Abstract
Long admired for informational role and recognition function in multidisciplinary science, DNA nanohybrids have been emerging as ideal materials for molecular nanotechnology and genetic information code. Here, we designed an optical machine-readable DNA icon on microarray, Avatar DNA, for automatic identification and data capture such as Quick Response and ColorZip codes. Avatar icon is made of telepathic DNA-DNA hybrids inscribed on chips, which can be identified by camera of smartphone with application software. Information encoded in base-sequences can be accessed by connecting an off-line icon to an on-line web-server network to provide message, index, or URL from database library. Avatar DNA is then converged with nano-bio-info-cogno science: each building block stands for inorganic nanosheets, nucleotides, digits, and pixels. This convergence could address item-level identification that strengthens supply-chain security for drug counterfeits. It can, therefore, provide molecular-level vision through mobile network to coordinate and integrate data management channels for visual detection and recording.
In 1996, Joe Davis created the first biological analog icon, called Microvenus, using recombinant DNA techniques to represent artistic information1,2. This insight into genetic art has challenged multidisciplinary scientists to develop digital-writing strategies and rapid-reading techniques of DNA language for next-generation data storage3,4,5,6,7,8,9,10. The lifespan of these devices are much greater than those of devices based on magnetic, electronic, or photonic phenomena11,12,13.
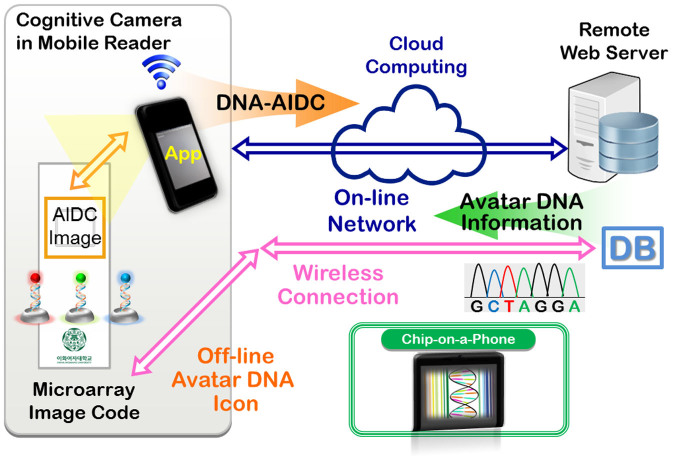
We describe the Avatar DNA that identifies DNA information using chip-on-a-phone, and convergence of this system with nano-bio-info-cogno (NBIC) technology for safe supply-chain management. Avatar DNA is an optical machine-readable hybrid icon fabricated on a biosensor through combination of biological DNA code with digital automatic identification and data capture (AIDC) image code, and therefore, it can connects specific information encoded in DNA base-sequence from an off-line AIDC image code to an on-line mobile computer system likes smartphone. Fig. 1 and Fig. S1 show how the digital DNA icons inscribed on a microarray chip can be automatically captured with a mobile phone camera, and how the information can be directly accessed by software. This technology employs fabricated DNA image codes for AIDC to rapidly transmit information to remote databases, including text messages, base-sequences, indexes, profiles, documents, and URLs.
Figure 1. Schematic illustration of the Avatar DNA-AIDC hybrid icon in chip-on-a-phone.
Results
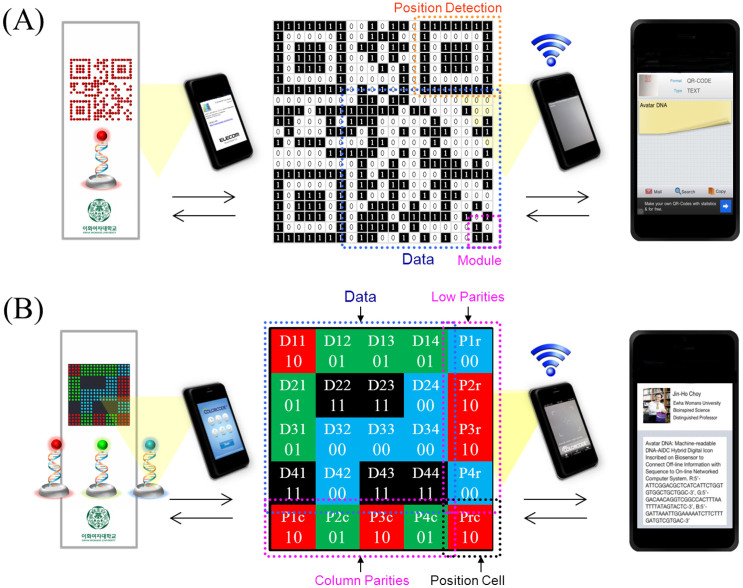
We successfully identified the DNA-AIDC hybrid icons using a smartphone in a laboratory environment. Two different AIDC image codes such as two-dimensional (2D) Quick Response (QR) and three-dimensional (3D) ColorZip were fabricated in spot-arrayed DNA chips, respectively. As shown in Fig. 2, the Avatar hybrid icons of biological DNA and digital AIDC image designed here are geometrically identical to the typical QR code and ColorZip code. Unique information can be represented in the DNA-AIDC hybrid icons through specific combination of DNA base-sequences and AIDC image patterns. In the each spot of microarray chips, different probe DNA with a specific base sequence are embedded to telepathically capture complementary target DNA code labelled with specific fluorophores. The fluorescently labelled DNA encoded functions as a photographical information emulsion. When these Avatar DNA icons were acquired by smartphone with application software, the DNA-QR icon was directly coupled with the text information of Avatar DNA, and the DNA-ColorZip icon was also linked to the information in the form of index including name card, definition, three different base-sequences, and website address.
