Abstract
We report a synergistic method using bioassay-directed liquid chromatography fractionation and time-of-flight mass spectrometry to guide and accelerate bioactive compound discovery. To steer purification and assays toward anticipated neutral lipid activators of a constitutive androstane receptor splice variant, a relative mass defect filter was calculated, based on the ratio of the mass defect to the measured ion mass, and used to reduce the number of candidate ion masses. Mass measurements often lack sufficient accuracy to provide unambiguous assignments of elemental compositions, and since the relative mass defect reflects fractional hydrogen content of ions, this value is largely determined by the hydrogen content of a compound’s biosynthetic precursors. A relative mass defect window ranging from 600–1000 ppm, consistent with an assortment of lipids, was chosen to assess the number of candidate ions in fractions of fetal bovine serum. This filter reduced the number of candidate ion m/z values from 1345 to 892, which was further reduced to 21 by intensity and isotope filtering. Accurate mass measurements from time-of-flight mass spectrometry and fragment ion masses generated using nonselective collision-induced dissociation suggested dioctyl phthalate as one of few neutral lipid constituents in the active fraction. The identity of this compound was determined to be di(2-ethylhexyl) phthalate using GC/MS, and it was ranked as a promising candidate for reporter assay screening.
Recent years have led to discoveries of numerous receptors and signaling mechanisms that regulate a wide variety of biological functions, but identification of potent bioactive compounds that activate these receptors involves lengthy and expensive efforts.1,2 Contributing to this are the need to screen an enormous number of compounds in both complex biological samples and combinatorial libraries for activity. Once a receptor ligand is identified, efforts to accelerate compound discovery often generate large libraries of compounds with structural similarity to known active substances, followed by activity screening.3,4 Complementary efforts have focused on reducing discovery time by improving technologies for bioactivity assays and compound fractionation, including development of high-throughput microfluidic assays,5 two-dimensional high-performance liquid chromatography (2-D HPLC), and microdialysis-HPLC methods.6
Despite these advances, discoveries of ligands that activate newly discovered receptors still depend on exhaustive and challenging bioassay-guided purification of active ligands followed by their identification based on nuclear magnetic resonance (NMR) and mass spectrometry (MS).7–9 There is a need for a rapid screening tool to direct fractionation or exclude fractions by preselecting specific compounds to minimize the number of bioassays that must be performed. Our aim in this study has been to develop and implement an efficient approach to use mass spectrometry data to guide bioassay-directed fractionation. In this specific case, we focused on isolation of a previously unknown component of fetal bovine serum (FBS) that activates a splice variant of constitutive androstane receptor (CAR2). Our approach exploits accurate measurements of ion masses and calculations of ion mass defects to distinguish likely candidates and guide this fractionation.
The importance of accurate ion mass measurements was established in 1920 by F. W. Aston.10 Using an early form of mass spectrometry, he determined the masses, within 1 part-per-thousand, of hydrogen, carbon, and oxygen to be 1.008, 12.000, and 16.000 Da, respectively. These measurements were the first to suggest that atomic masses could deviate from integer values. While accurate mass measurements are frequently used to confirm a suspected elemental composition, even measurements accurate to less than 1 ppm error often cannot provide unambiguous assignment of a unique elemental formula.11 Despite this shortcoming, accurate measurements of ion masses retain value even when mass measurement accuracy is insufficient to define elemental composition. One important concept, known as the mass defect, is defined as the deviation of an ion’s measured mass from its nominal mass, which is the truncated integer value. Most elements (oxygen and heavier) have a negative mass defect, or a mass deficiency, caused by increases of nuclear binding energy with increasing atomic number. Only the six elements preceding oxygen, except carbon, have a positive mass defect or sufficiency, but hydrogen’s is most notable at +7.825 mDa.12 Since many organic molecules have a multitude of hydrogen atoms, an ion’s mass defect is largely influenced by its hydrogen content.
