Significance
We report the ancestral roles of the A20 molecule as a dual-function enzyme in a basal chordate that adds and removes ubiquitin moieties to its target proteins. Moreover, we found amphioxus A20-binding inhibitors of NF-κB (ABINs) fulfilled ancestral roles in connecting the ubiquitin chain by competing with other ubiquitinated proteins. We further suggest that although ubiquitination is an ancient strategy in regulating immune signaling, taxon-specific regulating mechanism may occur in amphioxus. The emergence of A20 and ABINs adds information on the means by which amphioxus can effectively defend against pathogenic invasion in the absence of classical adaptive immunity.
Abstract
In the past decade, ubiquitination has been well documented to have multifaceted roles in regulating NF-κB activation in mammals. However, its function, especially how deubiquitinating enzymes balance the NF-κB activation, remains largely elusive in invertebrates. Investigating bbtA20 and its binding proteins, bbt A20-binding inhibitor of NF-κB (bbtABIN1) and bbtABIN2, in Chinese amphioxus Branchiostoma belcheri tsingtauense, we found that bbtABIN2 can colocalize and compete with bbt TNF receptor-associated factor 6 to connect the K63-linked polyubiquitin chains, whereas bbtABIN1 physically links bbtA20 to bbt NF-κB essential modulator (bbtNEMO) to facilitate the K48-linked ubiquitination of bbtNEMO. Similar to human A20, bbtA20 is a dual enzyme that removes the K63-linked polyubiquitin chains and builds the K48-linked polyubiquitin chains on bbt receptor-interacting serine/threonine protein kinase 1b, leading to the inhibition of NF-κB signaling. Our study not only suggests that ubiquitination is an ancient strategy in regulating NF-κB activation but also provides the first evidence, to our knowledge, for ABINs/A20-mediated inhibition of NF-κB via modifying the ubiquitinated proteins in a basal chordate, adding information on the stepwise development of vertebrate innate immune signaling.
Studies emerging in the past decade have revealed multifaceted roles of ubiquitin in receptor trafficking, signal transduction, and DNA damage response (1). In particular, it has been reported to be instrumental in regulating several steps in NF-κB signaling (2). For instance, when Toll-like receptor (TLR) signaling is triggered, the ubiquitin ligase activity of TNF receptor-associated factor 6 (TRAF6) was activated, leading to K63-linked polyubiquitination of targets including NF-κB essential modulator (NEMO) and TRAF6 itself (2). Because ubiquitination is a reversible posttranslational modification of proteins, NF-κB signaling is generally attenuated by deubiquitinating enzymes (DUBs), which “trim” ubiquitin chains from specific substrate proteins (3).
In mammals, one of the best-known DUBs involved in NF-κB signaling is A20 (also known as TNFAIP3), which belongs to the ovarian tumor (OTU) DUB family (4). A20 can restrict NF-κB signaling downstream of TNF receptor 1 (TNFR1), CD40, TLRs, NOD-like receptors, and the IL-1 receptor (5–8). Indeed, A20 is not only able to deubiquitinate substrates modified by K63-linked polyubiquitins, but can also induce their K48-linked polyubiquitination and degradation through its E3 activity. For example, A20 may regulate NF-κB activation by cleaving K63-linked polyubiquitin chains from receptor-interacting serine/threonine protein kinase 1 (RIP1), TRAF6, and/or NEMO through its deubiquitination activity dependent on its OTU domain (6, 7). Meanwhile, it also builds K48-linked polyubiquitin chains on RIP1 (7). Moreover, A20 can regulate NF-κB activation by supporting the degradation of the E2 enzymes UbcH5 and Ubc13, thereby inhibiting E3 activity that is dependent on these enzymes (8). Therefore, A20 interfaces with, and modifies, ubiquitinated protein substrates in multiple ways and acts through a variety of biochemical mechanisms to regulate NF-κB signals.
Although A20 can interact with TRAF2 and NEMO, some studies have shown that these interactions are not sufficient for the inhibition of NF-κB, leading to the identification of a gene family called A20-binding inhibitor of NF-κB (ABIN) (9). The ABIN family comprises three members, ABIN-1, ABIN-2, and ABIN-3. All of them share a conserved ubiquitin-binding domain (UBD) and are characterized as negative regulators of NF-κB signaling when overexpressed (10). For example, overexpressed ABIN-1 physically links A20 to NEMO to facilitate deubiquitination of NEMO (11). ABIN-2 blocks the interaction between RIP1 and NEMO, thus resulting in inhibition of NF-κB (12). ABIN-3 negatively regulates the LPS-induced NF-κB activation downstream of TRAF6 and upstream of the IκB kinase (IKK) (13).
Several lines of evidence have suggested that the K63-linked polyubiquitination is also crucial for the activation of Drosophila Relish (14). The ubiquitin chains in Drosophila immune deficiency (IMD) and caspase 8 homolog DREDD serve as scaffolds for the recruitment of TGF-β–activated kinase 1 (dTAK1) and dIKK complex, allowing DREDD-mediated proteolysis of Relish and the expression of Relish-dependent antimicrobial peptide genes (15, 16). Although homologs of cylindromatosis and ubiquitin-specific protease 36, two other important DUBs in mammalian NF-κB signaling, have been found to deubiquitinate dTRAF2 and dIMD, likely serving as a switch to deactivate the IMD pathway (17, 18), no A20 or ABINs have been reported in Drosophila and other invertebrates. Therefore, identifying the A20 and ABIN homologs and characterizing their roles in ubiquitination in the basal chordate amphioxus will help us not only to understand when and in what ways the ABINs and A20 appeared in classical NF-κB signaling, but also to characterize the inactivation of NF-κB by DUBs in invertebrates.
Results
Identification of Genes Involved in Ubiquitination in Amphioxus.
