placebo”, then there should be a Placebo Arm defined and a placebo intervention
These constraints are now in OCRe and can be applied to OCRe-compliant instances via XSD and programmatic constraints. Applying these constraints to ClinicalTrials.gov would improve the scientific accuracy of crucial information on study design and intervention model, and would also improve derivative projects like Aggregate Analysis of ClinicalTrials.gov (AACT) [77] and LinkedCT.
Another example of constraint checking is for reproducible research, in which third-party researchers use a study’s protocol, dataset, and statistical code to verify and reproduce published results [78]. Having study design information in OCRe would enable automated constraint checking and ontology-driven visualization and interpretation of the study in the context of other related studies (e.g., CTeXplorer [79]). As medical publishing transitions “beyond the PDF,” computable models of study design like OCRe may prove increasingly valuable.
4.1.3 Federated query using OCRe semantics
In our HSDB project, we used the Query Integrator (QI) from the Brinkley lab [80] to query HSDB_XSD-compliant XML databases hosted at three CTSA institutions. QI can issue Xquerys as well as SPARQL queries to ontologies in BioPortal. This allowed us to issue live federated queries that exploited both OCRe’s logical structure and SNOMED’s taxonomic hierarchies using a single query formalism. We were thus able to automatically ingest study data from institutional e-IRB-based protocol descriptions, ingest the entire ClinicalTrials.gov repository, perform inter-institutional ontology-based queries at high granularity, and identify inconsistencies in instance data using OCRe’s axioms [7]. This proof of demonstration showed the value of OCRe and the feasibility of our general-purpose federated query infrastructure.
4.2 Study Phenotype Matching
Several core clinical research use cases involve the matching of a study’s design features against a query, other studies, or patient data. To facilitate these matching tasks, we define a study’s “phenotype” as its PICO protocol elements. For example, consider a randomized trial that tests high dose versus low dose vitamin D supplementation for Primary Outcome Phenomenon Hyperlipidemia and Primary Outcome Variable LDL:HDL ratio with “HgbA1c > 6.5%” as one of the eligibility criteria. This study design can be represented in PICO as: Population = diabetic patients, Intervention = high dose vitamin D supplementation, Comparison Intervention = low dose vitamin D supplementation, Primary Outcome = hyperlipidemia, as measured via the variable LDL:HDL ratio, with Timing = 6 months.
4.2.1 Matching a study’s phenotype against a query
The study retrieval task can be construed as the matching of a PICO query against a study’s PICO phenotype, with the semantic distance between the two PICO vectors being a measure of relevance. If we consider each dimension of PICO as an axis in a vector space, a study’s PICO features define a subspace that we will call the study phenotype space Sstudy_phenotype(studyA). Retrieval relevance is then the semantic distance between Ssp(studyA) and Ssp(query). We can exploit the hierarchies in standard clinical vocabularies to generate a distance metric D(studyA, query). The lesser the distance, the more relevant Study A is to the query.
The relevance determination task divides into three match domains: 1) study population, 2) predictors (interventions and comparisons), and 3) outcomes and their assessment timing. These methods will generate P-, IC-, and O- similarity scores Dpop, Dpredictor, Doutcome for each comparison of a study to the query. The overall similarity of a study to the query, Dstudy_phenotype(study, query), will be computed as a function of the three underlying similarity scores. The rest of this section lays out our plan for using OCRe for this matching. We will assume that a PICO query will consist of three sets of query terms, one set for each P, IC and O domain.
Matching study population
A study’s population is most clearly defined in the eligibility criteria. (Baseline variables describe only selected features of the enrolled population). OCRe captures eligibility criteria as simple and compound statements in ERGO Annotation (3.3). After pre-processing and annotation, each study’s eligibility criteria will be a Boolean combination of noun phrases with constraints. For sake of simplicity, we will consider cases where the constrained noun phrases are coded as SNOMED terms or their extensions.
To match a study A’s eligibility criteria against the P of a PICO query, we first identify the study’s population space Spop(studyA). This space is defined as all eligibility criteria that are characterized by the SNOMED terms that are of a “clinical” semantic type (e.g., disease, symptom, demographics, labs). The population match distance Dpop(studyA, query) is the minimal semantic distance between Spop(studyA) and Spop(query). If Spop(studyA) coincides with Spop(query), then Dpop(studyA, query) = 0. Otherwise, Dpop(studyA, query) is defined algorithmically, as follows:
Step 1 – First, for each term in studyA, we find the eligibility term Tquery,n in the query that is semantically closest to it. Suppose studyA’s first eligibility criterion is annotated by the term “heart failure.” In the straightforward case, if the query includes a SNOMED term that is a synonym of heart failure, then Dcriterion1(studyA, query) = 0. Other cases are more complicated. If the query has a population term Tquery,2 “cardiomyopathy,” that term is a neighbor to studyA’s criterion “heart failure.” To calculate Dcriterion1(studyA, query), we can use a semantic distance function from NCBO Annotator [81,82] or Jiang et al. [83]. We take DcriterionA(studyA, query) to be the minimum of criterion1’s distances to any term Tquery,n in query. Thus, if all terms Tquery,n of query are far from those of studyA, then Dcriterion1(studyA, query) approaches 1 (i.e., no match).
