Abstract
Sulforaphane is a naturally occurring isothiocyanate in cruciferous vegetables. Sulforaphane inhibits histone deacetylases, leading to the transcriptional activation of genes including tumor suppressor genes. The compound has attracted considerable attention in the chemoprevention of prostate cancer. Here we tested the hypothesis that sulforaphane is not specific for tumor suppressor genes but also activates loci such as long terminal repeats (LTRs), which might impair genome stability. Studies were conducted using chemically pure sulforaphane in primary human IMR-90 fibroblasts and in broccoli sprout feeding studies in healthy adults. Sulforaphane (2.0 μM) caused an increase in LTR transcriptional activity in cultured cells. Consumption of broccoli sprouts (34, 68, or 102g) by human volunteers caused a dose dependent elevation in LTR mRNA in circulating leukocytes, peaking at more than a 10-fold increase. This increase in transcript levels was associated with an increase in histone H3 K9 acetylation marks in LTR 15 in peripheral blood mononuclear cells from subjects consuming sprouts. Collectively, this study suggests that sulforaphane has off-target effects that warrant further investigation when recommending high levels of sulforaphane intake, despite its promising activities in chemoprevention.
Keywords: Histone deacetylases, long terminal repeats, off target effects, sulforaphane
1. Introduction
Cruciferous vegetables such as broccoli and cauliflower contain a large number of glucosinolates including glucoerucin, glucoiberin, and glucoraphanin (1). When unheated vegetables are processed by chopping or chewing, the enzyme myrosinase is released from myrosin grains in myrosin cells and glucosinolates are released from adjacent S cells (2–4). Myrosinase catalyzes the hydrolytic removal of the glucose moiety in glucoraphanin, followed by non-enzymatic release of a hydrogen sulfate moiety and spontaneous rearrangement of the unstable intermediate to form the aliphatic isothiocyanate sulforaphane (SFN) (1). SFN has attracted considerable attention due to its putative role in cancer prevention (5).
Various, not mutually exclusive, mechanisms have been proposed to explain the chemopreventive activities of SFN. One theory is that SFN enhances drug-mediated cytotoxicity against cancer cells including cancer stem cells (6–8). The significance of these observations is not limited to the chemotherapy of cancer but extends to the prevention of cancer through enhancing cellular sensitivity to cell death signals in tumor initiation. SFN-dependent inhibition of anti-apoptotic NF-κB signaling pathways appear to play a major role in the elimination of abnormal cells (9, 10). A second theory is that SFN-dependent inhibition of histone deacetylases (HDACs) causes an increase in the expression of tumor suppressor genes such as p21 and Bax, leading to cell cycle arrest and apoptosis (11, 12). Evidence suggests that SFN inhibits class I and class II HDACs (13). The locus (gene) specificity of SFN is uncertain, despite a general consensus that chemoprevention needs to pursue gene-specific gene expression through the modulation of epigenetic marks in distinct genomic loci (14). Gene-specific epigenetic editing can, theoretically, be achieved by fusing enzymes or inhibitors to gene-specific DNA binding domains.
In this paper we tested the hypothesis that the inhibition of HDACs by SFN does not only de-repress tumor suppressor genes, but also has undesirable off-target effects, meditated be de-repression of genes other than tumor suppressor genes. Studies were conducted in both cell cultures and healthy adults to take advantage of the small inter-individual variation in cell cultures and to capture effects of biotransformation in human studies. As model loci for detecting off-target effects we used long-terminal repeats (LTRs), based on the following rationale.
LTRs make up about 8% of the human genome and at least 51 LTRs are transcriptionally competent (15). Repetitive elements such as LTRs pose a burden to genome stability, as their mobilization facilitates recombination between non-homologous loci, leading to chromosomal deletions and translocations (16, 17). Mobilization of LTR transposons is associated with 10% of all spontaneous mutations in mice (18). The transcriptional activity of LTRs is controlled by histone acetylation and other epigenetic marks; inhibition of HDACs leads to an increase in LTR transcription (19). De-repression of LTRs may impair genome stability through insertional mutagenesis, recombination events that cause translocations and other rearrangements, deregulation of genes in the host genome mediated by LTR promoter activity, and antisense effects if transcription extends into exon sequence downstream of the transposon (20).
