Introduction
Early evidence for a genetic etiology in eosinophilic esophagitis (EoE) came in the form of several epidemiological studies demonstrating a high prevalence of disease in specific genders and races, with nearly three-quarters of patients being male and almost all (~90%) being of European descent, respectively (1). Moreover, there is an increased disease risk among familial cases, which demonstrate a non-Mendelian inheritance pattern (2). Expression profiling from esophageal biopsies acquired during routine endoscopic procedure has provided incredible molecular insight into genetic dysregulation occurring within the inflamed esophagus. These transcriptional changes affect both coding and non-coding (microRNA) transcripts and underscore consistent, disease-specific alterations in the levels of select molecules expressed by activated immune cells and structural cells of the esophagus (3, 4). Importantly, these dysregulated transcripts and associated biological pathways represent potential targets for novel therapeutics and diagnostic methods (5).
In addition to the genetic elements, a role for environmental factors in EoE has been established through both clinical and basic research. Patients with EoE are often hypersensitized to multiple food antigens, making directed dietary modification one of the most effective therapies for EoE (6). Several early life exposures, including Cesarean birth, antibiotics, and formula feeding, have been identified to influence the risk of pediatric EoE (7). In addition, geographical location, industrialized environments, history of H. pylori infection, and seasonal variations in disease implicate environmental antigens (8-11). These clinical findings have been supported through multiple basic research studies showing that epidermal and pulmonary exposure to various antigens can induce EoE-like symptoms in mice (12-14). Emerging epigenetic data are now beginning to provide clues as to how these environmental factors may be intricately intertwined with the genetic dysregulation in EoE and thus act in a concerted fashion to affect disease pathophysiology.
Genetic variants
Several candidate gene approaches have identified a handful of genetic risk variants in EoE. For instance, a common single nucleotide variant (SNV) (minor allele frequency [MAF] = 0.25 in the HapMap (15) population of European descent) located in the 3′ untranslated of the chemokine (C-C motif) ligand 26 (CCL26) was overrepresented in EoE patients in both a case-control and a family-based analysis (4). Furthermore, two coding variants (R501X and 2282del4) in the epidermal barrier gene filaggrin (FLG), which is negatively regulated by IL-13 and is decreased in the esophageal mucosa of patients with EoE, associate with EoE risk (16). Lastly, in a small cohort of steroid-treated EoE patients, a genetic variant in the promoter of the transforming growth factor, beta 1 (TGFB1) gene, was associated with steroid unresponsiveness and correlated with increased TGF-β1-positive cells in the esophagus (17). The genetic link between the TGF-β pathway and EoE identified in this study is quite remarkable given the evidence demonstrating a high rate of EoE, other eosinophilic gastrointestinal disorders (EGID), and atopic disease in patients with connective tissue disorders (18) like Loeys-Dietz syndrome (LDS), which has been associated with variants in the TGF-β receptors 1 and 2 (19) (Table 1).
Table 1. Genetic and epigenetic modifications associated with EoE and related genetic disorders.
| Target | Modification | Potential biological effect | ||
|---|---|---|---|---|
| Genetic |
Disease risk
variants |
CCL26 | SNV in 3′UTR | Enhanced mRNA stability; increased expression |
| TGFB1 | SNV in promoter | Increased expression; non- responsiveness to topical steroid therapy |
||
| FLG | Nonsense and missense SNVs |
Loss of function; reduced barrier function |
||
| TSLP | SNVs in promoter region and introns |
Increased expression; correlates with esophageal levels of basophil and GMP- like cells |
||
| CRLF2 | Missense SNV | Male-specific association; Enhanced TSLP signaling |
||
| DSG1 | Missense SNVs in patients with SAM syndrome1 |
Loss of function; reduced epithelial integrity; increased IL5 and TSLP |
||
| TGFBR1/ | Missense SNVs in | Increased TGF-β signaling; | ||
| TGFBR2 | patients with LDS2 | elevated CD4+ Th2 cells | ||
| PTEN | Missense SNVs, insertions, and deletions in patients with PHTS3 |