Figure 2. Identification of the DNA-AIDC hybrid icons fabricated on a microarray chip on a smartphone.
(A) Digital images of DNA-DNA hybrids on DNA-chip (21 × 21 arrays in 16 mm2) are recognized as a two-dimensional binary quick-response code matrix and simultaneously coupled with the text of Avatar DNA. (B) Graphic image of three different DNA-DNA hybrids on DNA-chip (20 × 20 arrays in 20 mm2) captured as a three-dimentional ColorZip code matrix; the Avatar DNA icon is connected to a web server-based index to locate the information content. Inset images for DNA-QR and DNA-ColorZip hybrid codes were captured from the smartphone screen through real Avatar DNA chip experiments (Inset application execution image courtesy of ELECOM QR Code Reader for QR code and COLORCODE® V3.0 for ColorZip code. Inset face image was authorized by Jin-Ho Choy at Ewha Womans University, Republic of Korea).
The first model icon contained 21-by-21 modules with red-fluorescence dots arranged in a square grid on a white background (Fig. 2A). The DNA base-sequence was stored in a single DNA-QR code by automatically assigning binary number values to a 2D matrix through a logical QR code algorithm14. In our experiment, the icon in the form of Version 1 was simultaneously recognized as a QR code matrix of square dots mapped by binary digits of ones (red modules are perceived as black) and zeros (white background). The mobile reader likes smartphone simultaneously scanned the Avatar DNA-QR icon on the DNA chip by detecting its structural patterns, including position, module, and data area. The Avatar DNA information was then decoded through the sequence of black (B) and white (W) modules on a certain line, 1:0:1:1:1:0:1 (B:W:B:B:B:W:B), that passes the center of the detector from an omnidirectional angle. This could enable ultra-high-speed reading of the Avatar DNA QR code with the mobile chip-on-a-phone.
The second model icon contained a 25-subset array of 4-by-4 dots emitting 4 different colors (red, green, blue, and black) with a total of 20-by-20 modules (Fig. 2B). This icon was also able to be scanned as a ColorZip matrix that was mapped by color-filled 5-by-5 square cells. The 2-bit data can be expressed by one color cell, since each cell in the ColorZip code was encoded with one of four colors. Therefore, if the colors of the data cells D11, D12, D22, and D24 are encoded as red, green, black, and blue, the values are decoded as 10, 01, 11, and 00, respectively, according to the ColorZip code theory (International Standard ISO/IEC 18004:2006 Information technology — Automatic identification and data capture techniques — QR Code 2005 bar code symbology specification)15. In our experiment, the smartphone decoder used sampled pixels from each cell, and classified the colors of sampled pixels from the DNA chip into bitstream data.
These results can be explained by fact that whole spots of DNA chip, namely, 226 spots for the QR chip or 320 spots for the ColorZip chip, can be recognized as a single digital image code icon consisted of DNA pixels, DNA-QR code icon (226 pixels) and DNA-ColorZip code icon (320 pixels), respectively. It is worthy of note that, therefore, the Avatar DNA icon can be sufficiently identified with real DNA molecules at concentration of less than nanomolar-level (10−9 moles) through mobile phone camera, since DNA molecules with picomolar-level concentration were embedded in the each one spot of the microarray chip.
At a theoretical maximum for DNA, a single byte (or 8 bits) with 4 different bit combinations (00, 01, 10, and 11) is sufficient to store 4 base-sequences (A-T-C-G) in binary form without compression software. It has been reported that maximum data of 455 exabytes (EB, 1018 bytes) can be stored in one gram of single-strand DNA7. Most recently developed DNA-encoding algorithm using base-3 trits (0/1/2) can encode a single text character into 5 base-sequences without nucleotide repetition, which avoids homopolymers8. Data content can be from 0.338 to 1.58 bits per base, including error-correction redundancy, indexing, and parity check. Through such practical DNA encoding method, the 10 letters of ‘Avatar□DNA' (including the space denoted by □) were able to be encoded by 55 bits, cyber DNA along with 50-bases sequence, ‘CACAT AGATA CTCTG ACATC GAGAT CGACT ATGAT GACGA GTGCG TGTGC'. In our model, one probe DNA with 35-bases and three different probe DNA with 35-bases were fabricated inside the QR-DNA chip and for Colorzip-DNA chip, respectively, to translate the 10 letters, ‘Avatar□DNA' to DNA base-sequence.
