Significance
Nitrogen fixation is a process of conversion of atmospheric nitrogen to ammonia catalyzed by nitrogenase, which is quickly inactivated by oxygen. Cyanobacteria are a group of prokaryotes that perform oxygenic photosynthesis, and many cyanobacterial species have the ability to fix nitrogen. How nitrogen fixation is coordinated with oxygenic photosynthesis remains largely unknown. Here we report two transcriptional regulators, ChlR (chlorophyll regulator) and CnfR (cyanobacterial nitrogen fixation regulator), that activate the transcription of genes responsible for anaerobic chlorophyll biosynthesis and the nitrogen fixation genes, respectively, in response to low-oxygen conditions in Leptolyngbya boryana, a diazotrophic cyanobacterium lacking heterocysts.
Keywords: anoxia-induced expression, iron–sulfur cluster
Abstract
Leptolyngbya boryana (Plectonema boryanum) is a diazotrophic cyanobacterium lacking heterocysts. How nitrogen fixation is regulated in filamentous nonheterocystous cyanobacteria remains unclear. Here we describe a large 50-kb nitrogen fixation (nif) gene cluster in L. boryana containing 50 genes. This gene cluster contains 14 nif genes (nifBSUHDKVZT and nifPENXW), two genes encoding transcriptional regulators showing high similarity to ChlR (chlorophyll regulator) and PatB, three genes encoding ferredoxin, three genes encoding cytochrome oxidase subunits, and 28 genes encoding nif-related proteins and proteins with putative or unknown functions. Eleven mutants lacking one gene or a subset of genes were isolated. Five of them did not grow under diazotrophic conditions, including two mutants lacking the transcriptional regulators. Although the chlR homolog-lacking mutant showed a normal level of nitrogenase activity, various intermediates of chlorophyll biosynthesis were accumulated under micro-oxic conditions. The phenotype suggested that ChlR activates the expression of the genes responsible for anaerobic chlorophyll biosynthesis to support energy supply for nitrogen fixation. In another mutant lacking the patB homolog, no transcripts of any nif genes were detected under nitrogen fixation conditions, which was consistent with no activity. Constitutive expression of patB in a shuttle vector resulted in low but significant nitrogenase activity even under nitrate-replete conditions, suggesting that the PatB homolog is the master regulator of nitrogen fixation. We propose to rename the patB homolog as cnfR, after cyanobacterial nitrogen fixation regulator.
Nitrogen fixation is a process for converting atmospheric nitrogen (N2) to ammonia, the major bioavailable nitrogen (1). Together with industrial nitrogen fixation based on the Harber–Bosch process, biological nitrogen fixation supports the bioproductivity on the earth. The enzyme responsible for nitrogen fixation is nitrogenase, which consists of two separable components: the Fe protein and MoFe protein (2). The Fe protein, a homodimer of the NifH protein, plays a role as an ATP-dependent reductase component for the MoFe protein. A [4Fe-4S] cluster is held between the homodimeric interfaces of the Fe protein. On hydrolysis of ATP, an electron of the [4Fe-4S] cluster is transferred to the MoFe protein. The electrons from the Fe protein eventually reach the catalytic site, FeMo-cofactor (FeMo-co), via an intermediate cluster, the P-cluster, and the N2 molecule bound in the vicinity of FeMo-co is then reduced to ammonia. The metallocenters of the nitrogenase components are extremely vulnerable to oxygen. Once exposed to the air, both the components become irreversibly inactivated with half-lives in the order of several seconds to minutes (3). Thus, the process of nitrogen fixation requires an anoxic environment, and all nitrogen-fixing organisms have some mechanism for maintaining the diazotrophic cells anoxic to drive nitrogenase.
Cyanobacteria are photosynthetic prokaryotes that perform oxygenic photosynthesis similar to plants. Many cyanobacteria have the ability to fix nitrogen with nitrogenase (4). Unlike heterotrophic nitrogen-fixing bacteria, cyanobacteria have special mechanisms for protecting nitrogenase from oxygen, given that they evolve oxygen by oxygenic photosynthesis. Some filamentous cyanobacteria form heterocysts specialized for nitrogen fixation (5). The strategy of heterocyst differentiation is the spatial separation of nitrogenase from photosynthesis. On the deprivation of combined nitrogen compounds, approximately every 10th cell along the filament of vegetative cells differentiates to heterocysts within 20 h in the cyanobacterium Anabaena sp. PCC 7120 (Anabaena 7120). Heterocysts are characterized by the following properties: they have a reduced content of photosystem II, the oxygen-evolving complex (6); they develop special cell walls as a physical barrier for oxygen penetration from the environment (7); and they have high respiratory activity by cytochrome c oxidase to remove oxygen and produce ATP to support nitrogenase activity (8).
