Abstract
RAD51C is one of the RAD51 paralogs that plays an important role in DNA double-strand break repair by homologous recombination. Here, we identified and characterized OsRAD51C, the rice homolog of human RAD51C. The Osrad51c mutant plant is normal in vegetative growth but exhibits complete male and female sterility. Cytological investigation revealed that homologous pairing and synapsis were severely disrupted. Massive chromosome fragmentation occurred during metaphase I in Osrad51c meiocytes, and was fully suppressed by the CRC1 mutation. Immunofluorescence analysis showed that OsRAD51C localized onto the chromosomes from leptotene to early pachytene during prophase I, and that normal loading of OsRAD51C was dependent on OsREC8, PAIR2, and PAIR3. Additionally, ZEP1 did not localize properly in Osrad51c, indicating that OsRAD51C is required for synaptonemal complex assembly. Our study also provided evidence in support of a functional divergence in RAD51C among organisms.
Keywords: rice, meiosis, OsRAD51C, double-strand break repair, chromosome fragmentation
Introduction
The maintenance of genome stability is essential for cell and organism viability. Both environmental and endogenous DNA damaging agents, including ionizing radiation, chemicals, and spontaneous DNA breakage during DNA recombination and replication can cause DNA lesions (Ward, 1988; Friedberg et al., 1995; Olive, 1998; Flores-Rozas and Kolodner, 2000). Among these lesions, DNA double-strand breaks (DSBs) are one of the most disruptive forms of DNA damage (Jackson and Bartek, 2009). Unrepaired DSBs can lead to severe chromosomal aberrations that may ultimately result in apoptosis or aneuploidy. In response to the threats posed by DNA damage, organisms have evolved effective mechanisms for DSB repair, which include two major pathways: homologous recombination (HR) and non-homologous end joining (NHEJ). HR utilizes the sister chromatid as a template for new DNA synthesis, guaranteeing the delivery of stable genetic information to the next generation (Van Den Bosch et al., 2002; Sonoda et al., 2006). In contrast, NHEJ completes DSB repair by directly ligating the DNA break ends without using a homologous template. Therefore, due to this imprecise repair mechanism, inappropriate NHEJ is potentially mutagenic and can lead to chromosome anomalies (Lees-Miller and Meek, 2003; Lieber, 2008).
Meiotic recombination is initiated by SPO11-mediated DSBs. In Saccharomyces cerevisiae, Spo11 plays a major role in catalyzing the formation of DSBs via its topoisomerase-like transesterase activity (Keeney et al., 1997; Keeney, 2001). Recent studies indicate that the role of SPO11 in the initiation of meiotic recombination seems to be highly conserved, because homologs of the SPO11 gene have been found in a wide range of organisms (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; Hartung and Puchta, 2000; Romanienko and Camerini-Otero, 2000; Yu et al., 2010). Numerous genes involved in HR have been identified recently in various organisms. Rad51, a eukaryotic homolog of bacterial RecA protein, has DNA-dependent ATPase activity and facilitates strand exchange between homologous DNA molecules (Baumann and West, 1998). Rad51 is conserved from yeast to human in both structure and function (Bezzubova et al., 1993; Shinohara et al., 1993). In addition, five RAD51 paralog proteins (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) have also been studied in mammalian cells and are thought to play important roles in the process of recombination repair (Albala et al., 1997; Dosanjh et al., 1998; Liu et al., 1998; Pittman et al., 1998; Johnson et al., 1999; Pierce et al., 1999; Bleuyard et al., 2005). These proteins have 20–30% similarity and possess common functional domains (Miller et al., 2004). Biochemical and yeast two-hybrid analyses indicate that RAD51 paralogs form two distinct protein complexes: a RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex and a RAD51C-XRCC3 (CX3) complex (Masson et al., 2001; Miller et al., 2002; Wiese et al., 2002; Yokoyama et al., 2004). Consistent with this finding, a RAD51C-defective hamster cell line fails to form either of these two complexes, implying that RAD51C plays a prominent role in the HR processes (French et al., 2002).
The RAD51C gene was originally identified in humans as a new member of the RAD51 family by sequence similarity (Dosanjh et al., 1998). The first mammalian RAD51C-defective mutant was the RAD51C-deficient Chinese hamster cell line CL-V4B. CL-V4B displayed increasing numbers of spontaneous chromosomal aberrations and was highly hypersensitive to cross-linking agents such as mitomycin C (MMC) and cisplatin (Godthelp et al., 2002; Drexler et al., 2004). Subsequently, similar phenotypes were observed in both RAD51C-depleted chicken DT cells and HeLa cells (Takata et al., 2001; Lio et al., 2004). In addition, a RAD51C-like protein, Spn-D, has been identified in Drosophila, and shows 38% identity with human RAD51C. Intriguingly the spn-D mutant is insensitive to X-rays and methyl methanesulfonate, suggesting that Spn-D may play a role only in meiotic recombination rather than DNA damage repair in somatic cells (Abdu et al., 2003). In contrast, RAD51C is essential for early development in mammals, since deficiency leads to embryonic lethality in the mouse (Kuznetsov et al., 2009). With the help of the hypomorphic allele of the rad51c mutant, further study revealed that the rad51c mutation results in early prophase I arrest in males and broken chromosomes or aneuploidy at metaphase II in female mice (Kuznetsov et al., 2007). In the higher plant, Arabidopsis, the atrad51c mutant displayed complete sterility during flowering. In addition, it was sensitive to γ-radiation and cisplatin during vegetative development and demonstrated reduced HR frequencies and increased chromosome fragments in somatic cells (Abe et al., 2005; Li et al., 2005).