- Step 2 – If a population eligibility criterion is the disjunction or negation of population terms A, B, C, we define
Step 3 – Calculate overall Dpop(studyA, query), incorporating the distance between all criteria. At this step, we have a vector of N dimensions, where N is the number of population eligibility terms TA,n for studyA, and the values are the distances between each TA,n and its semantically closest corresponding term T (if any) in the query. We now need to collapse this vector to a single overall population distance Dpop(studyA, query). Ideally, we should not just sum the Euclidean distances of the vector because the population phenotypes are likely to be correlated. For example, studies enrolling “heart failure” patients are more likely to include patients with “hypertension”, such that two studies matching on both these criteria match less than a study matching on two clinically un-correlated criteria. However, this correlation problem may not be large enough to materially affect similarity determination for most researchers, so using the Euclidean distance measure is a reasonable first approximation.
Matching interventions/comparisons
In OCRe, a study’s predictor variables are described in the class outcome_analysis_specification. We can call a study’s space of predictors Spredictor(study). For queries, Spredictor(query) is defined by terms that are of semantic types that are appropriate for interventions, as defined for example, by Huang, et al. [84]. We can adapt the temporal sequence similarity scoring method developed by Lee [85,86] to define a distance metric Dpredictor(studyA, query) between a study and a query. In Lee’s method, a pattern of care refers to a set of temporally ordered, non-overlapping treatment courses. A treatment course is a single treatment or a set of treatments (called treatment regimen) that occurs over an interval period of time. A temporal sequence similarity score (T3S) between two treatment courses is defined by:
Constructing a similarity matrix, simE among the set of treatments E = {e1, e2, …,eN} based on distances derived from an ontology of such treatments;
Defining an Aligned Temporal Sequence Similarity Score (T3SA) for two patterns of care that have the same number of treatment courses, with T3SA taking into account differing durations and compositions of matched treatment courses;
Defining T3S for two non-aligned patterns of care as the T3SA of the overlap regions multiplied by a weight based on the ratio of the two patterns of care’s lengths.
There is nothing in Lee’s metric that limits its application to interventions. It can also apply to non-treatment predictors, such as exposures, to calculate Dpredictor(studyA, query). Additional methods are needed for matching on deeper predictor features for interventions and exposures (e.g., dose of a drug, indeterminate duration of a treatment or exposure event, e.g., “treat until the occurrence of X”).
Matching outcomes and their assessment times
Similarly, OCRe instances will define Soutcome(study) and the query will define Soutcome(query). To define the distance between these spaces, we first determine the “clinical” distance between the outcome variables (e.g., between “dull pain” and “burning pain”), then determine the “temporal” distance between the assessment times of “similar” variables (e.g., “3 months” and “6 months”). For each outcome variable in studyA, we pair it with an outcome term in the PICO query that is closest to it as defined by NCBO Annotator’s semantic distance function. We find the closest temporal distance between any pair of assessment timepoints for each outcome variable/term pair, and calculate the absolute difference in time duration since time zero, which is supposed to be stated in protocols [87–89]. We can explore heuristics for when outcomes are sufficiently similar semantically that calculating a temporal distance makes sense, and investigate alternative methods for calculating and capturing the temporal distance between two assessment timepoints (e.g., % of difference). Finally, we combine such time differences across multiple variable and timing pairs to calculate Doutcome(studyA, query). Methods for more complex matching (e.g., stroke documented by MRI vs. stroke coded as a discharge diagnosis) are beyond the scope of this paper.
Determining relevance to a query
After matching population, predictors, and outcomes between a study and a query, we calculate Dstudy_phenotype(study, query) as a function of (Dpop, Dpredictor, Doutcome). For browsing, the relative importance of population, predictor, and outcomes may vary from query to query. When a scientist is designing a new study, Dpredictor and Doutcome take on far more importance than Dpop. To quantify the overall match of a set of studies to a given query, we compute the average Dstudy_phenotype(study, query).
4.2.2 Matching a study’s phenotype against another study to determine similarity
As discussed in 2.5, interpreting clinical research for application to care or policy should involve interpreting a body of evidence. In systematic reviews, care must be taken that the studies included for synthesis are methodologically and clinically “similar” because heterogeneity contributes to statistical heterogeneity [30] and affects the applicability of the findings to patients. Thus, the notion of similarity is at the heart of evidence synthesis, yet there exists no formal conceptualization or metrics for this foundational notion.
Through the methods described above, though, OCRe and ERGO Annotation provide a foundation for computing similarity. Conceptually, relevance and similarity are closely related. Essentially, to find the similarity of studyA and studyB we can replace S(query) with S(studyB) and generate the same distance metrics as for relevance. The overall relevance of a set of studies against a query can then be generalized to the similarity between two sets of studies.
4.2.3 Matching a study’s phenotype against patient data
Eligibility determination can be seen from the perspective of a study or that of a subject. In the former case, the problem is to find the cohort of patients that may satisfy the eligibility criteria of a study. In the latter case, the problem is to identify those studies for which a person, whose state can be represented as a conjunction of predicates, may be eligible. We have argued [65] that the cohort identification task — screening a database of patient information to find potentially eligible participants — is best done with a conventional technology like relational databases, while the “study finding” task can be seen as a subsumption problem solvable using a description logic reasoner.