Many nitrogen-fixing cyanobacteria lack heterocysts. Such cyanobacteria include unicellular [Synechococcus (9, 10), Gloeothece (11), and Cyanothece (12)] and filamentous species [Trichodesmium (13) and Leptolyngbya (14)]. Nitrogen fixation in such species is temporally separated from oxygenic photosynthesis and regulated by circadian rhythms (9, 10, 15). In microbial mats in Yellowstone National Park, nitrogenase activity by some unicellular Synechococcus strains was detected only at dawn and sunset (16), supporting the circadian regulation of nitrogen fixation in field environments. In Gloeothece, nitrogen fixation is mainly regulated by circadian rhythm and modulated by other factors such as fixed nitrogen amounts and environmental O2 levels (11). Trichodesmium species are a group of major nitrogen fixers in the ocean. Almost all cells along with the trichome filaments fix nitrogen, mainly using a temporal separation mechanism under circadian control (17, 18). Cyanothece species performing nitrogen fixation under aerobic conditions have recently received attention (19). Extensive studies on transcriptomic and proteomic aspects conducted with the aim of understanding how these species combine nitrogen fixation and oxygenic photosynthesis in one cell have revealed dynamic metabolic changes resulting from complex regulatory mechanisms, including circadian control (15, 20, 21). However, the mechanisms by which genes responsible for nitrogen fixation are transcriptionally regulated under conditions that require nitrogen fixation in nonheterocystous cyanobacteria remain unknown.
Leptolyngbya boryana (formerly Plectonema boryanum) is a filamentous nonheterocystous nitrogen-fixing cyanobacterium that grows diazotrophically under micro-oxic conditions (14). Early studies of nitrogen fixation in L. boryana have indicated that the induction of nitrogenase requires both micro-oxic and nitrogen starvation conditions (14, 22, 23). Temporal separation of nitrogen fixation from photosynthesis has been observed in photosynthetic growth under micro-oxic conditions, suggesting the presence of a circadian control mechanism (24). The nucleotide sequence of nifH of L. boryana has been determined (25), and a transformation system for isolating a mutant lacking a targeted gene via electroporation has been established (26). Thus, L. boryana provides a promising model to investigate nitrogen fixation in cyanobacteria.
In the present study, we identified a large nitrogen fixation gene cluster in L. boryana strain dg5 that contains two genes encoding transcriptional regulators showing high similarity to ChlR (chlorophyll regulator) from Synechocystis sp. PCC 6830 (Synechocystis 6803) and to PatB from Anabaena 7120, respectively. Our results suggest that the ChlR homolog supports nitrogen fixation by the transcriptional activation of genes responsible for chlorophyll, heme, and bilin biosynthesis under low-oxygen conditions and that the PatB homolog, cyanobacterial nitrogen fixation regulator (CnfR), plays a role as the master transcriptional regulator for the nif genes in this gene cluster.
Results
Nitrogen Fixation Gene Cluster.
We determined the nucleotide sequence of genes encoding nitrogenase, as well as nitrogen fixation-related genes in L. boryana strain dg5. The nucleotide sequence of the 3.4-kb HindIII fragment containing nifUHD has been previously reported in L. boryana (25). Starting from this known sequence, genomic walking was performed both downstream of nifD and upstream of nifU (Fig. S1 and Table S1). The nucleotide sequence of a 50-kb region (50,712 bp) was determined. This gene region contains 50 genes and is divided into right and left parts at the intergenic region between nifP and nifB (Fig. 1 and Table S2). Right and left genes appear to be divergently transcribed from this intergenic region. The right part (Fig. 1 and Table S2, Nos. 30–50) contains the nitrogenase structural genes nifH, nifD, and nifK, which are flanked by four genes (nifB, fdxN, nifS, and nifU) and three genes (nifV, nifZ, and nifT) in upstream and downstream regions, respectively. A gene (orf534) encoding an XRE (xenobiotic response element) family transcriptional regulator is found in the opposite direction of nifVZT. The deduced amino acid sequence of this gene shows the highest similarity (58%) to that of patB from Anabaena 7120 (27). Downstream of patB, three genes encoding subunits of cytochrome c oxidase are found in the same direction as nifHDK. These three proteins show the highest similarity to those of the heterocyst-specific type oxidase subunits, CoxB2, CoxA2, and CoxC2, in Anabaena 7120 (8).
