Significance
UV damage in genomic DNA is identified by the human UV-damaged DNA-binding protein (UV-DDB). Recognition of DNA damage by UV-DDB serves to initiate global genomic nucleotide excision repair (NER) in humans. Recent work has revealed that UV-DDB dimerizes at sites of damage. This study demonstrates that prior to stable damage recognition, UV-DDB interrogates DNA for damage via a 3D diffusion mechanism coupled to the formation of multiple transient intermediates. Stable binding at sites of damage is achieved by dimerization of UV-DDB. This study also analyzed a disease-causing mutant of UV-DDB, which was found to slide on DNA, while retaining the ability to dimerize on DNA. These results enhance our understanding of damage recognition in NER in humans.
Keywords: DNA damage recognition, single-molecule tracking, DNA tightrope, human nucleotide excision repair
Abstract
How human DNA repair proteins survey the genome for UV-induced photoproducts remains a poorly understood aspect of the initial damage recognition step in nucleotide excision repair (NER). To understand this process, we performed single-molecule experiments, which revealed that the human UV-damaged DNA-binding protein (UV-DDB) performs a 3D search mechanism and displays a remarkable heterogeneity in the kinetics of damage recognition. Our results indicate that UV-DDB examines sites on DNA in discrete steps before forming long-lived, nonmotile UV-DDB dimers (DDB1-DDB2)2 at sites of damage. Analysis of the rates of dissociation for the transient binding molecules on both undamaged and damaged DNA show multiple dwell times over three orders of magnitude: 0.3–0.8, 8.1, and 113–126 s. These intermediate states are believed to represent discrete UV-DDB conformers on the trajectory to stable damage detection. DNA damage promoted the formation of highly stable dimers lasting for at least 15 min. The xeroderma pigmentosum group E (XP-E) causing K244E mutant of DDB2 found in patient XP82TO, supported UV-DDB dimerization but was found to slide on DNA and failed to stably engage lesions. These findings provide molecular insight into the loss of damage discrimination observed in this XP-E patient. This study proposes that UV-DDB recognizes lesions via multiple kinetic intermediates, through a conformational proofreading mechanism.
Unrepaired photoproducts in the genome arising from exposure to UV irradiation can be highly mutagenic and three pathways have evolved in mammalian cells to process these lesions, which include (i) global genomic repair, (ii) transcription-coupled repair, and (iii) translesion synthesis (1–5). During global genomic repair, cyclobutane pyrimidine dimers (CPDs) and pyrimidine(6–4)pyrimidone photoproducts [(6–4) photoproducts] are repaired by the nucleotide excision repair (NER) pathway that recognizes and excises bulky helix distorting lesions in the genome (6, 7). The recognition of CPD lesions in UV-damaged chromatin is mediated by UV-damaged DNA-binding protein (UV-DDB), composed of the tightly associated heterodimer of damage-specific DNA binding protein (DDB) 1 (p127) and DDB2 (p48) (5, 8). Following surveillance and CPD identification by UV-DDB, NER proceeds via lesion handover to XPC–hHR23B–centrin2 (XPC) followed by damage verification, helix opening and stabilizing of the repair intermediates, dual incision of the DNA in the context of the lesion, repair synthesis, and DNA ligation (7). In contrast to global genomic repair, transcription-coupled repair is initiated when CPD lesions in transcribed chromatin cause stalling of RNA polymerases (3). In mammalian NER, these two pathways converge after damage detection and are orchestrated by over 30 different gene products (9). Deficiencies in the molecular functions in seven of these NER proteins lead to various forms of the autosomal recessive disorder termed xeroderma pigmentosum (XP) (10). Finally, unrepaired, CPDs can be bypassed during DNA replication by specialized DNA polymerases, such as DNA polymerase η (pol η) (2). Mutations in the gene encoding pol η give rise to the eighth complementation group of XP, the XP variant phenotype.
Molecular defects in DDB2 lead to a slower loss of UV-induced photoproducts and presentation of the skin cancer prone XP complementation group E (XP-E) (11, 12). Recombinant DDB2 has been demonstrated to bind a variety of DNA structures including 6–4 photoproducts, abasic sites, and two base mismatches with remarkably high affinity and CPD lesions and cisplatin adducts with relatively lower affinity (13–15). Molecular analysis of XP-E patients revealed genetic defects in the DDB2 gene, which give rise to truncations, misfolding, or a modification of the DNA-binding interface of DDB2 (11, 12). In the case of the XP82TO patient, a lysine-to-glutamate point mutation at position 244 (K244E) was observed in DDB2, which results in significantly reduced DNA-binding activity and specificity for damage (11, 12).
In vivo, UV-DDB is constitutively associated with Cullin4A or 4B and RBX1, forming the CRL4DDB2 E3 ligase complex (16–18). In this complex, DDB2 is a DNA damage-recognition factor and functions as an adapter protein which targets the E3 ligase activity to sites of UV-induced photoproducts, promoting chromatin relaxation, and enabling access to subsequent repair factors (14, 18–20). A current working model for damage recognition in global genomic repair is that UV photoproducts are first recognized by UV-DDB (18, 21, 22). Stable binding of UV-DDB to sites of damage activates the ubiquitination activity of CRL4DDB2, which targets histones, primarily H2A, and enables nucleosome disassembly and subsequent recruitment of the XPC complex, which is also a ubiquitination substrate (18–24). Lesion handover between CRL4DDB2 and XPC is thought to be achieved by the autoubiquitination of DDB2 at lysines in the intrinsically disordered N terminus of DDB2 (18, 21). This modification of DDB2 serves to flag the repair factor for degradation (25).
We recently provided crystallographic and biophysical evidence for the dimerization of UV-DDB at sites of damage (26). The identification of this dimeric UV-DDB complex on DNA reveals a previously unanticipated complexity in damage recognition and raises several important questions in the initial damage recognition step of human GG-NER. In this study, we sought to answer: How do ∼180,000 molecules of UV-DDB (14) scan 3.2 billion bp of genomic DNA to find relatively rare lesions in DNA? How does UV-DDB interrogate the DNA to achieve remarkable specificity in damage discrimination? How does dimeric UV-DDB modulate the specificity of damage discrimination? How do mutations in the DNA-binding interface found in the K244E mutant of DDB2 influence the kinetics of DNA binding and damage recognition?
To better understand damage recognition by UV-DDB, we used a single-molecule DNA tightrope assay (27–30) to observe the real time interactions of quantum dot (QD)-conjugated wild-type (WT) UV-DDB or UV-DDB containing the K244E mutation in DDB2, with damaged DNA substrates with high temporal and spatial resolution. Observations of individual molecules reveal the presence of short-lived intermediates and heterogeneity in molecular properties that may be lost due to bulk averaging of the properties of an unsynchronized ensemble of molecules. We found that WT UV-DDB performs a 3D search to locate UV damage in DNA, whereas UV-DDB containing the K244E mutation in DDB2 slides on DNA. Unexpectedly, we identified multiple kinetic intermediates that participate in a complex kinetic cascade of damage recognition by WT UV-DDB. Here, we propose a working model wherein UV-DDB conformationally proofreads (31) DNA and uses dimerization as a strategy to enhance specificity of the damage recognition process.
Results
Visualizing the DNA Damage Search Mechanism of UV-DDB.