Several precedents for the utility of mass defects have been reported, including the development of the Kendrick scale, based on the deviation in mass from the mass of –CH2– groups, which were defined as “exactly 14 amu”.13 This approach has been particularly useful in characterization of petroleum products where homologs differing by the number of –CH2– groups are abundant.14 In a more recent development, Zhang and co-workers implemented an absolute mass defect filter to aid drug metabolite identification.15–17 Taking advantage of the observation that most modifications to a drug compound by metabolism change the mass defect by less than ±0.050 Da relative to the parent compound, their software filter removes all m/z values outside of this range, eliminating potentially interfering endogenous compounds from consideration. Since then, other researchers have used mass defect to aid identifications for other compound classes. Koulman and Volmer used mass defect to distinguish phosphatidyl choline and phosphatidyl ethanolamine lipids by assigning differences in m/z values to addition of different two-carbon units (–C2–, –C2H2–, and –C2H4–) within each class.18 Mass defects have also been used to identify peptides from other compounds in biological extracts.19 The concept of fractional mass, which represents the mass defect, has also been used in proteomics to distinguish peptides from non-peptide ions in analyses of complex digestion mixtures, with utility for on-the-fly selection of ions for tandem mass spectrometric (MS/MS) analysis.20,21
While the Kendrick mass defect, mass defect filters, and fractional mass have provided important steps toward identifying many unknown compounds, mass measurement alone is often inadequate to provide unambiguous elemental formulae. In this report, we exploit the notion that hydrogen atoms provide the major contribution to molecular mass defects, and suggest an approach based on filtering measured ion masses according to a relative mass defect in a manner that helps classify ions based upon the fraction of molecular mass accounted for by hydrogen atoms.
EXPERIMENTAL
Materials
Fetal bovine serum (FBS) was purchased from Invitrogen (Carlsbad, CA, USA). Activated charcoal/Dextran treated FBS was purchased from Hyclone (Logan, UT, USA). Di(2-ethylhexyl) phthalate (DEHP, CAS No. 117-81-7) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Di-n-octyl phthalate (DNOP, CAS No. 117-84-0) was purchased from Alfa Aesar (Ward Hill, MA, USA). Ammonium acetate was purchased from EMD Chemicals Inc. (Gibbstown, NJ, USA). Methanol, 2-propanol, and chloroform were all HPLC grade.
Avoidance of contamination
During sample preparation, glass containers were used whenever feasible, limiting exposure to plastics and minimizing contamination. In some cases, the availability of labware required the use of phthalate-free polypropylene plastics. All procedural blanks using polypropylene labware analyzed over 6 months yielded phthalate ester levels below our limit of detection (90 nM in extracts).
Extraction of lipids for GC/MS analysis
Lipids were extracted from 100 µL aliquots of serum samples in 15 mL polypropylene conical tubes using 3 mL of 2:1 methanol/chloroform (v/v). Tubes were vortexed for 15 s then placed in a hot water bath (50–55°C) for 15 min to inactivate and precipitate proteins. Samples were then centrifuged for 15 min at 3200 g and 25°C. The supernatant was removed and washed twice with 1 mL of 1 M aqueous KCl. Of the resulting lower organic layer 400 µL was added to a clean glass autosampler vial. After evaporation to dryness under a stream of nitrogen gas, the residue was redissolved in 200 µL of 1:1 100 mM aqueous ammonium acetate/2-propanol (v/v).
Extraction of lipids for discovery/fractionation
Serum lipids were extracted from 50 mL of FBS using a method adapted from Frost and Wells.22 Briefly, 50 mL of serum was mixed with 310 mL of 1:2:0.1 chloroform/methanol/1N aqueous H2SO4 (v/v/v). This mixture was incubated for 1 h at room temperature (RT) with mixing every 15 min, followed by addition of 1 volume of chloroform and 1 volume of 0.36 M aqueous H2SO4 containing 15 mM NaCl. The sample was then centrifuged at 25°C for 20 min at 4000 g. The lower organic phase was collected and the volume was reduced to about 17 mL using a rotary evaporator. The liquid residue was mixed 1:1 with chloroform, washed with water, and the lower organic phase was collected using a separatory funnel. The organic phase was transferred to a glass test tube and evaporated to approximately 3 mL under a stream of argon.