To reveal how ubiquitination functions in amphioxus NF-κB signaling, we conducted a systematic analysis of the ubiquitination-related genes in the amphioxus genome. Initially, the full-length cDNA of amphioxus ubiquitin with a ubiquitin domain was cloned. The derived 76 amino acids of amphioxus ubiquitin were 100% identical to those of human and rat ubiquitin. As in most invertebrate genomes, amphioxus possesses a single E1 with an ubiquitin-associated domain at its C terminus and two conserved motifs, the ATP-binding motif (GXGXXGCE) and the PXCTXXXP motif, which form thiolester with ubiquitin. All E2s except UbcH12 and Ube2S2 have been found in the amphioxus genome, especially that the UbcH5 family is clearly conserved. Proteins involved in E3 in amphioxus are comparable to those in mammals, including 389 putative RING finger-containing E3s, 25 homologous to the E6-AP carboxyl terminus E3s, 9 U-box E3s, and 69 plant homeodomain E3s (Table S1). Nearly 90 putative DUBs belonging to five families are encoded by the amphioxus genome, including 5 ubiquitin C-terminal hydrolases, 41 ubiquitin-specific proteases, 32 OTU proteases, 2 Josephins, and 12 JAB1∕MPN∕MOV34 metalloenzymes (Table S1). Moreover, some putative E3s and DUBs seem to be amphioxus-specific, because proteins with similar domain architectures could not be found in other species. For example, RING finger containing E3s have additional death effector domain (DED), and OTU containing DUBs have additional DED or death domain or leucin-rich repeats (Fig. S1). These comparative analyses imply that although the ubiquitination strategy is well conserved during evolution, the hierarchy of ubiquitin modification in amphioxus immune signaling pathways may not be the same as that in mammals.
Sequencing and Phylogenetic Analysis of bbtA20, bbtABIN1, and bbtABIN2.
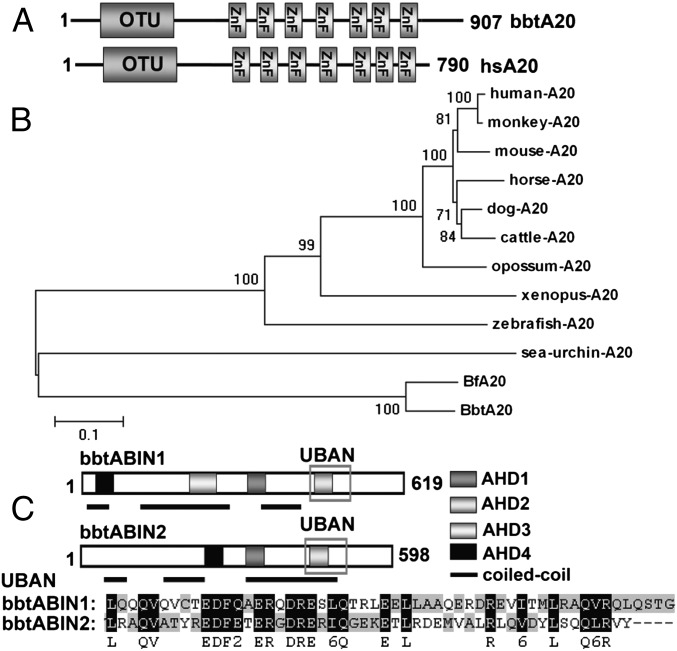
A20 is one of the most prominent and well-studied DUBs that regulate NF-κB signaling. To find molecular evidence for the roles of ubiquitination in amphioxus immune regulation, full-length cDNA of 2,701 bp was isolated from Chinese amphioxus, Branchiostoma belcheri tsingtauense, and designated bbtA20. This bbtA20 encodes a polypeptide of 907 amino acids with an N-terminal OTU domain and seven A20-type zinc fingers (ZnFs) at its C terminus (Fig. 1A). To gain information on the evolution of the A20 molecule, we searched several invertebrate genomes for A20 homologs and found a single sequence in the purple sea urchin (Strongylocentrotus purpuratus), but not in other lower invertebrates. A phylogenetic analysis based on the full-length sequences of A20 from the purple sea urchin, amphioxus, zebrafish, frog, and several mammalian species showed that all A20 proteins are evolved from a common ancestor and that A20 homologs in invertebrates are largely diverged from vertebrate A20 proteins (Fig. 1B).
Fig. 1.
Sequence and phylogenetic analyses of bbtA20 and bbtABINs. (A) Protein architecture of bbtA20 shows an N-terminal OTU domain and seven ZnFs at its C terminus. (B) Phylogenetic analysis of bbtA20. (C) Protein architecture shows that bbtABIN1 possesses all four AHD regions, whereas bbtABIN2 lacks AHD3. Both bbtABIN1 and bbtABIN2 contain a conserved UBAN domain. All A20 protein sequences were obtained from the GenBank database. Protein sequences were first aligned using ClustalX 1.83 and were manually corrected using GeneDoc. The neighbor-joining tree was obtained using MEGA4 with 1,000 bootstrap tests.
Because ABINs are key players in coupling with A20 to regulate the NF-κB pathway, two full-length cDNAs, designated bbtABIN1 and bbtABIN2, were obtained from B. belcheri tsingtauense. Sequence analysis indicated that bbtABIN1 encoded a 619-aa protein with 50–58% similarity to vertebrate ABIN-1, whereas bbtABIN2 encoded a 598-aa protein with 43–53% similarity to vertebrate ABIN-2. Similar to their mammalian counterparts, the previously described ABIN homology domain 1 (AHD1), AHD2, and AHD4 were found in both bbtABIN1 and bbtABIN2, and an AHD3 was also present in bbtABIN1 (Fig. 1C and Fig. S2A). Among four AHD regions, the most conserved was the AHD2 region, which comprised a UBD, also referred to as ubiquitin binding in ABIN and NEMO (UBAN) (Fig. 1C). Because mutations in the UBAN domain have been shown to lead to a loss of ubiquitin binding and impaired the NF-κB inhibitory potential of ABINs (19, 20), the conservation in this region indicates that bbtABINs may play roles similar to those of their mammalian counterparts in the ubiquitination process. Phylogenetic analysis further revealed bbtABIN1 to be the primitive form of vertebrate ABIN-1 and ABIN-3, whereas bbtABIN2 is the ortholog of vertebrate ABIN-2 (Fig. S2B).