Loss of function; hyperproliferation |
||
|
|
||||
| Transcriptome | CCL26 | Increased expression (esophagus) |
Promotes eosinophil trafficking into the esophagus |
|
| POSTN | Increased expression (esophagus) |
Increased eosinophil adhesion; promotes esophageal remodeling ; increased TSLP expression |
||
| DSG1 | Decreased expression (esophagus) |
Reduced barrier function; increased POSTN expression |
||
|
| ||||
| Epigenetic | Histones | H3 | Acetylated | Enhanced CCL26 promoter activity |
| H3 | Methylated (lysine 4) | Enhanced CCL26 promoter activity |
||
|
|
||||
| DNA | CCL26 | Hypomethylation in promoter region |
Enhanced CCL26 promoter activity |
|
|
|
||||
| MicroRNAs | miR-21 | Increased expression (esophagus) |
Skewed Th2 response; increased eosinophil survival |
|
| miR-223 | Increased expression (esophagus, blood) |
Increased eosinophil progenitors |
||
| miR-375 | Decreased expression (esophagus) |
Enhanced IL-13 transcriptional responses |
||
EoE was a comorbidity in one of three SAM syndrome patients
High prevalence of EoE and other EGlDs (n = 6) in 58 LDS patients
Significant enrichment of EGID in PHTS (odds ratio = 272; confidence interval 89-831, P < 10−4)
To identify disease risk variants in a more unbiased fashion, a genome-wide association study (GWAS) in which 351 EoE patients and 3,104 healthy controls were genotyped for over 550,000 common variants was performed. On chromosome 5q22, a single locus spanning the thymic stromal lymphopoietin (TSLP) and WD repeat domain 36 (WDR36) genes showed a significant association with EoE susceptibility (20). TSLP is a potent Th2-promoting cytokine involved in the development of multiple allergic diseases (21). Expression analyses showed increased TSLP in EoE and a genotypic effect of the top associated variant on TSLP expression, with patients carrying the risk allele having elevated TSLP expression (20). In addition, TSLP risk genotypes correlated with increased levels of basophils, which have a key role in promoting EoE-like disease in mice, as well as granulocyte-monocyte progenitor-like cells in the esophagus (14, 22).
A secondary candidate gene approach also identified variants within the TSLP locus that were significantly associated with EoE risk (23). In this study assessing over 700 variants in epithelial-derived genes linked to atopy, TSLP variants were the most significant genetic hits linked to EoE that, importantly, showed a stronger association with disease risk when compared to controls with atopic diseases (atopic dermatitis and asthma) (23). Moreover, a coding variant in the cytokine receptor-like factor 2 (CRLF2) gene, which encodes for the receptor for TSLP, showed a sex-specific association with EoE risk in males only (23). These cumulative data support aberrant regulation affecting the TSLP pathway as a specific genetic etiology in EoE. Given the established role of TSLP in the initiation of allergic diseases, the fact that WDR36 was not differentially expressed in EoE underscores TSLP as the most likely gene involved in driving the esophageal inflammatory responses in EoE. Importantly, however, variants in WDR36 have been linked with peripheral blood eosinophil levels and atopic asthma (24). Thus, further studies are needed to identify the precise causal variant(s) of EoE, as well as to fully investigate a potential non-esophageal role for WDR36 that may contribute to disease.
The EoE transcriptome
A total of 574 highly dysregulated esophageal genes, termed the EoE transcriptome, distinguishes EoE patients from healthy controls and, importantly, from patients with non-eosinophilic forms of esophagitis was identified (4). Despite the patchiness of EoE and phenotypic diversity within the EoE patients analyzed, the EoE transcriptome is surprisingly well-conserved across patient age, gender, atopic status, and non-familial relationship (4, 25). Indeed, a large-scale screen based on 94 signature EoE transcriptome genes has shown promise as a diagnostic tool capable of discriminating EoE patients from non-eosinophilic forms of esophagitis and patients with active EoE from those with EoE in remission (inactive EoE) (26). Notably, the cytokine IL-13 is capable of inducing an esophageal epithelial cell gene signature that represents 22% of the EoE transcriptome (27). We will discuss three key genes within the EoE transcriptome, their regulation by IL-13, and their influence on disease pathophysiology.