2. Methods and Materials
2.1. Cell cultures
Primary human IMR-90 lung fibroblasts from a female Caucasian were obtained from American Type Culture Collection (ATCC CCL-186; Manassas, VA). IMR-90 fibroblasts are primary human cells that have not been transformed or immortalized in any way. Therefore, the genetic make-up of IMR-90 fibroblasts is identical to that of human tissues, leaving no room for uncertainties regarding possible impacts of altered genetics as encountered in immortalized cell lines. Fibroblasts (from passages 32–36) were cultured in Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum, 0.1% sodium pyruvate, 100,000 U/L penicillin,100 mg/L streptomycin, and 0.1% non-essential amino acids (final concentrations). R,S-SFN was obtained from LKT Laboratories (St. Paul, MN) and was dissolved in DMSO to prepare a stock solution containing 40 mmol/l SFN. Aliquots were frozen at −20°C until use. SFN concentrations in cell culture media were adjusted to a final concentration of 2.0 μmol/l; controls were treated with solvent. Samples were collected at timed intervals.
2.2. Human feeding study
Eight apparently healthy adults (4 male, 4 female) participated in a broccoli sprout feeding study approved by the Institutional Review Board at the University of Nebraska-Lincoln. Exclusion criteria included pregnancy, smoking, self-reported health problems, and use of SFN supplements. Participants in this study included 6 Caucasians (2 Hispanics) and 2 Asians ages 19–30 years. Subjects were instructed not to consume cruciferous vegetables in the 48 hours leading up to the study and during the eight-hour period in which blood samples were collected. Each subject consumed three doses (34 g, 68 g, 102 g) of BroccoSprouts® broccoli sprouts from a local supermarket (HyVee, Lincoln, NE). Sprouts were consumed with a bagel and cream cheese (11). Each treatment was separated by a two week washout period and the order of doses was randomized. Thirty milliliters of blood were collected before sprout consumption (baseline, time 0 h) and at timed intervals (2, 4, and 8 h) after consumption. Peripheral blood mononuclear cells (PBMC) and plasma were purified using Histopaque and gradient centrifugation as described previously (21); aliquots were frozen at −80°C until analysis.
2.3. Quantitation of SFN
SFN was extracted and quantitated by HPLC as previously described (22). Briefly, 1 g of BroccoSprouts® was combined with 20 ml of water acidified with 0.1 M hydrochloric acid and homogenized for 10–15 seconds with a Tissue-Tearor model 985370 hand-held homogenizer (Biospec Products, Inc., Bartlesville, OK). Extracts were transferred to a 45°C water bath for 2 hours and then cooled to room temperature. The solution was extracted twice with 20 ml dichloromethane and the solvent phases were collected and combined. The solvent was dried through sodium sulfate and then loaded onto a Supelclean™ LC-Florisil® SPE column (Supelco). The column was washed with 3 ml ethyl acetate and SFN was collected with 3 ml methanol. Samples were passed through a 0.2 μm filter and stored at −20°C until analysis. Determination of SFN concentration was performed by injection on a Waters (Milford, MA) 600S HPLC system equipped with a VyDac C18 (Grace, Deerfield, IL) column and a Waters 2996 Photodiode Array detector. Separation was achieved with an isocratic flow rate of 1.0 ml/min using water and acetonitrile (30:70 v/v). A sample volume of 20 μl was injected.
2.4. Quantitative Real-Time PCR (qRT-PCR)
The abundance of p21 mRNA and LTR mRNA (transcript R/U5) was quantified by qRT-PCR as previously described using the cycle threshold method; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize for PCR efficiency (23, 24). PCR primer sequences for p21 were 5′-AGGCGGTTATGAAATTCACC-3′ (forward) and 5′-CCCTTCAAAGTGCCATCTG-3′ (reverse). LTR primer sequences were the same as previously reported (25). Note that the values for LTR mRNA represent the grand total of all transcriptionally active LTRs due to near-identical sequences in these repeats (25). Areas under the curves (AUCs) for LTR mRNA were calculated using the linear trapezoidal rule and were corrected for baseline levels (26).
2.5. Chromatin Immunoprecipitation (ChIP) assays
ChIP assays were performed as previously described (23). Antibody against K9-acetylated histone H3 (H3K9ac; ab10812) was purchased from Abcam (Cambridge, MA). Data were normalized for nucleosomal occupancy using an antibody to the C-terminus in histone H3. The enrichment of H3K9ac marks in LTR15 {nomenclature as per (15)} were quantified by qRT-PCR, using GAPDH as a control. PCR primer sequences were the same as in our previous studies (25). In case of ChIP assays, as opposed to mRNA quantification, individual LTRs can be distinguished by having one PCR primer anneal with sequences in the host genome adjacent to the LTR of interest.