The maximum data capacities of the Avatar DNA sensors can be expanded by combining different DNA base-sequence and logical system of AIDC code information at the genomic scale. When specific information including base-sequence is entered into the Avatar DNA-AIDC hybrid code, the arrangement of the QR code matrix can be logically expressed as one of diverse versions, ranging from Version 1 with 21-by-21 modules to Version 40 with 177-by-177 modules, for each row and column on the array chip sensors, depending on the data content to be inputted. Although the data capacity of original QR code is limited to 4,296 characters for alphanumeric data case, DNA-QR hybrid code can function both as a portable database and as an index to a remote database by variation their patterns to be inserted with DNA molecule. In a similar principle, the combination coding of color-code with DNA base-sequence can generate more than 17 billion patterns inside 5-by-5 cells of ColorZip-DNA matrix comprising of 4-by-4 data cells (D11−D44) and 9 alignment cells (P1r−P4c) along with parity (Prc). Considering the capacity for data storage, the amount and type of DNA information that can be stored on the ColorZip code icon are significantly larger than those of QR code icon. In our experiment for Avatar DNA ColorZip, we were able to input a maximum of 300 characters memo in the form of index, and afterward, the desired translation dataset for the original information was wirelessly read out.
The Avatar DNA icon could be useful in an institution with flexible data management using mobile networks. The concepts for future genomic-hybrid art will involve the self-assembly, cogno-manipulation, and engineering of DNA molecules16. Rationally designed DNA can contain specific information for a data archive, and can be used in place of analog images for mobile device-readable digital iconography.
Potential applications for Avatar DNA include health, safety, environment, and security, which are core missions in the U.S. Food and Drug Administration (FDA)17. One challenge is point-of-care (POC) diagnostics with an end-to-end risk management system for telemedical healthcare, to detect, monitor, and control hazardous diseases at resource-limited sites18,19. Without the isolation and transportation of bacteria to an infrastructure-rich laboratory, customized DNA-AIDC hybrids will be available soon for smartphones with special software, to interpret mutation rates in real-time, expediently transmit information via library networks, and prevent the spread of diseases.
Other safety and security applications include information hiding, data steganography, and applications related to supply-chain management at the point-of-sale (POS)12,20,21,22. Maintaining safety for medical products is currently a serious challenge23. The most recent counterfeiting incidents of Avastin, an injectable cancer medicine, highlighted the vulnerabilities in U.S. distribution chains for purchases of non-FDA-approved or unregulated drugs from a foreign supplier24. The counterfeit versions can occur anywhere along the chain, including the manufacturer, distributor, and supplier. These problems present significant risks to public health. It is important that inspection agencies improve supply-chain security and reevaluate current safety strategies within the global pharmaceutical market.
Discussion
The DNA-AIDC hybrid platform described here is a new infohybrid discipline that combines information technology and hybrid science, which can provide strong systems approach to supply-chain safety and security issue through comprehensive item-level identification based on a DNA molecule. Our approach involves NBIC convergence, where the main building blocks are interactive between boundaries with Avatar DNA. In a similar principle of NBIC convergence system defined from U.S. National Science Foundation25, our NBIC convergence is stands for:
Nanotechnology from nanosheets: Inorganic nanoparticles in the form of exfoliated metal hydroxide nanosheets with a chemical composition of [M2+1-xM3+x(OH)2]x+n (M2+ = divalent metal cations; M3+ = trivalent metal cations; 0.2 ≤ molar fraction of x ≤ 0.33) that originate from the 2D layered compounds with macromolecular anisotropic properties26,27. The ultrathin nanosheets have a thickness of ~1 nanometer, with a lateral size of several tens of nanometers. Nanosheets can be used for supramolecular hybrid assembly with functional substances like a DNA molecule at the molecular level to control cognitive responses to external stimuli.
Biotechnology from nucleotides: Rationally designed and fabricated DNA molecules consist of four digital nucleotides (A/T/C/G) with functional labels28,29. Lab-on-a-chip technology enables molecular sensing to read information stored in DNA and transmit the information for identification and communication30,31,32.
Information technology from digits: An applied computer system for software and hardware, including a digit algorithm and a mobile device that can interpret DNA information with logical data processing (encoding-encrypting-decrypting-decoding), is linked with wireless networking and tele-info communication.
Cognitive science from pixels: A cognitive process co-evolved with infocommunications using artificial intelligence and a smartphone equipped with an automatically focusing optical camera. This recognizes and acquires digital information images using a similar principle to that of the optic nervous system of the human eye.