Chemokine (C-C motif) ligand 26 (CCL26)
Expression of CCL26, which encodes the eosinophil chemoattractant eotaxin-3, was upregulated 53 fold in EoE, making it the most highly induced gene of the EoE transcriptome. CCL26 is believed to be the main driver for eosinophil recruitment into the esophagus, as the upregulation of CCL26 was unique among other closely related chemokines from the eotaxin family (CCL11 and CCL24) (4); however, other studies have indicated that CCL11 and CCL24 are induced at low levels in EoE (28, 29). The levels of CCL26 in patients with EoE correlated significantly with the esophageal levels of eosinophils and mast cells (4). The induction of CCL26 in EoE patients was determined to be largely due to the influence of IL-13 on esophageal epithelial cells, as CCL26 was also the most highly induced gene in IL-13-treated cells (279 fold when compared to untreated cells) (4, 27). Molecular analyses defined two STAT6 binding sites in the CCL26 promoter that were necessary for the induction of CCL26 by IL-13, as well as by IL-4 (30, 31). Furthermore, several co-activators, including poly-ADP ribosyl polymerase 14 (PARP14), have been shown to act upon the CCL26 locus. PARP14 was identified as a specific co-regulator of STAT6 signaling, and its overexpression in esophageal epithelial cells enhanced IL-13-induced CCL26 expression in a STAT6-dependent manner (32, 33). Finally, exposure of esophageal epithelial cells to acidic pH enhances eotaxin-3 release, providing a potential mechanism by which proton pump inhibitor (PPI) therapies could have some anti-inflammatory effects in EoE (16). A role for the eotaxins and their receptor CCR3 is supported by studies in mice that have shown attenuated eosinophil levels and/or tissue remodeling in eotaxin and/or CCR3-deficient mice (4, 34, 35).
Periostin (POSTN)
Periostin is a matricellular protein capable of interacting with multiple extracellular matrix molecules and cell surface receptors such as type 1 collagen and Notch1, respectively (36). Periostin is directly involved in regulating multiple cellular processes including cell migration and adhesion (36). Its influence on metastasis, tissue remodeling, and wound healing has made periostin a highly studied molecule in the context of various human diseases like cancer, asthma, and atopic dermatitis (37-39). A 47-fold induction of periostin mRNA was observed in EoE, making it the second most highly upregulated gene in the EoE transcriptome (4). Periostin protein was also increased in EoE, primarily localized within the lamina propria, indicating fibroblasts as the main cellular source of periostin induction (40). Indeed, TGF-β and IL-13 induced greater levels of periostin expression in esophageal fibroblasts than in esophageal epithelial cells (40). Using periostin-deficient mice, it was shown that periostin promotes allergic inflammatory responses in the lung and esophagus, in part through enhancing eosinophil adhesion (40). Notably, in skin keratinocytes, periostin can induce the expression of TSLP (37). These collective findings suggest a molecular loop between TGF-β, periostin, and TSLP, which act synergistically to drive the esophageal pathophysiology associated with EoE. It is notable that circulating periostin levels in patients with asthma help stratify patients into those that respond to biological therapeutics such as anti-IgE and anti-IL-13, and appear to identify eosinophilic asthmatic phenotypes, extending the significance of the eosinophil/periostin connection from EoE to other common atopic disorders (38, 41, 42).
Desmoglein 1 (DSG1)
Desmoglein 1 (DSG1) is a transmembrane molecule belonging to the family of desmosomal cadherins, which has an essential role in maintaining epithelial integrity through calcium-dependent intercellular adhesion. The focus on DSG1 as an etiological component in human disease stemmed from observations linking DSG1 alteration in various dermatological disorders, where epithelial integrity and barrier function is compromised (43, 44). In EoE, DSG1 mRNA is specifically downregulated in the esophageal mucosa of patients with active disease (27, 45). This specific decrease in DSG1 was shown to result from IL-13 stimulation of differentiated esophageal epithelial cells (45). Functionally, DSG1-deficeint esophageal epithelial cells exhibited greater cell dissociation, weaker adhesive properties, and reduced capacity to form an intact epithelial barrier (45). Moreover, the loss of DSG1 triggered epithelial gene expression changes reflective of those in EoE patient biopsies, including increased POSTN expression (45).