BbtA20, bbtABIN1, and bbtABIN2 Are the Target Genes of NF-κB.
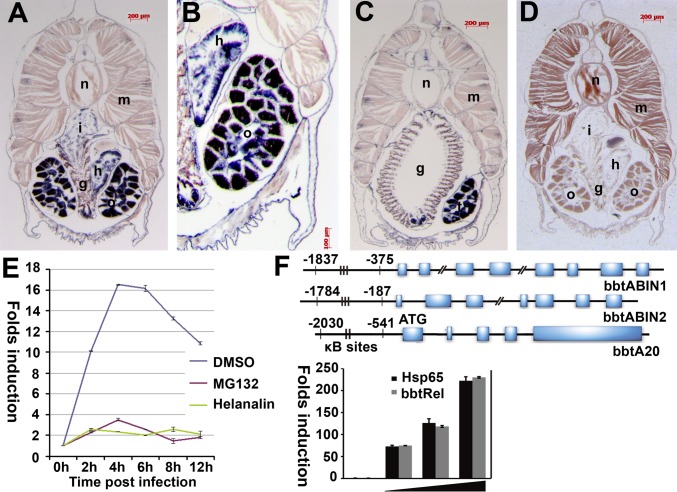
To shed further light on the relevance of bbtA20 and bbtABINs in amphioxus immunity, a section in situ hybridization was performed to determine their tissue distribution. Transcripts of bbtABIN2 are abundant in intestine, hepatic cecum, and gonad, but not in gill (Fig. 2 A–D). The expression of bbtABIN2 is similar to that of murine ABIN-2, being strongly expressed in homologous organs; the hepatic cecum of amphioxus is the counterpart of vertebrate liver. The expression pattern of bbtABIN1 is similar to bbtABIN2 (Fig. S3). Because the expression of bbtA20 in normal tissue is relatively weak and difficult to detect, we used RT-PCR to measure its expression and observed significant induction in 4 h following bacterial challenge (Fig. 2E). In our previous study, we demonstrated that the NF-κB–specific inhibitor helenalin can inspire the interaction between κB motif and bbtRel, a p65 homolog in amphioxus (21). Treatment with the helenalin before infection consistently suppressed the up-regulation of bbtA20 (Fig. 2E), suggesting that the expression of bbtA20 is tightly regulated by amphioxus NF-κB.
Fig. 2.
Expression patterns of bbtABIN2 and bbtA20. (A) Section in situ hybridization showed that transcripts of bbtABIN2 are abundant in intestine, hepatic cecum, and gonad. (B) Macroscopic view of the hybridization signals of bbtABIN2 in hepatic cecum and ovary. (C) Transcripts of bbtABIN2 are sparse in gill slits but abundant in testis. (D) Negative control of bbtABIN2 using the sense probe. g, gill slit; h, hepatic cecum; i, intestine; m, muscle; n, notochord; o, ovary; t, testis. Blue indicates strong hybridization, and dark brown indicates a weak signal. (E) RT-PCR analyses of expression patterns of bbtA20 after bacterial challenge in the presence or absence of the NF-κB inhibitor helenalin. Results were presented as “fold induction” of mRNA expression done in triplicate, using the 2-ΔΔCt method. Endogenous control for quantification was cytoplasmic β-actin. Values were considered significant at P < 0.05. (F) Sequence analysis indicated that all bbtABINs and bbtA20 possess several conserved κB-binding motifs in the promoter region. A reporter assay confirmed that the region containing two κB-binding sites upstream of the ATG of bbtA20 is essential for the binding to bbtRel. Hsp65 indicates the Homo sapiens p65. Reporter experiments were conducted in triplicate; vertical bars indicate mean ± SD. Data are representative of three independent experiments.
To investigate further whether the bbtABINs and bbtA20 are classical NF-κB target genes, 2-kb genomic sequences upstream of the ATG of bbtABIN1, bbtABIN2, and bbtA20 were obtained and subjected to the Transcription Element Search System (TESS) prediction program (www.cbil.upenn.edu/cgi-bin/tess/tess) to determine whether these regions contain NF-κB–binding motifs. Similar to their vertebrate homologs, the promoter regions of bbtABIN1, bbtABIN2, and bbtA20 contain several conserved κB-binding sites (Fig. 2F). Reporter assays showed that the sequence including two κB-binding sites of bbtA20 is essential for the interaction with bbtRel, further suggesting that bbtA20 is the target of classical NF-κB signaling (Fig. 2F).
Overexpressed bbtABIN2 Can Connect the K63-Linked Ubiquitin Chains by Competing with bbtTRAF6.