4.2.3.1 Cohort identification
The cohort identification task can be performed using OCRe and ERGO Annotations with these steps:
Transforming eligibility criteria from text into ERGO Annotation as described in 3.3.
Obtaining the local relational schema for patient data (e.g., a medication table containing drug names and codes, the dose and frequency of taking the drug, and the start and end times of the prescription)
Making assumptions regarding terminologies, e.g., problems are specified as ICD-9 codes;
Developing rules for assigning ERGO Annotation terms to tables in the patient data model (e.g., assigning terms whose UMLS semantic type was Disease or Syndrome to terms in the ProblemList table);
Creating a Hierarchy table that specified the UMLS terms as the ancestor(s) for each ICD-9 and drug code corresponding to a patient’s problems and medications.
Steps 2 through 5 are more fully described in [65]. We have extended this work to address eligibility criteria with temporal relationships. For the criterion “Bone marrow toxicity 3 weeks after end of chemotherapy”, for example, we would start with its OCRe representation in ERGO Annotation, decompose the representation to find noun phrases (as coded information), and add their corresponding tables in the patient data model to the FROM part of the query. We would use the Hierarchy table to map the terms in the database (e.g., codes for “neutropenia” or codes for specific drugs like “methotrexate” and “cytoxan”) and the terms in the criterion (e.g., code for “Bone marrow toxicity” or “Chemotherapy”). Finally, exploiting the structure of the timepoint constraint and using knowledge of the target SQL implementation, we would create an SQL query of the form:
SELECT * FROM ProblemList as p, Medication as m, Hierarchy as h
WHERE p.problem_name=h.descendant and h.ancestor= code for “Bone marrow toxicity” and
m.drug_name=h.descendant and h.ancestor = code for “Chemotherapy” and
p.time > DateAdd(week, 3, m.stop)
While the above SELECT query is manually crafted, our past experiences with templated expression languages suggest that automated generation of queries for common constraint patterns is very feasible [90].
4.2.3.2 Searching for Applicable Studies to Enroll In
Our earlier work showed how non-temporal ERGO Annotations can be transformed into OWL expressions [65]. To use a description logic reasoner to satisfy the “study finding” use case, we start by creating ERGO Annotations of all the eligibility criteria of the potential studies. For each study, the conjunction of the transformed OWL expressions for the inclusion criteria and the negation of the exclusion criteria approximates the characteristics of the study’s target population. A query for studies based on a set of patient characteristics (e.g., studies that include subjects who are pre-diabetic and have LDL below a certain threshold), formulated as an OWL expression, can be resolved by a description logic reasoner by finding all studies whose associated OWL eligibility expressions are subsumed by the query expression.
4.3 Evidentiary Reasoning
Our OCRe work to date has focused on modeling study design features to support data description and data sharing for scientific purposes. As a knowledge representation, however, OCRe can also provide value for knowledge-based decision support. To show this value, we are extending OCRe to include concepts related to statistical inference and structural and confounding biases, as well as developing computational methods for evidentiary reasoning to provide decision support for critiquing study designs. First, we explicitly model the notion of an inference as the result of a statistical analysis of dependent and independent variables. Each study design specifies many inferences. Next, inferences are at risk of bias (a systematic deviation away from the true value). The types of bias that each inference is at risk for depends on the study’s design type (from our study design typology, 3.2.1) and on the clinical content of the inference. We are extending OCRe with a typology of bias and constraining specific types of biases to specific study design types. Examples of high-level bias classes include selection, information, allocation, and detection bias [91]. In addition, we model the notion of a confounder as variables measured by the study protocol and used as covariates in inferential analyses.
This modeling of biases and confounding expressed in OCRe terms provides a first-ever ontological conceptualization of evidentiary reasoning, and is an important component for any informatics foundation for clinical research science.
5. Discussion
In this paper, we have shown that OCRe can capture detailed abstract study protocol information, which can yield numerous benefits along the entire scientific lifecycle of human studies. For the Design phase, OCRe-based models of the design of past and ongoing studies can help researchers retrieve, critique, and interpret the evidence base to more quickly iterate towards a worthwhile question to investigate. For Study Execution, the notion of matching study phenotypes to clinical phenotypes can assist with cohort identification and eligibility determination. For Study Interpretation, OCRe offers a common semantics for study protocol reporting, federated search and retrieval, and for semi-automated methods for evidentiary reasoning. Finally, for Study Application, OCRe-based study phenotypes can be useful for determining study’s applicability to patients with matching clinical phenotypes. The fact that we can describe how OCRe applies to such a broad range of applications is preliminary evidence of OCRe’s breadth, depth, and utility. Our ongoing program of research aims to demonstrate and evaluate OCRe’s ability to serve as a common informatics foundation for the science of clinical research.