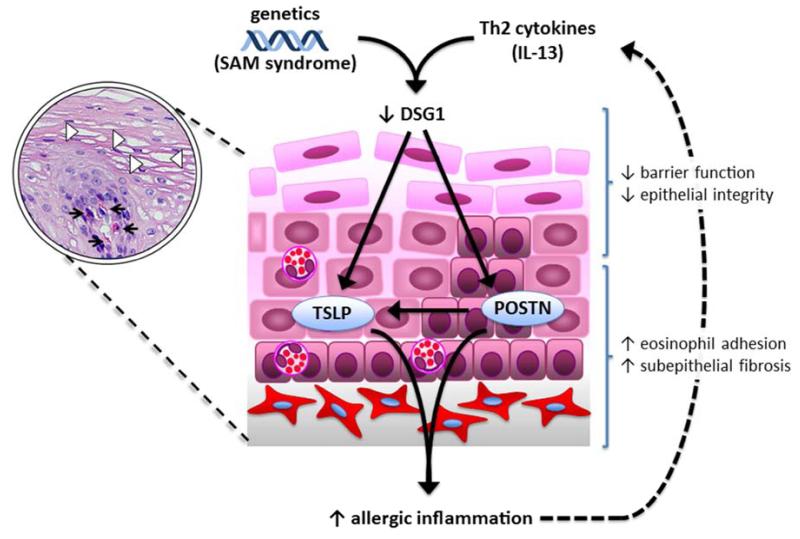
A key contributory role for DSG1 dysregulation in the allergic disease process was independently demonstrated in genetic studies that identified loss-of-function mutations in DSG1 in consanguineous individuals with severe atopic dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome. All three SAM patients analyzed had failure-to-thrive diagnoses and had multiple food allergies; notably, one patient also had an EoE diagnosis (46) (Table 1). Skin biopsies from SAM patients showed reduced DSG1 expression and acantholysis, while isolated keratinocytes showed increased expression of IL-5 and TSLP (46). Interestingly, an intronic mutation in DSG1 showed a suggestive association with EoE risk (20). Given this potential association and the findings in SAM syndrome, further investigation into EoE risk variants in DSG1 is warranted (Fig. 2).
Figure 2. Regulation of DSG1 promotes allergic inflammation.
Decreased DSG1 expression by Th2 cytokines (IL-13) or DSG1 coding variants (as in SAM syndrome) alter the levels of functional DSG1, leading to impaired barrier function and reduced epithelial integrity. DSG1 deficiency also induces the expression of periostin (POSTN), which can lead to enhanced eosinophil adhesion and subepithelial fibrosis, and TSLP, either directly or indirectly through periostin, culminating in a pro-inflammatory cycle.
Epigenetics
Epigenetics are the heritable phenotypic modifications that result from gene activation or repression through mechanisms that are independent of changes to the DNA sequence (47). Capable of being influenced by environmental stimuli, the epigenome lies at the crossroads of gene-environment interactions, placing it at the forefront for studying mechanisms underlying environmentally driven allergic inflammatory diseases. We will discuss in detail the current view of epigenetic regulation associated with EoE, which includes histone modification and DNA methylation, as well as posttranscriptional repression by microRNAs (miRNAs).
Histone modification and DNA methylation
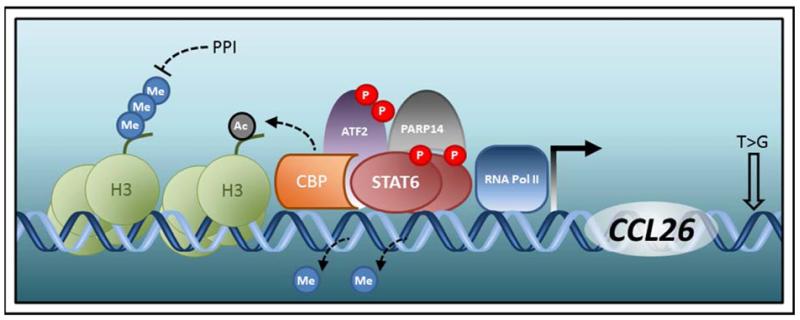
Posttranslational modification of histone tails is an epigenetic mechanism that can alter the accessibility to gene promoters located proximally or distally to the modified histone. These reversible modifications, the most prominent of which include acetylation and methylation, are made by diverse families of modifying enzymes in monomeric or even multimeric fashion to exposed amino acid residues (typically lysine) of the histone tail (48). The type of modification, the specific histone and position of the amino acid residue involved, and the degree to which the residue is modified can all influence the degree of gene activation or repression (49). Much of what little is known on the involvement of epigenetics in EoE has been attained from biochemical studies of the promoter of the leading EoE candidate gene, CCL26 (Figure 1). The mapping of two STAT6 binding sites roughly established the promoter proximal regions required for IL-13-induced transcriptional activation of CCL26 (30). Further analysis demonstrated a requirement for two co-activators, activating transcription factor 2 (ATF2) and the histone acetyltransferase cAMP-responsive element (CRE)-binding protein (CBP) (50). Chromatin immunoprecipitation (ChIP) assays indicated that STAT6, CBP, ATF2, and acetylated histone 3 bound within the same region of the CCL26 promoter in esophageal epithelial cells following IL-13 treatment (50). Thus, IL-13 induces the formation of a multi-protein complex on the CCL26 promoter that includes CBP, leading to increases in acetylated histone 3 and opening of the CCL26 promoter for additional transcriptional machinery. Interestingly, PPIs have been suggested to dampen the levels of trimethylated histone 3 lysine 4 (H3K4) and STAT6 bound to the CCL26 promoter, resulting in decreased eotaxin-3 expression (51). These findings could explain the emerging observation of PPI-responsive EoE, in which PPI therapy yields partial resolution of symptoms (52).