Fig. 1.
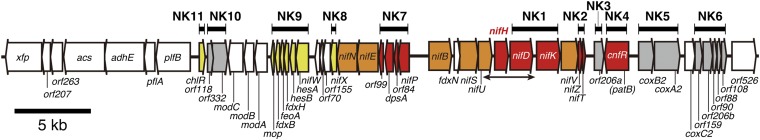
Gene organization of the 50-kb nitrogen fixation gene cluster from L. boryana. The thick horizontal bars above the gene organization indicate the coding regions that were replaced with the kanamycin cartridge in the NK mutants. Based on the phenotype of the NK mutants, the genes are color-coded as follows: red, genes essential for nitrogen fixation; orange, genes essential for nitrogen fixation (but with no experimental evidence in L. boryana); yellow, genes important for nitrogen fixation; gray, genes not essential for nitrogen fixation; white, no experimental evidence available to date. It should be noted that at least one of the genes (not yet specified in this study) is essential or important for nitrogen fixation in NK2, NK7, and NK9. The thin bar with arrowheads at both ends indicates the chromosomal 3.4-kb nifUHD region whose nucleotide sequence has been previously reported (25).
In the left part of the gene cluster (Fig. 1 and Table S2, Nos. 1–29), nifP (cysE) encoding serine O-acetyltransferase is located in the opposite direction and followed by nine genes: orf84, dpsA, orf99, nifE, nifN, nifX, orf155, orf77, and nifW. nifEN encodes the scaffold protein NifEN for the biosynthesis of FeMo-co. Another gene subcluster, hesAB, fdxH, feoA, fdxB, and mop, is found downstream of nifW. Three genes, modA, modB, and modC, encoding a molybdenum transporter are located downstream of mop. Three genes, orf332, orf118, and orf136, lie just downstream of modABC in the opposite direction. The amino acid sequence of orf136 shows 55% similarity to that of chlR from the nondiazotrophic cyanobacterium Synechocystis 6803 (28). Seven genes, pflB, pflA, adhE, acs, orf263, orf207, and xfp, most of which encode enzymes for anaerobic metabolism, are found in the leftmost part of the left region. Compared with other nonheterocystous strains, 30 genes from modC to orf206a in the central part of the gene cluster in L. boryana are conserved in the nif gene cluster of Cyanothece sp. ATCC 51142 in almost the same gene order, with the exception of a single inversion of the 12-kb region from dpsA to nifK (Fig. S2). A draft genome of L. boryana sp. PCC 6306, a different strain of L. boryana, was recently reported (29) as 1 of 54 diverse cyanobacterial species to analyze cyanobacterial phylogenetic relationship. We found that the 50-kb nif gene cluster of dg5 was identical to that of the draft genome of L. boryana sp. PCC 6306.
Isolation of 11 KO Mutants and Their Phenotypes.