The main difference between OCRe and other foundational models for clinical research informatics (e.g., BRIDG, CDISC, OBX, ClinicalTrials.gov XSD) is that OCRe takes a logic-oriented ontological modeling approach that mostly consists of logical statements about the design of human studies. The other models take an information model approach that more directly helps adopters achieve interoperability in data exchange and application development, but with less support for computational reasoning. OCRe also differs from other models in scope and domain of coverage. Compared to BRIDG, which focuses on operational support of protocol-driven regulated research, OCRe models study-design features more deeply for scientific purposes and covers both interventional and observational studies. BRIDG, however, models business processes and makes other distinctions necessary for supporting study execution and regulatory reporting of primarily interventional studies. CDISC’s Protocol Representation Model is derivative of BRIDG and the same comparisons apply. OBX models results data, which OCRe does not yet do, but has limited depth of modeling of study design features. ClinicalTrials.gov covers administrative and study design information, but its modeling of study design lacks depth and is rife with inconsistencies and ambiguities [7,76]. Overall, the BRIDG and CDISC information models are best for supporting study execution and regulatory reporting. The ClinicalTrials.gov model is important because ClinicalTrials.gov is the world’s largest collection of study design information. Many other data models exist for various systems, but to our knowledge none have the breadth, scope, and rigor of modeling that OCRe has for the explicit purpose of supporting the science of clinical research.
ERGO Annotation is an example of OCRe’s modeling power. This specialized part of OCRe tackles the difficult challenge of representing eligibility criteria, which often exhibit high semantic complexity. ERGO Annotation has a simple information model that makes distinctions regarding the structure of the criterion (e.g., existence of logical connectors, numerical comparisons) while leaving much of the semantics to noun phrases that are captured by terminological expressions. Other approaches for expressing clinical trial eligibility criteria such as Weng [43] and Milian [92] use more detailed information models and can use query templates for eligibility determination and for authoring criteria, while Huang’s approach uses Prolog rules as the model [93]. Weng’s EliXR is notable for using a data-driven approach to discover semantic patterns in a collection of textual eligibility criteria which are then organized into a semantic network with rich and fine-grained semantic knowledge. The detailed patterns developed by Milian can be parsed by NLP methods but the patterns are suitable only for specific clinical domains. In contrast to the above-mentioned approaches, ERGO Annotation can support OWL-based classification reasoning once ERGO Annotations are transformed into OWL expressions, as we have previously shown [65]. Our semi-automated approach is also more scalable than Huang’s and other similar approaches that use existing rule-based languages (such as Drools), which rely on manual crafting of rule criteria from the source text.
There are three other high-level ways in which our OCRe work is distinct. First, our work conceptualizes and renders study designs and eligibility criteria as structures to be represented, searched, visualized, analyzed, and reasoned about in their own right. This approach contrasts with the more common approach of focusing computational attention almost exclusively on results data [94,95] even though searching and analyzing study design information is a prerequisite for making sense of results data.
Second, while there have been heuristic and machine-learning approaches to determining the relevance of studies to a search query [96,97] many of the approaches require training a classifier against a hand-coded gold standard, which limits their scalability. In contrast, we exploit OCRe’s more granular modeling to directly match study features against a PICO query, and to define relevance matching more sharply than is possible using only article keywords. Our approach offers a general, scalable, high-throughput method for determining “PICO relevance”, which is of direct interest to clinical researchers. Moreover, the same approach is useful for cohort identification, eligibility determination, and similarity determination, such that once a study’s eligibility criteria are coded in ERGO Annotation, both the instance data and the methods can be reused across multiple use cases.
Finally, we know of no other group that is integrating the conceptualization of bias and clinical confounding with clinical research informatics models. This integration is crucial for enabling fluid support for a learning health system in which clinical research and methodologically rigorous evidence-based care are two sides of the same coin.
5.1 Limitations
OCRe continues to be a work in progress. The study design typology has been revised since its pilot evaluation and needs renewed validation. Our proposed matching methods have not been tested. Our work on modeling sources of bias is early and a proof of concept is still needed on using OCRe as a valid way to organize and index biases and clinical confounders for a wide range of study types. Above all, we have not yet modeled summary or individual-patient level results in OCRe, although the stubs for doing so are already present.
5.2 Current and future work
Our OCRe work is currently focused on three main objectives: 1) deployment into real use environments; 2) modeling, visualization, and computational methods for evidentiary reasoning; and 3) modeling of results data. OCRe is being incorporated into AHRQ’s Systematic Review Data Repository (SRDR) system, which is a workflow support system and study results repository [56]. Exploratory work is continuing on using OCRe as the common semantics for Cochrane Collaboration’s Central Register of Studies, and for data sharing between SRDR and the Cochrane Collaboration.
In the longer term, OCRe’s greatest value will be to underpin all five phases of the study lifecycle within a learning health system, such that study and clinical phenotypes can be matched and integrated during the processes of care. A step towards this vision is that OCRe will be used as the semantic foundation for evidence in the Open mHealth architecture for mobile health [98]. The goal is “embedded evaluation:” large and small-scale patient-centered studies can be conducted using mobile and sensing technology to “embed” the collection of evidence into daily life, where health prevention and chronic disease are most salient [60]. OCRe will be used to index software modules corresponding to data or functional elements of study designs (e.g., randomization, baseline characteristics), shared variable libraries, and protocol libraries. Our first project is using OCRe to inform the modular construction of a mobile-based system for n-of-1 studies for chronic pain in the PREEMPT project [99].
6. Conclusion
Now is an auspicious time for clinical research informatics to be grounded on an ontology for the science of clinical research. Technically, the science of ontologies is maturing [100] with tools like BioPortal [101]. Both US and European policies are spurring greater results reporting not just as PDFs but also directly into databases. Reproducible research and other initiatives are pushing for the sharing of protocol information as well as results data. As more clinical research data and information become shared and computable, OCRe provides well-motivated and well-formed semantics at the core of the science of clinical research to drive and integrate all phases of the study lifecycle. We have shown that with its breadth and richness, OCRe is a foundational informatics framework for clinical research and the application of evidence in a learning health system.