2.6. Statistics
Homogeneity of variances was tested using Bartlett’s Test. If variances were heterogeneous, data was log transformed before analysis. Data from IMR90 cell cultures were analyzed by using the Wilcoxon Signed-Rank Test. Data from PBMC experiments were analyzed using one-way analysis of variance (ANOVA) and Fisher’s LSD for posthoc comparisons (27) for gene expression data and Wilcoxon Signed-Rank Test for ChIP data. Differences were considered statistically significant if p<0.05. StatView 5.0.1 was used for conducting statistical analyses (SAS Institute; Cary, NC).
3. Results
3.1. LTR transcript levels increase after SFN treatment in IMR-90 fibroblast cultures
SFN de-repressed LTRs in IMR-90 fibroblasts (Fig. 1). A significant increase in LTR mRNA was detectable at t = 4 h, and peak values were achieved at t = 6 h compared with vehicle controls. LTR expression levels returned to baseline levels after 16 h of SFN treatment. The expression of the p21 tumor suppressor also increased in response to SFN treatment (Fig. 2), consistent with previous reports in BPH-1, LnCaP, and PC-3 prostate epithelial cells (28), but this increase in p21 mRNA was not statistically significant.
Fig. 1.
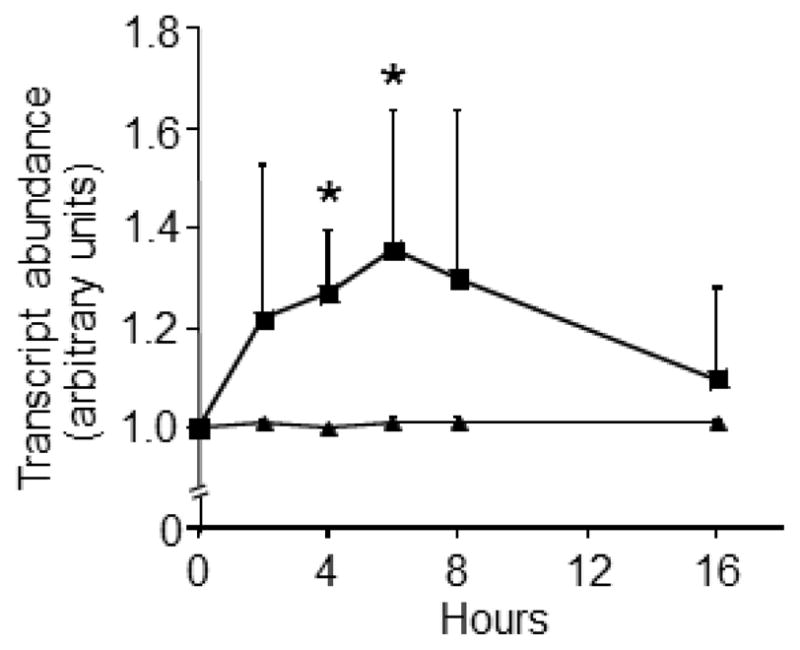
SFN increases LTR transcription in IMR-90 fibroblast cultures. Fibroblasts were treated with 2.0 μmol/l SFN (squares) or solvent (triangles). *Significantly different compared with vehicle control at the same collection time (p<0.05 by Wilcoxon Signed-Rank test, N=4–7).
Fig. 2.
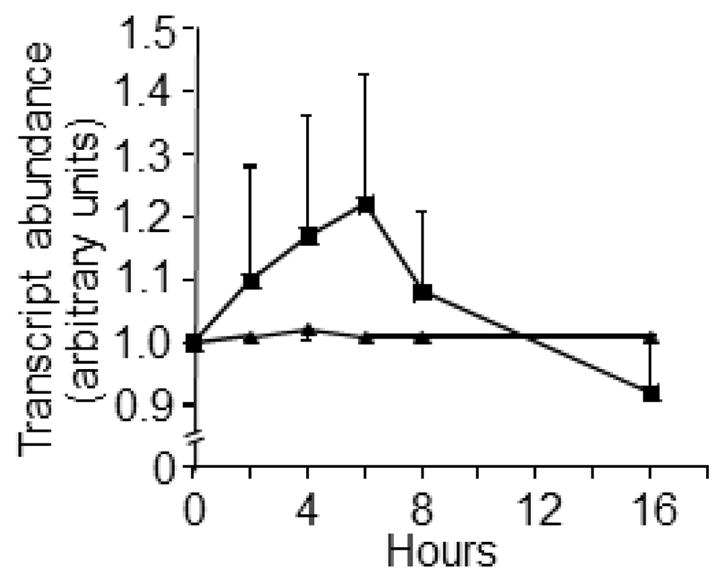
SFN had no significant effect on p21 transcription in IMR-90 fibroblast cultures. Fibroblasts were treated with 2.0 μmol/l SFN (squares) or solvent (triangles). *Significantly different compared with vehicle control at the same collection time (p<0.05 by Wilcoxon Signed-Rank test, N=4–7).