An increasing number of genetic and biochemical studies have provided further understanding of the precise role of RAD51C in HR. RAD51C exhibits an ATP-independent DNA strand exchange activity, DNA-stimulated ATPase activity, and ssDNA binding activity in vitro (Lio et al., 2003), suggesting it may participate in both early and late HR events. Consistent with this, it has been reported that RAD51C not only promotes RAD51 assembly in the early stages of recombination (French et al., 2002), but is also involved in branch migration and Holliday junction (HJ) resolution in mammalian cells (Liu et al., 2004). However, RAD51C does not directly interact with the human Holliday junction resolvase GEN1, which binds specifically to and promotes resolution of HJs (Ip et al., 2008; Rass et al., 2010). The role of RAD51C in the late stages of HR remains obscure. Furthermore, RAD51C can also facilitate CHK2 phosphorylation after irradiation and thereby promotes the transduction of damage signals (Badie et al., 2009).
A RAD51C homolog has recently been characterized in rice. It has been reported that OsRAD51C is required for both male and female gamete development, and its mutant exhibits sensitivity to DNA damaging agents (Kou et al., 2012). However, the specific functions of OsRAD51C in meiotic recombination, its spatial and temporal expression pattern, and its relationships with other determinant proteins in meiosis remain obscure. In this study, we identified another Osrad51c mutant showing a similar phenotype to that reported by Kou et al. By studying chromosome behavior in Osrad51c, we found that OsRAD51C is required for processing of DSBs, homologous pairing, and synaptonemal complex (SC) assembly. Furthermore, OsRAD51C shows a localization pattern in rice that is distinct from that of its homolog in mice. Our results therefore provide further clues to the role of OsRAD51C in meiosis.
Materials and methods
Plant materials
The rice (Oryza sativa L.) Osrad51c mutant, induced by 60Co γ-ray radiation, was isolated from the indica rice variety Zhongxian 3037. We then generated the Osrad51c crc1 double mutant by crossing heterozygous Osrad51c± and crc1± and selecting double mutants in the F2 progeny. The meiotic mutants used in this study, including Osrec8, pair2, pair3, and crc1, have been reported in previous work (Shao et al., 2011; Wang et al., 2011; Miao et al., 2013). All plants were cultivated in paddy fields under normal growth conditions.
Map-based cloning of OsRAD51C
The F2 mapping populations were constructed from a cross between heterozygous Osrad51c± and a japonica cultivar, Zhonghua11, and 898 sterile plants were selected for isolating the target gene. Sequence-tagged site (STS) markers were developed based on sequence differences between the indica variety 9311 and the japonica variety Nipponbare according to the data published on the NCBI website (http://www.ncbi.nlm.nih.gov). All primer sequences are listed in Supplemental Table 1.
RNAi analysis
For the RNAi construct, a 470-bp fragment of the OsRAD51C cDNA sequence was amplified using the primer pairs CR1 (Supplemental Table 1). RNAi vector construction and transformation were performed as previously described (Wang et al., 2009).
Antibody production
To generate antibody against OsRAD51C, a 210-bp fragment of OsRAD51C cDNA was amplified using primer CA1 (Supplemental Table 1). Vector construction, fusion peptide expression, and protein purification were performed as previously described (Wang et al., 2009). The fusion peptide was used to immunize mice to generate anti-OsRAD51C polyclonal antibody. The polyclonal antibodies to OsREC8, PAIR2, PAIR3, and ZEP1 have been described previously (Wang et al., 2010; Shao et al., 2011; Wang et al., 2012).
Meiotic chromosome preparation and cytological analysis
Young panicles were harvested and fixed in Carnoy's fixative solution and stored at −20°C. Microsporocytes at the appropriate meiotic stage were squashed and stained with acetocarmine. After washing with 45% acetic acid, the chromosome preparations were frozen in liquid nitrogen. After removing the coverslips, the slides were dehydrated through an ethanol series (70, 90, and 100%). Chromosomes were counterstained with 4,6-diamidinophenylindole (DAPI) in an antifade solution (Vector Laboratories, Burlingame, CA, USA). Images were captured under the ZEISS A2 fluorescence microscope with a micro CCD camera.
Immunofluorescence assays
Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 10–30 min at room temperature. Anthers were squashed on a slide in 1× PBS solution. After freezing in liquid nitrogen and quickly removing the coverslip, the slides were dehydrated through an ethanol series (70, 90, and 100%). The slides were then incubated in a humid chamber at 37°C for 4 h in different combinations of primary antibodies diluted 1:500 in TNB buffer (0.1M Tris-HCl, pH 7.5, 0.15M NaCl, and 0.5% blocking reagent). After three washes in 1× PBS, slides were incubated with the Texas-red-conjugated goat anti-rabbit antibody and fluorescein isothiocyanate-conjugated sheep anti-mouse antibody at 37°C for 2 h, and again washed three times in 1× PBS. Finally, the slides were counterstained with DAPI in an antifade solution. Images were captured by a fluorescence microscope.
Results
Morphological characterization of the Osrad51c mutant
A sterile mutant was isolated from the progeny of Zhongxian 3037 treated with γ-ray radiation, and named Osrad51c. The mutant plant was normal during vegetative growth, but exhibited complete spikelet sterility. To further investigate the fertility of the male gametes, we examined mature pollen viability in Osrad51c by staining with iodine potassium iodide solution (I2-KI). Only empty, shrunken and unshaped pollens were observed, indicating that microspores were inviable. When pollinated with wild-type pollen, the mutant plants still did not produce any seeds, demonstrating that megaspore development was also affected. These results suggest that the sterile phenotype in Osrad51c was caused by both male and female sterility.