Highlights.
A study’s lifecycle is design, execution, reporting, interpretation, application
The Ontology of Clinical Research (OCRe) models the study design of human studies
OCRe is a generic model applicable to multiple applications and clinical domains
ERGO Annotation represents the essential meaning of eligibility criteria
OCRe supports the entire study lifecycle and the science of clinical research
Acknowledgments
This publication was made possible by Grant Numbers LM-06780, RR026040, and UL1RR025005 (Johns Hopkins), UL1RR024143 (Rockefeller), UL1RR024131 (UCSF), UL1RR025767 (UTHSC-San Antonio) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
We thank the following for their contributions to the work described in this paper: Jim Brinkley, Todd Detwiler, Rob Wynden, Karl Burke, Meredith Nahm, Davera Gabriel, Herb Hagler, Richard Scheuermann, Swati Chakraborty, Shamim Mollah, Ed Barbour, Svetlana Kiritchenko, Berry de Bruijn, Joel Martin, Karla Lindquist, Tarsha Darden, Graham Williams, Margaret Storey.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clinical Research Informatics | AMIA. [[Accessed: 7/5/2013].]; http://www.amia.org/applications-informatics/clinical-research-informatics.
- 2.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG project: a technical report. Journal of the American Medical Informatics Association : JAMIA. 2008;15:130–7. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BRIDG. [[Accessed: 7/5/2013].]; http://www.bridgmodel.org/
- 4.Megan Kong Y, Dahlke C, Xiang Q, Qian Y, Karp D, Scheuermann RH. Toward an ontology-based framework for clinical research databases. J Biomed Inform. 2011;44:48–58. doi: 10.1016/j.jbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDISC. [[Accessed: 7/5/2013].]; http://www.cdisc.org/
- 6.Sim I, Carini S, Tu S, Wynden R, Pollock BH, Mollah SA, et al. The human studies database project: federating human studies design data using the ontology of clinical research. AMIA Summits Transl Sci Proc. 2010;2010:51–55. [PMC free article] [PubMed] [Google Scholar]
- 7.Sim I, Carini S, Tu SW, Detwiler LT, Brinkley J, Mollah SA, et al. Ontology-based federated data access to human studies information. AMIA Annu Symp Proc. 2012;2012:856–865. [PMC free article] [PubMed] [Google Scholar]
- 8.Katzan IL, Rudick RA. Time to integrate clinical and research informatics. Sci Transl Med. 2012;4:162fs41. doi: 10.1126/scitranslmed.3004583. [DOI] [PubMed] [Google Scholar]
- 9.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2:57cm29. doi: 10.1126/scitranslmed.3001456. [DOI] [PubMed] [Google Scholar]
- 10.Olsen L, Aisner D, McGinnis JM. The learning healthcare system: workshop summary. National Academies Press; Washington, D.C: 2007. [PubMed] [Google Scholar]
- 11.Richardson WS, Wilson MC, Nishikawa J, Hayward RS. The well-built clinical question: a key to evidence-based decisions. ACP J Club. 1995;123:A12–3. [PubMed] [Google Scholar]
- 12.Brian Haynes R. Forming Research Questions. In: Brian Haynes R, Sackett DL, Guyatt G, Tugwell P, editors. Clinical epidemiology: how to do clinical practice research. Lippincott Williams & Wilkins; Philadelphia: 2006. p. 496. [Google Scholar]
- 13.PICO Linguist. [[Accessed: 7/4/2013].]; http://babelmesh.nlm.nih.gov/pico.php.
- 14.Hernandez M, Falconer SM, Storey M, Carini S, Sim I. Synchronized tag clouds for exploring semi-structured clinical trial data. Proceedings of the 2008 conference of the center for advanced studies on collaborative research: meeting of minds; Ontario, Canada. 2008; pp. 42–56. [Google Scholar]
- 15.Brian Haynes R. Forming research questions. J Clin Epidemiol. 2006;59:881–886. doi: 10.1016/j.jclinepi.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PubMed Clinical Queries. [[Accessed: 7/5/2013].]; http://www.ncbi.nlm.nih.gov/pubmed/clinical?term=vitamind#clincat=Therapy,Narrow.
- 17.Hartling L, Bond K, Santaguida PL, Viswanathan M, Dryden DM. Testing a tool for the classification of study designs in systematic reviews of interventions and exposures showed moderate reliability and low accuracy. J Clin Epidemiol. 2011;64:861–871. doi: 10.1016/j.jclinepi.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Bekhuis T, Demner-Fushman D, Crowley RS. Comparative effectiveness research designs: an analysis of terms and coverage in Medical Subject Headings (MeSH) and Emtree. J Med Libr Assoc. 2013;101:92–100. doi: 10.3163/1536-5050.101.2.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohrig B, du Prel JB, Wachtlin D, Blettner M. Types of study in medical research: part 3 of a series on evaluation of scientific publications. Dtsch Arztebl Int. 2009;106:262–268. doi: 10.3238/arztebl.2009.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes DA, Schulz KF. An overview of clinical research: the lay of the land. Lancet. 2002;359:57–61. doi: 10.1016/S0140-6736(02)07283-5. [DOI] [PubMed] [Google Scholar]
- 21.What study designs should be included in an EPOC reviews and what should they be called. . [[Accessed: 7/4/2013].]; http://epocoslo.cochrane.org/sites/epocoslo.cochrane.org/files/uploads/WhatstudydesignsshouldbeincludedinanEPOCreviewsandwhatshouldtheybecalled.pdf.