Figure 1. Genetic and epigenetic regulation of the CCL26 locus.
Induction of CCL26 expression is initiated upon phosphorylation (P) of STAT6 by Th2 cytokines (IL-13 and IL-4). Complete activation of the CCL26 promoter by phosphorylated STAT6 is aided presumably by the opening of the CCL26 promoter region by DNA demethylation, CBP-mediated acteylation (Ac) of histone 3 (H3), and interaction with cofactors such as CBP, phosphorylated ATF2, and PARP14. A genetic variant (T>G) in the 3′ untranslated region of CCL26 is linked to EoE risk. Proton pump inhibitors (PPI) can silence CCL26 expression by removing trimethylated (Me) H3 from the CCL26 promoter region, potentially making the promoter inaccessible to phosphorylated STAT6 and RNA polymerase (Pol) II.
In addition to the epigenetic regulation by histone acetylation, the CCL26 promoter is also controlled by DNA methylation. DNA methylation occurs on cytosine nucleotides located within CpG (cytosine-guanine) dinucleotide motifs and, like other epigenetic marks, is dynamically regulated (53). Two CpG sites in the CCL26 promoter were identified as hypomethylated in esophageal epithelial cells derived from EoE patients (54). These data remarkably demonstrate the longevity of epigenetic marks in that they remain detectable even in cells cultured ex vivo through multiple rounds of cell division. The methylation status at one of the two CpG sites correlated with increased STAT6 binding to the CCL26 promoter and induction of CCL26 expression by IL-13 (54). Moreover, this CpG site flanks the CBP binding sequence, and its methylation prohibited the binding of CBP to the CCL26 promoter (54). Collectively, these findings suggest that a coordinated interaction involving DNA demethylation followed by histone acetylation occurs at the CCL26 promoter in response to IL-13.
MicroRNAs (miRNAs)
MicroRNAs (miRNAs) are short, noncoding RNAs that fine-tune the expression of target genes at the posttranscriptional level. MiRNAs act to repress translation and/or induce mRNA degradation through binding complementary “seed” sequences in the 3′ untranslated region of target mRNAs, forming double-stranded RNA molecules that are digested within the RNA-induced silencing complex (RISC) (55). Much like its coding counterparts composing the EoE transcriptome, a select set of miRNAs has been demonstrated to be dynamically altered in the esophageal mucosa of patients with EoE. The miRNA signature associated with EoE, which was distinct from both healthy controls and patients with chronic, non-eosinophilic forms of esophagitis, included 21 upregulated and 11 downregulated miRNAs (3).
Two of the most highly induced miRNAs in EoE, miR-21 and miR-223, also displayed the highest correlation with esophageal eosinophil levels in EoE patients (3). Both miRNAs have potentially significant functional implications in the pathophysiology of EoE. MiR-21 is upregulated in multiple mouse models of allergic lung inflammation (56) and has been implicated in promoting eosinophil survival (57). Induction of miR-223 in EoE showed significant correlations with the induction of the genes encoding Charcot-Leyden crystal protein (CLC), an eosinophil granule protein, and IL-5 (3), suggesting a role for miR-223 in eosinophil development. Indeed, in vivo studies demonstrated enhanced proliferation of eosinophil progenitors and a severe defect in eosinophil development from the bone marrow of miR-223-deficient mice (58). However, it remains unknown how the localized increase of miR-223 in the esophageal mucosa affects terminally differentiated eosinophils within the Th2 microenvironment.
