Abstract
While certain archaea appear to synthesize and/or metabolize fatty acids, the respective pathways still remain obscure. By analyzing the genomic distribution of the key lipid-related enzymes, we were able to identify the likely components of the archaeal pathway of fatty acid metabolism, namely, a combination of the enzymes of bacterial-type β-oxidation of fatty acids (acyl-CoA-dehydrogenase, enoyl-CoA hydratase, and 3-hydroxyacyl-CoA dehydrogenase) with paralogs of the archaeal acetyl-CoA C-acetyltransferase, an enzyme of the mevalonate biosynthesis pathway. These three β-oxidation enzymes working in the reverse direction could potentially catalyze biosynthesis of fatty acids, with paralogs of acetyl-CoA C-acetyltransferase performing addition of C2 fragments. The presence in archaea of the genes for energy-transducing membrane enzyme complexes, such as cytochrome bc complex, cytochrome c oxidase, and diverse rhodopsins, was found to correlate with the presence of the proposed system of fatty acid biosynthesis. We speculate that because these membrane complexes functionally depend on fatty acid chains, their genes could have been acquired via lateral gene transfer from bacteria only by those archaea that already possessed a system of fatty acid biosynthesis. The proposed pathway of archaeal fatty acid metabolism operates in extreme conditions and therefore might be of interest in the context of biofuel production and other industrial applications.
Keywords: biofuels, β-oxidation, halobacteria, methanogens, rhodopsin, bioenergetics
Introduction
In archaea, whose membrane lipids are built of isoprenoids (Koga and Morii, 2007), the role(s) of long-chain fatty acids and even their very presence remain controversial. On one hand, several thorough studies of archaeal membranes found no fatty acid-based lipids (Koga et al., 1993; Corcelli and Lobasso, 2006; Falb et al., 2008). On the other hand, palmitate, a C16 fatty acid, was shown to be crucial for the functional integrity of halorhodopsin and could be synthesized by cells of Halobacterium salinarum even when they were grown in a defined culture medium with no added fatty acids (Corcelli et al., 1996). The crystal structure of halorhodopsin from H. salinarum (Protein DataBank entry 1E12) showed three molecules of palmitate tightly bound between the monomers in the functionally active trimer (Kolbe et al., 2000). A fatty acid synthase (FAS) activity has been detected in a cell fraction from the archaeon H. salinarum as early as in 1971 (Pugh et al., 1971). Later, it was shown that cells of H. salinarum could grow on a synthetic medium with known composition (a mixture of amino acids, nucleotides, glycerol and inorganic salts) and produce functional bacteriorhodopsin and halorhodopsin (Gochnauer and Kushner, 1969; Oesterhelt and Krippahl, 1973; Helgersons et al., 1992). To exclude the possibility of the growth being supported by residual amounts of nutrients transferred from the previous rich medium, the cells have been even cultured in a synthetic medium for 9 passages (Gochnauer and Kushner, 1969). Hence, cells of H. salinarum appear to be capable of synthesizing long-chain fatty acids, in particular, palmitate, from C2-C3 precursors. For several other archaea, the ability to metabolize long-chain fatty acids has been reported (Falb et al., 2008; Slobodkina et al., 2009). For example, growth of Archaeoglobus fulgidus could be sustained by the oxidation of long-chain fatty acids (Khelifi et al., 2010). However, the respective enzymes have not been characterized.
In a recent paper, Lombard and coworkers (Lombard et al., 2012a) reported finding in various archaeal genomes homologs of the components of the bacterial fatty acid synthase II (FAS II), except for the acyl-carrier protein (ACP) and acyl-ACP synthase (see Rock and Cronan, 1996; Rock and Jackowski, 2002, and the Supplementary Materials for a description of FAS II components). Accordingly, they suggested that archaea are able to synthesize fatty acids by using these homologous components in combination with some functional analogues of ACP and acyl-ACP synthase (Lombard et al., 2012a). However, no such analogues of ACP and acyl-ACP synthase have ever been identified in archaea and the few homologs of bacterial FAS II components in archaeal genomes had a patchy distribution, insufficient to form a functional pathway in a single archaeal genome (see Table S1 of Lombard et al., 2012a). Thus, the problem of fatty acid biosynthesis in archaea remained unresolved, which prompted us to re-examine this issue.
We report here a genomic reconstruction of the archaeal fatty acid metabolism based on the Clusters of Orthologous Groups (COGs) approach (Tatusov et al., 1997; Koonin and Galperin, 2003). The results of this study suggest that many archaea are capable of synthesizing fatty acids by means of a chimeric pathway that includes bacterial-type enzymes of fatty acid β-oxidation in combination with (multiple paralogs of) the archaeal acetyl-CoA acetyltransferase, an enzyme of the mevalonate biosynthesis pathway. Accordingly, these results do not support the idea of an ancestral origin of fatty acid biosynthesis and, instead, suggest that most of the respective enzymes have been acquired from bacteria via lateral gene transfer.
COG-based pathway analysis
Phylogenomic reconstruction of archaeal fatty acid metabolism was based on the COGs approach (Tatusov et al., 1997; 2000; Koonin and Galperin, 2003), which improves accuracy of gene function prediction by distinguishing orthologous and paralogous genes, which is particularly important when the analyzed genomes come from different domains of life and the corresponding proteins show low sequence similarity (Makarova et al., 1999; 2007). The analysis of archaeal protein families relied on the assignments in the latest public release of the COG database (Tatusov et al., 2003), currently available at ftp://ftp.ncbi.nlm.nih.gov/pub/COG/COG/whog, the NCBI’s RefSeq database (Pruitt et al., 2012), and the recently updated version of archaeal COGs (Wolf et al., 2012), which is available at ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/arCOG. We also used the latest versions of the eggNOG, KEGG and MetaCyc databases (Caspi et al., 2012; Powell et al., 2012; Nakaya et al., 2013). Pathway details were taken from KEGG and the available literature data.