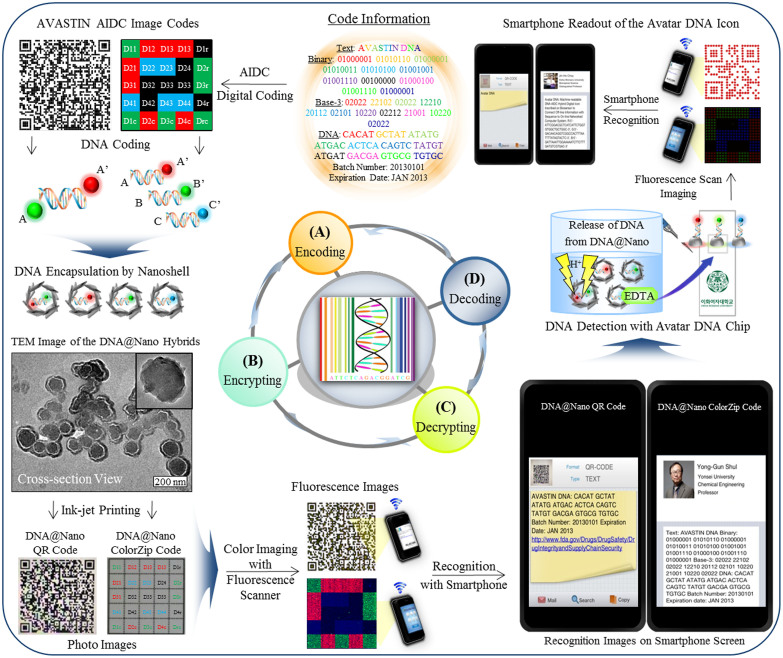
The Avatar DNA chip platform addresses the counterfeit incidents involving Avastin, an injectable cancer medicine. This technology is a practical application that strengthens security for supply-chain safety through a comprehensive item-level identification system. Proof-of-concept experiments were performed with a systems approach involving four primary steps: encoding, encrypting, decrypting, and decoding (Fig. 3).
Figure 3. Avatar DNA systems approach with NBIC convergence during proof-of-concept workflow on drug integrity and supply-chain security.
(A) Encoding generates the AVASTIN DNA-AIDC hybrid icons: the QR code, ColorZip code, and their corresponding DNA molecules are prepared to carry the information of AVASTIN drug, Text: AVASTIN DNA, Binary: 01000001 01010110 01000001 01010011 01010100 01001001 01001110 00100000 01000100 01001110 01000001, Base-3: 02022 22102 02022 12210 20112 02101 10220 02212 21001 10220 02022, DNA: CACAT GCTAT ATATG ATGAC ACTCA CAGTC TATGT ATGAT GACGA GTGCG TGTGC, Batch Number: 20130101, Expiration Date: JAN 2013. (B) Encrypting prints the DNA-AIDC hybrid tags: the fluorescence- and sequence-encoded DNA molecules with a size of 100 nm are encapsulated inside spherical nanocavity with a wall thickness of 10 nm reassembled with metal hydroxides nanosheets to form a DNA@Nano core-shell structure. Thus stabilized biological DNA is ink-jet printed safely from severe chemical resistant so that the dually coded DNA-AIDC hybrid tags are transmitted to a receiver. (C) Decrypting reads the fluorescent DNA-AIDC hybrid tags: after scanning the DNA-AIDC tags under fluorescence illumination, and then the resulting fluorescent DNA-AIDC images are recognized by smartphone camera with application software to read the pre-encoded information of AVASTIN drug (Inset face image was authorized by Yong-Gun Shul at Yonsei University, Republic of Korea). (D) Decoding identifies the DNA-AIDC hybrid icons: following hybridization experiment for detecting DNA within 1 second time at a concentration of 100 picomole, the resulting Avatar DNA icons on chip are fluorescently imaged by scanner, and then On/Off identified by smartphone readout.
The encoding step created phenotype DNA-AIDC icons that contained information about Avastin encoded in a specific DNA sequence (Fig. 3A). The integrity mark ‘AVASTIN DNA' was encoded with DNA in silico, and then the information was expressed as a QR or ColorZip code. The ‘AVASTIN DNA' binary code contains 88 digits with 11 bytes represented via ASCII characters, ‘01000001 01010110 01000001 01010011 01010100 01001001 01001110 00100000 01000100 01001110 01000001'. Using the base-3 (0/1/2) Huffman-compression coding method, the corresponding characters were converted to 55 trits, ‘02022 22102 02022 12210 20112 02101 10220 02212 21001 10220 02022'. Using a practical DNA encoding method8, the secured ‘AVASTIN DNA' code would be stored in the form of cyber DNA with 55 non-repeated base-sequences, ‘CACAT GCTAT ATATG ATGAC ACTCA CAGTC TATGT ATGAT GACGA GTGCG TGTGC'. This drug-integrity information was then displayed with mobile phone-recognizable image matrixes, including QR code (Version 9 with 53-by-53 modules) and ColorZip code (5-by-5 cells). When these DNA-AIDC models were generated, a 6-digit numeric batch number (20130101) and expiration date (3-letter month and 4-digit year, JAN 2013) were included according to FDA-approved packaging for Avastin.