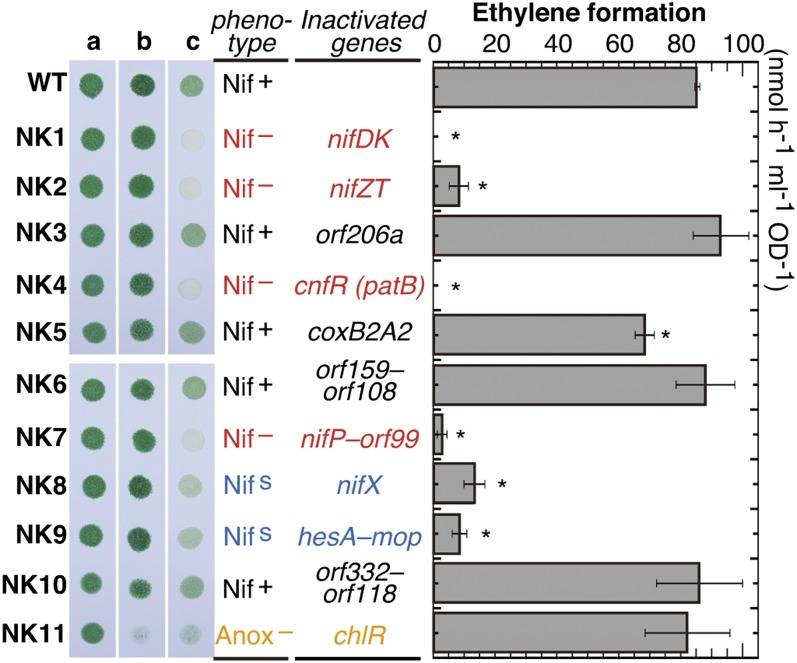
To determine whether the genes in this region are involved in nitrogen fixation, we isolated 11 mutants, NK1–NK11, lacking single or multiple genes (Figs. 1 and 2). We evaluated their growth under the three conditions (Fig. 2, lanes a–c) and their nitrogenase activity as acetylene reduction activity (Fig. 2, Right). The growth of all of the mutants was the same as that of the WT in nitrate-replete agar medium under aerobic conditions (Fig. 2, lane a). On the nitrate-replete agar medium under micro-oxic conditions, all of the mutants showed normal growth except for a chlR homolog-lacking mutant, NK11, which showed poor growth with abnormal bluish color. Under conditions for nitrogen fixation (nitrate-depleted/micro-oxic), the four mutants NK3, NK5, NK6, and NK10 grew normally like the WT (Nif+). These results suggested that all of the genes disrupted in these mutants are not important for nitrogen fixation.
Fig. 2.
Comparison of growth and acetylene reduction activity of the WT and transformants. L. boryana cells grown on BG-11 agar plates containing nitrate under aerobic conditions for 2 d were collected to adjust the cell density to an OD730 of 1.0, and aliquots (6 µL) were spotted onto new BG-11 agar plates containing nitrate (lanes a and b) or BG-110 without combined nitrogen (lane c), followed by growth under aerobic (lane a) or micro-oxic (lanes b and c) conditions for 4 d. Acetylene reduction activity was determined in the transformant cells. Cells grown under nitrate-replete conditions were incubated under nitrogen-fixing conditions for 12 h. Asterisks indicate values of ethylene formation rates that are statistically different from those of the WT (n = 3).
The growth of NK8 (∆nifX) and NK9 (∆hesAB-fdxH-feoA-fdxB-mop) was markedly slower than that of the WT under nitrogen-fixing conditions (NifS). This slow growth was accompanied by a marked decrease in acetylene reduction activity in NK8 (16%) and NK9 (10%). This phenotype suggested that NifX plays an important, although not essential, role in nitrogen fixation and that one or some genes disrupted in NK9 also take part in nitrogen fixation. All six genes, hesA, hesB, fdxH, feoA, fdxB, and mop, are probably involved in nitrogen fixation. Further experiments are in progress to ascertain the contribution of the individual genes for nitrogen fixation.
NK1 (∆nifDK), NK2 (∆nifZT), NK4 (∆patB), and NK7 (∆nifP-orf84-dpsA-orf99) were completely deficient in diazotrophic growth ability (Nif–), and NK11 (∆chlR) showed a growth defect under anoxic conditions (Anox–). The growth defect of NK1 in diazotrophic conditions was as expected. NK2 showed only ∼10% of acetylene reduction activity, suggesting that either or both of the small proteins, NifZ and NifT, are essential for the maximal activity of nitrogenase. Interestingly, similar to NK1, NK4 showed no acetylene reduction activity. The transcriptional regulator PatB homolog is probably essential for the induction of nif genes in response to the depletion of combined nitrogen (see below). NK7 lacks nifP, dpsA, and two unknown genes: orf84 and orf99. The deletion of the four genes caused a severe decrease (3%) in acetylene reduction activity. Despite growth defect under nitrogen fixation conditions, NK11 showed a normal level of acetylene reduction activity. The common growth defect under micro-oxic conditions and the normal level of acetylene reduction activity strongly suggested that the ChlR homolog (Orf136) is not directly involved in nitrogen fixation.