UV-DDB could use a number of different approaches to find DNA damage (SI Materials and Methods, section 1.1 and Fig. S1A), as reviewed in ref. 32, and single-molecule methods have been used to experimentally validate and visualize various protein search strategies (27, 28, 33–35). We have previously developed a DNA tightrope assay that enables the direct visualization of dynamics of QD-conjugated proteins on DNA (27–30). Briefly, in this assay, λ-DNA tightropes are strung-up between 5-μm poly-l-lysine–coated beads, which are deposited on a PEGylated coverslip (Fig. 1 A and B). Biomolecular interactions on DNA tightropes in the absence of buffer flow and surface interactions are visualized by oblique-angle fluorescence microscopy imaging (Fig. 1A). A schematic of the flow cell under oblique-angle illumination is shown in Fig. 1A and a YOYO-1–stained image of DNA obtained using oblique-angle fluorescence microscopy is shown in Fig. 1C.
Fig. 1.
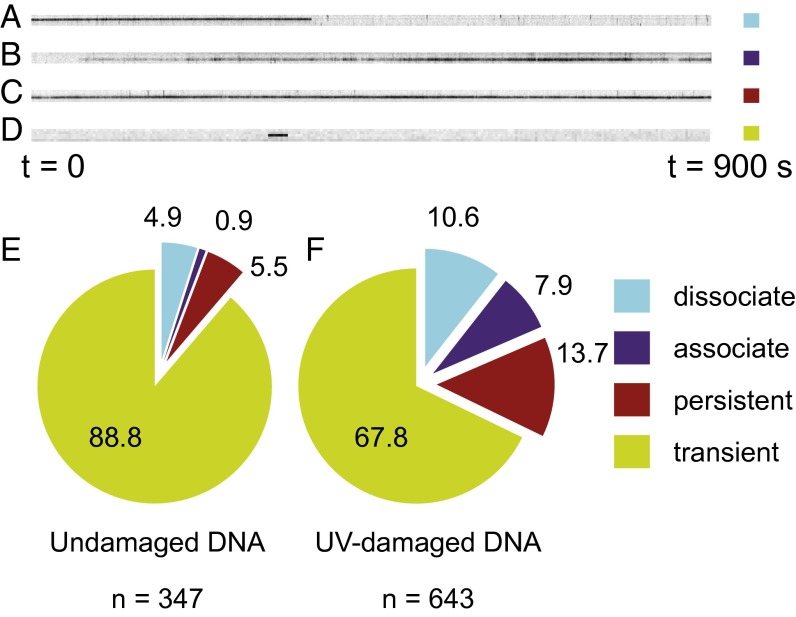
(A) Schematic of flow cell and microscope setup used in the DNA tightrope assay with oblique-angle illumination. (B) Schematic of 5-μm poly-l-lysine–coated beads deposited on a PEGylated glass surface with DNA tightropes elongated between them. (C) Oblique-angle fluorescence microscopy image of YOYO-1–stained λ-DNA tightropes (arrows) between poly-l-lysine–coated beads. Arrows indicate DNA between beads. (D) Image of QD UV-DDB binding to unstained DNA tightropes between beads (shown by green masks). Arrows indicate bound QD UV-DDB (Fig. S1 and Movie S1).
QD UV-DDB Performs a 3D Search on Undamaged as Well as UV-Damaged DNA.
To observe the interactions of UV-DDB with DNA, we conjugated UV-DDB to streptavidin-coated QDs (SA-QDs). These nanoparticles provide superior brightness and resistance to photobleaching compared with conventional fluorophores and fluorescent proteins (36, 37). To that end, we systematically tested three strategies for conjugating QDs to UV-DDB and proceeded with a His-Ab conjugation strategy that enabled us to conjugate UV-DDB to SA-QDs using a biotinylated penta-His antibody, while retaining DNA damage binding activity (SI Materials and Methods, section 1.2.3) (29). We incubated QD UV-DDB with undamaged λ-DNA tightropes or λ-DNA containing on average one lesion per 2,200 bp (SI Materials and Methods, section 2) and observed these interactions in a time window of 900 s. Imaging was performed in the absence of YOYO-1 dye to minimize potential double-strand breaks in the DNA tightropes (SI Materials and Methods, section 1.2.4) (38). We observed the binding of UV-DDB to DNA (Fig. 1D; undamaged DNA, Movie S1; UV-damaged DNA, Movie S2). For both undamaged and UV-damaged DNA, four classes of binding events were identified in a typical 900-s observation window (Fig. 2 A–D). These included molecules that (i) are present at the start of observation, but dissociate during observation (“dissociate” in Fig. 2A); (ii) associate during observation and are present at the end of observation (“associate” in Fig. 2B); (iii) are present both at the start and end of observation (“persistent” in Fig. 2C); and (iv) both associated and dissociated during observation (“transient” in Fig. 2D). In the majority of cases (>98%, n = 990 events), upon incubation with either undamaged or UV-damaged DNA, QD UV-DDB molecules associated with the DNA and did not show any sliding behavior (Movies S1 and S2) within the limits of our spatial and temporal resolutions (36 nm, ∼100 bp, 100 ms; SI Materials and Methods, section 1.3). Taken together, these data indicate that UV-DDB probes for DNA damage using a 3D search mechanism. Because these assays were performed in the absence of flow, we were able to observe macroscopic dissociation and rapid reassociation behavior of UV-DDB on separate DNA molecules, a phenomenon that is consistent with “jumping” (Fig. S2B and Movie S1), which we have reported previously for bacterial UvrA (28).
Fig. 2.
Kymographs of individual QD UV-DDB molecules that (A) dissociate from DNA during observation, (B) associate with DNA during observation, (C) are present during the entire observation window, and (D) both associates and dissociates during observation (transients). (E and F) Pie chart of percentage of each of the observations described above for undamaged DNA (n = 347) and UV-damaged DNA (n = 643), respectively (Fig. S2 and Movies S1 and S2).
UV Radiation-Induced DNA Photoproducts Shift the Binding Equilibrium to Longer-Lived States.
The presence of UV-induced photoproducts at a density of about one lesion per 2.2 kb of λ-DNA alters and greatly increases the binding of UV-DDB molecules (Fig. 2E, n = 347 for nondamaged DNA and n = 643 for damaged DNA). For example, UV damage caused a 2.5-fold increase in the molecules that persisted for all 900 s of observation time (Fig. 2 E and F). Furthermore, UV-induced damage caused an eightfold increase in UV-DDB molecules that associated onto the DNA during this 900-s observation window (Fig. 2 E and F). As expected, DNA damage decreases the percentage of transient UV-DDB molecules (Fig. 2D, those that bound and then dissociated) on UV-irradiated λ-DNA molecules (compare 67.8% vs. 88.8% for nondamaged DNA which was not UV irradiated, respectively, Fig. 2 E and F). These data reveal that across all these classes of binding events, UV-DDB associates with and persists on DNA for longer time periods when the DNA tightropes contain UV damage while showing lower transient behavior.
Single-Molecule Visualization Reveals Multiple Kinetic Intermediates of UV-DDB.
To further understand the heterogeneity in dwell times and to extract rate constants, we analyzed the kinetics of transient binding on both undamaged and UV-damaged DNA. Dissociation of UV-DDB complexes on DNA can be modeled as a first-order decay (Poisson) process from the DNA-bound intermediate state to the DNA-free state (SI Materials and Methods, section 3.1.1). With the objective of quantifying this process, we performed a cumulative residence time distribution (CRTD) analysis (SI Materials and Methods, section 3.1.2) (39). The CRTD may be interpreted as a type of survival curve representing the fraction of the population of DNA-bound proteins remaining on DNA as a function of time. Fitting the CRTD to a Poisson process (T ≡ exp{−kdt}) yields the dissociation rate constant (kd) and consequently the mean lifetime (τ = kd−1) of particles dissociating from the DNA. For systems comprised of multiple intermediates, the number of terms fit to the CRTD reveals the number of measurable intermediates (SI Materials and Methods, section 3.1.2 and Table S1).