3.2. Human consumption of broccoli sprouts increases LTR expression and histone acetylation
The expression of LTRs increased dose-dependently in response to consumption of broccoli sprouts, as judged by the AUCs for LTR mRNA in PBMC (Fig. 3). The AUCs for LTR transcripts in subjects consuming 34 g, 68 g, and 102 g sprouts were 1.29 ± 2.38, 5.54 ± 5.26, and 13.06 ± 23.68 arbitrary units, respectively, compared with baseline. The increase was significantly greater for the 68 g and 102 g doses compared with the 34 g dose (p<0.05, N=8). The AUCs for p21 mRNA in subjects consuming 34 g, 68 g, and 102 g sprouts were 0.80 ± 1.43, 0.73 ± 0.90, 2.46 ± 3.00 arbitrary units, respectively, compared with baseline; however the differences were not statistically significant (p>0.05, N=8). Note that the level of SFN in Broccosprouts® was 18 mg/g fresh weight.
Fig. 3.
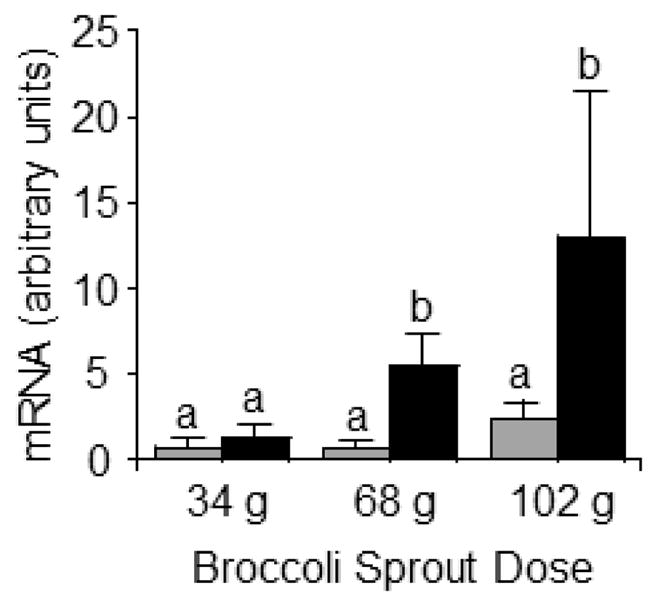
AUC for LTR (black bars) and p21 (gray bars) expression increases in a dose-dependent manner following three broccoli sprout doses in healthy adults. Statistical significance determined by ANOVA with Fisher’s LSD used for posthoc comparisons. a,bColumns not sharing the same letters are significantly different for the same treatment group (SFN vs. control; p<0.05, N=8).
The de-repression of LTRs was associated with an increased abundance of H3K9ac marks in LTR15 at t = 4 h compared with t = 0 h (Fig. 4A). The magnitude of the effect depended on the amount of broccoli sprouts that was consumed; meaningful increases in H3K9ac enrichment at t = 4 h were observed when sprout doses exceeded 34 g (Fig. 4B).
Fig 4.
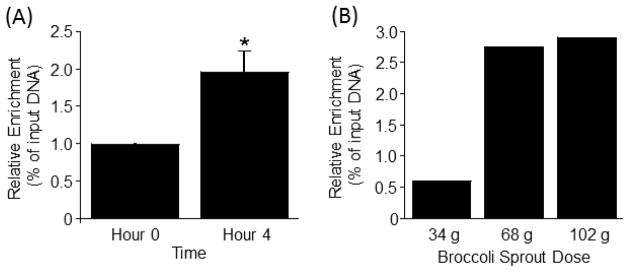
(A) Broccoli sprout consumption increases enrichment of H3K9ac mark in LTR15 in PBMCs from human subjects consuming 102 g broccoli sprouts. *Significantly different than controls as per Wilcoxon Signed-Rank test (p<0.05, N=8). (B) Dose-response curve for the H3K9ac mark in LTR15 in PBMCs from a representative subject.