Characterization of NK11 (ΔchlR).
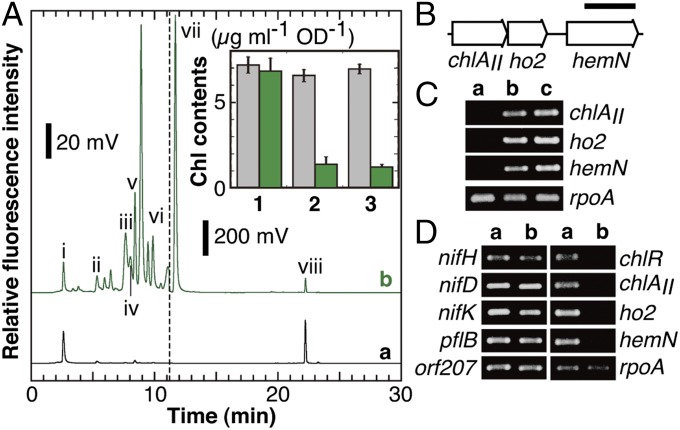
chlR encodes a MarR-type transcriptional activator for the low-oxygen induction of a small operon, chlAII-ho2-hemN, in Synechocystis 6803 (28). The ∆chlR mutant of Synechocystis 6803 shows poor growth under micro-oxic conditions with a very low level of Chl because the chlAII operon is not expressed and Chl biosynthesis is delayed in the steps of oxygen-dependent reactions. The three genes chlAII, ho2, and hemN encode Mg-protoporphyrin IX monomethylester (MPE) cyclase, heme oxygenase 2, and coproporphyrinogen III oxidase, respectively, and all of the enzymes complement the activities of the three oxygen-dependent reactions that are normally catalyzed by ChlAI (30), HO1 (31), and HemF (32), respectively, under micro-oxic conditions. To confirm the function of the chlR-homolog, we determined the Chl content and analyzed the pigment composition by HPLC. The Chl content of NK11 cells incubated under micro-oxic conditions for 7 d was only ∼20% that of the WT, in contrast to the normal content under aerobic conditions (Fig. 3A, Inset). This marked decrease in the Chl content is consistent with the abnormal blue color derived from phycobiliproteins (28) exhibited by colonies of NK11. As shown in the HPLC profile (Fig. 3A), NK11 cells accumulated various intermediates of Chl biosynthesis. This phenotype was substantially the same as that of ∆chlR of Synechocystis 6803 (28, 33).
Fig. 3.
Phenotype analysis of NK11. (A) WT and NK11 cells were incubated under micro-oxic conditions for 7 d. Aliquots (100 µL with an OD730 of 100) of the cell suspension were mixed with 900 µL of ice-cold methanol for HPLC analysis. The elution of pigments was monitored by fluorescence emission at 630 nm with excitation at 400 nm (a, WT; b, NK11). The elution profiles from 0 to 11.2 min are enlarged 10 times to emphasize small peaks (see the scale bars). Peaks i to viii were previously assigned as coproporphyrin III (i), Mg-protoporphyrin IX (ii), divinyl protochlorophyllide (iii), monovinyl protochlorophyllide (iv), MPE (v), protoporphyrin IX (vi), demetallated MPE (vii), and Chl a (viii), respectively. (Inset) Comparison of Chl levels of the WT and NK11. WT (gray bars) and NK11 (green bars) cells were grown in BG-11 agar plates containing nitrate (lanes 1 and 2) or BG-110 without combined nitrogen (lane 3) under aerobic (lane 1) or micro-oxic (lanes 2 and 3) conditions (n = 3). (B) The gene organization of the three genes, chlAII, ho2, and hemN, forming a small gene cluster. (Scale bar, 1 kb.) (C) Transcript profiles by RT-PCR for the three genes. WT cells were grown under nitrate-replete/aerobic (lane a), nitrate-replete/micro-oxic (lane b), and nitrate-depleted/micro-oxic (lane c) conditions, and RNA was extracted from the cells. rpoA was used as an internal control. (D) Transcript profiles by RT-PCR for nifHDK and the six genes in WT and NK11 cells. WT (lane a) and NK11 (lane b) cells were grown under nitrogen-fixing conditions, and RNA was extracted from the cells.