An analysis of the genes involved in lipid metabolism (functional group I in the COG database) identified in the genome of H. salinarum strain NRC-1 genes for the four key enzymes of the bacterial pathway of β-oxidation of fatty acids (see Figure S1): acyl-CoA dehydrogenase (ACD or FadE, COG1960, EC 1.3.1.8, 1.3.8.1, 1.3.8.7, 1.3.8.8, or 1.3.8.9), enoyl-CoA hydratase (ECH or FadB_1, COG1024, EC 4.2.1.17), and 3-hydroxyacyl-CoA dehydrogenase (HDH or FadB_2, COG1250, EC 1.1.1.35), and 3-ketoacyl-CoA thiolase (BKL or FadA, COG0183, EC 2.3.1.16). Each of these genes was found in H. salinarum genome in several paralogous copies (6, 3, 2 and 3, respectively, see Table 1). In contrast, the KEGG scheme of fatty acid degradation (http://www.genome.jp/kegg-bin/show_pathway?mapno=00071&org_name=hal) indicated the presence of 3-ketoacyl-CoA thiolase but not of any other enzyme of this pathway. These data demonstrated that the issue of archaeal fatty acid metabolism remained unresolved and was worthy of a comprehensive analysis.
Table 1.
Potential enzymes of fatty acid β-oxidation and membrane energy-converting complexes in archaea
| Organism name | Steps of β-oxidation | Biosynthetic | Rhodo | Complex III | Complex IV | |||||||
| 1a | | 1 | | 2 | | 3 | | thiolases | psin | subunits | subunits | |||||
| ACDa | ACD | ECH | HDH | BKL | BKL | |||||||
| HpaB | FadE | FadB1 | FadB2 | FadA | AcaB | CytB | Rieske | CoxA | CoxB | CoxC | ||
| Crenarchaeota | COG | COG | COG | COG | COG0183 | COG | COG | COG | COG | COG | COG | |
| 2358 | 1950 | 1024 | 1250 | Bact. | Arch. | 5524 | 1290 | 0723 | 0843 | 1522 | 1845 | |
| Acidilobus saccharovorans 345 15 | – | 4 | 3 | 1 | 1 | 4 | – | 1 | – | – | – | – |
| Aeropyrum pernix K1 | – | 5 | 3 | 1 | 1 | 4 | – | 1 | 2 | 2 | 2 | 1 |
| Desulfurococcus kamchatkensis 1221 n | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Desulfurococcus mucosus DSM 2162 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Hyperthermus butylicus DSM 5456 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Ignicoccus hospitalis KIN4 1 | 1 | – | 1 | 1 | 1 | 1 | ||||||
| Ignisphaera aggregans DSM 17230 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Pyrolobus fumarii 1A | 1 | – | 1 | 1 | 1 | 1 | ||||||
| Staphylothermus hellenicus DSM 12710 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Staphylothermus marinus F1 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Thermosphaera aggregans DSM 11486 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| Acidianus hospitalis W1 | 1 | 3 | 2 | 2 | 1 | 1 | – | 2 | 2 | 4 | 1 | – |
| Metallosptiaera sedula DSM 5348 | 4 | 6 | 6 | 4 | 1 | 7 | – | 3 | 4 | 6 | 4 | 1 |
| Sulfolobus solfataricus P2 | 3 | 9 | 8 | 4 | 2 | 8 | – | 3 | 3 | 3 | 2 | 1 |
| Caldivirga maquilingensis IC 167 | – | 2 | 2 | 1 | 1 | – | 2 | 2 | 1 | 1 | – | |
| Pyrobaculum aerophiium IM2 | 1 | 4 | 3 | 2 | 5 | – | 1 | 1 | 2 | 2 | 1 | |
| Thermofilum pendens Hrk 5 | – | – | – | – | 1 | – | – | – | – | – | – | |
| Thermoproteus neutrophilus V24Sta | 1 | 3 | 1 | 1 | 1 | |||||||
| Thermoproteus tenax Kra 1 | 1 | 4 | 3 | 2 | 5 | – | 1 | 1 | – | – | – | |
| Thermoproteus uzoniensis 768 20 | 1 | 4 | 3 | 2 | 5 | – | 2 | 2 | 1 | 1 | 1 | |
| Vulcanisaeta moutnovskia 768 28 | 2 | 7 | 6 | 2 | 2 | 5 | ||||||
| Euryarchaeota | ||||||||||||
| Archaeoglobus fulgidus DSM 4304* | 3 | 14 | 9 | 10 | 3 | 12 | – | – | – | – | 2 | – |
| Archaeoglobus profundus DSM 5631 | – | 1 | – | – | – | 2 | – | – | – | – | – | – |
| Archaeoglobus veneficus SNP6 | – | 1 | – | – | 1 | 1 | ||||||
| Ferroglobus placidus DSM 10642 | 1 | 9 | 6 | 5 | 2 | 7 | – | – | – | – | 2 | – |
| Halalkalicoccus jeotgali B3 | – | 12 | 12 | 5 | 2 | 3 | – | 2 | 2 | 2 | 1 | 1 |
| Haloarcula marismortui ATCC 43049 | – | 10 | 7 | 5 | 3 | 4 | 6 | 3 | 3 | 4 | 4 | 3 |
| Halobacterium salinarum R1 | – | 6 | 3 | 2 | 1 | 2 | 4 | 2 | 1 | 2 | 2 | 1 |
| Haloferax volcanii DS2 | – | 7 | 7 | 4 | 1 | 3 | – | 4 | 3 | 3 | 3 | 2 |
| Halogeometricum borinquense DSM 11551 | – | 8 | 7 | 4 | 1 | 4 | – | 3 | 2 | 4 | 3 | 2 |
| Halomicrobium mukohataei DSM 12286 | – | 6 | 3 | 3 | 1 | 2 | 3 | 3 | 2 | 4 | 2 | 1 |
| Halopiger xanaduensis SH 6 | – | 6 | 1 | 2 | 2 | 2 | 2 | – | 1 | 4 | 3 | 3 |
| Haloquadratum walsbyi DSM 16790 | – | 1 | 1 | 1 | – | 3 | 3 | 2 | 1 | 1 | 1 | 1 |
| Halorhabdus utahensis DSM 12940 | – | – | 2 | 1 | – | 1 | 2 | 3 | 2 | 1 | 1 | 1 |
| Halorubrum lacusprofundi ATCC 49239 | – | 7 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | 2 | 2 |
| Haloterrigena turkmenica DSM 5511 | – | 18 | 16 | 10 | 4 | 12 | 1 | – | 1 | 4 | 3 | 3 |
| Natrialba magadii ATCC 43099 | – | 14 | 9 | 7 | 3 | 5 | 3 | – | 1 | 3 | 3 | 1 |
| Natronomonas pharaonis DSM 2160* | – | 12 | 8 | 5 | 3 | 5 | 4 | – | – | 3 | 3 | 1 |
| Methanothermus fervidus DSM 2088 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanobrevibacter smithii ATCC 35061 | – | – | – | 1 | – | – | – | – | – | – | – | |
| Methanosphaera stadtmanae DSM 3091 | – | – | – | 1 | – | – | – | – | – | – | – | |
| Methanothermobacter thermautotrophicus | ||||||||||||