Fractionation of lipid extracts (Process 1)
From the extracts obtained from the first extraction protocol above, a neutral lipid fraction was generated using a 1000 mg BondElut Jr. NH2 solid-phase extraction (SPE) column (Varian Inc., Palo Alto, CA, USA) using sequential elution with various solvents as described.23 The hexane fraction was evaporated to dryness under argon and redissolved in 200 µL of hexane before the second stage of fractionation, which employed a 1 mL Bakerbond SPE silica gel column (J.T. Baker Inc., Phillipsburg, NJ, USA), eluting with an isocratic mobile phase of 95:5 hexane/2-propanol (v/v). A 10 mL volume of this mobile phase was used to elute the sample, which was further fractionated using a modified method developed by Murphy.24 In this third stage of fractionation, the silica SPE column eluant was evaporated to dryness under argon and dissolved in 1 mL of hexane. A 100 µL aliquot of this solution was then applied to a µPorasil silica analytical HPLC column (3.9 × 300 mm, 10 µm; Waters Corp., Milford, MA, USA) and fractionated by isocratic separation using a HPXL solvent delivery system (Rainin, Oakland, CA, USA). The mobile phase consisted of 1.08% 2-propanol and 0.09% acetic acid in hexane. Column temperature was maintained at 55°C and the flow rate at 0.6 mL/min while fractions were collected every minute over 30 min. Fractions were then dried under nitrogen gas then dissolved in 200 µL of 1:1 100 mM aqueous ammonium acetate/2-propanol (v/v) for mass spectrometric analysis.
Reporter assay
Bioassay-guided fractionation of substances with CAR2 ligand activity was guided through use of a cell-based luciferase reporter assay based on cytomegalovirus (CMV) promoter and human liver CAR2 as previously described.25 All DNA transfections were performed in a 48-well format using COS-1 cells and a CMV-CAR2 plasmid. Luciferase assays were performed using the Dual-Luciferase reporter assay system (Promega, Madison, WI, USA) and a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA, USA).
Flow injection analysis-MS and HPLC/MS of serum fractions
Aliquots (10 µL) of individual fractions isolated from FBS were analyzed using flow injection analysis (FIA) on a LCT Premier time-of-flight mass spectrometer (Waters Corp.) using electrospray ionization in both positive ion mode (ESI+) and negative ion mode (ESI−) and V mode ion path. The sample cone was held at 25 V, and the ESI capillary at 3200 V. Spectrum acquisition covered m/z 100–1500 with a spectrum accumulation time of 0.2 s. Multiplexed nonselective collision-induced dissociation (CID) was performed using quasi-simultaneous acquisition of spectra with three different aperture 1 settings: 15, 40, and 65 V.26,27 FIA analyses of samples employed a 100% methanol mobile phase flowing at 0.2 mL/min supplied by Prominence LC-20AD (Shimadzu Corp.) HPLC solvent delivery modules.
Individual fractions from FBS from Process 1 showing activity in the reporter assay were fractionated further (Process 2) using a Hypersil GOLD (Thermo Scientific) C18 column (2.1 × 50 mm, 1.9 µm particle size). Each FBS fraction was divided further into six new fractions (at 4.9, 5.5, 6.5, 10, 15, and 20 min) using a 20 min ternary gradient with a flow of 0.2 mL/min. Solvent A (10 mM NH4OAc), solvent B (methanol) and solvent C (2-propanol) were programmed using linear gradients as follows: initial=99% A/1% B, linear to 100% B at 1 min, then linear to 95% B/5% C at 11 min, then to 50% B/50% C at 13 min, and 100% C at 14 min. After holding at 100% C for 1 min the gradient was returned to initial conditions for the remaining 5 min.