Isolation and identification of the OsRAD51C gene in rice
To identify the mutated gene, a map-based cloning approach was carried out in an F2 population, which was constructed by crossing the heterozygous Osrad51c± to a japonica cultivar Zhonghua11. The OsRAD51C gene was initially mapped to the long arm of chromosome 1 and further narrowed to a 19-kb region. Within this region, one candidate gene (Os01g0578000) was identified and annotated as a DNA repair and recombination protein. Phylogenetic analysis revealed that it is a rice homolog of human RAD51C that has been previously reported by Kou et al. (2012). Sequencing revealed an A-to-T single base mutation within the ninth exon of the OsRAD51C gene, which leads to premature protein termination. To further confirm that OsRAD51C was the gene responsible for the observed phenotype, we generated transgenic rice plants in which OsRAD51C transcription was downregulated using the RNAi approach. As predicted, the transgenic plants exhibited complete pollen and spikelet sterility, providing confirmation that the phenotype of Osrad51c is caused by mutation of the OsRAD51C gene.
Chromosome behaviors in the Osrad51c mutant
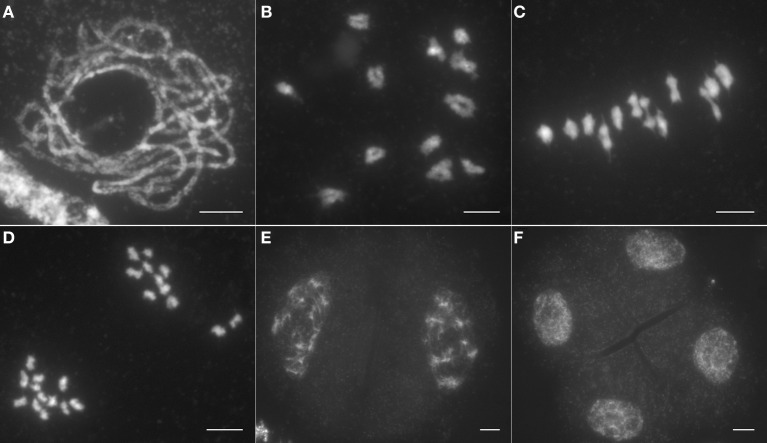
To ascertain the causes of the sterile phenotype in Osrad51c, we investigated meiotic progression in pollen mother cells (PMCs) from both wild-type plants and the Osrad51c mutant. In the wild-type, representative meiotic stages were observed: chromosomes began to condense and appeared as long thin threads at leptotene; pairing and initial synapsis occurred between homologous chromosomes at the zygotene stage; homologs became tightly associated and fully synapsed during pachytene (Figure 1A). After further chromosome condensation at diplotene, 12 short bivalents were clearly observed at diakinesis (Figure 1B), and these aligned on the equatorial plate during metaphase I (Figure 1C). From anaphase I to telophase I, homologous chromosomes separated from each other and migrated to opposite poles (Figure 1D). In meiosis II, sister chromatids separated and tetrads were generated (Figures 1E,F).
Figure 1.
Male meiosis in wild type. (A) Pachytene; (B) Diakinesis; (C) Metaphase I; (D) Anaphase I; (E) Dyad; (F) Tetrad. Bars, 5 μm.
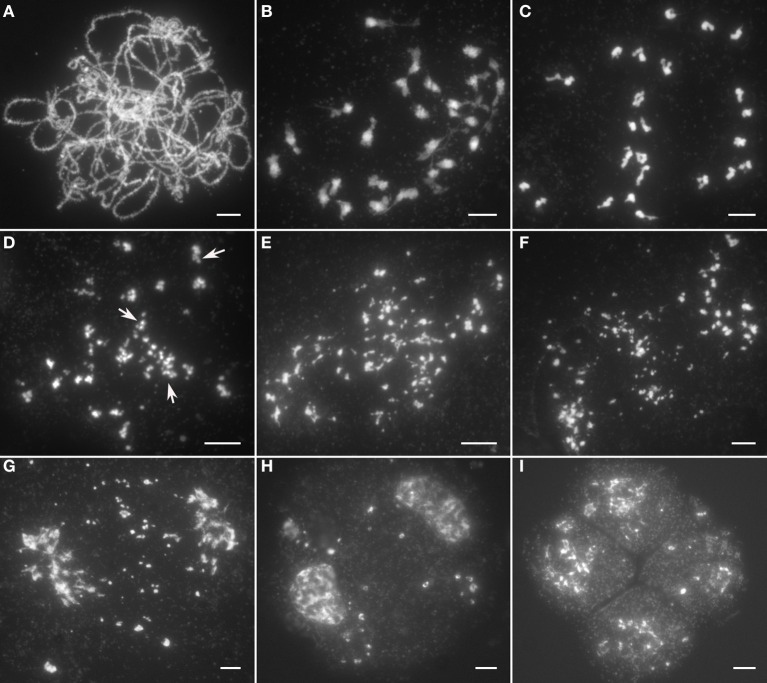
Chromosomal behavior at leptotene in Osrad51c was quite similar to the wild-type. However, obvious defects in meiotic chromosomes were first observed at zygotene, when Osrad51c chromosomes remained as single threads and did not pair up with their homologs. Fully synapsed homologs were never observed at the pachytene stage in Osrad51c mutants (Figure 2A). At the diakinesis stage, following further condensation, 24 irregularly shaped univalents were observed in the PMCs (Figure 2B). At early metaphase I, the univalents did not align along the equatorial plate and remained scattered throughout the nucleus (Figure 2C). Meanwhile, a few small pieces which had broken away from the main chromosome were detected for some univalents (Figure 2D, arrows). Subsequently, the number of chromosomal fragments increased markedly in a short time, and became randomly distributed throughout the entire nucleus (Figure 2E). Despite the massive genome fragmentation, the meiotic cell cycle still entered into anaphase I, with the chromosomes asynchronously migrating toward the two opposite poles and acentric chromosomal fragments becoming randomly scattered in the nucleus (Figure 2F). At telophase I, when the major chromosomes arriving at the poles started to decondense, many lagging fragments could still be seen excluded from the nuclei (Figure 2G). They finally led to the formation of numerous micronuclei in the subsequent dyads and tetrads (Figures 2H,I). We therefore concluded that the sterility of the Osrad51c mutant is caused by defective homologous chromosome pairing and massive chromosomal fragmentation. Similar abnormalities were also observed in OsRAD51C RNAi plant meiocytes.