| Delta H | ||||||||||||
| Methanocaldococcus jannaschii DSM 2661 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanococcus aeolicus Nankai 3 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanothermococcus okinawensis IH1 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanotorris igneus Kol 5 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanocella paludicola SANAE | – | – | – | – | – | – | – | – | – | – | – | |
| Methanocorpusculum labreanum Z | – | – | – | – | – | – | – | – | – | – | – | |
| Methanoculleus marisnigri J R1 | – | – | – | – | – | – | – | 1 | – | – | – | |
| Methanoplanus petrolearius DSM 11571 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanoregula boonei 6A8 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanosphaerula palustris E1 9c | – | – | – | – | – | – | – | – | – | – | – | |
| Methanospirillum hungatei JF 1 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanococcoides burtonii DSM 6242 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanohalobium evestigatum Z 7303 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanohaiophilus mahii DSM 5219 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanosaeta thermophila PT | – | – | – | – | – | – | – | – | – | – | – | |
| Methanosalsum zhilinae DSM 4017 | – | – | – | – | – | – | – | – | – | – | – | |
| Methanosarcina barkeri Fusaro | – | – | – | – | – | – | – | – | – | – | 1 | |
| Methanopyrus kandleri AV19 | – | – | – | – | – | – | – | – | – | – | – | |
| Pyrococcus furiosus DSM 3638 | – | – | – | – | – | – | – | – | – | – | – | |
| Thermococcus sibiricus MM 739 | – | – | – | – | – | – | – | – | – | – | – | |
| Picrophilus torridus DSM 9790 | – | 6 | 2 | 2 | 2 | 2 | – | 1 | 1 | 1 | 1 | – |
| Thermoplasma acidophilum DSM 1728 | – | 7 | 5 | 3 | 2 | 3 | – | 2 | 2 | – | – | – |
| Aciduliprofundum boonei T469 | – | – | – | – | – | 1 | – | – | – | – | – | – |
| other archaea | ||||||||||||
| Korarchaeum cryptofilum | – | 1 | 1 | 1 | 1 | 1 | – | 1 | 1 | – | – | – |
| Nanoarchaeum equitans | ||||||||||||
| Cenarchaeum symbiosum | 1 | – | 1 | 1 | – | 2 | – | 1 | 1 | 1 | 1 | – |
| Nitrosopumilus maritimus | 1 | – | 1 | 1 | – | 2 | – | 1 | 1 | 1 | 1 | – |
The organisms are ordered according to their taxonomic positions (see Table S1). Two organisms shown to metabolize fatty acids (Falb et al., 2008; Khelifi et al., 2010) are marked with asterisks. The columns represent the numbers of paralogous genes within each respective COG (Tatusov et al., 2003; Wolf et al., 2012), or their absence, indicated with a dash, in the corresponding genome. For COG0183 (3-ketothiolases, acetyl-CoA C-acetyltransferases, and related enzymes), the two columns show the numbers of genes in the “bacterial” and “archaeal” parts of the tree (Fig. 1). Grey shading indicates those genomes that encode only a single “archaeal” form member of COG0183. The enzyme names are abbreviated as follows: ACD, acyl-CoA-dehydrogenase; ECH, enoyl-CoA hydratase; HDH, 3-hydroxyacyl-CoA dehydrogenase; BKL, β-ketothiolase. The prediction that members of COG2368 function as alternative acyl-CoA-dehydrogenases, ACDa, has not been experimentally verified (see text for details).
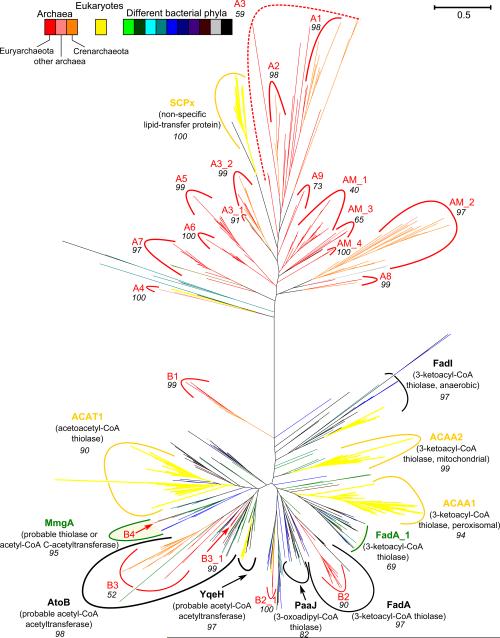
An analysis of the archaeal COGs and their genome neighborhoods (Tables 1 and S1) in 69 species representing the key lineages of archaea (listed in Table S2) revealed the presence of co-occurring genes encoding three enzymes of β-oxidation of fatty acids, ACD, ECH, and HDH in approximately half of the analyzed archaeal genomes. These enzyme sequences showed highly significant similarity to the respective bacterial enzymes and were typically assigned to the same COG. In contrast, β-ketothiolases (COG0183) were found in every archaeal genome except for the highly reduced genome of Nanoarchaeum equitans; in some organisms they were encoded by a single gene but often were found in several paralogous forms (Table 1). Such a biased distribution of various β-ketothiolase homologs could to be due to their involvement in several substantially different reactions (pathways). Indeed, in addition to the 3-ketoacyl-CoA thiolase, COG0183 includes another enzyme of the thiolase superfamily (Pereto et al., 2005; Jiang et al., 2008), archaeal acetyl-CoA C-acetyltransferase (EC 2.3.1.9). This latter enzyme catalyzes the first condensation reaction in the mevalonate pathway of the biosynthesis of C5 isoprenoid precursors (for a review see (Miziorko, 2011). Since mevalonate pathway is essential for the synthesis of archaeal isoprenoid lipids, at least one member of COG0183 is present in complete genomes of all free-living archaea. Remarkably, the genomes that encode the three β-oxidation enzymes (ECH, HDH and ACD) invariably encode more than one member of COG0183 (Table 1). To sort out the respective proteins and trace their evolutionary relationships, we have constructed a maximum likelihood phylogenetic tree for the potential β-ketothiolases and inspected it in order to figure out the likely functions of these enzymes.