The chlAII-ho2-hemN operon is conserved in the draft genome sequence of L. boryana stain dg5 (Fig. 3B). As observed in Synechocystis 6803, the three genes were only expressed in cells grown under micro-oxic conditions (Fig. 3C). Under nitrate-depleted micro-oxic conditions, no transcripts were detected for the chlAII-ho2-hemN operon in NK11, whereas the transcripts for nifHDK were normally detected as in the WT (Fig. 3D). We also examined whether the pflB subcluster (pflBA, adhE, acs, orf263, orf207, and xfp) located next to chlR is regulated by ChlR. The seven genes, including pflB and orf207 as representatives of the pflB subcluster (Fig. 3D), were expressed in NK11 as observed in the WT, indicating that ChlR does not regulate their expression. These results indicated that orf136 is the chlR ortholog in L. boryana.
Characterization of NK4 (ΔpatB).
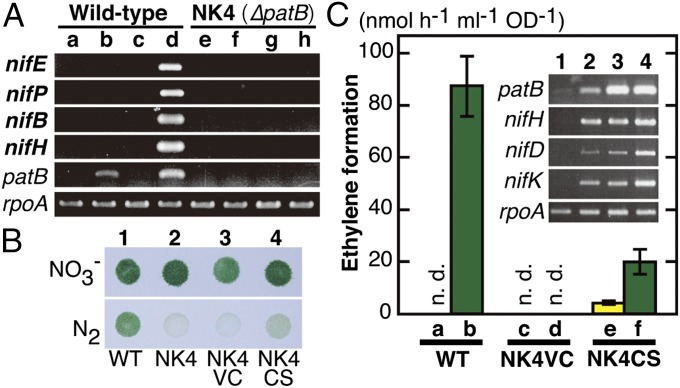
NK4 showed the most prominent Nif– phenotype comparable to NK1 (Fig. 2). To examine whether the PatB-homologous protein regulates the expression of nif genes, we compared the transcript profiles between the WT and NK4 (Fig. 4A and Fig. S3). No transcripts for any nif genes were detected in NK4 cells incubated under nitrogen-fixing conditions, an observation that explains why NK4 showed no detectable nitrogenase activity (Fig. 2). The expression of nine genes from chlR to xfp in the leftmost part and orf526 was not affected by the loss of the patB homolog. This result suggests that the PatB homolog activates the transcription of the nif genes in this cluster. RT-PCR in the WT indicated that the patB homolog was predominantly expressed under nitrate-depleted conditions, irrespective of aerobic or micro-oxic conditions (Fig. 4A).
Fig. 4.
Transcript levels in the WT and NK4 and complementation analyses. (A) Transcript profiles by RT-PCR. WT (lanes a–d) and NK4 (lanes e–h) cells grown on BG-11 agar plates under aerobic conditions were transferred to new BG-11 agar plates containing nitrate (lanes a, c, e, and g) or BG-110 (lanes b, d, f, and h), followed by incubation under aerobic (lanes a, b, e, and f) or micro-oxic (lanes c, d, g, and h) conditions for 6 h. RNA was extracted from the cells to prepare cDNA, and the transcript levels were estimated by RT-PCR. (B) NK4VC and NK4CS are NK4 derivatives harboring the shuttle vectors pPBH202 (vector control) (38) and pPBHPatBS containing strep-patB under the control of the T5 promoter, respectively. Cells of the WT (lane 1), NK4 (lane 2), NK4VC (lane 3), and NK4CS (lane 4) grown on nitrate-replete BG-11 agar plates under aerobic conditions were spotted on new nitrate-replete BG-11 agar plates (NO3–) or BG-110 (N2), followed by incubation under micro-oxic conditions for 4 d. (C) Acetylene reduction activity was determined in the transformant cells. Cells grown under nitrate-replete conditions were incubated under nitrate-replete (yellow bars; lanes a, c, and e) and nitrogen-fixing (green bars; lanes b, d, and f) conditions for 12 h. n.d., not detected. (Inset) The transcript levels of patB and nifHDK were estimated by RT-PCR. WT (lanes 1 and 2) and NK4CS (lanes 3–4) cells grown on BG-11 agar plates under aerobic conditions were transferred to new BG-11 agar plates containing nitrate (lanes 1 and 3) or BG-110 (lanes 2 and 4) under micro-oxic conditions for 12 h. The transcript levels were estimated by RT-PCR as shown in A.