Conversely, miR-375 is the most repressed miRNA in the EoE-associated signature. Mechanistically, IL-13 can downregulate miR-375 in cultured esophageal epithelial cells (59). Exogenous expression of miR-375 in esophageal epithelial cells modulated the levels of several immunomodulatory genes at baseline and after IL-13 stimulation, indicating a unique role for miR-375 in the regulation of IL-13-induced transcriptional responses (59). Interestingly, the interaction between epithelial-derived miR-375 and IL-13 has also been observed in the intestine with strikingly different results, where IL-13-treatment of intestinal epithelial cells enhanced miR-375 expression (60). MiR-375 was also shown to regulate goblet cell differentiation through targeting of Kruppel-like factor 5 (KLF5) and induce TSLP expression (60). Taken together, these findings suggest that miRNA dysregulation affects multiple inflammatory processes connected to EoE.
The miRNA signature associated with EoE exhibited near-complete reversibility (27 out of 32 dysregulated miRNAs, or 84%) during disease remission induced by swallowed fluticasone therapy (3). Notably, similar levels of normalization were observed in an independent, longitudinal EoE cohort analyzed both pre- and post-steroid therapy, where only 32 out of the 377 miRNAS analyzed, or less than 9%, remained dysregulated (61). However, one miRNA, miR-675 was significantly elevated in fluticasone-responsive EoE remission patients compared to unresponsive EoE patients (3). Several miRNAs, including miR-146a, miR-146b, and miR-223 were identified as dysregulated in plasma samples from patients with active EoE (3). Together, these data implicate miRNAs as potential biomarkers for EoE diagnosis and steroid responsiveness.
Summary
In summary, gene expression profiling of patient tissue and screening for disease risk variants have taken unbiased approaches to reveal many of the critical molecular pathways underlying EoE pathogenesis. While these pathways continue to undergo rigorous investigation, new research into the epigenetic modification of immunoregulatory genes like CCL26 and a dysregulated miRNA signature in EoE add additional layers to the molecular entities governing the transcriptome of the inflamed esophageal mucosa. In addition, there has been increasing recognition of EoE associated with a number of Mendelian disorders such as SAM syndrome (DSG1), connective tissue disorders (TGFBR1/2 mutations), and PTEN hamartoma tumor syndromes (PHTS) (62) (Table 1). Although many challenges exist and much work remains, dissecting the genetic and epigenetic factors of EoE and related genetic disorders represents a promising area for translational research aimed at novel therapies, noninvasive diagnostics, and biomarkers for therapy response.
Synopsis.
Eosinophilic esophagitis (EoE) is a complex, polygenic disorder driven by both genetic predisposition and environmental exposures. Due to the recent emergence of EoE as a bona fide global health concern, there is a paucity of available therapeutic and diagnostic options. However, rapid progress has been made in an effort to rectify this lack and to improve our understanding of the etiological factors driving EoE. Here, we will highlight key advances in elucidating the genetic (and epigenetic) components involved in EoE.
Key Points.
Eosinophilic esophagitis is a complex, polygenic disorder
Disease risk variants and an altered esophageal transcriptional profile underlie the genetic etiology of eosinophilic esophagitis
Emerging epigenetic modifications link environmental exposures to the genetic dysregulation in eosinophilic esophagitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sherrill JD, Rothenberg ME. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J Allergy Clin Immunol. 2011;128:23–32. doi: 10.1016/j.jaci.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–1059. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 3.Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, Franciosi JP, Putnam PE, Eby M, Martin LJ, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–1075. e1069. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner BS, Book W, Busse WW, Butterfield J, Furuta GT, Gleich GJ, Klion AD, Lee JJ, Leiferman KM, Minnicozzi M, et al. Workshop report from the National Institutes of Health Taskforce on the Research Needs of Eosinophil-Associated Diseases (TREAD) J Allergy Clin Immunol. 2012;130:587–596. doi: 10.1016/j.jaci.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagalwalla AF, Shah A, Li BU, Sentongo TA, Ritz S, Manuel-Rubio M, Jacques K, Wang D, Melin-Aldana H, Nelson SP. Identification of specific foods responsible for inflammation in children with eosinophilic esophagitis successfully treated with empiric elimination diet. J Pediatr Gastroenterol Nutr. 2011;53:145–149. doi: 10.1097/MPG.0b013e31821cf503. [DOI] [PubMed] [Google Scholar]