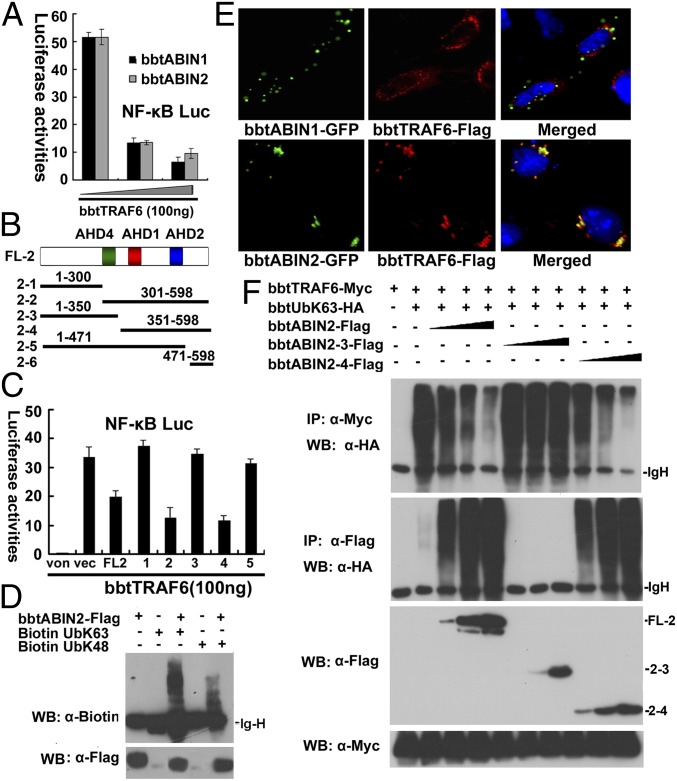
To reveal the potential cellular roles of bbtABINs and bbtA20 in the activation of NF-κB signaling, we first performed reporter assays to measure the modulation of NF-κB by these proteins. All bbtABIN1, bbtABIN2, and bbtA20 can attenuate the TNF-α–induced NF-κB activation in a dose-dependent manner in HEK 293T cells (Fig. S4A). Moreover, bbtTRAF6 mediated NF-κB activation, and secretion of TNF-α can be inhibited by gradient bbtABINs, indicating that bbtABINs may play inhibitory roles in NF-κB activation by targeting TRAF6 or molecules downstream of TRAF6 (Fig. 3A and Fig. S4B). To reveal whether the inhibitory role of bbtABINs is dependent on the ubiquitin-binding activity, several truncated mutants for bbtABIN2, designated bbtABIN2-1 to bbtABIN2-6, were constructed (Fig. 3B). Overexpression of the vectors containing AHD2 domain significantly reduced the bbtTRAF6-mediated NF-κB activation (Fig. 3C). A further in vitro ubiquitin-binding assay suggested that bbtABIN2 binds K63-linked, as well as K48-linked, polyubiquitin chains and displays a preference for K63-linked polyubiquitin chains (Fig. 3D). These results implied that the UBAN in the AHD2 region is not only structurally but also functionally conserved in connecting ubiquitin chains and involved in the regulation of the NF-κB pathway.
Fig. 3.
K63-linked ubiquitinated bbtTRAF6 is the target of bbtABIN2. (A) Luciferase reporter assays showed that both bbtABIN1 and bbtABIN2 can attenuate the NF-κB activation mediated by bbtTRAF6 in a dose-dependent manner in HEK 293T cells. Reporter experiments were conducted in triplicate; vertical bars indicate mean ± SD. Data are representative of three independent experiments. (B) Vectors of bbtABIN2 used in this study. (C) Luciferase reporter assays showed that truncated mutants containing the AHD2 region of bbtABIN2 can inhibit the bbtTRAF6-mediated activation of NF-κB. FL2 indicates the WT bbtABIN2; 1–6 indicate bbtABIN2-1 to bbtABIN2-6. von indicates the blank control; vec indicates the pcDNA3.1 vector. (D) In vitro binding assays showed that bbtABIN2 can bind both K63-linked and K48-linked ubiquitin chains. WB, Western blot. (E) Confocal analysis showed that bbtABIN2, but not bbtABIN1, can colocalize with bbtTRAF6. Original magnification 400×. (F) Both WT bbtABIN2 and the truncated mutant bbtABIN2-4 can bind to K63-linked polyubiquitin chains by competing with bbtTRAF6, whereas bbtABIN2-3, which lacks the UBAN domain, cannot. The abbreviation bbtUb indicates the WT amphioxus ubiquitin, whereas bbtUbK63 indicates the amphioxus ubiquitin mutant with substitution of arginine for all lysine residues except the lysine at position 63. All of the co-IP (IP) assays were done at least twice.
To reveal whether bbtABINs can target ubiquitinated bbtTRAF6, coimmunoprecipitation (co-IP) assays were conducted, and results showed that bbtTRAF6 can interact with bbtABIN1 and bbtABIN2, but not with bbtA20 (Fig. S4C). Immunostaining further showed that bbtABIN2, but not bbtABIN1, can colocalize with bbtTRAF6 (Fig. 3E), indicating that the interaction between bbtABIN1 and bbtTRAF6 may require some still unidentified proteins. In vivo ubiquitination assays showed that both K63-linked and K48-linked polyubiquitin chains can be built on bbtTRAF6 (Fig. S4D). However, when bbtTRAF6 was coexpressed with bbtABIN2, but not with bbtABIN1 or bbtA20, lower efficacy of K63-linked ubiquitination was observed (Fig. S4E). In vivo ubiquitination assays further showed that when bbtTRAF6 was coexpressed with WT bbtABIN2 or bbtABIN2-4, but not with bbtABIN2-3, which lacks a UBAN domain, lower efficacy of K63-linked ubiquitination of bbtTRAF6 but enhanced K63-linked ubiquitination of bbtABIN2 was observed (Fig. 3F and Fig. S4F). Taken together, the overexpressed bbtABIN2 can inhibit the bbtTRAF6-mediated activation of NF-κB by competing with bbtTRAF6 for the K63-linked polyubiquitin chains.
BbtABIN1 Physically Linked bbtA20 to bbtNEMO to Facilitate the Degradation of bbtNEMO.
Integrating ubiquitination into NF-κB activation requires NEMO, which not only displays affinity for polyubiquitin chains, but is also modified posttranslationally by a complex set of reactions involving ubiquitin. We cloned a NEMO homolog from B. belcheri tsingtauense, designated bbtNEMO. Phylogenetic analysis confirmed that bbtNEMO is the common ancestor of vertebrate NEMO and optineurin, which is a Golgi-associated NEMO homolog that plays a role in TNFR1 signaling, indicating that the two genes were produced by duplication when invertebrates evolved into vertebrates (Fig. S2C).