The tree (Figure 1, see also Figure S2) revealed a clear split between mostly bacterial and archaeal sequences, with the few archaeal branches (B1 through B4) in the bacterial part representing clear-cut cases of lateral gene transfer from bacteria to archaea. Within the archaeal part, one group of sequences (clades AM_1 through AM_4) contained those members of COG0183 that are present in single copies in the majority of archaeal genomes (shaded grey in Table 1), with separate groupings of euarchaeal (AM_1) and crenarchaeal (AM_2) sequences. Analysis of the genomic neighborhoods of these genes showed that they were frequently located next to the genes that code for 3-hydroxy-3-methylglutaryl CoA synthase and hydroxymethylglutaryl-CoA reductase, enzymes of the mevalonate pathway (Table S1). This observation suggested that β-ketothiolases from clades AM_1 through AM_4 indeed function in this pathway.
Figure 1. Schematic view of the phylogenetic tree for β-ketothiolases (acetoacetyl-CoA acyltransferases, COG0183).
Separate clades are labelled with the names of the experimentally characterized genes from model organisms. The names of E. coli genes are in black, names of B. subtilis genes are in green, names of human genes are in yellow. The branches of tree are colored according to the taxonomical identity of the source organisms; the color code is shown in the upper left corner. Clades with archaeal sequences are indicated by red arks or red arrows and named from A1 to AM_4 and from B1 to B4. The numbers underneath the clade labels indicate the reliability of respective branches (as tested using SH-like statistics implemented in PhyML). The bar in the top right corner represents the scale for the branch lengths (the expected number of substitutions per site). The tree has been constructed based on conserved blocks (total 227 positions) from 511 sequences. The sequences were aligned with Muscle (Edgar, 2004) and the alignment was manually edited using GeneDoc (Nicholas et al., 1997). The phylogenetic tree was constructed by PhyML (Guindon and Gascuel, 2003) with the SPR algorithm of tree construction. The tree was visualized with MEGA5 program (Tamura et al., 2011). The complete version of this tree is available in the Supplementary Materials as Figure S2.
Other groups of sequences (clades A1 through A9 on Figure 1) contain proteins that are encoded only in some archaeal genomes, usually in large paralogous groups. Their genes almost always co-occur with the genes of aforementioned three proteins of the β-oxidation pathway (Table 1) and are often adjacent to them (Table S1), forming predicted operons. This observation makes it increasingly likely that these enzymes participate in the same pathway.
Table 1 also shows that the correlation between the presence of three β-oxidation enzymes and multiple β-ketothiolases is not absolute. Four out of the 117 inspected archaeal genomes (Ignicoccus hospitalis, Pyrolobus fumarii, Cenarchaeum symbiosum and Nitrosopumilus maritimus) encode ECH and HCD, along with two paralogs of β-ketothiolase, but do not have the genes for ACD (COG1960). A PSI-BLAST search (Altschul et al., 1997) identified members of COG2368 (annotated as ‘Aromatic ring hydroxylase’) as distant homologs of FadE that could potentially have the acyl-CoA dehydrogenase activity and therefore functionally replace the bacterial ACD. Indeed, an experimentally characterized member of this COG was shown to have 4-hydroxybutyryl-CoA dehydratase activity, and also performed isomerization of the double bond (Muh et al., 1996). Archaeal members of COG2368 appear only in the genomes with other β-oxidation genes (Table 1) and appear to be plausible candidates for non-orthologous gene displacement of FadE-like acyl-CoA dehydrogenase. In two of these four organisms, C. symbiosum and N. maritimus, COG2368 members are encoded next to the genes for the NDP-forming acyl-CoA synthetase (Sanchez et al., 2000), see Table S3. Therefore, COG2368 members are listed in Table 1 as ACDa (predicted alternative ACD).
Hence, in all inspected archaeal genomes, ECHs, HDHs, and either FadE-like acyl-CoA-dehydrogenases (ACD) or members of COG2368 (ACDa) co-occur with additional thiolases, either of bacterial type or paralogs of archaeal acetyl-CoA acetyltransferase. The correlation is significantly positive at any level of sampling, with the Z-score ≥ 14.6. Thus, almost a half of 69 archaeal genomes shown in Table 1 are potentially capable of fatty acid metabolism, which might be important for survival of these organisms in their natural habitats.
A search for additional enzymes that co-occurred with the β-oxidation enzymes (see Table S4) identified three more COGs that, in principle, could be involved in archaeal fatty acid metabolism. COG1607 unifies various thioesterases, including the acyl-CoA hydrolase YciA from E. coli (Kuznetsova et al., 2005). COG1804 includes members of an additional family of CoA transferases, such as crotonobetainyl-CoA:carnitine CoA-transferase CaiB (Heider, 2001). Finally, COG2030 includes MaoC-like R-specific enoyl-CoA dehydratases (whereas the β-oxidation pathway includes S-enoyl-CoA dehydratase) (Fukui et al., 1998). However, none of the enzymes listed in Table S4 appears to be able to catalyze the addition or cleavage of C2 fragments.
Could fatty acids be synthesized by reversing the β-oxidation pathway?
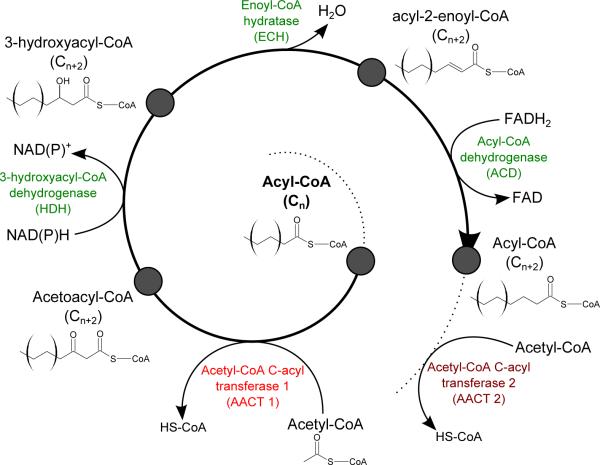
In principle, the reactions of the fatty acid β-oxidation pathway are reversible, which begs the question of whether the same enzymes could be used for synthesizing fatty acids from C2 precursors. In fact, this reverse mode of operation of β-oxidation enzymes from Escherichia coli has been recently realized in the context of biofuel production, yielding a pathway of carbon chain elongation (Dellomonaco et al., 2011; Clomburg et al., 2012). In particular, thiolase AtoB was shown to catalyze condensation of two acetyl-CoA molecules, while thiolase FadA was appeared to operate on longer substrates (Clomburg et al., 2012). A similar arrangement could function in archaea, where the first step, synthesis of acetoacetyl-CoA, would be shared with the mevalonate pathway and carried out by the universal archaeal acetyl-CoA C-acetyltransferase, whereas its various paralogs would catalyze subsequent steps of fatty acid chain elongation. The proposed pathway of archaeal fatty acid synthesis is shown in Figure 2. The large number of paralogs of acetyl-CoA C-acetyltransferase in some archaeal genomes (Tables 1 and S1) may reflect their specificity to hydrocarbon chains of particular length, as shown in Figure 2.