Figure 2.
Male meiosis in the Osrad51c mutant. (A) Pachytene; (B) Diakinesis; (C) Early metaphase I; (D) Middle metaphase I; (E) Late metaphase I; (F) Anaphase I; (G) Telophase I; (H) Dyad; (I) Tetrad. Bars, 5 μ m.
DSB formation is a prerequisite for chromosome fragmentation in Osrad51c
To further clarify whether chromosome fragmentation resulted from unrepaired DSBs, we generated Osrad51c crc1 double mutants. CRC1, a component of the central region of the SC in rice, is required for meiotic chromosome pairing and recombination, and DSB formation is completely abolished in the absence of CRC1 (Miao et al., 2013). In the crc1 single mutant, homologous chromosome pairing and synapsis were defective and 24 univalents were seen at diakinesis and metaphase I (Figures 3A,B). However, in the Osrad51c crc1 double mutant, chromosomal behavior was very similar to that in crc1. No DNA fragments were detected; instead, only 24 intact univalents were observed at both diakinesis and metaphase I in Osrad51c crc1 (Figures 3C,D). The massive chromosomal fragmentation seen in Osrad51c was completely suppressed by the defective CRC1. We therefore concluded that failure of meiotic DSB repair is responsible for the massive chromosomal fragmentation in Osrad51c.
Figure 3.
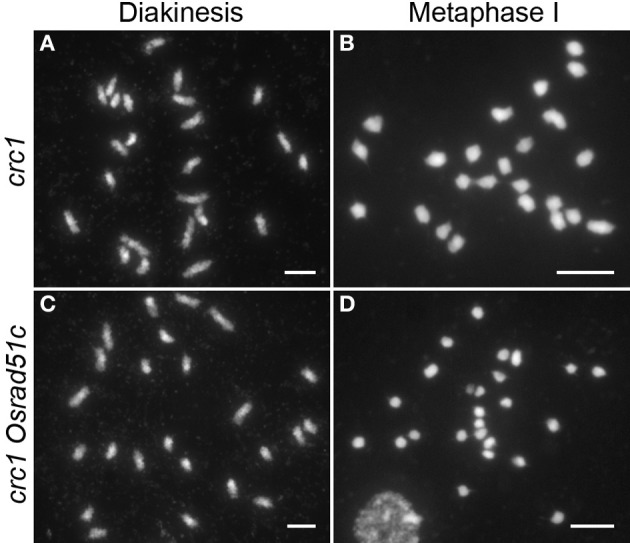
Male meiosis in crc1 (A,B) and crc1 Osrad51c (C,D). (A,C) Diakinesis; (B,D) Metaphase I. Bars, 5 μm.
OsRAD51C localizes to meiotic chromosomes from leptotene to early pachytene
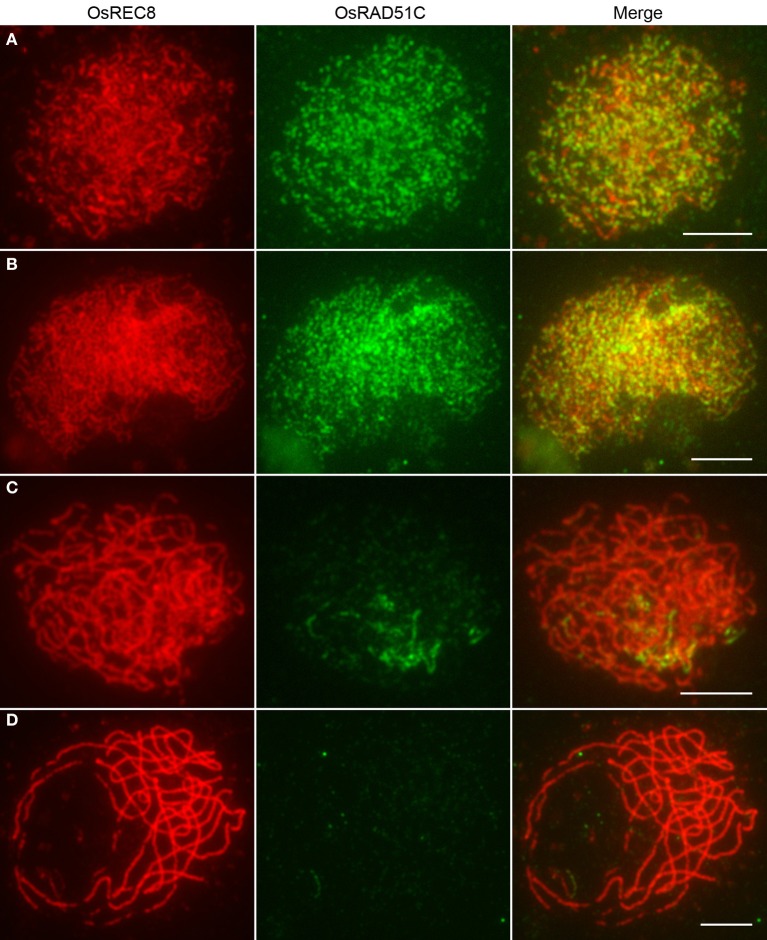
In order to investigate the spatial and temporal localization patterns of OsRAD51C protein during meiosis, immunofluorescence assays were performed using polyclonal antibodies against OsRAD51C and OsREC8, from mice and rabbits, respectively. REC8, a key component of the sister chromatid cohesion complex, is required for axial element formation and homolog pairing. OsREC8 is first detectable as a diffuse signal during premeiotic interphase and gradually elongates along the entire length of the chromosomes from leptotene to pachytene. The signal eventually disappears at metaphase I. Consequently, OsREC8 has frequently been used as a chromosome marker in prophase I (Shao et al., 2011). The OsRAD51C signals were first detected as punctuate foci at leptotene (Figure 4A). The number of these foci gradually increased and reached a maximal level at zygotene (Figure 4B). Subsequently, OsRAD51C protein began to rapidly dissociate from the chromosomes. Once meiosis entered early pachytene, only a few residual signals could be seen (Figure 4C). At late pachytene, the OsRAD51C had completely disappeared from the chromosomes and was not detected in subsequent stages (Figure 4D). We therefore conclude that OsRAD51C plays a role in homologous chromosome pairing and DSB repair from leptotene to early pachytene in rice meiosis.