- 7.Jensen ET, Kappelman MD, Kim H, Ringel-Kulka T, Dellon ES. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;57:67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 8.Almansa C, Krishna M, Buchner AM, Ghabril MS, Talley N, DeVault KR, Wolfsen H, Raimondo M, Guarderas JC, Achem SR. Seasonal distribution in newly diagnosed cases of eosinophilic esophagitis in adults. Am J Gastroenterol. 2009;104:828–833. doi: 10.1038/ajg.2008.169. [DOI] [PubMed] [Google Scholar]
- 9.Spergel JM, Book WM, Mays E, Song L, Shah SS, Talley NJ, Bonis PA. Variation in prevalence, diagnostic criteria, and initial management options for eosinophilic gastrointestinal diseases in the United States. J Pediatr Gastroenterol Nutr. 2011;52:300–306. doi: 10.1097/MPG.0b013e3181eb5a9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurrell JM, Genta RM, Dellon ES. Prevalence of esophageal eosinophilia varies by climate zone in the United States. Am J Gastroenterol. 2012;107:698–706. doi: 10.1038/ajg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dellon ES, Peery AF, Shaheen NJ, Morgan DR, Hurrell JM, Lash RH, Genta RM. Inverse association of esophageal eosinophilia with Helicobacter pylori based on analysis of a US pathology database. Gastroenterology. 2011;141:1586–1592. doi: 10.1053/j.gastro.2011.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayapudi M, Mavi P, Zhu X, Pandey AK, Abonia JP, Rothenberg ME, Mishra A. Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2010;88:337–346. doi: 10.1189/jlb.0110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope SM, Fulkerson PC, Blanchard C, Akei HS, Nikolaidis NM, Zimmermann N, Molkentin JD, Rothenberg ME. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J Biol Chem. 2005;280:13952–13961. doi: 10.1074/jbc.M406037200. [DOI] [PubMed] [Google Scholar]
- 14.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, Benitez AJ, Ruymann KR, Muir AB, Hill DA, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013;19:1005–1013. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International HapMap, C The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, Buckmeier BK, Jameson SC, Greenberg A, Kaul A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aceves SS, Newbury RO, Chen D, Mueller J, Dohil R, Hoffman H, Bastian JF, Broide DH. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65:109–116. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, Collins MH, Putnam PE, Franciosi JP, von Tiehl KF, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132:378–386. doi: 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, Chichester K, Myers L, Halushka MK, Oliva-Hemker M, Wood RA, Dietz HC. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med. 2013;5:195ra194. doi: 10.1126/scitranslmed.3006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, Gober L, Kim C, Glessner J, Frackelton E, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler SF. The role of thymic stromal lymphopoietin (TSLP) in allergic disorders. Curr Opin Immunol. 2010;22:795–799. doi: 10.1016/j.coi.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siracusa MC, Saenz SA, Tait Wojno ED, Kim BS, Osborne LC, Ziegler CG, Benitez AJ, Ruymann KR, Farber DL, Sleiman PM, et al. Thymic stromal lymphopoietin-mediated extramedullary hematopoiesis promotes allergic inflammation. Immunity. 2013;39:1158–1170. doi: 10.1016/j.immuni.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, Franciosi JP, Kushner JP, Abonia JP, Assa’ad AH, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:160–165. e163. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 25.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, Putnam PE, Jameson SC, Assa’ad AH, Konikoff MR, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–629. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, Franciosi JP, Garza JM, Kaul A, King EC, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–1299. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, Collins MH, Putnam PE, Wells SI, Rothenberg ME. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Lucendo AJ, De Rezende L, Comas C, Caballero T, Bellon T. Treatment with topical steroids downregulates IL-5, eotaxin-1/CCL11, and eotaxin-3/CCL26 gene expression in eosinophilic esophagitis. Am J Gastroenterol. 2008;103:2184–2193. doi: 10.1111/j.1572-0241.2008.01937.x. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya B, Carlsten J, Sabo E, Kethu S, Meitner P, Tavares R, Jakate S, Mangray S, Aswad B, Resnick MB. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38:1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber JC. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005;37:2559–2573. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Hoeck J, Woisetschlager M. Activation of eotaxin-3/CCLl26 gene expression in human dermal fibroblasts is mediated by STAT6. J Immunol. 2001;167:3216–3222. doi: 10.4049/jimmunol.167.6.3216. [DOI] [PubMed] [Google Scholar]