Figure 2. Proposed scheme of archaeal fatty acid biosynthesis.
The synthesis of fatty acids in archaea is proposed to proceed through essentially the same steps as in FAS I or FAS II mechanisms (see Figure S3), but by using CoA instead of the acyl-carrier protein, as in the β-oxidation of fatty acids (see Figure S1). Enzymes that catalyze the reduction (HDH and ACD) and dehydratation (ECH) steps are proposed to be the same as those involved in the bacterial β-oxidation pathway, whereas initiation of the cycle and fatty acid chain elongation are proposed to be catalyzed by various paralogs of the archaeal acetyl-CoA C-acetyltransferase. As discussed in the text, the presence of several paralogs of the latter enzyme in many archaeal genomes (denoted here as AACT1 and AACT2) might reflect their specificity towards chains of particular lengths. As an example, enzymes of Aeropyrum pernix K1, proposed to catalyze the respective steps, are indicated by their genome locus tags (GenBank accession no. BA000002).
Distribution of membrane energy-converting enzymes
In addition to the above-mentioned COG1607 (YciA), COG1804 (CaiB), and COG2030 (MaoC), a search for genes that frequently co-occur with genes of fatty acid β-oxidation genes and multiple paralogs of acetyl-CoA C-acetyltransferase revealed a correlation between the presence in archaeal genomes of these genes and the genes encoding certain membrane energy-converting proteins, namely various rhodopsins and the key membrane subunits of the cytochrome bc complex and the cytochrome c oxidase (Tables 1 and S4). Specifically, genes of these energy-converting enzymes were only found in those archaea that had the β-oxidation pathway genes and multiple paralogs of acetyl-CoA C-acetyltransferase, but not the other way around. This was quite remarkable, as no such correlation has been observed in bacteria (data not shown). We could not come up with any explanation of why the presence of cytochrome bc complex, the cytochrome c oxidase and rhodopsins would correlate with the presence of the fatty acid degradation pathway. On the other hand, there was an easy explanation for the co-occurrence of these enzymes and the fatty acid biosynthesis pathway. Indeed, membrane subunits of rhodopsin, cytochrome bc complex and the cytochrome c oxidase often incorporate fatty acid chains as essential structural elements (Gomez and Robinson, 1999; Sedlák and Robinson, 1999; Kolbe et al., 2000; Shinzawa-Itoh et al., 2007; Hasan et al., 2011). Therefore, proper functioning of these membrane enzymes would depend on the ability of the organisms to synthesize fatty acids, at least in free-living archaea that cannot acquire these fatty acids from the surrounding milieu.
Archaeal fatty acids metabolism: specific or bacterial-like?
The fundamental difference between the fatty acid biosynthesis pathways of bacteria and archaea has been suggested back in 1971 from an observation that fatty acid synthesis in the cell fraction of Halobacterium cutirubrum (currently H. salinarum) did not change after the addition of bacterial ACP (Pugh et al., 1971), while the preparations from E. coli showed an almost tenfold increase in activity. A recent work (Lombard et al., 2012a) did not find in archaeal genomes genes of eukaryotic fatty acid synthase (FAS I, see Smith, 1994; Maier et al., 2008, for reviews) but reported finding certain components of the bacterial fatty acid synthase (FAS II). Our analysis has shown that some archaea indeed encode orthologs of certain components of the bacterial FAS II. However, with two exceptions that are discussed below, these components have a patchy distribution, incompatible with a functional fatty acid biosynthesis pathway (see Table S2). These results are consistent with the idea that archaea use a system of fatty acid synthesis that is substantially different from the bacterial one.
Fatty acid metabolism in methanogens?
The above analysis could help sorting out the often controversial data on the presence of fatty acids in archaea. For example, a single, never confirmed report claimed the presence of fatty acid-based lipids in diverse methanogenic archaea; this report was based on identification of fatty acids in membrane fractions that were obtained from archaeal cells grown on yeast extract (Gattinger et al., 2002). These fatty acid-based lipids were identified in the yeast extract used in the growth medium, although at the concentration about 25 times less than that observed in frozen archaeal cells. In our opinion, these data cannot be considered as conclusive evidence for the ability of these methanogens to synthesize fatty acids. Since the genomes of methanogens do not seem to contain genes of β-oxidation of fatty acids (Table 1), fatty acid-containing hydrophobic constituents of the yeast extract would selectively accumulate in the membranes of these methanogens upon consumption of the other components of the extract. Furthermore, fatty acids of organic waste, being weak uncouplers (Korshunov et al., 1998), suppress the growth of methanogenic archaea within well-studied syntrophic bacterial-archaeal consortia that are widely used for production of methane from such waste (Worm et al., 2010). Accordingly, sustainability of such microbial systems depends on vigorous consumption of fatty acids by bacteria, which prevents their accumulation in the media. Apparently, methanogens cannot deal with long-chain fatty acids.
Curiously, in an early study, an addition of bacterial ACP to the cell fraction of Methanobacterium thermoautotrophicum (currently Methanothermobacter thermautotrophicus) led to a three-fold increase in the yield of fatty acids (Pugh and Kates, 1994). This observation has been interpreted as evidence for the presence of a bacterial-type FAS in this methanogen (Pugh and Kates, 1994; Lombard et al., 2012a). However, the genome of M. thermoautotrophicus does not encode any components of FASII that could interact with the added bacterial ACP (Table S2). Therefore, the reasons for the effect observed by Pugh and Kates (1994) remain obscure, unless their cell extract contained residual amounts of bacterial FAS components.
Accumulation from yeast extract might also account for the reported presence of long-chain fatty acids (Carballeira et al., 1997) and long-chain alcohols (Nishihara et al., 2000) in membrane fractions of the archaeon Pyrococcus furiosus which also lacks β-oxidation enzymes (Table 1). It is noteworthy that the most recent study of the total lipids of P. furiosus found no fatty acids or fatty acid-based lipids (Lobasso et al., 2012).