- 22.Carini S, Pollock BH, Lehmann HP, Bakken S, Barbour EM, Gabriel D, et al. Development and evaluation of a study design typology for human research. AMIA Annu Symp Proc. 2009;2009:81–85. [PMC free article] [PubMed] [Google Scholar]
- 23.Evidence-Based Medicine Working Group, Evidence-based medicine. A new approach to teaching the practice of medicine. JAMA. 1992;268:2420–2425. doi: 10.1001/jama.1992.03490170092032. [DOI] [PubMed] [Google Scholar]
- 24.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. J Biomed Inform. 2004;37:108–119. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Dechartres A, Charles P, Hopewell S, Ravaud P, Altman DG. Reviews assessing the quality or the reporting of randomized controlled trials are increasing over time but raised questions about how quality is assessed. Journal of clinical epidemiology. 2011;64:136–44. doi: 10.1016/j.jclinepi.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Assessing the Risk of Bias of Nonrandomized Studies of Interventions or Exposures: Further Development of the RTI Item Bank - Abstract - Draft | AHRQ Effective Health Care Program. [[Accessed: 7/7/2013].]; http://effectivehealthcare.ahrq.gov/index.cfm/search-for-guides-reviews-and-reports/?productid=956&pageaction=displayproduct.
- 27.Greenland S, O’Rourke K. On the bias produced by quality scores in meta-analysis, and a hierarchical view of proposed solutions. Biostatistics (Oxford, England) 2001;2:463–71. doi: 10.1093/biostatistics/2.4.463. [DOI] [PubMed] [Google Scholar]
- 28.Guyatt G, Rennie D, Meade MO, Cook DJ. Users’ Guides to the Medical Literature: A Manual for Evidence-Based Clinical Practice. 2 [Google Scholar]
- 29.Statistics notes | BMJ. [[Accessed: 7/4/2013].]; http://www.bmj.com/bmj-series/statistics-notes.
- 30.Higgins J, Green S. Cochrane Handbook for Systematic Reviews of Interventions [updated March 2011]. Version 5.1.0. The Cochrane Collaboration; 2011. [Google Scholar]
- 31.CEBM > EBM Tools > Critical Appraisal > Overview. [Accessed: 7/5/2013]; http://www.cebm.net/index.aspx?o=1157.
- 32.Lehmann HP. Bayesian computer-based approach to the physician’s use of the clinical research literature. 1992 [Google Scholar]
- 33.Rennels GD, Shortliffe EH, Stockdale FE, Miller PL. A computational model of reasoning from the clinical literature. Comput Methods Programs Biomed. 1987;24:139–149. doi: 10.1016/0169-2607(87)90025-3. [DOI] [PubMed] [Google Scholar]
- 34.CDISC SDTM. [[Accessed: 7/5/2013].]; http://www.cdisc.org/sdtm.
- 35.Stuart C. JAMA feature: Most clinical trials fail to add meaningful evidence 2012 September 21, 2012- May 1, 2012 [Google Scholar]
- 36.Altman DG. The scandal of poor medical research. BMJ. 1994;308:283–4. doi: 10.1136/bmj.308.6924.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinson BC, Anderson MS, de Vries R. Scientists behaving badly. Nature. 2005;435:737–738. doi: 10.1038/435737a. [DOI] [PubMed] [Google Scholar]
- 38.Bruno R, Mercier F, Claret L. Model-Based Drug Development in Oncology: What/’s Next? Clin Pharmacol Ther. 2013;93:303–305. doi: 10.1038/clpt.2013.8. [DOI] [PubMed] [Google Scholar]
- 39.van Herick A, Schuetz, Alperin, Bullano, Balu, Gandhi S. The impact of initial statin treatment decisions on cardiovascular outcomes in clinical care settings: estimates using the Archimedes Model. ClinicoEconomics and Outcomes Research. 2012:337. doi: 10.2147/CEOR.S35487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nammuni K, Pickering C, Modgil S, Montgomery A, Hammond P, Wyatt JC, et al. Design-a-trial: a rule-based decision support system for clinical trial design. Knowledge-Based Syst. 2004;17:121–129. [Google Scholar]
- 41.Richesson RL, Andrews JE. Clinical Research Informatics. Springer; 2012. [Google Scholar]
- 42.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010;2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 43.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(Suppl 1):i116–24. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuggia M, Besana P, Glasspool D. Comparing semi-automatic systems for recruitment of patients to clinical trials. Int J Med Inf. 2011;80:371–388. doi: 10.1016/j.ijmedinf.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 45.WMA Declaration of Helsinki. [Accessed: 7/5/2013];Ethical Principles for Medical Research Involving Human Subjects. http://www.wma.net/en/30publications/10policies/b3/
- 46.Williams RJ, Tse T, Harlan WR, Zarin DA. Registration of observational studies: is it time? CMAJ. 2010;182:1638–1642. doi: 10.1503/cmaj.092252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godlee F. Clinical trial data for all drugs in current use. BMJ. 2012:345. doi: 10.1136/bmj.e7304. [DOI] [PubMed] [Google Scholar]
- 48.Trials | Instructions for Authors. [[Accessed: 7/6/2013].]; http://www.trialsjournal.com/authors/instructions.