- 32.Goenka S, Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc Natl Acad Sci U S A. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamurthy P, Sherrill JD, Parashette K, Goenka S, Rothenberg ME, Gupta S, Kaplan MH. Correlation of increased PARP14 and CCL26 expression in biopsies from children with eosinophilic esophagitis. J Allergy Clin Immunol. 2013 doi: 10.1016/j.jaci.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125:1419–1427. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Kudo A. Periostin in fibrillogenesis for tissue regeneration: periostin actions inside and outside the cell. Cell Mol Life Sci. 2011;68:3201–3207. doi: 10.1007/s00018-011-0784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, Toda S, Inagaki N, Kurihara Y, Hayashida S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012;122:2590–2600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, Shikotra A, Carter R, Audusseau S, Hamid Q, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–654. e610. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong GS, Lee JS, Park YY, Klein-Szanto AJ, Waldron TJ, Cukierman E, Herlyn M, Gimotty P, Nakagawa H, Rustgi AK. Periostin cooperates with mutant p53 to mediate invasion through the induction of STAT1 signaling in the esophageal tumor microenvironment. Oncogenesis. 2013;2:e59. doi: 10.1038/oncsis.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, Stringer K, Abonia JP, Molkentin JD, Rothenberg ME. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–296. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, Harris JM, Scheerens H, Wu LC, Su Z, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 42.Hanania NA, Wenzel S, Rosen K, Hsieh HJ, Mosesova S, Choy DF, Lal P, Arron JR, Harris JM, Busse W. Exploring the effects of omalizumab in allergic asthma: an analysis of biomarkers in the EXTRA study. Am J Respir Crit Care Med. 2013;187:804–811. doi: 10.1164/rccm.201208-1414OC. [DOI] [PubMed] [Google Scholar]
- 43.Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, Bonafe JL, Wilkinson J, Taieb A, Barrandon Y, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 44.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat Med. 2000;6:1275–1277. doi: 10.1038/81385. [DOI] [PubMed] [Google Scholar]
- 45.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, Kemme KA, Costello MS, Mingler MK, Blanchard C, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2013 doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, Koetsier JL, Gat A, Goldberg I, Bergman R, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–1248. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286:13193–13204. doi: 10.1074/jbc.M110.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Cheng E, Huo X, Yu C, Zhang Q, Pham TH, Wang DH, Spechler SJ, Souza RF. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS One. 2012;7:e50037. doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dohil R, Newbury RO, Aceves S. Transient PPI responsive esophageal eosinophilia may be a clinical sub-phenotype of pediatric eosinophilic esophagitis. Dig Dis Sci. 2012;57:1413–1419. doi: 10.1007/s10620-011-1991-5. [DOI] [PubMed] [Google Scholar]
- 53.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 54.Lim E, Rothenberg ME. Demethylation of the Human Eotaxin-3 Gene Promoter Leads to the Elevated Expression of Eotaxin-3. The Journal of Immunology. 2013 doi: 10.4049/jimmunol.1302454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 56.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu TX, Lim EJ, Itskovich S, Besse JA, Plassard AJ, Mingler MK, Rothenberg JA, Fulkerson PC, Aronow BJ, Rothenberg ME. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS One. 2013;8:e59397. doi: 10.1371/journal.pone.0059397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu TX, Lim EJ, Besse JA, Itskovich S, Plassard AJ, Fulkerson PC, Aronow BJ, Rothenberg ME. MiR-223 deficiency increases eosinophil progenitor proliferation. J Immunol. 2013;190:1576–1582. doi: 10.4049/jimmunol.1202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu TX, Lim EJ, Wen T, Plassard AJ, Hogan SP, Martin LJ, Aronow BJ, Rothenberg ME. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol. 2012;5:388–396. doi: 10.1038/mi.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo-Russo S, Avnit-Sagi T, Cojocaru G, Zreik F, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 61.Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, Brodsky AS, Resnick MB. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS One. 2012;7:e40676. doi: 10.1371/journal.pone.0040676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henderson CJ, Ngeow J, Collins MH, Martin LJ, Putnam PE, Abonia JP, Marsolo K, Eng C, Rothenberg ME. Increased prevalence of eosinophilic gastrointestinal disorders (EGID) in pediatric PTEN hamartoma tumor syndromes (PHTS) J Pediatr Gastroenterol Nutr. 2013 doi: 10.1097/MPG.0000000000000253. [DOI] [PMC free article] [PubMed] [Google Scholar]