Fatty acid metabolism in haloarchaea
The case of the H. salinarum, which is apparently capable of synthesizing long-chain fatty acids, is more complex. The ability of H. salinarum to oxidize long-chain fatty acids has never been demonstrated, despite the presence of a bacterial-type β-ketothiolase in the genome (Table 1). In fact, long-chain fatty acids have been shown to suppress the growth of H. salinarum (Gonzalez, 2009). A recent study demonstrated activated expression of the H. salinarum genes for β-oxidation pathway (ECH, HDH, ACD) under the conditions of a low-salt stress, 2.6 M NaCl as compared to the optimal NaCl concentration of 4.3 M (Leuko et al., 2009). The genes for two β-ketothiolases, VNG0678G and VNG0931G, which belong to the archaeal clades A5 and A7, respectively (Figures 1 and S2), were also upregulated under these conditions, as were the Bop and CoxA genes that encode bacteriorhodopsin and cytochrome c oxidase subunit I (Leuko et al., 2009). It is tempting to suggest that this entire set of co-expressed enzymes is responsible for energy conservation under stress conditions, when newly synthesized long-chain fatty acids are needed to stabilize the membrane-bound energy-transducing enzymes.
That would mean, however, a reversal of the typical roles of bacterial- and archaeal-type β-ketothiolases in H. salinarum. If the typical archaeal β-ketothiolases, VNG0678G and VNG0931G, are involved in fatty acid biosynthesis, this leaves the bacterial-type β-ketothiolase VNG2063G to participate in the mevalonate synthesis pathway, in agreement with the proposal of (Falb et al., 2008). In fact, the mevalonate pathway in halobacteria is atypical since it seems to begin with the condensation of acetyl-CoA with yet unknown C2 compound (possibly a derivative of lysine) into acetoacetyl-CoA (Ekiel et al., 1986; Falb et al., 2008). The proposed recruitment of the bacterial-type β-ketothiolase for this reaction, implying a change in its substrate specificity, could explain why the cells of H. salinarum lack the ability to oxidize long-chain fatty acids (Gonzalez, 2009).
In contrast to other archaea, genomes of two haloarchaeal species, Halogeometricum borinquense and Haloterrigena turkmenica, encode almost complete sets of FAS II genes. The former contains genes for ACP, ACP synthase, 3-oxoacyl-ACP synthase, ACP-S-malonyl-transferase, and 3- oxoacyl-ACP reductase, whereas the latter encodes all these enzymes as well as enoyl-ACP reductase (Table S2). However, neither organism encodes 3-hydroxyacyl-ACP dehydratase (FabA) or acetyl-CoA carboxylase. As a result, it is not clear whether either of these organisms has a fully functional bacterial-type FAS II. Haloarchaea are known for the high extent of genes acquired by lateral gene transfer from bacteria (Makarova et al., 2007; Nelson-Sathi et al., 2012; Wolf et al., 2012), so the presence of certain bacterial FAS genes in certain haloarchaea is hardly surprising.
Evolutionary considerations
The organization of the archaeal fatty acid biosynthesis pathway, if any, is important for resolving a long-standing evolutionary problem of the origin of biological membranes and, more specifically, the nature of the membrane organization in the last common ancestor of bacteria and archaea (LUCA). Earlier studies made a clear distinction between the archaea with their isoprenoid-based lipids, synthesized via mevalonate pathway, and bacteria and eukaryotes, who have fatty acids-based phospholipid membranes (Koga et al., 1998; Boucher et al., 2004). Since the components of the mevalonate pathway, as well as isoprenoids, are found in bacteria and eukaryotes, the prevailing opinion, until recently, has been that LUCA could synthesize only isoprenoid-based lipids (Benner et al., 1989; Smit and Mushegian, 2000; Mulkidjanian and Galperin, 2010; Dibrova et al., 2012; 2013). The recent papers by Lombard and co-workers challenged this notion by proposing that modern archaea possess a version of the bacterial FAS II (Lombard et al., 2012a). This led to an important evolutionary conjecture that the fatty acid biosynthesis pathway has already been present in LUCA and therefore that LUCA could have had modern-type, two-tail fatty acid lipids (Lombard et al., 2012b; 2012c). While the likely nature of LUCA’s membranes is outside the scope of this report, we would like to note here that the great majority of archaea encode only non-specific components of bacterial FAS II that do not constitute a functional pathway (see Table S2, as well as Table S1 of Lombard et al., 2012a). The majority of bacterial-type fatty acid metabolism enzymes found in archaea represent likely cases of lateral gene transfer and are not relevant to the status of LUCA.
The remarkable presence of the membrane energy-converting enzymes only in those organisms that also encode the proposed fatty acid biosynthesis pathway (Table 1) can also be analyzed in evolutionary terms. There is growing evidence that proton-translocating energy-converting complexes, such as cytochrome bc complex and cytochrome c oxidase, have evolved in bacteria and have been acquired by archaea via lateral gene transfer (Hemp and Gennis, 2008; Hemp et al., 2012; Dibrova, 2013; Dibrova et al., 2013). If so, these energy-transducing enzymes could be acquired only by those archaea that were able to synthesize fatty acids. Exploitation of these enzymes by archaea would have additionally required the presence of advanced, proton tight membranes. As discussed elsewhere, different branches of archaea utilize different means to make their ether-based membranes proton-tight (Mulkidjanian et al., 2008; 2009).
Concluding remarks
This work provides evidence suggesting that the archaeal pathway of fatty acids metabolism combines bacterial-like enzymes of β-oxidation and archaea-specific paralogs of the acetyl-CoA C-acetyltransferase. The core set of three enzymes with reversible ACD, ECH, and HDH activities could be involved in either synthesis or oxidation of long-chain fatty acids. Coupling of these enzymes with archaeal acetyl-CoA C-acetyltransferase would then yield a synthetic pathway, with various paralogs of the acetyl-CoA C-acetyltransferase likely to be involved in the addition of C2 fragments to the growing carbon chain. A combination of the core set with the archaeal homologs of the bacterial β-ketothiolase, as found in some genomes (Tables 1 and S1) is expected to result in β-oxidation of fatty acids. Indeed, both archaea that have been shown to be able to oxidize long-chain fatty acids, A. fulgidus and Natronomonas pharaonis (Falb et al., 2008; Khelifi et al., 2010), contain bacterial-type β-ketothiolases in their genomes. In several archaea, members of COG2368 could potentially replace the missing acyl-CoA-dehydrogenases. In some organisms, members of COG2030 and COG1680 could also be involved, as substrate-specific enoyl-CoA dehydratases and acyl-CoA hydrolases, respectively.