Figure 4.
Dual immunolocalization of OsREC8 (red) and OsRAD51C (green) in wild type PMCs. (A) Leptotene; (B) Zygotene; (C) Mid prophase I; (D) Late pachytene. Bars, 5 μm.
OsRAD51C is required for normal SC assembly
The formation of SCs accompanies meiotic recombination and thus serves as a cytological marker of mature HR. To determine whether SC formation was affected in Osrad51c, we further investigated the localization of ZEP1, PAIR2, and PAIR3. PAIR2, the homolog of S. cerevisiae HOP1 and Arabidopsis ASY1, which associates with the axial element/lateral element (AE/LE), is required for homologous chromosome synapsis during early prophase I in rice (Nonomura et al., 2006). ASY1 foci also serve as a marker to detect chromosome axis formation both in Arabidopsis and barley (Chelysheva et al., 2005; Wang et al., 2010; Higgins et al., 2012). PAIR3 is an AE/LE associated protein, which is essential for homologous pairing, normal recombination, and SC assembly (Wang et al., 2011). ZEP1, a transverse filament protein, is the central element of the SC (Wang et al., 2010). These three proteins were used to monitor the assembly of SCs in Osrad51c meiocytes. We found that PAIR2 and PAIR3 in Osrad51c behaved in the same manner as in the wild-type. Both of them were co-localized and distributed along the chromosomes (Figure 5B), indicating that the axial elements of SCs were able to assemble completely in the absence of OsRAD51C. In the wild-type prophase I, ZEP1 signals were first visible as punctuate foci at leptotene, then quickly elongated to form linear signals along the entire chromosomes at early pachytene. However, in Osrad51c meiocytes, SC extension was blocked, and only short discontinuous linear signals were observed at pachytene (Figure 5A), suggesting the failure of SC assembly in Osrad51c. Together, although the formation of AEs appeared normal, the central element of the SC was not completely assembled, demonstrating that mature SC formation was disrupted in the absence of OsRAD51C.
Figure 5.
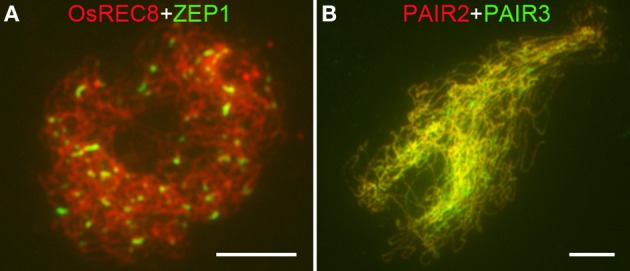
Dual immunolocalization of several meiotic elements in Osrad51c PMCs. (A) OsREC8 (red) and ZEP1 (green) signals at early pachytene; (B) PAIR2 (red) and PAIR3 (green) signals at early pachytene. Bars, 5 μ m.
OsRAD51C localization relies on OsREC8, PAIR2, and PAIR3, but not on ZEP1
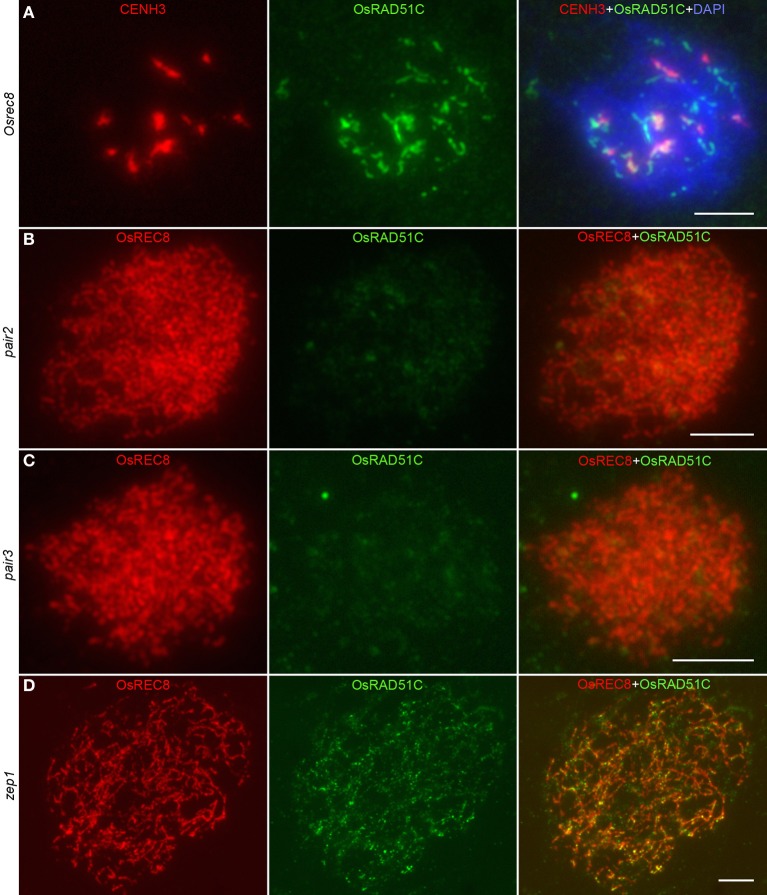
To further examine OsRAD51C function in rice meiosis, we investigated the recruitment of OsRAD51C in Osrec8, pair2, pair3, and zep1 mutants by immunofluorescence. In Osrec8, we employed CENH3 as a centromere marker to indicate the PMCs. Unlike the discrete dot-like signals at zygotene in the wild-type, OsRAD51C signals were gathered into bigger and brighter aggregates in Osrec8 (Figure 6A), implying that correct loading of OsRAD51C requires OsREC8. We then examined the location of OsRAD51C proteins in the other three mutants using anti-OsREC8 antibody as a chromosome marker. We failed to detect any OsRAD51C signals in both the pair2 and pair3 mutants (Figures 6B,C), suggesting that OsRAD51C loading also relies on PAIR2 and PAIR3. However, fine punctuate foci of OsRAD51C protein were distributed normally along the chromosomes in zep1 zygotene meiocytes (Figure 6D), indicating that defective ZEP1 does not affect OsRAD51C loading. We therefore concluded that normal localization of OsRAD51C depends on OsREC8, PAIR2, and PAIR3, rather than ZEP1.