To examine whether constitutive expression of the patB homolog induces the nif genes even under nitrogenase-repressed conditions, we constructed a shuttle vector carrying a fusion gene encoding Strep-tagged PatB-homolog under the control of an Escherichia coli T5 promoter, and transformed NK4 to isolate NK4CS (Fig. 4 B and C). NK4CS restored diazotrophic growth under nitrogen-fixing conditions, although the growth was slower than that of the WT (Fig. 4B). In addition, ∼20% activity of that in the WT was restored in NK4CS (Fig. 4C, lane f). Furthermore, low but significant nitrogenase activity was detected in NK4CS, although the cells grew in the presence of nitrate (Fig. 4C, lane e). The transcript levels of the patB homolog and nifHDK are shown in the Inset to Fig. 4C. The transcript levels of the patB homolog (lanes 3 and 4) were much higher than that of WT under nitrate-depleted conditions (lane 2), and their levels were similar in both nitrate-replete and nitrate-depleted conditions, indicating that the T5-promoter regulation brought about overexpression of the patB homolog in NK4CS. The nifHDK transcripts, as representatives of the nif genes, were higher than those of WT under nitrate-depleted conditions (compare lanes 4 and 2). Furthermore, the nifHDK transcripts were detected in the nitrate-replete conditions in almost the same levels as those of WT under nitrate-depleted conditions (compare lanes 3 and 2), which was consistent with nitrogenase activity in nitrate-replete conditions (lane e), albeit at a relatively low level. These results strongly suggested that the PatB homolog is the master transcriptional regulator for nitrogen fixation in L. boryana.
Discussion
Cyanobacteria have developed various regulatory mechanisms to drive nitrogenase together with oxygenic photosynthesis during evolution. Heterocyst formation is the most elaborate mechanism for separating the nitrogen fixation process spatially from vegetative photosynthetic cells (5). However, molecular genetic analysis of nitrogen fixation in heterocystous strains is often complicated by a large number of genes for heterocyst differentiation and the synthesis of polysaccharide and glycolipid layers, which are not directly associated with the nitrogen fixation process itself. Therefore, nonheterocystous strains provide better systems for focusing only on nitrogen fixation. Among nonheterocystous cyanobacteria, some Cyanothece strains have been studied by a combination of transcriptomic and proteomic analyses (20, 21). However, the lack of an efficient transformation system has hindered the identification of the detailed mechanism underlying the regulation of nitrogen fixation (34). In the present study, we isolated 11 mutants with an efficient transformation system using the nonheterocystous strain L. boryana. A significant effect on diazotrophic growth was observed in seven of these mutants. This result demonstrated that L. boryana is a very useful model cyanobacterium for investigating the regulatory mechanism of nitrogen fixation in molecular biological aspects. Because multiple genes were disrupted in most mutants, studies are ongoing to identify the genes that are responsible for the Nif– or NifS phenotype using a complementation system with a shuttle vector.
NK11 showed a unique phenotype: poor growth under micro-oxic conditions (Anox–) and normal activity of nitrogenase, distinct from other Nif– mutants (Fig. 2). The growth defect confirmed the critical role of a continuous supply of tetrapyrrole pigments for photosynthesis and respiration to support nitrogenase activity under micro-oxic conditions (Fig. 5). The phenotype of NK11 strongly suggested that orf136 is the L. boryana ortholog of the Synechocystis 6803 chlR gene (28). The diazotrophic growth defect despite of normal nitrogenase activity in NK11 can be explained as follows. Cells used for the acetylene reduction assay were precultured in nitrate-replete media under aerobic conditions, and the cells were exposed to nitrogen-fixing conditions (nitrate-depleted/micro-oxic) for just 12 h before the assay. The contents of preexisting Chl, bilins, and heme in the cells were probably high enough to produce and drive nitrogenase in such a short time. However, the contents of Chl, bilins, and heme were decreased due to dilution by cell divisions and eventually became too low to support nitrogenase and various cellular activities resulting in growth defect.
Fig. 5.