The encrypting step encapsulated artificial DNA molecules conjugated with red, green, and blue color fluorophores inside a supramolecular inorganic nanobody (DNA@Nano), and placed them behind the generated AIDC patterns in the printed form of a packaging label to be affixed onto the drug vials (Fig. 3B). A naked DNA molecule is easily destroyed in unfavorable environmental conditions33,34, and synthetic DNA molecules should be protected within adequate hosts. Inorganic nanoscale structures were superior to biological hosts such as microorganisms, because biological hosts may induce mutations or damage to the embedded DNA11,34,35,36. Therefore, we encapsulated each DNA molecule in the duplex form of 35 bases conjugated with different fluorophores (100 nm in diameter) into a spherical inorganic nanoshell with a wall thickness of 10 nm. This inorganic shell was a hermetic diffusion barrier that protected the supramolecular assembly and the DNA molecule from endonucleases, oxidative stress induced by free radicals, and high-temperature in a colloidal suspension (see the supplementary information). Therefore, the artificial DNA stabilized inside the robust nanobody could be used to encode the ‘AVASTIN DNA' security mark, which could be printed with the desired AIDC labels onto a piece of transparent adhesive paper. The DNA-QR code pattern was produced with one kind of DNA@Nano diluted with a black water-based ink, and used 100 ppm DNA. The DNA-ColorZip code matrix was printed with a series of three different kinds of DNA@Nano diluted with an aqueous solution at the DNA concentration of 100 ppm containing polymeric wetting agent.
The decrypting step performed capture and reading of AVASTIN DNA-AIDC images based on a smartphone reader at authentication nodes for timely oversight (Fig. 3C). If this DNA-AIDC was packaged as part of FDA-approved labeling into all legitimate products, they could be effectively tracked and traced to monitor sales records and protect against counterfeiting. We obtained proof-of-principle by automatically reading scanned fluorescent images for both DNA-QR and DNA-ColorZip codes using a smartphone camera and a conventional application, typically achieved in a non-destructive manner. The scan displays the hidden drug-integrity DNA code on the screen of the smartphone. The fluorescent AIDC images also serve as sentinels to tag a specific index of secured DNA information (similar to the file header in magnetic tape) for later retrieval and identification. The off-line DNA-AIDC tag can retrieve authentic information by directly connecting to an on-line database library linked to the URL of the Office of Drug Security, Integrity, and Recalls of the U.S. FDA, to browse and cross-check drug integrity codes against a reference database. It is strongly recommended that users check the intermediate results carefully before accepting the species identity of any item for constructing dynamic logistics.
In the decoding step (Fig. 3D), we collected the artificial AVASTIN DNA molecules from DNA@Nano hybrids by dissolving the nanoshell with an acidic solution in the presence of EDTA. Using prior knowledge of the DNA base-sequences for both DNA-QR and DNA-ColorZip codes, DNA identification proceeds through complementary DNA-DNA hybrid reactions on each DNA-chip without an amplification procedure like a polymerase chain reaction. The resulting DNA-AIDC image icons on the microarray were automatically recognized by a smartphone camera with application software, which identified the secured integrity as ‘Avatar DNA' on the screen. These final DNA icons can provide further information through the phone-based communication system for scientific notification, public services, user guides, advertising, and multimedia content. Therefore, the NBIC convergence strategy could sufficiently verify the forensic proof (AVASTIN DNA@Nano hybrid) to upstream and downstream users for a seamless supply chain protected from counterfeits.
The Avatar DNA icon was detected with the camera of chip-on-a-phone using application software to address safety and security issues in supply-chain management by converging with NBIC science and technology. The 2D Avatar DNA-QR and 3D Avatar DNA-ColorZip code images made of telepathic DNA-DNA hybrids on a microarray could be spontaneously readable to wirelessly communicate specific off-line information with an on-line network system. The Avatar DNA chip-on-a-phone platform is a burgeoning new field for visual detection and recording. Therefore, it can provide molecular-level strategic vision to coordinate and integrate convergence channel programs. The systems approach will profoundly influence the safety of the drug supply chain. This approach can be expanded for a broad variety of applications using multidisciplinary NBIC convergence combined with biological logic gates, electric DNA pedigrees, cloud-based DNA big data management, DNA-Fi networks, or chip-on-a-phone equipped with a handheld single-DNA-molecule sequencer.