GC/MS of phthalate ester standards and extracts of FBS
Gas chromatography/mass spectrometry analyses for phthalate esters were performed using a 6890 gas chromatograph coupled to a model 5973 mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). Samples were injected onto a DB-5-MS column (30 m × 0.25 mm, 0.25 µm film) using pulsed splitless mode, helium carrier gas at 43 cm/s, and an initial injector temperature of 280°C. The column was held initially at 50°C, then programmed to 240°C (35°C/min) then to 300°C (6°C/min) with a 5 min hold. Electron ionization (70 eV) was employed, and spectra were acquired over m/z 40–500 at 3.14 spectra/s.
RESULTS AND DISCUSSION
Bioassay-directed fractionation
Previous experiments conducted in our laboratories discovered CAR2 activation when using cell culture media containing FBS, but treatment of FBS with activated charcoal/Dextran to remove various low molecular weight compounds eliminated this activity. Fractionation of lipid extracts of untreated and treated FBS using isocratic elution HPLC on a normal-phase silica column generated two sets of 30 fractions per sample, all of which were assayed for CAR2 activity. Fraction 17 from untreated FBS exhibited CAR2 activity, while the corresponding fraction 17 from treated FBS remained inactive. Based on the retention characteristics of the normal-phase LC solvent system, it was proposed that an endogenous neutral lipid, suspected to be a steroid based on known CAR ligands, was present in FBS and was responsible for CAR2 activation.
To identify constituents in the active fraction, FIA-MS was performed on active fraction 17 and inactive fractions 15, 16, 18, and 19. Each summed spectrum, in both positive and negative mode, contained between 975 and 1085 peaks. After background subtraction, between 298 and 408 peaks remained, with more than 90% of the peaks below m/z 700. In the negative ion mode spectra, many peaks had a mass defect consistent with that of a lipid, and the two most prominent peaks, at m/z 255 and 283, corresponded to hexadecanoate and octadecanoate, respectively. Comparisons of spectra for the various fractions failed to reveal distinguishable differences between biologically active and inactive fractions, suggesting the compound of interest was of low abundance relative to other inactive fraction constituents. Therefore, a method was needed to distinguish bioactive substances from other compounds present in the sample by reducing spectrum complexity and accelerating recognition of neutral lipid candidates. The scheme employed in this study, as depicted in Fig. 1, is driven by bioassay-directed LC fractionation of the active fractions, with fractions screened using MS and ranked in priority in part using a relative mass defect data filter.
Figure 1.
Work flow for discovery and identification of bioactive compounds.
The untreated FBS fraction with greatest activity was fractionated further using reversed-phase LC on an ultra-performance C18 column with a ternary gradient. Owing to concerns that some neutral lipids might not yield protonated molecules during electrospray, ammonium acetate was added to the mobile phase to enhance the possibility of forming [M+NH4]+ ions for such compounds.