Figure 6.
The localization of OsRAD51C at leptotene in different meiotic mutants of rice. (A) Osrec8; (B) pair2; (C) pair3; (D) zep1. The CENH3 signals (red) indicating the PMC in Osrec8, and the OsREC8 signals (red) indicating the PMCs in pair2, pair3, and zep1. OsRAD51C signals in green. Chromosomes stained with DAPI (blue). Bars, 5μ m.
Discussion
The role of OsRAD51C in rice meiosis
Meiotic recombination is initiated by the deliberate introduction of DSBs. Repair of these SPO11-mediated DSBs is a complicated process, involving multiple proteins via various pathways. RAD51C is known to play an important role in DSB repair by HR. In mammalian cells, RAD51C participates in branch migration and Holliday junction resolution (Liu et al., 2004). RAD51C mutation in cell lines results in spontaneous chromosomal aberrations and hypersensitivity to DNA damaging agents (Godthelp et al., 2002; Drexler et al., 2004). In addition, disruption of RAD51C always leads to embryonic lethality in mammals (Shu et al., 1999; Deans et al., 2000; Kuznetsov et al., 2007).
In contrast, RAD51C mutation in higher plants generally leads to a sterile phenotype, without affecting vegetative growth. Previous studies in Arabidopsis demonstrated that AtRAD51C is required for HR and SPO11-dependent DSB repair. The Atrad51c-1 mutant shows chromosomal fragmentation, which can be suppressed in the spo11-1 atrad51c-1 double mutant (Abe et al., 2005; Li et al., 2005). Moreover, AtRAD51C-deficient plants are also sensitive to DNA damaging factors (Bleuyard et al., 2005). In this study, considerable numbers of chromosome fragments were also detected from metaphase I in Osrad51c, suggesting that DSBs were formed but not properly repaired. As rice CRC1 mutation inhibits DSB formation (Miao et al., 2013), we employed crc1 instead of spo11-1 to generate the Osrad51c crc1 double mutant. The massive chromosomal fragmentation phenotype in Osrad51c was completely suppressed by the deletion of CRC1, indicating that OsRAD51C likely acts downstream of CRC1 and plays an important role in the process of DSB repair.
In mice, RAD51C foci could still be visualized at the pachytene stage, when they were clearly observed as one or two distinct foci associated with each paired chromosome. Interestingly, the staining pattern was similar to that of MLH1, which serves as a marker of chiasmata at the late pachytene/diplotene stage. In addition, in Mlh1-deficient mice, the RAD51C foci were markedly reduced (Liu et al., 2007). These results suggest that RAD51C foci may represent the sites of crossovers, and provide support for the role of RAD51C in the late stages of HR. In our study, OsRAD51C signals were first observed at leptotene as punctuate foci; these increased in number and reached a peak at zygotene. However, in contrast to the findings in mice, the signal had completely disappeared at pachytene. The differences in the distribution pattern of RAD51C between mammals and plants suggest that functional differentiation has taken place during evolution.
Timing of the appearance of unrepaired chromosomal fragments
Several studies on mutants with chromosome fragmentation have shown that the fragments appear at different stage of meiosis. In the Arabidopsis Atxrcc3 mutant, chromosomal fragments first appear at diplotene, while in the atrad51 mutant they occur at diakinesis. Another study demonstrated that mutation in rice RAD51C led to the appearance of chromosomal fragments at pachytene (Bleuyard and White, 2004; Li et al., 2004; Puizina et al., 2004; Kou et al., 2012). However, in the present study, the chromosomal fragments were first observed at early metaphase I. A possible reason for this discrepancy is that the unrepaired fragment of chromatids were stuck together by sister chromatid cohesion. It is recognized that cohesion between sister chromatids depends on the multi-protein complex known as the cohesin complex. In meiosis, the cohesin along sister chromatid arms is dissolved during the metaphase I to anaphase I transition, while centromeric cohesion is maintained by shugoshin proteins until meiosis II (Xiong and Gerton, 2010). Generally, DSBs are generated during leptotene and are ultimately processed into crossovers and non-crossovers at zygotene. The sister chromatid arms are held together during this process by cohesin until they are gradually released at metaphase I. We therefore suggest that this is why we did not observe DNA fragments during early prophase I in the Osrad51c mutant. As the meiotic cell cycle proceeded, uneven chromosome condensation occurred, and chromosomes with irregular shape were found at diakinesis and early metaphase I. Following the degradation of cohesin on the chromosome arms, acentric chromosome fragments could be seen randomly distributed in the nucleus. Finally, we speculate that due to differences in chromosome size and cohesin distribution patterns, or even methods of chromosome preparation, chromosome fragments may seem to appear at different meiotic stages in different studies (Luo et al., 2014).