The CRTDs describing the data for transient binding (Fig. 2D) to undamaged DNA and damaged DNA are presented in Fig. 3 A and B, respectively. Binding events that persisted for the entire 900-s observation window were necessarily excluded from this analysis. Examination of these CRTDs on undamaged DNA revealed the presence of three kinetic intermediates with mean lifetimes over three orders of magnitude (0.8, 8.1, and 113 s; Table 1). Surprisingly, we detected similar kinetics for transiently bound UV-DDB on UV-damaged DNA. However, as described below, we found that UV damage enriched the population of the longest-lived intermediates on DNA indicating that more UV-DDB bound with higher affinity on DNA containing UV damage. We found that all of the features of each of the CRTDs were best described when three exponentials terms corresponding to three decay processes were used (SI Materials and Methods, sections 3.1.1, 3.1.2, and 3.2; log-log representation Fig. S3 A and B; log-linear representation Fig. S3 C and D). We denote these three independent Poisson processes that describe the dissociation kinetics of transiently bound UV-DDB as T1,ud, T2,ud, and T3,ud (Table 1 and SI Materials and Methods, sections 3.1.1, 3.1.2, and 3.2). Analysis of the dwell times, τ, of UV-DDB on UV-damaged DNA also revealed three kinetic intermediates which dissociate from DNA according to three Poisson processes: T1,d, T2,d, and T3,d (Table 1 and SI Materials and Methods, sections 3.1.1, 3.1.2, and 3.2). It is important to note that the detection of decay processes in the CRTD analysis of transient molecules is limited by the time resolution, size of the time window of acquisition, and the probability of occurrence of the decay process. Given that the time resolution of acquisition (0.1 s) is of the order of the mean lifetime of UV-DDB molecules dissociating according to the T1 process (0.3s, 0.8s), the apparent differences between T1,ud and T1,d processes may be biologically indistinguishable. It is noteworthy that the T1 process identified here does not correspond to nonproductive collisions that are found to occur on an ∼100× faster time scale (∼5 ms) (35).
Fig. 3.
CRTD plots for transient UV-DDB (Fig. 2D) dissociating from (A) undamaged DNA and (B) UV-damaged DNA with experimental data in black circles and triple exponential fits (red). (C and D) Pie chart showing the percentage of molecules participating in each of the three Poisson processes (Table 1) observed in the transient binding events (Fig. 2D) observed for binding to undamaged DNA (n = 307) or UV-damaged DNA (n = 436), respectively. (E) Estimated survival functions for dissociating molecules (Fig. 2A) from undamaged DNA (blue, n = 16) and UV-damaged DNA (red, n = 67) (Fig. S3 and Table 2).
Table 1.
Estimates for the amplitudes and dissociation rate constants for transient molecules
| Analysis of transient molecules | ||||||||||
| T1 | T2 | T3 | ||||||||
| DNA type | N | a1 | kd,1, s−1 | τ1, s | a2 | kd,2, s−1 | τ2, s | a3 | kd,3, s−1 | τ3, s |
| Undamaged DNA | 307 | 0.79 | 1.22 | 0.8 | 0.39 | 1.23 × 10−1 | 8.1 | 0.07 | 8.85 × 10−3 | 113.0 |
| Lower bound of 95% CI | 0.76 | 1.13 | 0.8 | 0.37 | 1.07 × 10−1 | 7.2 | 0.05 | 4.50 × 10−3 | 75.8 | |
| Upper bound of 95% CI | 0.81 | 1.31 | 0.9 | 0.41 | 1.40 × 10−1 | 9.3 | 0.09 | 1.32 × 10−2 | 222.1 | |
| Damaged DNA | 436 | 1.11 | 3.03 | 0.3 | 0.25 | 1.25 × 10−1 | 8.0 | 0.21 | 7.89 × 10−3 | 126.7 |
| Lower bound of 95% CI | 1.03 | 2.78 | 0.3 | 0.24 | 1.09 × 10−1 | 7.1 | 0.20 | 7.13 × 10−3 | 115.4 | |
| Upper bound of 95% CI | 1.19 | 3.28 | 0.4 | 0.27 | 1.41 × 10−1 | 9.2 | 0.22 | 8.66 × 10−3 | 140.4 | |
N represents the total number of observed counts. ai represent the coefficient of the exponential terms obtained from the fits. kd,i represent the dissociation rate constants and τi represent the mean lifetimes. The lower and upper bounds corresponding to the 95% confidence intervals (CI) are presented for the population of transient molecules observed in this study.
To better understand how the presence of photoproducts in the DNA changes the extent of UV-DDB binding, we first calculated the fraction of the population dissociating from each DNA substrate according to the decay processes identified above (Table 1). We then compared the relative fractions of the three kinetic intermediates represented on each DNA substrate (Fig. 3 C and D and SI Materials and Methods, section 3.3). In the population of molecules bound to undamaged DNA, 58% were found to dissociate with a lifetime of τ1,ud (0.8 s) compared with 57% for UV-damaged DNA with a lifetime of τ1,d (0.3 s). Similarly, 35% were found to dissociate from undamaged DNA compared with 23% for UV-damaged DNA (1.5-fold higher) with a lifetime of τ2 (8 s) and 7% of the molecules dissociated with a lifetime of τ3 (120 s) compared with 20% for binding to UV-damaged DNA (2.9-fold higher; Fig. 3 C and D). These data indicate that a greater fraction of UV-DDB dissociates according to the T2 process on undamaged DNA compared with that on UV-damaged DNA. This suggests that the T2 process is a damage verification process which facilitates the dissociation of weakly bound UV-DDB molecules on DNA.
These data also indicate that the fraction of molecules that do not dissociate according to the T2 process, participate in a much slower, T3 process. We found that a 2.9-fold greater fraction of UV-DDB dissociates, according to the T3 process, from UV-damaged DNA compared with undamaged DNA. These results indicate that the T3 process represents the slow dissociation of a tightly bound kinetic intermediate of UV-DDB, which is represented to a greater extent on UV-damaged DNA.
Further evidence for the presence of long-lived complexes was the class of observations that are present at the beginning of data acquisition and dissociate during observation (Fig. 2A). We performed a CRTD analysis for this class of particles and obtained the survival curves presented in Fig. 3E for undamaged DNA (n = 16, blue) and damaged DNA (n = 67, red). We found that this population dissociated from undamaged λ-DNA according to the T2 process (τ2,ud = 11.7 s) and a distinct T4 process (τ4,ud = 446.2 s; Table 2 and SI Materials and Methods, section 3.5). Additionally, we found that UV-DDB also dissociated from UV-damaged λ-DNA according to a T4 process with τ4,d = 336.7 s (Table 2 and SI Materials and Methods, section 3.5).
Table 2.