Methods
Avatar DNA-AIDC hybrid icon
Capture (Probe) DNA with 50 base-sequences were prepared from Integrated DNA Technologies, USA. One capture DNA [A] was used for the DNA-QR code. Three different capture DNA were used for the DNA-ColorZip code as follows: 1) capture DNA [A] (5′-NH2-TTT TTT TTT TTT TTT GCC AGC AGC CAC ACC AGA ATG ATG AGC GTC CGA AT-3′); 2) capture DNA [B] (5′-NH2-TTT TTT TTT TTT TTT GAG TAC TAT AAA ATT AAA GTG GCC GAC CTG TTG TC-3′); and 3) capture DNA [C] (5′-NH2-TTT TTT TTT TTT TTT GTC ACG ACA TCA AAG AAG ATT TTT CCA ATT TAA TC-3′). The Avatar DNA chips were custom-fabricated by Genomictree Inc., Korea. In the DNA-QR code, there were 226 spots within 21 × 21 arrays per block (4 mm × 4 mm = 16 mm2 area), and the spot diameter was 140 μm with 40-μm pitch between spots. In the DNA-ColorZip code, there were 320 spots within 20 × 20 arrays per block (4.5 mm × 4.5 mm = 20.25 mm2 area), and the spot diameter was 140 μm with 80-μm pitch between spots. As-prepared capture DNA were spotted using an OmniGrid™ Microarrayer (GeneMachines, San Carlos, CA) onto silanized glass slides. The fluoroprobe-labeled target DNAs with 35 complementary base-sequences were synthesized by Integrated DNA Technologies. One red-color target DNA, [A′-Cy5], was used in the DNA-QR code. The following three different target DNAs were used in the DNA-ColorZip hybrid code icon: 1) red-color target DNA, [A′-Cy5] (5′-Cy5-ATT CGG ACG CTC ATC ATT CTG GTG TGG CTG CTG GC-3′); 2) green-color target DNA, [B′-Cy3] (5′-Cy3-GAC AAC AGG TCG GCC ACT TTA ATT TTA TAG TAC TC-3′); and 3) blue-color target DNA, [C′-Alexa350] (5′-Alexa350-GAT TAA ATT GGA AAA ATC TTC TTT GAT GTC GTG AC-3′). All target DNA samples (10 nM) were suspended in 80 μl of hybridization buffer (3 × SSC, 0.3% SDS, 30% formamide). The hybridization mixtures were directly introduced to each Avatar DNA-chip. After incubating at 42°C for 30 min, thus hybridized chips were washed with gentle agitation to eliminate non-specific binding as follows: 5 min at 42°C in 2 × SSC and 0.1% SDS, 5 min at room temperature in 0.1 × SSC and 0.1% SDS, and 2 min at room temperature in 0.1 × SSC (twice). The Avatar DNA icon images were visualized with a fluorescence scanner (Typhoon 9400, Amersham Biosciences Inc., USA) with software (typhoon scanner control) at appropriate sensitivity levels of the photomultiplier (PMT), at 488 nm for Alexa 350, 532 nm for Cy3, and 633 nm for Cy5. The results were analyzed by ImageQuant software. After acquisition chip images by scanner instrument, the chip data images were then recognized and captured by smartphone (iPhone touch 5th generation, Apple Inc., USA) using either QR code application (ELECOM QR Code Reader) or a ColorZip code application (COLORCODE® V3.0). The distance between the fluorescence images and the smartphone reader was approximately 20 cm using a typical light source in a laboratory environment. Finally, thus decoded images from the Avatar DNA icon chip were directly captured on the smartphone screen for figure display.
Avatar DNA NBIC convergence system for drug supply chain management
In the encoding step, the secured text ‘AVASTIN DNA' was encoded with a DNA base-sequence in silico, and information was then generated in the format of a QR code or ColorZip code. The 11 characters (including one space) of ‘AVASTIN DNA' were represented as the binary code via ASCII characters. Using the base-3 (0/1/2) Huffman-compression coding method, the 88 bits of characters were then converted to 55 trits. Using a practical DNA-encoding method8, the text, AVASTIN DNA, was then translated to 55 base-sequences. The encoded AVASTIN DNA information, including the batch number, expiration date, and URL of the Drug Integrity and Supply Chain Security of the U.S. FDA (http://www.fda.gov/Drugs/DrugSafety/DrugIntegrityandSupplyChainSecurity), was stored in the DNA-image hybrid code matrix for Version 9 QR code (53 × 53 modules) using a conventional QR-code generator program (Formtec Design Pro 8), and for ColorZip code (5 × 5 cells) using a generator program for ColorZip name cards available at http://www.colorzip.co.kr.