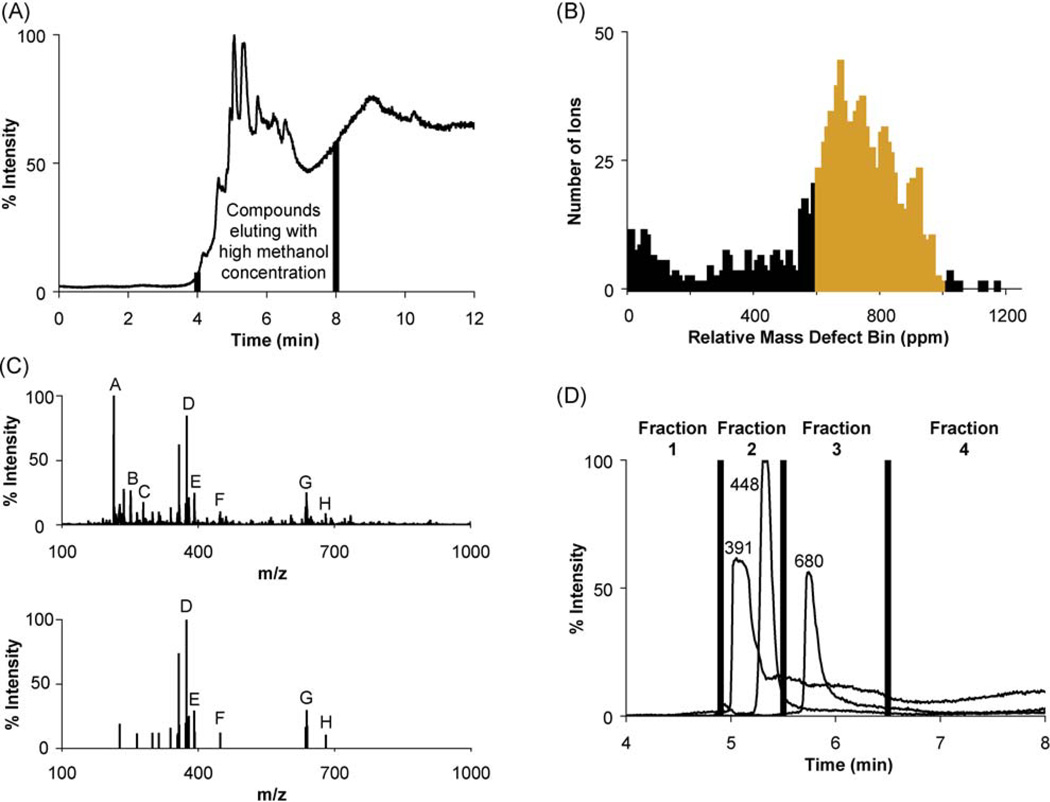
Based on previous experiments, the retention time range from 4 to 8 min in the reversed-phase HPLC separation corresponds to compounds that elute in high methanol concentration (Fig. 2(A)). Spectra were then summed over this retention time range using a 0.025 Da mass window and a 0.1% intensity filter followed by lock mass correction using m/z 214.0896 (protonated N-butyl benzenesulfonamide, a common source contaminant). This process generated a list of 1345 ions of discrete m/z values.
Figure 2.
(A) Total ion chromatogram from the LC separation of the active fraction from Process 1 (fraction 17), highlighting the retention time window selected for summation of mass spectra. (B) Histogram representing the frequency of relative mass defects for the 1345 discrete m/z values from summing all mass spectra from retention times 4–8 min. Highlighted in orange is the range of RMD values of interest, 600–1000 ppm, targeting likely neutral lipids. (C) Two summed spectra obtained from the m/z values in the selected range in (B). The top spectrum presents all m/z values from this range while the bottom spectrum presents the 21 remaining m/z values after intensity filtering and deisotoping. Peak A = m/z 214, B = m/z 251, C = m/z 279, D = m/z 374, E = m/z 391, F = m/z 448, G = m/z 638, H = m/z 680. (D) Extracted ion chromatograms depicting the second fractionation scheme (Process 2) in which the three ion m/z values of interest are distributed among two collected fractions.
To narrow the list of m/z values to those candidates most consistent with an expected neutral lipid, we turned our attention to compounds rich in hydrogen content. Hydrogen is unusual among elements in having a large positive mass defect. For quick recognition of ions with high hydrogen content, we calculated a relative mass defect (RMD) as follows: , measured in ppm
Values of RMD provide a measure of elemental composition analogous to % H determined by combustion and gravimetric analysis, as has been a long-established technique for structure confirmation. The RMD shows substantial correlation with % H, and the former can be useful for grouping ions of similar structural classes because they share similar % H content. Compounds with large RMD values have a greater percent of hydrogen and a smaller percent of heavier elements with negative or neutral mass defects such as phosphorus and oxygen. For example, alkanes have relative mass defects greater than 1000 ppm, membrane lipids and steroids often lie between 600–1000 ppm, sugars such as sucrose between 300–400 ppm, and organic acids such as citric acid have RMDs less than 300 ppm. Examples are illustrated in Fig. 3. As per the Kendrick mass defect, no heteroatom-containing organic compound should yield a RMD value greater than 1200 ppm. In these cases, the mass defect should be treated as negative, or arising from multiple charging.