Estimates for the amplitudes and dissociation rate constants for dissociating molecules
| Analysis of dissociating molecules | ||||||||
| T4 | T2 | |||||||
| DNA type | N | a4 | kd,4, s−1 | τ4, s | N | a2 | kd,2, s−1 | τ2, s |
| Undamaged DNA | 11 | 0.71 | 2.24 × 10−3 | 446.2 | 5 | 0.42 | 0.85 | 11.7 |
| Lower bound of 95% CI | 0.62 | 1.78 × 10−3 | 370 | 0.28 | 0.01 | 6.5 | ||
| Upper bound of 95% CI | 0.79 | 2.70 × 10−3 | 562.1 | 0.60 | 0.15 | 63.3 | ||
| Damaged DNA | 67 | 1.02 | 2.97 × 10−3 | 336.7 | ||||
| Lower bound of 95% CI | 0.99 | 2.79 × 10−3 | 317.5 | |||||
| Upper bound of 95% CI | 1.06 | 3.15 × 10−3 | 358.6 | |||||
N represents the total number of observed counts. ai represent the coefficient of the exponential terms obtained from the fits. kd,i represent the dissociation rate constants and τi represent the mean lifetimes. The lower and upper bounds corresponding to the 95% CIs are presented for the population of dissociating molecules observed in this study.
WT UV-DDB Is Persistent on UV-Damaged DNA and Slides at High Ionic Strength.
As mentioned above, we detected a population of molecules that persisted during the entire observation window of 900 s (Fig. 2C and Movie S2), with some individual molecules persisting for up to 90 min. Importantly, these persistent molecules represent a distinct, stable complex that does not dissociate according to any of the T1, T2, T3, or T4 processes described here (SI Materials and Methods, section 4.1). UV damage caused a 2.5-fold increase in these persistent UV-DDB molecules. Furthermore, these persistent molecules exhibited a salt-dependent mobility and are not irreversible aggregates of UV-DDB on DNA (SI Materials and Methods, section 4.2, Fig. S4, and Movie S3).
UV-DDB Colocalizes and Is Persistent at Sites of Lesions.
We posited that long-lived UV-DDB molecules on UV-damaged λ-DNA represented UV-DDB bound to sites of photoproducts. Because UV-DDB has previously been demonstrated to bind DNA containing an abasic site (AP site) analog with high affinity (14), we engineered long DNA substrates containing defined AP sites every 2 kb, for use in the DNA tightrope assay (SI Materials and Methods, section 5.1) (40). To identify the site of the lesion in these long DNA substrates, we engineered a biotin modification near the AP site. This enabled us to mark the site of the lesion using SA-QDs and test the hypothesis that UV-DDB is long-lived at sites of lesions in these DNA damage arrays. Upon incubation with SA-QDs, we were able to observe QD arrays on these substrates marking sites of introduced biotins in the proximity of the AP sites (APbiodT) (Fig. 4A). The pair-wise inter-QD distances were consistent with integral multiples of the linearized plasmid length of 0.65 μm (Fig. 4B). This distribution reflects the probability of occurrence of pairs of QDs with discrete numbers of plasmid lengths between them. This skewed distribution arises from the distribution of DNA substrate lengths, as well as the lower number of pairs of QDs with large number of plasmid lengths between them on any given DNA molecule. To investigate whether long-lived UV-DDB molecules bound DNA at sites of DNA damage, we performed dual-color experiments involving the incubation of QD UV-DDB conjugates with QD-conjugated APbiodT DNA tightropes. We found long-lived UV-DDB molecules that colocalized to sites of damage (n = 22; Fig. 4C, Movie S4, and SI Materials and Methods, section 5.2) and persisted through the entire observation window. Significantly, this general method of creating long DNA substrates with site-specific modifications provides new opportunities for studying site-specific interactions of DNA repair proteins in the DNA tightrope platform.
Fig. 4.
(A) Oblique-angle fluorescence microscope image of array of QDs on a DNA tightrope of a long DNA molecule containing a defined abasic site analog with a proximal biotin marking the site of the lesion. (Scale bar: 1 μm.) (B) Histogram of pairwise distances between QDs marking sites of the introduced lesion. (C) Kymograph of UV-DDB (red) colocalized to sites of defined lesion (green) (one pixel = 46 nm along y axis) (Movie S4). Here, data in the red channel was collected at 10 fps whereas data in the green channel was collected at 0.3 fps. (D and E) Dual-color kymographs of the UV-DDB dimer on UV-damaged DNA (Movie S5) and APbiodT substrate, respectively (one pixel = 46 nm along the y axis) (Fig. S5 and Movies S4 and S5).
Dimeric UV-DDB Is Persistent at Sites of Damage.
Previously we identified that UV-DDB dimerizes on DNA (26). We next probed whether the long-lived intermediates detected in this study represented dimeric UV-DDB. To address the nature of UV-DDB stoichiometry, we incubated UV-DDB molecules which had been separately labeled with two differently colored QDs, together with UV-damaged DNA tightropes. We observed colocalization of both colors representing dimerization of WT UV-DDB (Movie S5 and Fig. 4D). This colocalization did not exhibit a dependence on the choice of QD conjugate. In addition, we found that 13 (72%) of 18 observations of dual-colored dimeric UV-DDB complexes (compared with a total of 47 single-colored persistent UV-DDB) persisted during the entire observation window of 900 s, indicating that UV-DDB dimers are long-lived on UV-damaged DNA. Additionally, long-lived UV-DDB dimers were also observed on APbiodT substrates (n = 18 dual-colored complexes and n = 67 single-colored persistent UV-DDB; Fig. 4E). Control EMSA experiments revealed that UV-DDB binds to biodT containing substrates essentially the same as undamaged DNA substrates (SI Materials and Methods, section 5.3 and Fig. S5C), confirming that the colocalization observed in these experiments indeed reflects dimeric UV-DDB at abasic sites in the DNA.
UV-DDB Containing the K244E Mutant of DDB2 Dimerizes and Slides on DNA.
To gain insight into the structural nature of the complex binding kinetics of WT UV-DDB, we turned our attention to an XP-causing mutant of UV-DDB containing the K244E mutation in DDB2 [UV-DDB (K244E)] found in the XP82TO patient. This UV-DDB (K244E) variant contains a mutation in a crucial DNA-binding residue in DDB2, which greatly reduces its affinity for DNA and its specificity for damage (14, 24). We probed the DNA-binding ability of recombinant UV-DDB (K244E) in a pull-down experiment (see SI Materials and Methods, section 6.1 for details) and found that, consistent with a previous report (24), UV-DDB (K244E) lacks the ability to discriminate UV-induced damage in DNA while retaining strong end binding (SI Materials and Methods, section 6.1 and Fig. S6A). This finding was further confirmed by atomic force microscopy (AFM) experiments in which UV-DDB (K244E) was incubated with undamaged DNA (Fig. 5 A and B and SI Materials and Methods, section 6.2). UV-DDB (K244E) possesses residual DNA-binding activity mostly at DNA ends, exhibiting a threefold preference for binding to two DNA molecules over a single DNA molecule (SI Materials and Methods, section 6.2).
Fig. 5.
AFM images of UV-DDB (K244E) bound to (A) 517-bp DNA, or (B) plasmid DNA demonstrating that UV-DDB (K244E) dimerizes on DNA, 300 nm × 300 nm × 2 nm. (C) Kymograph of K244E sliding on DNA. (Vertical scale bar: 1 μm; horizontal scale bar: 5 s.) (D) Plot of anomalous diffusive exponent vs. diffusivity for sliding UV-DDB (K244E) (Movie S6). (E) Histogram of UV-DDB (K244E) volumes on DNA (n = 171) (Fig. S6 and Movie S6).