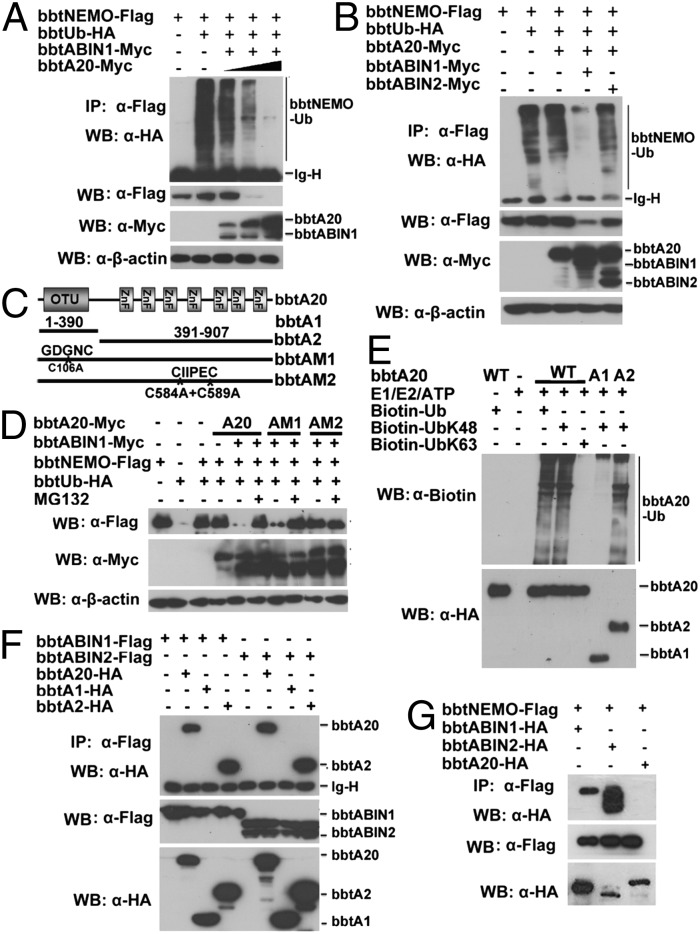
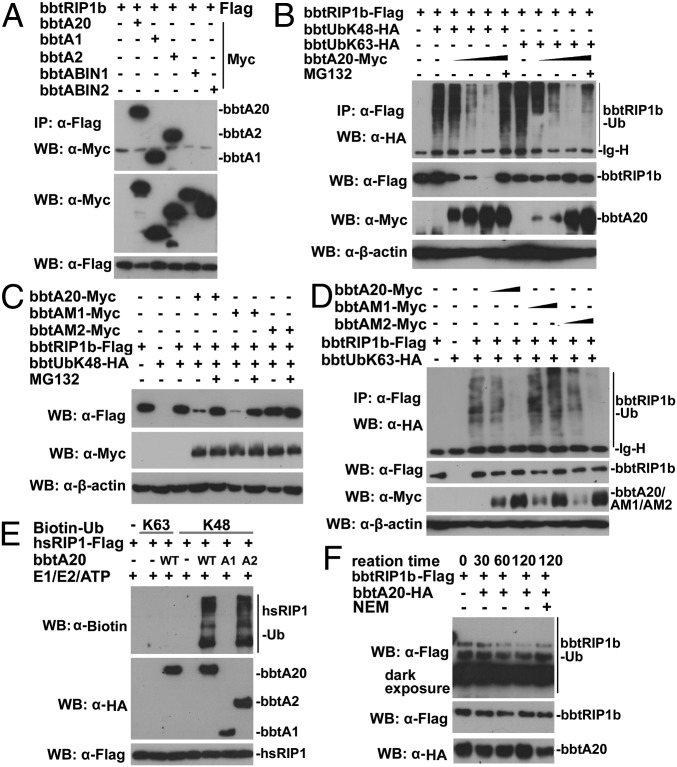
Both in vivo and in vitro ubiquitination assays indicated that K63-linked and K48-linked polyubiquitin chains can be built on bbtNEMO (Fig. S5 A and B). Alone, bbtABIN1, bbtABIN2, and bbtNEMO could not promote deubiquitination or degradation of bbtNEMO (Fig. S5C). However, when bbtNEMO was coexpressed with bbtABIN1 and gradient bbtA20, the degradation of bbtNEMO was observed in a dose-dependent manner (Fig. 4A). Such degradation is dependent on the ZnFs of bbtA20 and did not occur when bbtNEMO was coexpressed together with bbtA20 and bbtABIN2 (Fig. 4B and Fig. S5D). With the use of ubiquitin mutants with only K48 or K63 available for polymerization, the overexpressed bbtA20 can mediate the proteasome-dependent degradation of bbtNEMO but cannot remove the K63-linked ubiquitin chains on bbtNEMO (Fig. S5E). Because the DUB activity of human A20 is mediated by the catalytic cysteine at position 103 (C103) within the OTU domain, whereas the E3 activity is dependent on the ZnF4 motif of A20, two point-mutated constructs of bbtA20 (designed bbtAM1 and bbtAM2) and two truncated mutants (designed bbtA1 and bbtA2) were constructed for further analyses. The C106 within the OTU domain of bbtA20 was replaced as alanine in bbtAM1, whereas the C584 and C589 in ZnF4 were double-mutated into alanine in bbtAM2 (Fig. 4C). Notably, WT bbtA20 and bbtAM1, but not bbtAM2, attenuate the degradation of bbtNEMO, suggesting that bbtA20-mediated K48-linked ubiquitination was functionally linked and dependent on its ZnF4 (Fig. 4D). In vitro ubiquitination assays further confirmed that bbtA20 can function as an E3 to catalyze the K48-linked, but not the K63-linked, polyubiquitin chains (Fig. 4E). Such E3 activity is dependent on the C-terminal ZnFs of bbtA20 (Fig. 4E). To reveal whether bbtABIN1 serves as an adaptor to link bbtA20 with bbtNEMO, co-IP assays were performed. Results showed that bbtA20 can interact with bbtABIN1 and bbtABIN2 via its ZnFs (Fig. 4F). Moreover, bbtNEMO interacts with bbtABINs, but not with bbtA20 (Fig. 4G). The ZnF4 mutant or OTU mutant did not affect such interaction (Fig. S5F). Thus, we proposed that when bbtA20 interacts with bbtABIN1 through the ZnFs, it is linked to bbtNEMO to promote the degradation of bbtNEMO through a proteasome-dependent pathway, which is dependent on its ZnF4.
Fig. 4.