Figure 3.
The relationship between relative mass defect and % hydrogen for an assortment of organic compounds. Selected for by the dashed line is the range of interest, from 600 to 1000 ppm.
Since a relative mass defect filter can narrow the list of candidate ions to those with targeted hydrogen content, lipids were selected by eliminating all m/z values except those with a RMD between 600 and 1000 ppm. Figure 2(B) depicts a histogram of the calculated RMDs for all 1345 m/z values. Ions with mass defects outside this range, including those introduced during the preparative HPLC steps in Process 1, were judged to be unlikely neutral lipid candidates and were excluded from further consideration. The remaining m/z values were filtered by relative ion abundance, with the highest 10% selected, before a final filtering to remove m/z values corresponding to 13C isotopes. The resulting spectrum list contained 21 candidate ion m/z values (Fig. 2(C)). Of these values, three represented extracted ion chromatograms with well-defined peaks, m/z 391, 448, and 680. The other 18 values were excluded because their extracted ion chromatograms had a similar profile to the total ion chromatogram, suggesting their presence was from the HPLC solvent system or the column, not the FBS sample.
Further fractionation (Process 2) of the active fraction, 17, from Process 1 allowed for the m/z value candidates to be collected among different fractions. A total of six fractions were collected using reversed-phase fractionation with an ultra-performance C18 column and the ternary gradient described earlier, with candidates m/z 391 and 448 in fraction 2 and m/z 680 in fraction 3 (Fig. 2(D)). All Process 2 fractions were divided in half, with one aliquot used for FIA-MS analysis and the other used for bioassay analysis towards CAR2, finding fractions 2 and 3 active.
Identification of bioactive compound
Positive mode ESI FIA-MS analysis was conducted for each of the six C18 Process 2 fractions. Comparison of spectra from the active fractions (2 and 3) to spectra from inactive fractions (1, 4, 5, and 6) discovered a dominant ion, at m/z 391, present only in the active fractions. To identify this compound, the accurate mass, m/z 391.2836, was selected from the mass spectrum (fraction 2) and submitted to an elemental composition calculator. With the tolerance set to 10 ppm, the range of double-bond equivalents (DBE) set to −1.5 to 20, and the range of potential elements set to 0–50 carbon, 0–200 hydrogen, 0–4 nitrogen, and 0–25 oxygen, three potential formulae were displayed, two of which were eliminated owing to the low likelihood that this neutral lipid would contain nitrogen atoms. The only non-nitrogen-containing formula, (m/z 391.2843, 1.8 ppm error), was present in fractions 2 and 3, and was selected as the most likely candidate based on both mass measurement and isotope pattern matching.
The use of multiplexed in-source CID allows for analyses similar to MSE on a time-of-flight mass spectrometer.28,29 This is advantageous over common MS/MS experiments in that varying degrees of fragmentation are generated without the need for predetermined or data-dependent selection of the precursor ion mass. This technique is performed by switching the voltage on aperture 1, a focusing lens in the transit region between the source and analyzer. For fraction 2, at the low voltage setting of 15 V, the ion at m/z 391 dominates the spectrum, but at 65 V, fragment ions at m/z 279, 167, and 149 dominate the spectrum. Follow-up LC/MS analyses of the active fraction confirmed that these three fragments co-eluted with m/z 391. These fragment ion masses are consistent with published EI spectra of dioctyl phthalate suggesting that m/z 391 is protonated dioctyl phthalate.30
Three dioctyl phthalate isomers, differing only in the aliphatic ester group, are commonly used as plasticizers (Fig. 4); branched-chain isomers di(2-ethylhexyl) phthalate and diisooctyl phthalate, and the straight-chain ester di-n-octyl phthalate. However, owing to similarities in the physical properties of the isomers, as well as expected similarities in fragmentation, the initial LC/MS analyses did not distinguish which isomer was present. Therefore, established GC/MS techniques were used to distinguish the isomer content.31,32
Figure 4.