4. Discussion
To the best of our knowledge, this is the first report suggesting possible off target effects of SFN in the de-repression of LTRs in both primary cells and human feeding studies. The increase in LTR mRNA abundance was not only statistically significant but also appears to be of biological importance, considering that the AUC for LTR mRNA increased by more than 10 times for the highest dose of broccoli sprouts. The doses tested in cell cultures and feeding studies are nutritionally relevant as levels of SFN tested in cell culture were based on previous results in human feeding studies (29). Our observations were corroborated by an increase in the abundance of H3K9ac marks in LTRs, consistent with their de-repression. Note that SFN also increased the expression of the tumor suppressor gene p21, albeit to a smaller extent than previously reported (30). The comparably small increase in p21 expression in response to SFN might be due to us using nutritionally meaningful levels, whereas other studies typically concentrations of at least 15 μmol/l SFN (9, 31–33).
Our observations are important for human health, considering the roles of LTRs in impairing genome stability (see Introduction) and the widespread availability of SFN-containing foods and supplements. The sprouts used in this study, Broccosprouts®, are widely available in the United States; the lowest dose that was tested, 34 g, is about one serving according to product labeling. One serving of broccoli sprouts may contain levels of SFN sufficient to de-repress LTRs. SFN-containing supplements are also available to consumers over the counter. To the best of our knowledge, there are no data available for the consumption of SFN supplements in the United States, but supplement use in general is fairly common. For example, approximately 50% of all Americans and 64–81%, of cancer survivors take nutritional supplements (34). Of the many SFN supplements on the market, wide ranges of doses are available. One product, BroccoMax® is a supplement labeled as having 30 mg sulforaphane glucosinolate per serving, providing a dose of SFN similar to the dose tested in this study. One could assume with a reasonable level of confidence that consuming such doses of SFN on a regular basis could elicit effects exceeding those seen in this study where single doses were administered. Note that we do not dispute the potential benefits of SFN in the chemoprevention of cancer (6–8, 11, 12). However, we propose that SFN needs to be considered in the context of gene-specific editing (14).
There are some uncertainties that need to be addressed in future studies. First, the effects of SFN on the de-repression of LTRs (and p21) are transient, and it remains to be determined whether a de-repression that last a few hours is sufficient to elicit biologically meaningful effects. Second, while the de-repression of LTRs was quantitatively important, it remains to be seen whether the observed increase in LTR expression is sufficient to impair genome stability.
Collectively, this study suggests that sulforaphane has off-target effects that warrant further investigation when recommending high levels of sulforaphane intake, despite its promising activities in chemoprevention.
Footnotes
Funding Source: This work is supported by the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945 and DK077816.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56(1):5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 2.Luthy B, Matile P. The mustard oil bomb: Rectified analysis of the subcellular organisation of the myrosinase system. Biochemie und Physiologie der Pflanzen. 1984;179(1–2):5–12. [Google Scholar]
- 3.Andreasson E, Jorgensen LB, Hoglund A-S, Rask L, Meijer J. Different myrosinase and idioblast distribution in arabidopsis and Brassica napus. Plant Physiol. 2001;127(4):1750–63. doi: 10.1104/pp.010334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, Hedrich R. Identification of a new glucosinolate-rich cell type in arabidopsis flower stalk. Plant Physiol. 2000;124(2):599–608. doi: 10.1104/pp.124.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhoeven DT, Goldbohm RA, van Poppel G, Verhagen H, van den Brandt PA. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol Biomarkers Prev 1996. 1996 Sep 1;5(9):733–48. [PubMed] [Google Scholar]