To obtain dynamic information describing the binding of UV-DDB (K244E) to DNA, we incubated QD UV-DDB (K244E) with undamaged λ-DNA in the DNA tightrope assay. We found that overall binding of UV-DDB (K244E) was diminished and in stark contrast to the WT UV-DDB, 80% (79 of 99 observations) of UV-DDB (K244E) molecules exhibited sliding behavior. Next we wanted to identify whether UV-DDB (K244E) was able to stably colocalize to sites of damage. We incubated the mutant protein with DNA tightropes containing defined abasic site analog lesions (APbiodT) and found that UV-DDB (K244E) was also found to slide on APbiodT DNA without exhibiting obvious long-lived pausing behavior (Movie S6 and Fig. 5C). This finding is consistent with the findings from the pull-down experiment which indicate that UV-DDB (K244E) lacks the ability to stably associate with DNA damage (Fig. S6A). Of the sliding UV-DDB (K244E) molecules, we calculated diffusion constants for molecules that were found to slide on DNA for an observation window of at least 60 s. Sliding QD UV-DDB (K244E) exhibited heterogeneity in its diffusive behavior spanning three orders of magnitude (Fig. 5D and SI Materials and Methods, section 6.3). Importantly, the diffusive behavior of UV-DDB (K244E) on DNA (both undamaged λ-DNA and APbiodT) was found to have a mean diffusion constant of 0.11 ± 0.2 μm2⋅s−1 (mean ± SD), with an anomalous diffusive exponent (α) of 0.5 ± 0.22 (mean ± SD) (n = 31, SI Materials and Methods, section 6.3).
Unexpectedly, the DNA tightrope assay provided further insight into the interaction of UV-DDB (K244E) with DNA. We identified rare events that suggest that dimerization of UV-DDB (K244E) is DNA dependent and proceeds via random collisions of UV-DDB (K244E) molecules on DNA (Movie S7 and Fig. S6C). This observation prompted us to examine the stoichiometry of UV-DDB (K244E) bound to DNA. We have previously used AFM to identify the stoichiometry of WT UV-DDB bound to DNA using a calibration curve relating the AFM volume of the complex to its molecular weight (SI Materials and Methods, section 6.5 and Fig. S6D) (26). Volume analysis of DNA-bound UV-DDB (K244E) revealed a peak at 564.3 ± 10.1 nm3 corresponding to a molecular weight of 388.6 ± 11.8 kDa (mean ± SD), consistent with that of dimeric UV-DDB (K244E) bound to DNA (n = 171, Fig. 5E). These data demonstrate that a mutation in the DNA-binding interface of DDB2 does not inhibit dimerization of UV-DDB. However, in contrast to WT UV-DDB (26), dimeric UV-DDB (K244E) is more likely to be stabilized on two DNA molecules in a damage-independent manner (SI Materials and Methods, section 6.2). In summary, our study indicates that UV-DDB (K244E) retains the ability to dimerize on DNA but lacks specificity for lesions and consequently slides on DNA.
Discussion
In this study, we used single-molecule techniques to understand the kinetics of damage recognition by UV-DDB in the initial step of NER. We identified that UV-DDB consisting of DDB1 and DDB2 performs a 3D search for damage sites in DNA. Using DNA substrates containing UV-induced photoproducts or AP sites, we discovered a complex kinetic pathway of damage recognition by UV-DDB that culminates in the formation of long-lived dimers of UV-DDB [(DDB1-DDB2)2] at sites of damage. Specifically, we found that UV photoproducts or abasic sites (i) increased the total number of stably bound UV-DDB molecules, (ii) decreased the number of transient UV-DDB molecules that associate and dissociate from DNA, (iii) increased the number of UV-DDB molecules that associate and persist on DNA, and (iv) substantially increased the number of persistent dimers of UV-DDB on DNA. Additionally, we studied the stoichiometry and dynamics of the XP causing K244E mutant of DDB2 on DNA and identified that UV-DDB (K244E) dimerizes and slides on DNA but does not stably associate to damaged sites.
Damage Recognition Is a Multistep Kinetic Cascade Culminating in UV-DDB Dimerization.
The use of our single-molecule tightrope platform permitted the observation of previously undetected kinetic intermediates in the process of damage recognition by UV-DDB. We found evidence for five, progressively stable kinetic intermediates, four of which were transient and the fifth was found to be persistent during the 900-s observation window (SI Materials and Methods, section 3.4). Some of the slower decay processes detected here are consistent with previously available bulk estimates (SI Materials and Methods, section 7) and likely represent dissociation of bound UV-DDB from lesions in DNA (15, 17, 26). Likewise, some of the long-lived intermediates on undamaged DNA probably represent UV-DDB bound to spontaneous damage arising from depurination on commercially available λ-DNA, which is recognized by UV-DDB (14).
Dual-color fluorescence microscopy experiments revealed that dimeric UV-DDB persists on damaged DNA tightropes. Previous AFM experiments revealed that four of five WT UV-DDB complexes on DNA consisted of dimeric WT UV-DDB bound to a single DNA molecule (26). In contrast, 76% of dimeric UV-DDB (K244E) complexes on DNA were bound to two molecules of DNA (Fig. 5 A and B and SI Materials and Methods, section 6.2). These findings indicate that the presence of two DNA molecules in the dimeric UV-DDB DNA complex is a sufficient but not necessary requirement for dimerization.
We predict that these long-lived dimers of UV-DDB at sites of lesions could inhibit the progression of NER if these highly stable UV-DDB dimers are not actively dismantled. In support of this hypothesis, introduction of excess recombinant UV-DDB to in vitro reconstituted NER reactions resulted in inhibition of repair of (6–4) photoproducts (8, 9). In addition, in vivo studies of fluorescently tagged UV-DDB binding have reported immobile binding of UV-DDB to DNA for up to 4 h in XP-A cells (41). In a study involving siRNA knockdown of Cullin4A, fluorescently tagged UV-DDB exhibited delayed disappearance from CPD foci in HeLa cells (42). Inhibition of the proteasome using MG132 also resulted in inhibited recruitment of XPC to sites of CPD lesions in mammalian cells (43). We believe that the highly stable, persistent, dimeric UV-DDB complexes detected in our studies represent a distinct species on the kinetic pathway to recognize damage with high specificity and affinity.
Damage Recognition Involves Dynamic Conformational Changes in both UV-DDB and DNA.