Structures of three isomers of dioctyl phthalate: (DEHP) di(2-ethylhexyl) phthalate, (DIOP) diisooctyl phthalate, and (DNOP) di-n-octyl phthalate.
A standard solution of both DEHP and DNOP was tested and compared to a sample of untreated FBS. Figure 5 depicts the GC/MS results for the analysis, showing that only DEHP is present in the FBS sample, confirming it as the bioactive compound as it is present in both chromatograms and DNOP is absent from the serum extract sample. These results were confirmed by bioassay testing of the individual isomers with a cell-based reporter assay, yielding an EC50 value of 0.21 µM DEHP and no DNOP activity.33
Figure 5.
GC/MS extracted ion chromatograms for m/z 149 for phthalate ester standards and extract of FBS. The peaks eluting at 12.22 and 13.92 min correspond to DEHP and DNOP, respectively, confirming the presence of DEHP in extracts of FBS. (A) 5 µM DEHP and 5 µM DNOP standards, (B) extract of 100 µL of FBS.
CONCLUSIONS
Relative mass defect filtering offers a rapid way to screen lists of ion masses for compounds with fractional hydrogen content characteristic of specific compound classes or common biosynthetic origins. This approach accelerates processing of ion mass information, and allows grouping of compounds based on hydrogen content even when their absolute mass defects differ. Such is the case when the molecular masses within a class span a wide range. Coupling the use of relative mass defects with bioassay-directed LC fractionation offers a quick alternative to traditional bioactive compound discovery, focusing on targeted compound classes, and requiring fewer processing steps compared to the traditional cycle of fractionation, bioassay, and characterization. Calculations of relative mass defects could be performed on-the-fly to guide data-dependent MS/MS analyses, if implemented in instrument control software. Furthermore, this approach facilitates mining of LC/MS or GC/MS data for nontarget metabolite analyses, where a substantial fraction of detected signals are often unassigned to compound classes or elemental formulae.
Acknowledgements
This work was supported, in part, by the National Institutes of Health National Institute of General Medical Sciences [Grant GM066411 to CJO]; by the Intramural Research program of the National Institutes of Health National Institute of Environmental Health Sciences; by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Contract N01-DK-7-0004/HHSN267200700004c]; by the Michigan Agricultural Experiment Station; and by Michigan State University. The authors gratefully acknowledge assistance from Beverly Chamberlin of the Michigan State University Mass Spectrometry Facility.
REFERENCES
- 1.DiMasi JA, Hansen RW, Grabowski HG, Lasagna L. J. Health Econ. 1991;10:107. doi: 10.1016/0167-6296(91)90001-4. [DOI] [PubMed] [Google Scholar]
- 2.DiMasi JA, Hansen RW, Grabowski HG. J. Health Econ. 2003;22:151. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 3.Fauchère J-L, Boutin JA, Henlin J-M, Kucharczyk N, Ortuno J-C. Chemom. Intell. Lab. Syst. 1998;43:43. [Google Scholar]
- 4.Ertl P, Roggo S, Schuffenhauer A. J. Chem. Inf. Modeling. 2007;48:68. doi: 10.1021/ci700286x. [DOI] [PubMed] [Google Scholar]
- 5.Young SM, Curry MS, Ransom JT, Ballesteros JA, Prossnitz ER, Sklar LA, Edwards BS. J. Biomol. Screen. 2004;9:103. doi: 10.1177/1087057103262335. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, Kong L, Li X, Chen X, Guo M, Zou H. J. Chromatogr. B. 2004;812:71. doi: 10.1016/j.jchromb.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 7.Wise A, Jupe SC, Rees S. Annu. Rev. Pharmacol. Toxicol. 2004;44:43. doi: 10.1146/annurev.pharmtox.44.101802.121419. [DOI] [PubMed] [Google Scholar]