BbtA20 was physically linked to bbtNEMO to facilitate the K48-linked ubiquitination of bbtNEMO. (A) Cotransfection of bbtNEMO with bbtA20 and bbtABIN1 results in the degradation of bbtNEMO dependent on the dose of bbtA20. (B) Cotransfection of bbtNEMO with bbtA20 and bbtABIN1, but not with bbtA20, alone or together with bbtABIN2, results in the degradation of bbtNEMO. (C) Vectors of bbtA20 used in this study. (D) In vivo ubiquitination showed that point mutations within bbtA20 ZnF4, but not within bbtA20 OTU, attenuate the degradation of bbtNEMO. (E) In vitro analyses showed that bbtA20 can function as an E3 to catalyze the K48-linked ubiquitination, which is dependent on its ZnFs. (F) Transfected WT and the ZnFs only of bbtA20 bind bbtABIN1 and btABIN2. (G) Co-IP assays showed that both bbtABIN1 and bbtABIN2, but not bbtA20, can directly interact with bbtNEMO. All co-IP experiments were done at least twice.
BbtA20 Is a Dual Enzyme to Deubiquitinate K63-Linked Polyubiquitin Chains and Induce K48-Linked Polyubiquitin Chains on bbtRIP1b.
Although bbtA20 alone could not interact with bbtTRAF6 or bbtNEMO, it can interact with bbtRIP1b, which has been demonstrated to play similar roles to its human counterpart in HEK 293T cells (22). Both the OTU domain and ZnFs are responsible for the interaction (Fig. 5A), and the ZnF4 mutant and OTU mutant did not affect such interaction (Fig. S5F). In vitro ubiquitination assays showed that both K63-linked and K48-linked polyubiquitin chains can be built on bbtRIP1b (Fig. S6A). With the use of ubiquitin mutants with only K48 (bbtUbK48) available for polymerization, overexpressed bbtA20 led to the proteasome-dependent degradation of bbtRIP1b in a dose-dependent manner (Fig. 5B). However, in the presence of bbtUbK63, ubiquitination of low efficacy, but not degradation, of bbtRIP1b was observed when bbtA20 was cotransfected (Fig. 5B). When coexpressed with WT bbtA20 or bbtA2 (ZnFs only), bbtRIP1b was found to be degraded in a dose-dependent manner (Fig. S6B). Moreover, mutations within the bbtA20 ZnF4 markedly diminished the degradation of bbtRIP1b through a proteasome-dependent pathway (Fig. 5C), whereas mutation of the C106 within the bbtA20 OTU markedly attenuated deubiquitination of the K63-linked ubiquitin chains on bbtRIP1b (Fig. 5D). These observations are confirmed by further in vitro ubiquitination analyses, which showed that both bbtA20 and bbtA2 can catalyze the K48-linked ubiquitination of bbtRIP1b and human RIP1 (Fig. 5E and Fig. S6C). Meanwhile, in vitro deubiquitination assays indicated that the bbtA20 can remove the K63-linked polyubiquitin chains on bbtRIP1b dependent on its OTU domain (Fig. 5F and Fig. S6D). Thus, bbtA20 is a dual-function enzyme to remove and add ubiquitin chains on bbtRIP1b. Moreover, the molecular basis by which A20 performs this dual function is well conserved during evolution.
Fig. 5.
BbtA20 is a dual enzyme in removing the K63-linked ubiquitin chains and adding the K48-linked ubiquitination on bbtRIP1b. (A) Co-IP assays assessing the direct interaction between bbtRIP1b and bbtA20 and identifying both OTU and ZnFs of bbtA20 are responsible for the interaction. (B) Cotransfection of bbtA20 promotes bbtRIP1b degradation in HEK 293T cells. Such degradation could be inhibited by addition of MG132. Moreover, cotransfection of bbtA20 removes the K63-linked polyubiquitin chains from bbtRIP1b in a dose-dependent manner. (C) Cotransfection of WT bbtA20 or OTU mutant (bbtAM1), but not ZnF4 mutant (bbtAM2), results in the proteasome-dependent degradation of bbtRIP1b. (D) Cotransfection of WT bbtA20 or bbtAM2, but not bbtAM1, results in a low level of K63-linked ubiquitination, but not degradation, of bbtRIP1b in a dose-dependent manner. (E) In vitro ubiquitination assays showed that both WT bbtA20 and bbtA2 (the ZnFs only) can function as E3 to catalyze the K48-linked ubiquitination of hsRIP1b. (F) In vitro deubiquitination assays indicated that bbtA20 can deubiquitinate the K63-linked ubiquitinated bbtRIP1b. All co-IP experiments were done at least twice.
Discussion
Ubiquitination Is an Ancient Strategy in Regulating NF-κB Signaling, but the A20- and ABIN-Mediated Attenuation of NF-κB Originated in the Basal Deuterostome.
Ubiquitination is thought to be universal in eukaryotes, and its activity has been shown to be prevalent in various eukaryotic species (23). Therefore, it is not surprising to observe that such a posttranslational process is highly conserved in amphioxus, not only because amphioxus possesses comparable gene modules relating to ubiquitin-mediated signaling but also because the protein architecture and sequences of many key molecules involved in the process are well conserved in amphioxus, including the 100% identity of ubiquitin among species. Together with other studies, including those in vertebrates, plants, and Drosophila, our research clearly demonstrated that ubiquitination of TRAF6, RIP1, and NEMO is essential for the activation of amphioxus NF-κB, further indicating that ubiquitination is an ancient strategy in regulating inflammation and other immune processes.