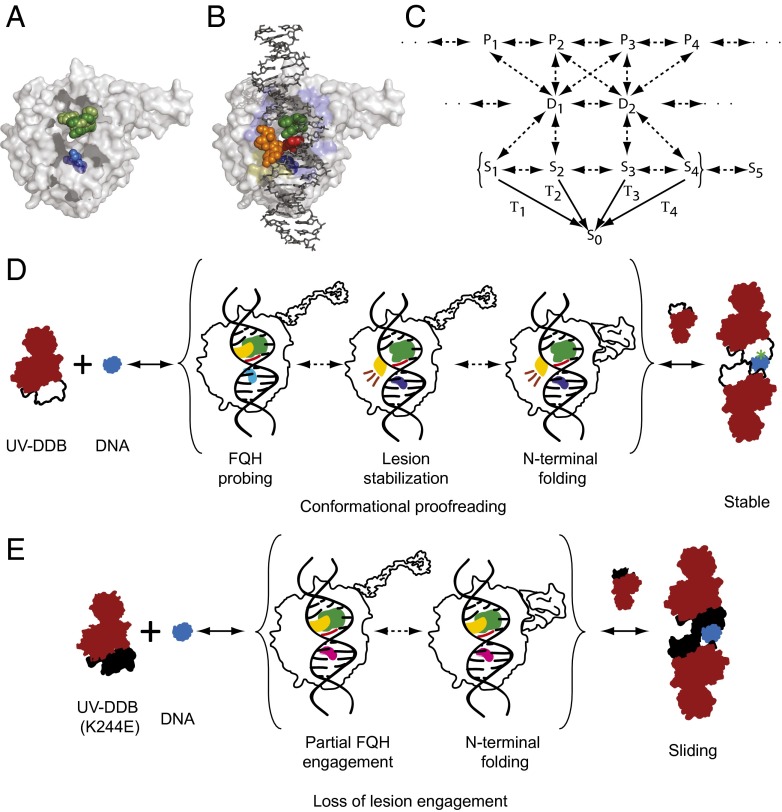
What might be the physical basis of the heterogeneity observed in the lifetimes of the repair intermediates? Crystal structures of UV-DDB in the apo and DNA damage-bound forms give insight into this question [Protein Data Bank (PDB) ID codes 3EI1 (44), 4A0A, 4A0K, (18) and 4E54 (26)]. The protein in the apo state [PDB ID code 3EI4 (18)] upon binding to damaged DNA undergoes an FQH hairpin (F334-Q335-H336) transition which probes the major groove of the DNA for the presence of damage [PDB ID code 4E45 (26); Fig. 6 A and D and Movie S8] (18, 26, 45). At sites of damage, this conformational change in DDB2 is accompanied by the base flipping of the damaged bases in DNA (consisting of the photoproducts in the case of UV damage, alternatively the abasic site and the adjacent 3′ base; Fig. 6 B and D) to an extrahelical conformation and stabilization in the lesion binding pocket of DDB2 (PDB ID codes 3EI1, 4A0A, and 4A0K) (18, 44). Stable damage recognition is thought to induce folding of the intrinsically disordered N terminus to form an α-paddle structure, which along with the β-wing, forms a winged helix structure upon DNA binding (PDB ID code 4E54; Fig. 6D) (18, 26). Damage recognition may thus be considered to progress along a reaction coordinate that describes a series of dynamically interconverting structural intermediates. Some of the highly transient, short-lived binding intermediates observed in this work might reflect abortive attempts at damage recognition by UV-DDB. These species may correspond to metastable intermediates that participate in varying extents of lesion engagement, failing to stabilize at sites of lesions. Indeed, previous work has demonstrated that the assembly and disassembly of subunits of large macromolecular complexes such as the spliceosome proceeds via a kinetic pathway, which rejects nonproductive subcomplexes along the reaction coordinate (46).
Fig. 6.
(A) Molecular model of DDB2 showing conformations of the FQH motif (green) and K244 (blue) in the apo (light) and DNA-bound (dark) states [PDB ID codes 3EI4 (45) and 4E54 (26)]. (B) DNA-binding interface of DDB2 illustrating the pinning of the undamaged base adjacent (red) to the lesion (orange) between the FQH motif (green) and K244 (dark blue) (PDB ID code 4E54). The molecular recognition scheme presented in C shows a small, illustrative subset of interconverting protein {P} and DNA {D} conformers which form intermediates {S}, whose decay rates are measured in this work (species in brackets). (D) Model for damage recognition and conformational proofreading by WT UV-DDB where green represents the FQH hairpin, blue represents K244, and yellow represents the lesion. This model highlights some of the known conformations of UV-DDB in the apo state and bound to DNA damage. (E) Model for sliding behavior of K244 on DNA, where magenta represents the glutamate in K244E DDB2 and other colors as described above (Movie S8).
K244 Is Required for Damage Recognition.
Specific damage recognition depends on K244 switching its conformation in the apo form to the DNA-bound form (Fig. 6A). We have demonstrated that the DDB2 K244E mutant supports UV-DDB dimerization but slides on DNA and fails to stably engage lesions. This finding suggests that processing of damaged DNA is contingent upon the successful sandwiching of the undamaged base 3′ to the two damaged bases, between the FQH hairpin and K244. The dimerization of UV-DDB (K244E) observed in our AFM experiments probably occurs by rapid 3D diffusion of one UV-DDB (K244E) molecule colliding with another, the DNA-bound UV-DDB (K244E) (Movie S7). Such a dimer may form a topologically constrained complex on the DNA, which is not actively engaged in a damage detection conformation (Fig. 6E). The subdiffusive nature of the UV-DDB (K244E) sliding indicates that this complex performs a constrained Brownian walk on the DNA, suggesting that although mutant DDB2 cannot bind to DNA damage to form persistent complexes, it has several strong interactions with the DNA, such as helix probing with the FQH motif (29). This sliding state may arise from either a conformational change in the monomer of UV-DDB (K244E) that is capable of sliding, or from dimeric UV-DDB (K244E) that fails to stably engage the lesion.
Conformational Proofreading Is a Candidate Mechanism for Damage Recognition.
As evidenced from the crystal structures, both UV-DDB and the DNA undergo a series of concerted conformational changes that ensure successful damage recognition [PDB ID codes 3EI4 (45), 3EI1 (18), 4A0A, 4A0K (18), and 4E54 (26)]. This problem of damage recognition by UV-DDB falls under a category of problems in molecular recognition which use conformational changes in the target and/or the ligand to achieve highly specific recognition in a noisy environment. This mechanism termed “conformational proofreading” (31) uses a structural mismatch between the protein-binding pocket and the ligand such that binding of the correct ligand facilitates a conformational change in the protein which stabilizes the binding, whereas the incorrect ligand is unlikely to allow this conformation change and is therefore rejected. This mechanism of conformational proofreading describes specific molecular recognition in the absence of energy consumption and is an alternative to kinetic proofreading (31, 47).
A working model for damage recognition by WT UV-DDB is presented here (Fig. 6 C and D and SI Materials and Methods, section 3.4). Each of the intermediate states detected here, {Si}, can be considered to be comprised of a protein configuration {Pi} and a DNA configuration {Di} (SI Materials and Methods, section 3.4). Here, the apo protein and the DNA interconvert between ensembles of conformers {Pi} and {Di}. Collisions between the protein and DNA result in the formation of interconvertible repair intermediates {Si}, some of which are relatively stable and observable (such that their decay to the DNA-free state {S0} is measurable). We propose that during the initial stages of damage recognition by WT UV-DDB, target specificity arises from the ability of the repair intermediate to cross energy barriers between the various states, whereas additional affinity arises from the dimerization of WT UV-DDB, which then locks the repair factor to the site of damage. The formation of dimers of mutant UV-DDB (K244E) on DNA suggests that this dimerization depends on the residence time of monomeric UV-DDB on DNA, independent of its specific association with damage. The observation of sliding UV-DDB (K244E) on DNA containing lesions suggests that mutation of the critical K244 leads to an inability of the mutant DDB2 to conformationally proofread the DNA for damage.
Jumping as a Mechanism for Target Search: Implications for Search.