- 6.Kallifatidis G, Labsch S, Rausch V, Mattern J, Gladkich J, Moldenhauer G, et al. Sulforaphane increases drug-mediated cytotoxicity toward cancer stem-like cells of pancreas and prostate. Mol Ther. 2011;19(1):188–95. doi: 10.1038/mt.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rausch V, Liu L, Kallifatidis G, Baumann B, Mattern J, Gladkich J, et al. Synergistic activity of sorafenib and sulforaphane abolishes pancreatic cancer stem cell characteristics. Cancer Res 2010. 2010 Jun 15;70(12):5004–13. doi: 10.1158/0008-5472.CAN-10-0066. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010 May 1;16(9):2580–90. doi: 10.1158/1078-0432.CCR-09-2937. Epub 2010/04/15.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon DO, Kim MO, Kang SH, Choi YH, Kim GY. Sulforaphane suppresses TNF-α-mediated activation of NF-κB and induces apoptosis through activation of reactive oxygen species-dependent caspase-3. Cancer Lett. 2009;274(1):132–42. doi: 10.1016/j.canlet.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Heiss E, Herhaus C, Klimo K, Bartsch H, Gerhäuser C. Nuclear factor κB is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem 2001. 2001 Aug 24;276(34):32008–15. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- 11.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007 Feb;232(2):227–34. Epub 2007/01/30.eng. [PMC free article] [PubMed] [Google Scholar]
- 12.Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006 Mar;20(3):506–8. doi: 10.1096/fj.05-4785fje. Epub 2006/01/13.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res 2004. 2004 Aug 15;64(16):5767–74. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 14.de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res 2012. 2012 Nov 1;40(21):10596–613. doi: 10.1093/nar/gks863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res. 2006;34(9):e67. doi: 10.1093/nar/gkl335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005 Feb 23;24(4):800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004 Mar 12;303(5664):1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 18.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002 Dec 5;420(6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 19.Brunmeir R, Lagger S, Simboeck E, Sawicka A, Egger G, Hagelkruys A, et al. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 2010;6(4):e1000927. doi: 10.1371/journal.pgen.1000927. Epub 2010/05/06.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997 Aug;13(8):335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 21.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol Cell Physiol. 1998;275:C382–C8. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 22.Campas-Baypoli ON, Sanchez-Machado DI, Bueno-Solano C, Ramirez-Wong B, Lopez-Cervantes J. HPLC method validation for measurement of sulforaphane level in broccoli by-products. Biomedical chromatography: BMC. 2010 Apr;24(4):387–92. doi: 10.1002/bmc.1303. Epub 2009/08/04.eng. [DOI] [PubMed] [Google Scholar]
- 23.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011;22:328–33. doi: 10.1016/j.jnutbio.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–8. doi: 10.1006/meth.2001.1262. eng. [DOI] [PubMed] [Google Scholar]
- 25.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, et al. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J Nutr. 2008;138:2316–22. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zempleni J, Galloway JR, McCormick DB. Pharmacokinetics and utilization of orally and intravenously administered riboflavin in healthy humans. Am J Clin Nutr. 1996;63:54–66. doi: 10.1093/ajcn/63.1.54. [DOI] [PubMed] [Google Scholar]
- 27.Abacus Concepts. StatView. Berkeley, CA: Abacus Concepts Inc; 1996. [Google Scholar]
- 28.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 2006. 2006 Apr 1;27(4):811–9. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin Chim Acta. 2002 Feb;316(1–2):43–53. doi: 10.1016/s0009-8981(01)00727-6. Epub 2001/12/26.eng. [DOI] [PubMed] [Google Scholar]
- 30.Chew YC, Adhikary G, Wilson GM, Xu W, Eckert RL. Sulforaphane induction of p21(Cip1) cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent. J Biol Chem. 2012 May 11;287(20):16168–78. doi: 10.1074/jbc.M111.305292. Epub 2012/03/20.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, et al. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000 Mar 1;60(5):1426–33. Epub 2000/03/23.eng. [PubMed] [Google Scholar]
- 32.Fimognari C, Nusse M, Cesari R, Iori R, Cantelli-Forti G, Hrelia P. Growth inhibition, cell-cycle arrest and apoptosis in human T-cell leukemia by the isothiocyanate sulforaphane. Carcinogenesis. 2002 Apr;23(4):581–6. doi: 10.1093/carcin/23.4.581. Epub 2002/04/19.eng. [DOI] [PubMed] [Google Scholar]
- 33.Singh SV, Srivastava SK, Choi S, Lew KL, Antosiewicz J, Xiao D, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J Biol Chem 2005. 2005 May 20;280(20):19911–24. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 34.Velicer CM, Ulrich CM. Vitamin and mineral supplement use among US adults after cancer diagnosis: a systematic review. J Clin Oncol. 2008 Feb 1;26(4):665–73. doi: 10.1200/JCO.2007.13.5905. Epub 2008/02/01.eng. [DOI] [PubMed] [Google Scholar]