At least eight types of ubiquitin chain exist, and individual linkages affect distinct cellular processes. It is clear that E3s and DUBs display specificity for substrates and ubiquitin chain types (19, 20). Because amphioxus has many E3s and DUBs with unique domain architectures, and because key molecules involved in immune regulation by targeting ubiquitination are not conserved during evolution, it is reasonable to speculate that the underlying mechanism of ubiquitination regulation of NF-κB signaling may differ among species. This hypothesis has been partially demonstrated between Drosophila and Homo sapiens, because the involved targets of the polyubiquitin chains and their specialized E3s and DUBs differ in immune signaling (2, 14). Here, we present further evidence that the well-known A20- and ABIN-mediated regulation of NF-κB signaling may be widely present in deuterostomes, but not in protostomes. In response to stimulation with proinflammatory cytokines, mammalian A20 mediates its inhibitory function in a complex with other regulatory proteins, including ring finger protein 11 (RNF11) and itchy E3 ubiquitin protein ligase (ITCH), which are both ubiquitin ligases, and Tax1-binding protein 1 (TAX1BP1), which is an adaptor protein (24, 25). By comparative analyses, we found that ITCH, RNF11, and TAX1BP1 are also present in amphioxus genome, but not in Drosophila. Moreover, all of them have similar domain architectures with their human counterparts, implying the conservation of A20 ubiquitin-editing complex in amphioxus (Fig. S7). Thus, due to the unique evolutionary position of amphioxus, studies on the ubiquitination-related proteins and enzymes in amphioxus should help us to understand how these molecules evolved to participate in the regulation of inflammation when invertebrates evolved into vertebrates.
Ancestral A20 Has Dual Functions in Adding or Removing Ubiquitin from Target Proteins to Regulate the NF-κB Activity.
The first identified target of mammalian A20, acting within the so-called “A20 ubiquitin editing complex,” has been shown to be ubiquitinated RIP1 (7).The OTU domain of A20 functions as a DUB to remove activated K63-linked polyubiquitin chains from RIP1, whereas the A20 ZnF domain functions as an ubiquitin ligase, adding K48-linked polyubiquitin chains to RIP1 and targeting the protein for proteasomal degradation (7). Similar to human A20, the bbtA20 ZnF domain can function as an ubiquitin ligase, adding K48-linked polyubiquitin chains to bbtRIP1b, whereas the bbtA20 OTU domain can remove the K63-linked polyubiquitin chains on bbtRIP1b. Thus, when it first emerged, the ancestral A20 molecule appears to have been a dual-function enzyme that added and removed ubiquitin moieties to its target proteins, which is demonstrated for the first time, to our knowledge, in an invertebrate species.
Except for targeting RIP1, mammalian A20 can also form a complex with TRAF6 or NEMO within minutes of ligand binding to the receptor, leading to their deubiquitination and the inhibition of IKK via a noncatalytic mechanism (6). To affect NEMO ubiquitination, A20 required ABIN-1, which is an A20-binding protein and recognizes NEMO via a two-step mechanism: Its UBAN domain interacts with the polyubiquitinated chains of NEMO, and its NEMO-binding domain directly contacts NEMO (11). Similar to human A20, to affect bbtNEMO ubiquitination, bbtA20 required bbtABIN1 to serve as a scaffold protein. However, unlike human A20, bbtA20 did not bind directly with bbtNEMO. Also unlike human A20, bbtA20 did not promote the deubiquitination of bbtNEMO but led to the degradation of bbtNEMO via its ZnF domain. It is important to note that bbtA20 is a classical NF-κB targeting gene and is induced within 2 h of NF-κB activation. Thus, we propose that in a negative feedback loop, amphioxus NF-κB up-regulates bbtA20, which can not only remove K63-linked polyubiquitin chains from bbtRIP1b but also promote the degradation of bbtRIP1b and bbtNEMO, leading to the failure of NF-κB activation as the balancer to attenuate excessive inflammation. In addition to bbtABIN1, the bbtA20-mediated degradation of bbtNEMO may still require other proteins, because an in vitro ubiquitination assay with the use of purified bbtA20 from HEK 293T cells as an E3 candidate did not show the K48-linked ubiquitination of bbtNEMO, even in the presence of purified bbtABIN1. Thus, further studies on amphioxus RNF11, ITCH, and TAX1BP1 should help us to understand how the A20 ubiquitin-editing protein complex is formed and functions in amphioxus.
UBAN Domain Provides the Molecular Basis for ABINs to Play Inhibitory Roles in NF-κB Signaling.
Although overexpressed ABIN-2 plays inhibitory roles in NF-κB activation, ABIN-2–deficient bone marrow-derived macrophages showed no defect in NF-κB signaling but had diminished ERK activation (26). It is unlikely that the association with ERK signaling is the sole function of ABIN-2, because this role does not require the conserved UBD of ABIN-2 and the binding of ABIN-2 to A20 is also not involved. Here, we demonstrated that bbtABIN2 can colocalize and interact with bbtTRAF6. When bbtABIN2 is bound to bbtTRAF6, it can compete with bbtTRAF6 for the K63-linked ubiquitin chains, leading to the attenuation of NF-κB signaling. Such competition is dependent on the UBAN domain of bbtABIN2, which can directly bind with K63-linked polyubiquitin chains in vitro. Due to the lack of any amphioxus cell lines and the difficulties of performing genetic analysis in amphioxus embryos, genetic or endogenous evidence on whether bbtABIN2 can target NF-κB in vivo could not be obtained. However, our study at least provides primitive evidence that the conserved UBAN domain and ubiquitin-binding activity represent the molecular basis by which ABINs perform their inhibitory roles in immune-related signaling, which should help us to understand the importance of ABIN-2 ubiquitin-binding activity in immune responses in mammals.
In addition to connecting A20 to NEMO to affect NF-κB signaling, as an ubiquitin sensor, a growing number of references have implicated ABIN-1 in embryonic development and in chronic inflammatory and autoimmune diseases, such as psoriasis, rheumatoid arthritis, and lupus nephritis (27–29). These studies have related the roles of ABIN-1 in anti-inflammation to its UBD, likely by targeting the TLR-signaling pathways that control NF-κB and MAPK activity (27–30). In this study, both bbtABIN1 and bbtABIN2 proteins, when expressed individually, were found to localize in cytoplasm and form unique punctuated spherical structures of various sizes, suggesting that bbtABIN1 and bbtABIN2 may form complexes with other unidentified proteins. Moreover, both bbtABIN1 and bbtABIN2 are classical NF-κB target genes and are abundant in the amphioxus digestive system, which is presumed to be the primary line of immune defense in amphioxus. Thus, further characterization of the relationships between bbtABINs and other ubiquitinated proteins should help us to define the ancestral roles of ABINs in anti-inflammation and to understand how these roles evolved when invertebrates evolved into vertebrates.