The role of UV-DDB in vivo is to recognize photoproducts in genomic DNA. The organization of genomic DNA into nucleosomes and other chromatin higher-order structures may occlude sites of damage and serve as a barrier to DNA damage search. How might the recognition of these lesions proceed inside living cells? Our work provides direct evidence for a 3D search mechanism for DNA damage search by UV-DDB. The interaction of a small population of UV-DDB in real time was consistent with jumping from one λ-DNA molecule to another in our single-molecule DNA tightrope assay (Movie S1). Of the total DNA-bound WT UV-DDB molecules observed, less than 2% showed perceptible linear diffusion. Importantly, because our spatial resolution is about 100 bp, we cannot rule out short-range 1D sliding (SI Materials and Methods, section 1.3). These findings are consistent with a recent report on the promoter search used by Escherichia coli RNA polymerase, which posits that target search is favored via a 3D diffusion mode over a facilitated mode of diffusion in a concentration-dependent manner (35, 48). Recently it has been demonstrated that nucleosomal core particles containing site-specific UV photoproducts exhibit spontaneous unwrapping of the DNA, providing access to damaged DNA (49). Rapid sampling of exposed DNA by a 3D search mechanism minimizes the need to overcome obstacles to 1D sliding, such as other DNA-binding proteins and higher-order chromatin organization. Confinement of UV-DDB in higher-order structures of chromatin may confer the ability to repeatedly sample nucleosomal DNA for damage. Such a 3D search mechanism coupled with short-range (<100-bp) diffusion may serve as an effective strategy to interrogate nucleosomal, as well as linker DNA in chromatin. Because UV-DDB exists in about 1.8 × 105 copies per human cell nucleus (translating to an in vivo concentration of the order of a few hundred nanomolars) (14), we propose that UV-DDB rapidly surveys the genome using 3D diffusion and proofreads the DNA for damaged bases in discrete kinetic steps in an excess of undamaged DNA. How the presence of histones and the configuration of DNA in nucleosomes affects the kinetics of binding remains to be investigated.
Our previous work revealed that full-length human UV-DDB dimerizes on DNA at sites of damage via the N terminus of DDB2 (26). Here, we demonstrated that these dimers of UV-DDB are long-lived on DNA, with residence times greater than 900 s. However, the residence of UV-DDB on DNA inside living cells is likely determined by posttranslational modifications of the N terminus of DDB2, which is involved in this dimerization. Autoubiquitination at lysines in the N terminus of DDB2 (18) may disrupt its secondary structure (26) and dismantle the highly stable UV-DDB dimer observed in this work. Recent reports have identified that the N terminus of DDB2 is also PARylated in vivo in response to UV damage, resulting in stabilization of UV-DDB on damaged chromatin (50, 51). How posttranslational modifications such as phosphorylation (52), SUMOylation (53), ubiquitylation (21), and PARylation (50, 51), as well as interacting partners such as XPC and XPA (21, 54, 55), may regulate the dimerization and lifetimes of the kinetic intermediates identified in this work need to be further investigated.
Our work reveals that even a relatively simple step of damage binding is highly regulated even in the absence of protein partners and supports the hypothesis that multiple layers of damage recognition and verification are needed before the final commitment to repair DNA is made (56). Previous work has suggested that damage verification in NER proceeds via an ATP-dependent kinetic proofreading mechanism performed by the XPD helicase in TFIIH (57, 58). Here, we propose that before kinetic proofreading by TFIIH, damage recognition by UV-DDB proceeds via the formation of multiple repair intermediates in a kinetic cascade, using a mechanism which resembles conformational proofreading (31). Further, we hypothesize that conformational proofreading is a common feature of damage recognition in the absence of energy consumption and is also used by XPC to discriminate damage. In this regard, it is interesting to note that conformational and kinetic proofreading mechanisms have been found to operate together for highly specific recognition of homologous sequences during homologous recombination (59, 60). We believe that this synergy of damage detection mechanisms is required for the successful navigation of the complex kinetic and thermodynamic landscape of DNA damage recognition, which achieves high specificity by rejecting nonoptimal repair intermediates. Future studies will help reveal whether the combination of proofreading mechanisms is a universal feature of DNA damage recognition.
Materials and Methods
Biological Reagents.
UV-DDB was purified as described previously (26). Various strategies were explored for QD conjugation and these are described in detail in SI Materials and Methods, section 1. The QD conjugation strategy described in ref. 29 was used to conjugate UV-DDB (His-DDB1/DDB2) to the penta-His–biotinylated conjugated (Qiagen; His-Ab). SA-QDs (Invitrogen) were conjugated to penta-His–biotinylated conjugated (Qiagen) in a molar ratio of 1:5 for 20 min at room temperature (RT). Following this, the His-Ab QD conjugates were incubated with UV-DDB so as to obtain a final molar ratio of UV-DDB:His-Ab:QD = 1:5:1.
DNA Tightrope Assay.
The DNA tightrope assay was performed as described before (28). Custom flow chambers were constructed essentially as described before (28). Poly-l-lysine hydrobromide [MW (molecular weight) > 300,000; Wako Pure Chemicals]-coated silica beads (5 μm; Polysciences Inc.) were deposited on a PEGylated (mPEG-succinimidyl valerate, MW 5,000; Laysan Bio, Inc.) glass coverslip in a flow chamber. Following this step, DNA substrates were elongated between beads using the protocol developed previously. Imaging was performed in either low-salt buffer [150 mM NaCl, 50 mM Hepes 7.5, 100 mM DTT, and 1 mg/mL BSA (Roche)] or high-salt buffer [1 M NaCl, 50 mM Hepes 7.5, 100 mM DTT, 1 mg/mL BSA (Roche)] with 0.8 nM of either QD UV-DDB or QD UV-DDB (K244E). Experiments on undamaged and UV-damaged DNA were performed with bacteriophage λ-DNA (New England Biolabs). UV damage was introduced into bacteriophage λ-DNA by irradiating the DNA with UV-C at a dose of 20 J⋅m−2 and was quantified using qPCR. UV-damaged DNA substrates contained on average one photoproduct in 2,200 bp of DNA (see SI Materials and Methods, section 2 for details). DNA substrates containing defined lesions were prepared by introducing a defined lesion in a plasmid as described before (40) and were linearized before tandem ligation to obtain long DNA substrates as described in the SI Materials and Methods, section 5.
Oblique-Angle Fluorescence Imaging.
Oblique-angle fluorescence imaging was performed using a Nikon Ti eclipse base with a 100× TIRF objective with 1.45 N.A. Flow cells were illuminated by a 488-nm laser. Emissions from QDs were separated using emission filters (Chroma) mentioned here: 655 nm (640/20 or 700/75), 705 nm (700LP or 700/75), 605 nm (600/50), 585 nm (600/50) and 565 nm (535/50) and 520 nm using a (520/40). Images were acquired using Nikon Elements Ar (4.11.00) with a temporal resolution of 100 ms with a laser power of 1–2 mW at the back focal plane of the objective using an Andor Neo sCMOS camera. Lifetime measurements were performed with the Qdot 655 nm streptavidin conjugate.
Data Analysis.
Movies captured with NIS-Elements Ar software were exported as a stack of Tagged Image File Format (TIFF) files. TIFF files were further analyzed using ImageJ software (http://imagej.nih.gov/ij/). Kymographs were generated for particles and lifetimes were measured from these kymographs as described previously (28). Diffusion constants were calculated from kymographs as described in SI Materials and Methods, sections 4.2 and 6.3.
Pull-Down Experiment.
Details of DNA substrates and conditions of the pull-down experiment are presented in SI Materials and Methods, section 6.1.
AFM.