The predicted pathway of fatty acid synthesis still needs to be experimentally verified. However, targeting of the respective genes by knock-out mutations might be complicated by the lethality of these mutations. For example, disruption of the fatty acid biosynthesis in H. salinarum would likely affect halorhodopsin and other fatty acid-dependent membrane enzymes.
Until now, successful biotechnological attempts to reverse the system of β-oxidation for producing biofuels centered on bacterial enzymes and, specifically, on enzymes from E. coli (Dellomonaco et al., 2011; Clomburg et al., 2012). The enzymes of the proposed archaeal fatty acid biosynthesis pathway from Sulfolobus solfataricus and other extremophiles can be expected to operate at higher temperatures and to remain stable at lower pH values than the corresponding mesophilic enzymes. For example, oxidation of fatty acids by A. fulgidus has been shown to proceed at 70°C (Khelifi et al., 2010), while the cells of Geoglobus ahangari were capable of β-oxidizing fatty acids at temperatures as high as 85°C (Kashefi et al., 2002). Thus, archaeal enzymes of fatty acid metabolism might be more suitable for biofuel production and other industrial applications than the respective bacterial enzymes.
Supplementary Material
Acknowledgements
We thank Drs. Kira S. Makarova, Yuri I. Wolf, and Eugene V. Koonin for their interest in this work and many helpful suggestions and Dr. Jonathan Lombard for useful discussion. This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-Mu-1285/1-10, DFG-436-RUS 113/963/0-1), the Russian Government (№ 02.740.11.5228), the Deutscher Akademischer Austausch Dienst (Ostpartnerschaften-Program), and the COST Action CM0902 of the EU (A.Y.M) and the NIH Intramural Research Program at the National Library of Medicine (M.Y.G).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zheng Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST - A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Ellington AD, Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci USA. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, Kamekura M, Doolittle WF. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol Microbiol. 2004;52:515–527. doi: 10.1111/j.1365-2958.2004.03992.x. [DOI] [PubMed] [Google Scholar]
- Carballeira NM, Reyes M, Sostre A, Huang H, Verhagen MF, Adams MW. Unusual fatty acid compositions of the hyperthermophilic archaeon Pyrococcus furiosus and the bacterium Thermotoga maritima. J Bacteriol. 1997;179:2766–2768. doi: 10.1128/jb.179.8.2766-2768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clomburg JM, Vick JE, Blankschien MD, Rodríguez-Moyá M, Gonzalez R. A synthetic biology approach to engineer a functional reversal of the β-oxidation cycle. ACS Synth Biol. 2012;1:541–554. doi: 10.1021/sb3000782. [DOI] [PubMed] [Google Scholar]
- Corcelli A, Lobasso S, Colella M, Trotta M, Guerrieri A, Palmisano F. Role of palmitic acid on the isolation and properties of halorhodopsin. Biochim Biophys Acta. 1996;1281:173–181. doi: 10.1016/0005-2736(96)00007-7. [DOI] [PubMed] [Google Scholar]
- Corcelli A, Lobasso S. Characterization of lipids of halophilic Archaea. In: Rainey FA, Oren A, editors. Methods in Microbiology, Extremophiles. Academic Press; 2006. pp. 585–613. [Google Scholar]
- Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- Dibrova DV, Chudetsky MY, Galperin MY, Koonin EV, Mulkidjanian AY. Role of energy in the emergence of biology from chemistry. Orig Life Evol Biosph. 2012;42:459–468. doi: 10.1007/s11084-012-9308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrova DV. Phylogenomic analysis of energy converting enzymes. 2013. p. 259. PhD thesis. Universität Osnabrück, Osnabrück.
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP, Mulkidjanian AY. Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim Biophys Acta. 2013;1827:1407–1427. doi: 10.1016/j.bbabio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiel I, Sprott GD, Smith IC. Mevalonic acid is partially synthesized from amino acids in Halobacterium cutirubrum: a 13C nuclear magnetic resonance study. J Bacteriol. 1986;166:559–564. doi: 10.1128/jb.166.2.559-564.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S, et al. Metabolism of halophilic archaea. Extremophiles. 2008;12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Shiomi N, Doi Y. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol. 1998;180:667–673. doi: 10.1128/jb.180.3.667-673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinger A, Schloter M, Munch JC. Phospholipid etherlipid and phospholipid fatty acid fingerprints in selected euryarchaeotal monocultures for taxonomic profiling. FEMS Microbiol Lett. 2002;213:133–139. doi: 10.1111/j.1574-6968.2002.tb11297.x. [DOI] [PubMed] [Google Scholar]
- Gochnauer MB, Kushner DJ. Growth and nutrition of extremely halophilic bacteria. Can J Microbiol. 1969;15:1157–1165. doi: 10.1139/m69-211. [DOI] [PubMed] [Google Scholar]
- Gomez B, Jr., Robinson NC. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- Gonzalez O. Reconstruction, modeling & analysis of haloarchaeal metabolic networks. 2009. p. 202. PhD thesis, Ludwig-Maximilians-Universität, München. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, Ryan CM, Whitelegge JP, Cramer WA. Conservation of lipid functions in cytochrome bc complexes. J Mol Biol. 2011;414:145–162. doi: 10.1016/j.jmb.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J. A new family of CoA-transferases. FEBS Lett. 2001;509:345–349. doi: 10.1016/s0014-5793(01)03178-7. [DOI] [PubMed] [Google Scholar]
- Helgersons S, Siemsens S, Dratz E. Enrichment of bacteriorhodopsin with isotopically labeled amino acids by biosynthetic incorporation in Halobacterium halobium. Can J Microbiol. 1992;38:1181–1185. [Google Scholar]
- Hemp J, Gennis RB. Diversity of the heme–copper superfamily in archaea: Insights from genomics and structural modeling. In: Schäfer G, Penefsky HS, editors. Results and Problems in Cell Differentiation. Springer-Verlag; Berlin: 2008. pp. 1–31. [DOI] [PubMed] [Google Scholar]
- Hemp J, Laura AP, Gennis RB. The heme-copper oxidoreductase superfamily: Diversity, evolution and ecology. Biochim Biophys Acta. 2012;1817:S107–S108. [Google Scholar]
- Jiang C, Kim SY, Suh DY. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol Phylogenet Evol. 2008;49:691–701. doi: 10.1016/j.ympev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Kashefi K, Tor JM, Holmes DE, Gaw Van Praagh CV, Reysenbach A-L, Lovley DR. Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int J Syst Evol Microbiol. 2002;52:719–728. doi: 10.1099/00207713-52-3-719. [DOI] [PubMed] [Google Scholar]