Materials and Methods
In Vivo Ubiquitination and Deubiquitination Assays.
HEK 293T cells were transfected with the indicated constructs in figures, and they were pretreated with 25 μM MG-132 to block bbtA20-mediated degradation of bbtRIP1b and bbtNEMO. Cells were collected, lysed, and then incubated with primary antibodies [4 μg of anti-FLAG M2 (Sigma–Aldrich) or 4 μg of anti-Myc (Sigma–Aldrich)] at 4 °C overnight, followed by a 3-h incubation with Protein G Plus beads (Roche). Samples were washed and prepared for Western blot analysis.
In Vitro Ubiquitination Assays.
Flag-tagged bbtNEMO, Flag-tagged Homo sapiens RIP1 (hsRIP1), Flag-tagged bbtRIP1b, HA-tagged bbtA20, HA-tagged bbtA1, HA-tagged bbtA2, and HA-tagged bbtABIN2 proteins were purified from HEK 293T cells as described in SI Materials and Methods. For in vitro ubiquitination of bbtNEMO and bbtRIP1b, ubiquitination assays were performed in 50-μL reaction volumes and contained the following components as indicated: 1.5 μg of N-terminal biotinylated Ub (Boston Biochem), 4 μL of conjugation fraction A (containing predominantly E1 and E2 enzymes; Boston Biochem), 4 μL of conjugation fraction B (containing predominantly E3 enzymes), 5 μL of 10× reaction buffer [300 mM Hepes (pH 7.2), 20 mM ATP, 50 mM MgCl2, 2 mM DTT], and 1 μg of purified Flag-tagged bbtNEMO or Flag-tagged bbtRIP1b. For in vitro bbtNEMO, bbtRIP1b, and hsRIP1 ubiquitinated by bbtA20, the reactions described above were performed, with the exception that 1 μg of purified HA-tagged bbtA20, HA-tagged bbtA1, or HA-tagged bbtA2 was used instead of conjugation fraction B. For autoubiquitination of bbtA20 and its truncated mutants, reactions including 1 μg of purified HA-tagged bbtA20, HA-tagged bbtA1, or HA-tagged bbtA2; 1.5 μg of N-terminal biotinylated Ub; 4 μL of conjugation fraction A; 4 μL of conjugation fraction B; and 5 μL of 10× reaction buffer were performed. Reactions were incubated at 30 °C for 1 h with agitation by Mini LabRoller (Labnet) and subsequently prepared for immunoblot analysis.
Further details regarding genomic analysis, animals and reagents, gene cloning and vector construction, section in situ analysis, acute immune challenge and RT-PCR analysis, immunofluorescence imaging, luciferase reporter assays, co-IP assays, protein purifications and FLAG or HA elutions, in vitro deubiquitination assays, and ubiquitin chain-binding and pull-down assays can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Project 2013CB835303 of the National Basic Research Program (973) and Project 2012AA092281 of the State High-Tech Development Project (863), as well as Key Project 30730089 and Project 31270018 of the National Nature Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.B. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KF006935 (bbtABIN1), KF006936 (bbtABIN2), and KF006937 (bbtA20)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321187111/-/DCSupplemental.
References
- 1.Liu YC, Penninger J, Karin M. Immunity by ubiquitylation: A reversible process of modification. Nat Rev Immunol. 2005;5(12):941–952. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7(8):758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8(7):501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma A, Malynn BA. A20: Linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12(11):774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289(5488):2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5(10):1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 7.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 8.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327(5969):1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145(7):1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verstrepen L, Carpentier I, Verhelst K, Beyaert R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem Pharmacol. 2009;78(2):105–114. doi: 10.1016/j.bcp.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Mauro C, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281(27):18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 12.Liu WK, et al. The inhibitor ABIN-2 disrupts the interaction of receptor-interacting protein with the kinase subunit IKKgamma to block activation of the transcription factor NF-kappaB and potentiate apoptosis. Biochem J. 2004;378(Pt 3):867–876. doi: 10.1042/BJ20031736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wullaert A, et al. LIND/ABIN-3 is a novel lipopolysaccharide-inducible inhibitor of NF-kappaB activation. J Biol Chem. 2007;282(1):81–90. doi: 10.1074/jbc.M607481200. [DOI] [PubMed] [Google Scholar]
- 14.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7(11):862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 15.Paquette N, et al. Caspase-mediated cleavage, IAP binding, and ubiquitination: Linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37(2):172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R, et al. The role of ubiquitination in Drosophila innate immunity. J Biol Chem. 2005;280(40):34048–34055. doi: 10.1074/jbc.M506655200. [DOI] [PubMed] [Google Scholar]
- 17.Thevenon D, et al. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6(4):309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Xue L, et al. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Wagner S, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27(26):3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 20.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S, et al. The archaic roles of the amphioxus NF-κB/IκB complex in innate immune responses. J Immunol. 2013;191(3):1220–1230. doi: 10.4049/jimmunol.1203527. [DOI] [PubMed] [Google Scholar]
- 22.Li J, et al. Functional conservation and innovation of amphioxus RIP1-mediated signaling in cell fate determination. J Immunol. 2011;187(8):3962–3971. doi: 10.4049/jimmunol.1100816. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16(8):1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shembade N, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9(3):254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 25.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 2009;28(5):513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papoutsopoulou S, et al. ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat Immunol. 2006;7(6):606–615. doi: 10.1038/ni1334. [DOI] [PubMed] [Google Scholar]
- 27.Caster DJ, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. J Am Soc Nephrol. 2013;24(11):1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208(6):1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, et al. A20-binding inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein β activation and protects from inflammatory disease. Proc Natl Acad Sci USA. 2011;108(44):E998–E1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457(7231):906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.