In the encrypting step, model DNA molecules labeled with fluorophores were encapsulated inside a layered double hydroxide (LDH) nanoshell matrix, and they were printed onto a packaging label in the form of generated QR code and ColorZip code patterns. First, model DNAs were prepared in the form of a 35-base-long duplex with complementary base-sequences using typical protocols. One DNA-duplex with both green and red fluorophores (Cy3-AA′-Cy5) was prepared for the AVASTIN DNA-QR code. Three different DNA-duplexes (red, AA′-Cy5; green, BB′-Cy3; and blue, CC′-Alexa350) were prepared for the AVASTIN DNA-ColorZip code. The model DNAs were each encapsulated inside an LDH nanoshell matrix using the exfoliation-reassembling method as previously reported11. Each DNA solution containing 150 μM of Cy3-AA′-Cy5, AA′-Cy5, BB′-Cy3, and CC′-Alexa350 was dissolved in a mixed solution of decarbonated water and formamide, and then dropped into a diluted colloidal suspension of LDH nanosheets (125 μM) at ambient temperature with gentle shaking under N2 atmosphere. The prepared DNA core@Nano shell hybrid was immediately freeze-dried. The desired AVASTIN DNA code labels drawn in the encoding step were printed with a size of 2.0 cm × 2.0 cm onto a piece of transparent adhesive paper (Formtec barcode label LQ-3104) using a heat-transfer inkjet printer (HP Deskjet D4160). For the AVASTIN DNA-QR code, the colloidal solution of DNA@Nano, at a DNA weight content of 100 ppm, was made with a black ink solution (pH 7.2) squeezed from the original cartridge (No. 98) in an HP Deskjet printer. For the AVASTIN DNA ColorZip code, ink-free aqueous solutions of each DNA@Nano, at a DNA weight content of 100 ppm, were made with 0.4% (w/v) of Surfynol 440 surfactant (Air Products), which was necessary for reliable printing20. A 10-ml solution of each DNA@Nano was then loaded into a blank inkjet cartridge compatible with the desktop inkjet printer. Once printed, the labels were dried for 10 min at room temperature.
In the decrypting step, the fluorescence of the AVASTIN DNA code was visualized using a fluorescence scanner set for 488 nm for Alexa350, 532 nm for Cy3, and 633 nm for Cy5 using appropriate PMT sensitivity levels. The results were analyzed with ImageQuant software. Thus obtained fluorescent images were then recognized using a either a QR-code application (ELECOM QR Code Reader) or a ColorZip-code application (COLORCODE® V3.0) downloaded on a smartphone (iPhone touch 5th generation, Apple Inc., USA), and finally were captured for data display.
In the decoding step, the DNA from DNA@Nano was collected by thawing and dissolving the LDH matrix from the printed code labels. The DNA in the LDH nanoshell was then released by dissolving LDH in a slightly acidic solution at pH 4 along with the addition of ethylenediaminetetraacetic acid disodium salt (EDTA) for 15 min at ambient temperature. Each collected DNA hybridization mixture was then directly detected with the corresponding Avatar DNA-chip using typical protocols. Finally, Avatar DNA-chips were read by smartphone and captured to display their information on screen.
Author Contributions
D.H.P. designed the original idea and performed experiments and wrote the manuscript. C.J.H. partly assisted experiments. Y.G.S. contributed to the discussion and important insights. J.H.C. conceived the original idea, supervised the whole study, and made critical comments on the manuscript.
Supplementary Material
Avatar DNA Nanohybrid System in Chip-on-a-Phone
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (2005-0049412 and R31-2008-000-10010-0).
References
- Davis J. Microvenus. Art J. 55, 70–74 (1996). [Google Scholar]
- Davis J., Boyd D., O'Reilly H. & Wieczorek M. Art and Genetics. eLS 10.1002/9780470015902.a0005868, 1 (2006). [Google Scholar]
- Clelland C. T., Risca V. & Carter B. Hiding messages in DNA microdots. Nature 399, 533–534 (1999). [DOI] [PubMed] [Google Scholar]
- Bancroft C., Bowler T., Bloom B. & Clelland C. T. Long-Term Storage of Information in DNA. Science 293, 1763–1765 (2001). [DOI] [PubMed] [Google Scholar]
- Wong P. C., Wong K. K. & Foote H. Organic Data Memory Using the DNA Approach. Comm. ACM 46, 95–98 (2003). [Google Scholar]
- Choy J. H., Oh J. M., Park M., Sohn K. M. & Kim J. W. Inorganic-Biomolecular Hybrid Nanomaterials as a Genetic Molecular Code System. Adv. Mater. 16, 1181–1184 (2004). [Google Scholar]
- Church G. M., Gao Y. & Kosuri S. Next-Generation Digital Information Storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- Goldman N. et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J. & Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008). [DOI] [PubMed] [Google Scholar]
- Chen P. et al. Gold nanoparticles for high-throughput genotyping of long-range haplotypes. Nat. Nanotechnol. 6, 639–644 (2011). [DOI] [PubMed] [Google Scholar]
- Park D. H., Kim J. E., Oh J. M., Shul Y. G. & Choy J. H. DNA Core@Inorganic Shell. J. Am. Chem. Soc. 132, 16735–16736 (2010). [DOI] [PubMed] [Google Scholar]
- Oh J. M., Park D. H. & Choy J. H. Integrated bio-inorganic hybrid systems for nano-forensics. Chem. Soc. Rev. 40, 583–595 (2011). [DOI] [PubMed] [Google Scholar]
- Paunescu D., Fuhrer R. & Grass R. N. Protection and Deprotection of DNA—High-Temperature Stability of Nucleic Acid Barcodes for Polymer Labeling. Angew. Chem. Int. Ed. 52, 4269–4272 (2013). [DOI] [PubMed] [Google Scholar]
- Han S. et al. Lithographically encoded polymer microtaggant using high-capacity and error-correctable QR code for anti-counterfeiting of drugs. Adv. Mater. 24, 5924–5929 (2012). [DOI] [PubMed] [Google Scholar]
- Cheong C. et al. Pictorial Image Code: A Color Vision-based Automatic Identification Interface for Mobile Computing Environments. Eighth IEEE Workshop on Mobile Computing Systems and Applications, 23–28 10.1109/HotMobile.2007.8 (2007). [Google Scholar]
- Zala K. Poetry in the genes. Nature 458, 35 (2009). [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration, Strategic Priorities 2011 – 2015 Responding to the Public Health Challenges of the 21st Century (Updated 4/20/2011), www.fda.gov/downloads/AboutFDA/ReportsManualsForms/Reports/UCM252092.pdf accessed January 2014.