- 49.Annals of Internal Medicine | Information For Authors | Research/Publication Ethics. [[Accessed: 7/4/2013].]; http://annals.org/public/authorsinfo.aspx#research-publication-ethics.
- 50.Eichler HG, Abadie E, Breckenridge A, Leufkens H, Rasi G. Open clinical trial data for all? A view from regulators. PLoS Med. 2012;9:e1001202. doi: 10.1371/journal.pmed.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murad M, Montori V. Synthesizing evidence: Shifting the focus from individual studies to the body of evidence. JAMA. 2013;309:2217–2218. doi: 10.1001/jama.2013.5616. [DOI] [PubMed] [Google Scholar]
- 52.Barton S. Which clinical studies provide the best evidence? The best RCT still trumps the best observational study. BMJ. 2000;321:255–256. doi: 10.1136/bmj.321.7256.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS Med. 2010;7:e1000326. doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsafnat G, Dunn A, Glasziou P, Coiera E. The automation of systematic reviews. BMJ. 2013;346:f139. doi: 10.1136/bmj.f139. [DOI] [PubMed] [Google Scholar]
- 55.RevMan | The Cochrane IMS. [[Accessed: 7/6/2013].]; http://ims.cochrane.org/revman.
- 56.SRDR - Systematic Review Data Repository | Home. [[Accessed: 7/6/2013].]; http://srdr.ahrq.gov/
- 57.OpenMeta[Analyst] -- CEBM @ Brown. [[Accessed: 7/6/2013].]; http://www.cebm.brown.edu/open_meta.
- 58.Fiore LD, Brophy M, Ferguson RE, D’Avolio L, Hermos JA, Lew RA, et al. A point-of-care clinical trial comparing insulin administered using a sliding scale versus a weight-based regimen. Clin Trials. 2011;8:183–195. doi: 10.1177/1740774511398368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kravitz RL, Paterniti DA, Hay MC, Subramanian S, Dean DE, Weisner T, et al. Marketing therapeutic precision: Potential facilitators and barriers to adoption of n-of-1 trials. Contemp Clin Trials. 2009;30:436–445. doi: 10.1016/j.cct.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Chen C, Haddad D, Selsky J, Hoffman JE, Kravitz RL, Estrin DE, et al. Making sense of mobile health data: an open architecture to improve individual- and population-level health. J Med Internet Res. 2012;14:e112. doi: 10.2196/jmir.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salathé M, Freifeld CC, Mekaru SR, Tomasulo AF, Brownstein JS. Influenza A (H7N9) and the Importance of Digital Epidemiology. N Engl J Med. 2013 doi: 10.1056/NEJMp1307752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rector AL, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Appl Ontol. 2009;4:51–69. [Google Scholar]
- 63.HL7: Reference Information Model (RIM) Downloads. [[Accessed: 7/6/2013].]; http://www.hl7.org/implement/standards/rim.cfm.
- 64.Code of Federal Regulations | HHS.gov. [[Accessed: 7/6/2013].]; http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.html#46.102.
- 65.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011;44:239–250. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen JF. Maintaining knowledge about temporal intervals. Commun ACM. 1983;26:832–843. [Google Scholar]
- 67.Chung GY, Coiera E. A study of structured clinical abstracts and the semantic classification of sentences. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing; Prague, Czech Republic. 2007; pp. 121–128. [Google Scholar]
- 68.Hara K, Matsumoto Y. Extracting clinical trial design information from MEDLINE abstracts. New Gen Comput. 2007;25:263–275. [Google Scholar]
- 69.Xu R, Garten Y, Supekar KS, Das AK, Altman RB, Garber AM. Extracting subject demographic information from abstracts of randomized clinical trial reports. Stud Health Technol Inform. 2007;129:550–554. [PubMed] [Google Scholar]
- 70.Chung GY. Towards identifying intervention arms in randomized controlled trials: extracting coordinating constructions. J Biomed Inform. 2009;42:790–800. doi: 10.1016/j.jbi.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 71.Hansen M, Rasmussen N, Chung G. Extracting number of trial participants from abstracts of randomized controlled trials. Proceedings of Tromsø, Telemedicine and eHealth Conference; 2008; [DOI] [PubMed] [Google Scholar]
- 72.Demner-Fushman D, Lin J. Knowledge Extraction for Clinical Question Answering: Preliminary Results. Proceedings of AAAI Workshop on Question Answering in Restricted Domains; Pittsburgh, PA. 10 July 2005; 2005. pp. 1–9. [Google Scholar]
- 73.Kiritchenko S, de Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC Med Inform Decis Mak. 2010;10 doi: 10.1186/1472-6947-10-56. 56-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tao J, Sirin E, Bao J, McGuinness DL. Extending OWL with Integrity Constraints | Tetherless World Constellation. Proceedings of Description Logics Workshop; May 4–7, 2010.2010. [Google Scholar]
- 75.Stardog: Documentation. [[Accessed: 7/8/2013].]; http://stardog.com/docs/
- 76.Roumiantseva D, Carini S, Sim I, Wagner TH. Sponsorship and design characteristics of trials registered in ClinicalTrials.gov. Contemp Clin Trials. 2013;34:348–355. doi: 10.1016/j.cct.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 77.AACT Database. [[Accessed: 7/6/2013].]; https://www.ctti-clinicaltrials.org/project-topics/clinical-trials.gov/aact-database.