- Khelifi N, Grossi V, Hamdi M, Dolla A, Tholozan JL, Ollivier B, Hirschler-Rea A. Anaerobic oxidation of fatty acids and alkenes by the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus. Appl Environ Microbiol. 2010;76:3057–3060. doi: 10.1128/AEM.02810-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Nishihara M, Morii H, Akagawa-Matsushita M. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol Rev. 1993;57:164–182. doi: 10.1128/mr.57.1.164-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Kyuragi T, Nishihara M, Sone N. Did archaeal and bacterial cells arise independently from noncellular precursors? A hypothesis stating that the advent of membrane phospholipid with enantiomeric glycerophosphate backbones caused the separation of the two lines of descent. J Mol Evol. 1998;46:54–63. doi: 10.1007/pl00006283. [DOI] [PubMed] [Google Scholar]
- Koga Y, Morii H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. Computational Approaches in Comparative Genomics. Kluwer Academic; Boston: 2003. Sequence - Evolution - Function; p. 461. [PubMed] [Google Scholar]
- Korshunov SS, Korkina OV, Ruuge EK, Skulachev VP, Starkov AA. Fatty acids as natural uncouplers preventing generation of O2− and H2O2 by mitochondria in the resting state. FEBS Lett. 1998;435:215–218. doi: 10.1016/s0014-5793(98)01073-4. [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Sanders SA, Reinking J, Savchenko A, Arrowsmith CH, et al. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol Rev. 2005;29:263–279. doi: 10.1016/j.femsre.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Leuko S, Raftery MJ, Burns BP, Walter MR, Neilan BA. Global protein-level responses of Halobacterium salinarum NRC-1 to prolonged changes in external sodium chloride concentrations. J Proteome Res. 2009;8:2218–2225. doi: 10.1021/pr800663c. [DOI] [PubMed] [Google Scholar]
- Lobasso S, Lopalco P, Angelini R, Vitale R, Huber H, Muller V, Corcelli A. Coupled TLC and MALDI-TOF/MS analyses of the lipid extract of the hyperthermophilic archaeon Pyrococcus furiosus. Archaea. 20122012:957852. doi: 10.1155/2012/957852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J, Lopez-Garcia P, Moreira D. An ACP-independent fatty acid synthesis pathway in archaea: implications for the origin of phospholipids. Mol Biol Evol. 2012a;29:3261–3265. doi: 10.1093/molbev/mss160. [DOI] [PubMed] [Google Scholar]
- Lombard J, Lopez-Garcia P, Moreira D. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012b;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- Lombard J, Lopez-Garcia P, Moreira D. Phylogenomic investigation of phospholipid synthesis in archaea. Archaea. 2012c;2012:630910. doi: 10.1155/2012/630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh U, Cinkaya I, Albracht SP, Buckel W. 4-Hydroxybutyryl-CoA dehydratase from Clostridium aminobutyricum: characterization of FAD and iron-sulfur clusters involved in an overall non-redox reaction. Biochemistry. 1996;35:11710–11718. doi: 10.1021/bi9601363. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Galperin MY, Makarova KS, Wolf YI, Koonin EV. Evolutionary primacy of sodium bioenergetics. Biol Direct. 2008;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Galperin MY, Koonin EV. Co-evolution of primordial membranes and membrane proteins. Trends Biochem Sci. 2009;34:206–215. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Galperin MY. Evolutionary origins of membrane proteins. In: Frishman D, editor. In Structural Bioinformatics of Membrane Proteins. Springer; Vienna: 2010. pp. 1–28. [Google Scholar]
- Nakaya A, Katayama T, Itoh M, Hiranuka K, Kawashima S, Moriya Y, et al. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41:D353–357. doi: 10.1093/nar/gks1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr., Deerfield DW., II GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- Nishihara M, Nagahama S, Ohga M, Koga Y. Straight-chain fatty alcohols in the hyperthermophilic archaeon Pyrococcus furiosus. Extremophiles. 2000;4:275–277. doi: 10.1007/s007920070013. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D, Krippahl G. Light inhibition of respiration in Halobacterium halobium. FEBS Lett. 1973;36:72–76. doi: 10.1016/0014-5793(73)80339-4. [DOI] [PubMed] [Google Scholar]
- Pereto J, Lopez-Garcia P, Moreira D. Phylogenetic analysis of eukaryotic thiolases suggests multiple proteobacterial origins. J Mol Evol. 2005;61:65–74. doi: 10.1007/s00239-004-0280-8. [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh EL, Wassef MK, Kates M. Inhibition of fatty acid synthetase in Halobacterium cutirubrum and Escherichia coli by high salt concentrations. Can J Biochem. 1971;49:953–958. doi: 10.1139/o71-138. [DOI] [PubMed] [Google Scholar]
- Pugh EL, Kates M. Acylation of proteins of the archaebacteria Halobacterium cutirubrum and Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1994;1196:38–44. doi: 10.1016/0005-2736(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- Rock CO, Jackowski S. Forty years of bacterial fatty acid synthesis. Biochem Biophys Res Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- Sanchez LB, Galperin MY, Müller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming) J Biol Chem. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- Sedlák E, Robinson NC. Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry. 1999;38:14966–14972. doi: 10.1021/bi9914053. [DOI] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodkina GB, Kolganova TV, Querellou J, Bonch-Osmolovskaya EA, Slobodkin AI. Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2009;59:2880–2883. doi: 10.1099/ijs.0.011080-0. [DOI] [PubMed] [Google Scholar]
- Smit A, Mushegian A. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 2000;10:1468–1484. doi: 10.1101/gr.145600. [DOI] [PubMed] [Google Scholar]
- Smith S. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 1994;8:1248–1259. [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct. 2012;7:46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm P, Muller N, Plugge CM, Stams AJM, Schink B. Syntrophy in Methanogenic Degradation. In: Hackstein JHP, editor. In Microbiology Monographs: (Endo)symbiotic Methanogenic Archaea. Springer-Verlag; Berlin: 2010. pp. 143–173. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.