Samples for AFM imaging in air were prepared by incubating 25 nM nondamaged 517-bp DNA with 50 nM UV-DDB (K244E) for 10 min at RT. Imaging conditions and deposition are described in SI Materials and Methods, section 6.4.
Supplementary Material
Acknowledgments
We thank Greg Gibson for help with troubleshooting related to imaging; Arthur S. Levine for support; Neil Kad for critically reading the manuscript; Guillermo Romero, Marcel Bruchez, and Roger Hendrix for helpful discussions; Jacob Piehler for providing the biotinylated trisnitrilotriaceticacid compound; and Chunwei Du and Peggy Hsieh for assistance with pSCW01. This work was made possible through the generous support of the University of Pittsburgh Cancer Institute Cancer Center Support Grant P30 CA 047904 and from National Institutes of Health Grant 1R01ES019566.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323856111/-/DCSupplemental.
References
- 1.Sugasawa K. Multiple DNA damage recognition factors involved in mammalian nucleotide excision repair. Biochemistry (Mosc) 2011;76(1):16–23. doi: 10.1134/s0006297911010044. [DOI] [PubMed] [Google Scholar]
- 2.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571(1-2):19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 5.Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5(4):737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aboussekhra A, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80(6):859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 7.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106(2):253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 8.Wakasugi M, et al. Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J Biol Chem. 2001;276(18):15434–15440. doi: 10.1074/jbc.M011177200. [DOI] [PubMed] [Google Scholar]
- 9.Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 10.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3 Pt 2):785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J Biol Chem. 1996;271(40):24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 12.Rapić-Otrin V, et al. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum Mol Genet. 2003;12(13):1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- 13.Iwai S, et al. Benzimidazolium triflate-activated synthesis of (6-4) photoproduct-containing oligonucleotides and its application. Nucleic Acids Res. 1999;27(11):2299–2303. doi: 10.1093/nar/27.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittschieben BO, Iwai S, Wood RD. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem. 2005;280(48):39982–39989. doi: 10.1074/jbc.M507854200. [DOI] [PubMed] [Google Scholar]
- 15.Reardon JT, et al. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J Biol Chem. 1993;268(28):21301–21308. [PubMed] [Google Scholar]
- 16.Groisman R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113(3):357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 17.Luijsterburg MS, et al. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci. 2007;120(Pt 15):2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- 18.Fischer ES, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147(5):1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Kapetanaki MG, et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. 2006;103(8):2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero-Santoro J, et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68(13):5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- 21.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121(3):387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Lan L, et al. Monoubiquitinated histone H2A destabilizes photolesion-containing nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase. J Biol Chem. 2012;287(15):12036–12049. doi: 10.1074/jbc.M111.307058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Takedachi A, Saijo M, Tanaka K. DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku and contributes to the recruitment of XPA. Mol Cell Biol. 2010;30(11):2708–2723. doi: 10.1128/MCB.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapić-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30(11):2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh JI, et al. Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair. Proc Natl Acad Sci USA. 2012;109(41):E2737–E2746. doi: 10.1073/pnas.1110067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes CD, et al. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Res. 2013;41(9):4901–4912. doi: 10.1093/nar/gkt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39(17):7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014;42(4):2493–2504. doi: 10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savir Y, Tlusty T. Conformational proofreading: The impact of conformational changes on the specificity of molecular recognition. PLoS ONE. 2007;2(5):e468. doi: 10.1371/journal.pone.0000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kad NM, Van Houten B. Dynamics of lesion processing by bacterial nucleotide excision repair proteins. Prog Mol Biol Transl Sci. 2012;110:1–24. doi: 10.1016/B978-0-12-387665-2.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15(8):768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 34.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264(2):675–678. [PubMed] [Google Scholar]
- 35.Wang F, et al. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat Struct Mol Biol. 2013;20(2):174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5(9):763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 37.Bruchez MP. Quantum dots find their stride in single molecule tracking. Curr Opin Chem Biol. 2011;15(6):775–780. doi: 10.1016/j.cbpa.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tycon MA, Dial CF, Faison K, Melvin W, Fecko CJ. Quantification of dye-mediated photodamage during single-molecule DNA imaging. Anal Biochem. 2012;426(1):13–21. doi: 10.1016/j.ab.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 39.Kastantin M, Schwartz DK. Identifying multiple populations from single-molecule lifetime distributions. ChemPhysChem. 2013;14(2):374–380. doi: 10.1002/cphc.201200838. [DOI] [PubMed] [Google Scholar]
- 40.Geng H, et al. In vitro studies of DNA mismatch repair proteins. Anal Biochem. 2011;413(2):179–184. doi: 10.1016/j.ab.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alekseev S, et al. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol Cell Biol. 2008;28(24):7402–7413. doi: 10.1128/MCB.01108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Mahdy MA, et al. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281(19):13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 43.Wang QE, et al. Cellular ubiquitination and proteasomal functions positively modulate mammalian nucleotide excision repair. Mol Carcinog. 2005;42(1):53–64. doi: 10.1002/mc.20065. [DOI] [PubMed] [Google Scholar]
- 44.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135(7):1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyal E, Yang LW, Bahar I. Anisotropic network model: Systematic evaluation and a new web interface. Bioinformatics. 2006;22(21):2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 46.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331(6022):1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman LJ, Mumm JP, Gelles J. RNA polymerase approaches its promoter without long-range sliding along DNA. Proc Natl Acad Sci USA. 2013;110(24):9740–9745. doi: 10.1073/pnas.1300221110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan MR, Smerdon MJ. UV damage in DNA promotes nucleosome unwrapping. J Biol Chem. 2010;285(34):26295–26303. doi: 10.1074/jbc.M110.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pines A, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199(2):235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robu M, et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci USA. 2013;110(5):1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Q, et al. The p38 mitogen-activated protein kinase augments nucleotide excision repair by mediating DDB2 degradation and chromatin relaxation. J Biol Chem. 2008;283(47):32553–32561. doi: 10.1074/jbc.M803963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuge M, et al. SUMOylation of damaged DNA-binding protein DDB2. Biochem Biophys Res Commun. 2013;438(1):26–31. doi: 10.1016/j.bbrc.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Wakasugi M, et al. Physical and functional interaction between DDB and XPA in nucleotide excision repair. Nucleic Acids Res. 2009;37(2):516–525. doi: 10.1093/nar/gkn964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otrin VR, McLenigan M, Takao M, Levine AS, Protić M. Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J Cell Sci. 1997;110(Pt 10):1159–1168. doi: 10.1242/jcs.110.10.1159. [DOI] [PubMed] [Google Scholar]
- 56.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: Decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10(7):673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 57.Reardon JT, Sancar A. Thermodynamic cooperativity and kinetic proofreading in DNA damage recognition and repair. Cell Cycle. 2004;3(2):141–144. [PubMed] [Google Scholar]
- 58.Mathieu N, Kaczmarek N, Naegeli H. Strand- and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc Natl Acad Sci USA. 2010;107(41):17545–17550. doi: 10.1073/pnas.1004339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Vlaminck I, et al. Mechanism of homology recognition in DNA recombination from dual-molecule experiments. Mol Cell. 2012;46(5):616–624. doi: 10.1016/j.molcel.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Sagi D, Tlusty T, Stavans J. High fidelity of RecA-catalyzed recombination: A watchdog of genetic diversity. Nucleic Acids Res. 2006;34(18):5021–5031. doi: 10.1093/nar/gkl586. [DOI] [PMC free article] [PubMed] [Google Scholar]