- 78.Peng RD, Dominici F, Zeger SL. Reproducible epidemiologic research. Am J Epidemiol. 2006;163:783–789. doi: 10.1093/aje/kwj093. [DOI] [PubMed] [Google Scholar]
- 79.Hernandez ME, Carini S, Storey MA, Sim I. An interactive tool for visualizing design heterogeneity in clinical trials. AMIA Annu Symp Proc. 2008:298–302. [PMC free article] [PubMed] [Google Scholar]
- 80.Brinkley JF, Detwiler LT. Structural Informatics Group, A query integrator and manager for the query web. J Biomed Inform. 2012;45:975–991. doi: 10.1016/j.jbi.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shah NH, Musen MA. UMLS-Query: a perl module for querying the UMLS. AMIA Annu Symp Proc. 2008:652–656. [PMC free article] [PubMed] [Google Scholar]
- 82.Lee WN, Shah N, Sundlass K, Musen M. Comparison of ontology-based semantic-similarity measures. AMIA Annu Symp Proc. 2008:384–388. [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang R, Gan M, He P. Constructing a gene semantic similarity network for the inference of disease genes. BMC Syst Biol. 2011;5(Suppl 2) doi: 10.1186/1752-0509-5-S2-S2. S2-0509-5-S2-S2. Epub 2011 Dec 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annu Symp Proc. 2006:359–363. [PMC free article] [PubMed] [Google Scholar]
- 85.Lee WN, Das AK. Local alignment tool for clinical history: temporal semantic search of clinical databases. AMIA Annu Symp Proc. 2010;2010:437–441. [PMC free article] [PubMed] [Google Scholar]
- 86.Lee W. Evaluating clinical practice variation using a knowledge-based temporal sequence alignment framework. 2012 [Google Scholar]
- 87.PCORI Methodology Standards. [[Accessed: 7/7/2013].]; http://www.pcori.org/assets/PCORI-Methodology-Standards1.pdf.
- 88.Chan A, Tetzlaff JM, Altman DG, Laupacis A, Gøtzsche PC, Krleža-Jerić K, et al. SPIRIT 2013 Statement: Defining Standard Protocol Items for Clinical Trials. Annals of Internal Medicine. 2013;158:200–207. doi: 10.7326/0003-4819-158-3-201302050-00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan AW, Tetzlaff JM, Gotzsche PC, Altman DG, Mann H, Berlin JA, et al. SPIRIT 2013 explanation and elaboration: guidance for protocols of clinical trials. BMJ. 2013;346:e7586. doi: 10.1136/bmj.e7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tu SW, Campbell JR, Glasgow J, Nyman MA, McClure R, McClay J, et al. The SAGE Guideline Model: achievements and overview. J Am Med Inform Assoc. 2007;14:589–598. doi: 10.1197/jamia.M2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delgado-Rodríguez M, Llorca J. Bias. Journal of Epidemiology and Community Health. 2004;58:635–641. doi: 10.1136/jech.2003.008466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Milian K, Teije A. In: Towards Automatic Patient Eligibility Assessment: From Free-Text Criteria to Queries. Peek N, Marín Morales R, Peleg M, editors. Springer; Berlin Heidelberg: 2013. pp. 78–83. [Google Scholar]
- 93.Huang Z, Teije A, Harmelen F. In: Rule-Based Formalization of Eligibility Criteria for Clinical Trials. Peek N, Marín Morales R, Peleg M, editors. Springer; Berlin Heidelberg: 2013. pp. 38–47. [Google Scholar]
- 94.Kaiser J. Making clinical data widely available. Science. 2008;322:217–218. doi: 10.1126/science.322.5899.217. [DOI] [PubMed] [Google Scholar]
- 95.Sharing Clinical Research Data: A Workshop - Institute of Medicine. [[Accessed: 7/6/2013].]; http://www.iom.edu/Activities/Research/SharingClinicalResearchData/2012-OCT-04.aspx.
- 96.Wallace BC, Trikalinos TA, Lau J, Brodley C, Schmid CH. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-55. 55-2105-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification. Journal of the American Medical Informatics Association : JAMIA. 2006;13:206–19. doi: 10.1197/jamia.M1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Estrin D, Sim I. Open mHealth Architecture: An Engine for Health Care Innovation. Science. 2010;330:759–760. doi: 10.1126/science.1196187. [DOI] [PubMed] [Google Scholar]
- 99.Kravitz RL. N-of-1 Trials Using mHealth in Chronic Pain, 1R01NR013938-01 [Google Scholar]
- 100.Musen MA, Noy NF, Shah NH, Whetzel PL, Chute CG, Story MA, et al. The National Center for Biomedical Ontology. Journal of the American Medical Informatics Association : JAMIA. 2012;19:190–5. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39:W541–5. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]