- 8.He W, Miao FJ-P, Lin DC-H, Schwandner RT, Wang Z, Gao J, Chen J-L, Tian H, Ling L. Nature. 2004;429:188. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 9.Byers JA. J. Chem. Ecol. 1992;18:1603. doi: 10.1007/BF00993233. [DOI] [PubMed] [Google Scholar]
- 10.Aston FW. Nature. 1920;105:617. [Google Scholar]
- 11.Kind T, Fiehn O. BMC Bioinform. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross JH. Mass Spectrometry: A Textbook. New York: Springer; 2004. [Google Scholar]
- 13.Kendrick E. Anal. Chem. 1963;35:2146. [Google Scholar]
- 14.Hughey CA, Hendrickson CL, Rodgers RP, Marshall AG, Qian K. Anal. Chem. 2001;73:4676. doi: 10.1021/ac010560w. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Zhang D, Ray K. J. Mass Spectrom. 2003;38:1110. doi: 10.1002/jms.521. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Zhu M, Ray KL, Ma L, Zhang D. Rapid Commun. Mass Spectrom. 2008;22:2082. doi: 10.1002/rcm.3585. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Ma L, Zhang D, Ray K, Zhao W, Humphreys WG, Skiles G, Sanders M, Zhang H. Drug Metab. Dispos. 2006;34:1722. doi: 10.1124/dmd.106.009241. [DOI] [PubMed] [Google Scholar]
- 18.Koulman A, Volmer DA. Proc. 57th ASMS Conf. Mass Spectrometry and Allied Topics; MP; May 30–June 4, 2009; Philadelphia, Pennsylvania. p. 242. [Google Scholar]
- 19.Toumi ML, Desaire H. Proc. 57th ASMS Conf. Mass Spectrometry and Allied Topics; ThP; May 30–June 4, 2009; Philadelphia, Pennsylvania. p. 495. [Google Scholar]
- 20.Dodds ED, An HJ, Hagerman PJ, Lebrilla CB. J. Proteome Res. 2006;5:1195. doi: 10.1021/pr050486o. [DOI] [PubMed] [Google Scholar]
- 21.Kirchner M, Timm W, Fong P, Wangemann P, Steen H. Bioinform. 26:791. doi: 10.1093/bioinformatics/btq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost SC, Wells MA. Arch. Biochem. Biophys. 1981;211:537. doi: 10.1016/0003-9861(81)90488-4. [DOI] [PubMed] [Google Scholar]
- 23.Kaluzny MA, Duncan LA, Merritt MV, Epps DE. J. Lipid Res. 1985;26:135. [PubMed] [Google Scholar]
- 24.Murphy EJ, Rosenberger TA, Horrocks LA. J. Chromatogr. B: Biomed. Sci. App. 1996;685:9. doi: 10.1016/0378-4347(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 25.Auerbach SS, DeKeyser JG, Stoner MA, Omiecinski CJ. Drug Metab. Dispos. 2007;35:428. doi: 10.1124/dmd.106.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu L, Jones AD, Last RL. Plant J. 2010;61:579. doi: 10.1111/j.1365-313X.2009.04083.x. [DOI] [PubMed] [Google Scholar]
- 27.Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL. Plant J. 2010;62:391. doi: 10.1111/j.1365-313X.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bateman KP, Castro-Perez J, Wrona M, Shockcor JP, Yu K, Oballa R, Nicoll-Griffith DA. Rapid Commun. Mass Spectrom. 2007;21:1485. doi: 10.1002/rcm.2996. [DOI] [PubMed] [Google Scholar]
- 29.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li G-Z, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Anal. Chem. 2005;77:2187. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 30.Yinon J. Org. Mass Spectrom. 1988;23:755. [Google Scholar]
- 31.Bove JL, Dalven P. Int. J. Environ. Anal. Chem. 1981;10:189. [Google Scholar]
- 32.Bove JL, Dalven P, Kukreja VP. Int. J. Environ. Anal. Chem. 1978;5:189. [Google Scholar]
- 33.DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Mol. Pharmacol. 2009;75:1005. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]