Meiotic DSB repair requires a proper structural platform
The cohesin complexes not only function in mediating sister chromatid cohesion but have also been shown to be required for DSB repair (Birkenbihl and Subramani, 1992; Sjögren and Nasmyth, 2001; Atienza et al., 2005). REC8 is a meiosis-specific component of the cohesin complex, and is involved in homologous pairing and recombination and monopolar orientation (Klein et al., 1999; Brar et al., 2006; Shao et al., 2011). REC8 is also a master regulator of chromatin structure and is required for AE formation (Pasierbek et al., 2001; Xu et al., 2005). AEs play a central role in establishing the leptotene chromosome and provide a structural platform for subsequent meiotic events. PAIR2 and PAIR3, two AE-associated proteins in rice, are involved in homologous pairing and recruiting recombination factors onto chromosomes. Although the exact relationship of PAIR2 and PAIR3 with AE proteins is still unclear, we speculate that they may be constituents of a structure that associates with AE, based on previous studies (Nonomura et al., 2006; Wang et al., 2011).
In this study, we found that normal loading of OsRAD51C was partially disrupted in the Osrec8 mutant. However, in pair2 and pair3 mutants, almost no signals were detectable during the zygotene stage, suggesting that PAIR2 and PAIR3 provide a structural platform for the recombination intermediates. Once the platform is destroyed, all other recombination proteins then fail to be loaded normally. Thus, we hypothesize that DSB processing requires a proper structural chromosome platform mainly composed of AE proteins. It is also possible that DSBs in pair2 and pair3 are repaired using sister chromatids as templates.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Ministry of Sciences and Technology of China (2011CB944602 and 2012AA10A301), and the National Natural Science Foundation of China (31230038 and 31170288).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00167/abstract
The list of primers used in this study.
References
- Abdu U., González-Reyes A., Ghabrial A., Schüpbach T. (2003). The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics 165, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., et al. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139, 896–908 10.1104/pp.105.065243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albala J., Thelen M., Prange C., Fan W., Christensen M., Thompson L., et al. (1997). Identification of a novel human RAD51 homolog, RAD51B. Genomics 46, 476–479 10.1006/geno.1997.5062 [DOI] [PubMed] [Google Scholar]
- Atienza J. M., Roth R. B., Rosette C., Smylie K. J., Kammerer S., Rehbock J., et al. (2005). Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368 10.1158/1535-7163.MCT-04-0241 [DOI] [PubMed] [Google Scholar]
- Badie S., Liao C., Thanasoula M., Barber P., Hill M., Tarsounas M. (2009). RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J. Cell Biol. 185, 587–600 10.1083/jcb.200811079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., West S. (1998). Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23, 247–251 10.1016/S0968-0004(98)01232-8 [DOI] [PubMed] [Google Scholar]
- Bezzubova O., Shinohara A., Mueller R., Ogawa H., Buerstedde J. (1993). A chicken RAD51 homologue is expressed at high levels in lymphoid and reproductive organs. Nucleic Acids Res. 21, 15770–11580 10.1093/nar/21.7.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R. P., Subramani S. (1992). Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20, 6605–6611 10.1093/nar/20.24.6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard J. Y., Gallego M. E., Savigny F., White C. I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41, 533–545 10.1111/j.1365-313X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Bleuyard J.-Y., White C. I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23, 439–449 10.1038/sj.emboj.7600055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar G. A., Kiburz B. M., Zhang Y., Kim J.-E., White F., Amon A. (2006). Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature 441, 532–536 10.1038/nature04794 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., Belcram K., et al. (2005). AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118, 4621–4632 10.1242/jcs.02583 [DOI] [PubMed] [Google Scholar]
- Deans B., Griffin C. S., Maconochie M., Thacker J. (2000). Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 19, 6675–6685 10.1093/emboj/19.24.6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A. M. (1998). Meiotic recombination in C.elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Dosanjh M., Collins D., Schild D., Fan W., Lennon G., Albala J., et al. (1998). Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res. 26, 1179–1184 10.1093/nar/26.5.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler G., Rogge S., Beisker W., Eckardt-Schupp F., Zdzienicka M., Fritz E. (2004). Spontaneous homologous recombination is decreased in Rad51C-deficient hamster cells. DNA Repair 3, 1335–1343 10.1016/j.dnarep.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H., Kolodner R. (2000). Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 25, 196–200 10.1016/S0968-0004(00)01568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C., Masson J., Griffin C., O'Regan P., West S., Thacker J. (2002). Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 277, 19322–19330 10.1074/jbc.M201402200 [DOI] [PubMed] [Google Scholar]
- Friedberg E., Walker G., Siede W. (1995). DNA Repair and Mutagenesis. Washington, DC: ASM Press [Google Scholar]
- Godthelp B., Wiegant W., Van Duijn-Goedhart A., Sch Rer O., Van Buul P., Kanaar R., et al. (2002). Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 30, 2172–2182 10.1093/nar/30.10.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Puchta H. (2000). Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 1548–1554 10.1093/nar/28.7.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Perry R. M., Barakate A., Ramsay L., Waugh R., Halpin C., et al. (2012). Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24, 4096–4109 10.1105/tpc.112.102483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S., Rass U., Blanco M., Flynn H., Skehel J., West S. (2008). Identification of Holliday junction resolvases from humans and yeast. Nature 456, 357–361 10.1038/nature07470 [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Liu N., Jasin M. (1999). Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401, 397–399 10.1038/43932 [DOI] [PubMed] [Google Scholar]
- Keeney S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Topics Dev. Biol. 52, 1–53 10.1016/S0070-2153(01)52008-6 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., Nairz K., et al. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- Kou Y., Chang Y., Li X., Xiao J., Wang S. (2012). The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J. Exp. Bot. 63, 5323–5335 10.1093/jxb/ers190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S., Haines D., Martin B., Sharan S. (2009). Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 69, 863–872 10.1158/0008-5472.CAN-08-3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S., Pellegrini M., Shuda K., Fernandez-Capetillo O., Liu Y., Martin B., et al. (2007). RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 176, 581–592 10.1083/jcb.200608130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller S. P., Meek K. (2003). Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85, 1161–1173 10.1016/j.biochi.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., et al. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10596–10601 10.1073/pnas.0404110101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yang X., Lin Z., Timofejeva L., Xiao R., Makaroff C., et al. (2005). The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138, 965–976 10.1104/pp.104.058347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R. (2008). The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 10.1074/jbc.R700039200 [DOI] [PubMed] [Google Scholar]