- Wang S. et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip 11, 3411–3418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Isikman S. O., Mudanyali O. Greenbaum A. & Ozcan A. Optical imaging techniques for point-of-care diagnostics. Lab Chip 13, 51–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas III S. W. et al. Infochemistry and infofuses for the chemical storage and transmission of coded information. Proc. Natl. Acad. Sci. USA 106, 9147–9150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M. A. et al. InfoBiology by printed arrays of microorganism colonies for timed and on-demand release of messages. Proc. Natl. Acad. Sci. USA 108, 16510–16514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H. & Choy J. H. Emerging Strategies in Infohybrid Systems. Eur. J. Inorg. Chem. 2012, 5145–5143 (2012). [Google Scholar]
- U.S. Food and Drug Administration, FDA Conducts Preliminary Review of Agency's Diversion and Counterfeit Criminal Case Information, September 2011, www.fda.gov/downloads/Drugs/DrugSafety/DrugIntegrityandSupplyChainSecurity/UCM272150.pdf accessed January 2014.
- U.S. Food and Drug Administration, Counterfeit Version of Avastin in U. S. Distribution, July 10. 2012, www.fda.gov/Drugs/DrugSafety/ucm291960.htm accessed January 2014.
- Roco M. C. & Bainbridge W. S. Converging Technologies for Improving Human Performance: Nanotechnology, Biotechnology, Information Technology, and Cognitive Science, National Science Foundation (2003) www.wtec.org/ConvergingTechnologies/Report/NBIC_report.pdf accessed January 2014.
- Choy J. H. Intercalative route to heterostructured nanohybrid. J. Phys. Chem. Solids 65, 373–383 (2004). [Google Scholar]
- Ruiz-Hitzky E., Aranda P., Dardera M. & Ogawa M. Hybrid and biohybrid silicate based materials: molecular vs. block-assembling bottom–up processes. Chem. Soc. Rev. 40, 801–828 (2011). [DOI] [PubMed] [Google Scholar]
- Park S. J., Taton T. A. & Mirkin C. A. Array-based electrical detection of DNA with nanoparticle probes. Science 295, 1503–1506 (2002). [DOI] [PubMed] [Google Scholar]
- Wu N. et al. “Molecular Threading” and tunable molecular recognition on DNA origami nanostructures. J. Am. Chem. Soc. 135, 12172–12175 (2013). [DOI] [PubMed] [Google Scholar]
- Lim D. K., Cui M. H. & Nam J. M. Highly stable, amphiphilic DNA-encoded nanoparticle conjugates for DNA encoding/decoding applications. J. Mater. Chem. 21, 9467–9470 (2011). [Google Scholar]
- Pei H., Liang L., Yao G., Li J., Huang Q. & Fan C. Reconfigurable Three-Dimensional DNA Nanostructures for the Construction of Intracellular Logic Sensors. Angew. Chem. Int. Ed. 51, 9020–9024 (2012). [DOI] [PubMed] [Google Scholar]
- Chen R. et al. DNA based identification of medicinal materials in Chinese patent medicines. Sci Rep. 2, 958; 10.1038/srep00958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993). [DOI] [PubMed] [Google Scholar]
- Yin R., Zhang D., Song Y., Zhu B. Z. & Wang H. Potent DNA damage by polyhalogenated quinones and H2O2 via a metal-independent and Intercalation-enhanced oxidation mechanism. Sci. Rep. 3, 1269; 10.1038/srep01269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunescu D., Puddu M., Soellner J. O., Stoessel P. R. & Grass R. N. Reversible DNA encapsulation in silica to produce ROS-resistant and heat-resistant synthetic DNA ‘fossils'. Nat. Protoc. 8, 2440–2448 (2013). [DOI] [PubMed] [Google Scholar]
- Gibson D. G. et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 329, 52–56 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Avatar DNA Nanohybrid System in Chip-on-a-Phone