- Lio Y., Mazin A., Kowalczykowski S., Chen D. (2003). Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 278, 2469–2478 10.1074/jbc.M211038200 [DOI] [PubMed] [Google Scholar]
- Lio Y., Schild D., Brenneman M., Redpath J., Chen D. (2004). Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J. Biol. Chem. 279, 42313–42320 10.1074/jbc.M405212200 [DOI] [PubMed] [Google Scholar]
- Liu N., Lamerdin J., Tebbs R., Schild D., Tucker J., Shen M., et al. (1998). XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1, 783–793 10.1016/S1097-2765(00)80078-7 [DOI] [PubMed] [Google Scholar]
- Liu Y., Masson J., Shah R., O'Regan P., West S. (2004). RAD51C is required for Holliday junction processing in mammalian cells. Science 303, 243–246 10.1126/science.1093037 [DOI] [PubMed] [Google Scholar]
- Liu Y., Tarsounas M., O'Regan P., West S. C. (2007). Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J. Biol. Chem. 282, 1973–1979 10.1074/jbc.M609066200 [DOI] [PubMed] [Google Scholar]
- Luo Q., Li Y., Shen Y., Cheng Z. (2014). Ten years of gene discovery for meiotic event control in rice. J. Genet. Genomics 41, 125–137 10.1016/j.jgg.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Masson J., Tarsounas M., Stasiak A., Stasiak A., Shah R., McIlwraith M., et al. (2001). Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15, 3296–3307 10.1101/gad.947001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Hayashi-Hagihara A. (1998). Mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932–2942 10.1101/gad.12.18.2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C., Tang D., Zhang H., Wang M., Li Y., Tang S., et al. (2013). CENTRAL REGION COMPONENT1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25, 2998–3009 10.1105/tpc.113.113175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., Sawicka D., Barsky D., Albala J. (2004). Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 32, 169–178 10.1093/nar/gkg925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., Yoshikawa D., McConnell I., Clark R., Schild D., Albala J. (2002). RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem. 277, 8406–8411 10.1074/jbc.M108306200 [DOI] [PubMed] [Google Scholar]
- Nonomura K.-I., Nakano M., Eiguchi M., Suzuki T., Kurata N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119, 217–225 10.1242/jcs.02736 [DOI] [PubMed] [Google Scholar]
- Olive P. (1998). The role of DNA single-and double-strand breaks in cell killing by ionizing radiation. Radiat. Res. 150, 42–51 10.2307/3579807 [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. (2001). A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349–1360 10.1101/gad.192701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A., Johnson R., Thompson L., Jasin M. (1999). XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman D. L., Weinberg L. R., Schimenti J. C. (1998). Identification, characterization, and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics 49, 103–111 10.1006/geno.1998.5226 [DOI] [PubMed] [Google Scholar]
- Puizina J., Siroky J., Mokros P., Schweizer D., Riha K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978 10.1105/tpc.104.022749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U., Compton S. A., Matos J., Singleton M. R., Ip S. C., Blanco M. G., et al. (2010). Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 24, 1559–1569 10.1101/gad.585310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 10.1016/S1097-2765(00)00097-6 [DOI] [PubMed] [Google Scholar]
- Shao T., Tang D., Wang K., Wang M., Che L., Qin B., et al. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156, 1386–1396 10.1104/pp.111.177428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Matsuda Y., Ushio N., Ikeo K., Ogawa T. (1993). Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat. Genet. 4, 239–243 10.1038/ng0793-239 [DOI] [PubMed] [Google Scholar]
- Shu Z., Smith S., Wang L., Rice M. C., Kmiec E. B. (1999). Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53-/- background. Mol. Cell. Biol. 19, 8686–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren C., Nasmyth K. (2001). Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. (2006). Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029 10.1016/j.dnarep.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki M., Tachiiri S., Fukushima T., Sonoda E., Schild D., et al. (2001). Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21, 2858–2866 10.1128/MCB.21.8.2858-2866.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Bosch M., Lohman P., Pastink A. (2002). DNA double-strand break repair by homologous recombination. Biol. Chem. 383, 873–892 10.1515/BC.2002.095 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Wang M., Lu J., Yu H., Liu J., et al. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122, 2055–2063 10.1242/jcs.049080 [DOI] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Miao C., Hu Q., et al. (2012). The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8:e1002809 10.1371/journal.pgen.1002809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Qin B., Li M., et al. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22, 12–19 10.1091/mbc.E10-08-0667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang K., Tang D., Wei C., Li M., Shen Y., et al. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430 10.1105/tpc.109.070789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. (1988). DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 35, 95–125 10.1016/S0079-6603(08)60611-X [DOI] [PubMed] [Google Scholar]
- Wiese C., Collins D., Albala J., Thompson L., Kronenberg A., Schild D. (2002). Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 30, 1001–1008 10.1093/nar/30.4.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong B., Gerton J. L. (2010). Regulators of the cohesin network. Annu. Rev. Biochem. 79, 131–153 10.1146/annurev-biochem-061708-092640 [DOI] [PubMed] [Google Scholar]
- Xu H., Beasley M. D., Warren W. D., Van Der Horst G. T., McKay M. J. (2005). Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 10.1016/j.devcel.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Yokoyama H., Sarai N., Kagawa W., Enomoto R., Shibata T., Kurumizaka H., et al. (2004). Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 32, 2556–2565 10.1093/nar/gkh578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Wang M., Tang D., Wang K., Chen F., Gong Z., et al. (2010). OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119, 625–636 10.1007/s00412-010-0284-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The list of primers used in this study.