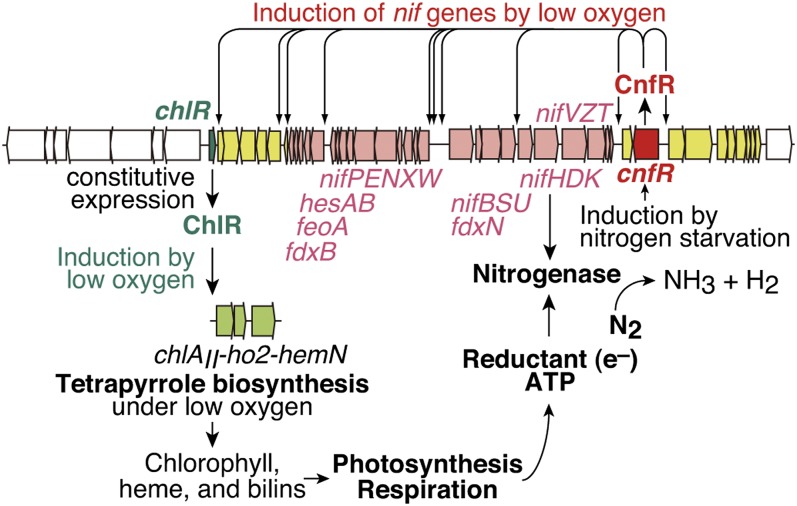
A working model of ChlR and CnfR in the regulatory circuits of the nif gene cluster in L. boryana. chlR (shown in green) is expressed in a constitutive manner, and the ChlR protein is converted to an active form on exposure to low-oxygen environments, and it activates the expression of the small gene cluster chlAII-ho2-hemN (shown in light green) in the other chromosomal locus. Thus, the three newly synthesized enzymes, ChlAII, HO2, and HemN, bypass the oxygen-dependent reactions in tetrapyrrole biosynthesis to maintain a constant supply of Chl, heme, and bilin pigments even under micro-oxic conditions. These pigments support activities of photosynthesis and respiration to supply reductants and ATP in an efficient manner to drive nitrogenase. cnfR (shown in red) is induced in response to nitrogen starvation, and the CnfR protein is converted to an active form when the oxygen level becomes low enough to drive nitrogenase. Following this, the active form of CnfR activates the transcription of the nif genes (shown in pink) to produce active nitrogenase. Some genes shown in yellow are also regulated by some other regulatory mechanisms while they are still under the control of CnfR.
The PatB-homologous protein (534 amino acid residues) has the N-terminal 2 Cys motifs (CxxCxxCxxxCP and CxxCx8CxxxC) that are found in a bacterial-type ferredoxin to hold two [4Fe-4S] clusters and the DNA-binding motif in the C-terminal portion. This architecture is conserved in all PatB homologs in cyanobacteria. In Anabaena 7120, patB is specifically expressed in heterocysts, and a patB-lacking mutant showed poor growth under nitrogen-fixing conditions that was accompanied by abnormal heterocyst differentiation pattern in the form of frequent multiple contiguous heterocysts (35). In L. boryana, the patB homolog is expressed in response to the deprivation of combined nitrogen (Fig. 4A). No transcripts for the nif genes were detected in NK4 (Fig. 4A and Fig. S3), a finding consistent with the total loss of acetylene reduction activity (Fig. 2). Even under nitrate-replete conditions, significant nitrogenase activity was detected in NK4CS (Fig. 4C, lane e), which is an NK4-derivative carrying an extra strep-patB gene in the plasmid. These results strongly suggest that the patB homolog encodes the master transcriptional activator for nitrogen fixation in L. boryana. We propose to rename the patB homolog as cnfR, after cyanobacterial nitrogen fixation regulator.
The constitutive overexpression of cnfR in the shuttle vector resulted in partial complementation of diazotrophic growth and nitrogenase activity (Fig. 4 B and C). A possibility that an N-terminal fusion with Strep-tag (Strep-CnfR) may lower the activity of CnfR can be excluded because the transcript levels of nifHDK were almost the same as those of WT or even higher in NK4CS (Fig. 4C, Inset). Two possibilities may be considered for the partial complementation. One is a polar effect caused by the kanamycin-resistance gene (KmR) cartridge. Because the KmR cartridge does not carry any terminator sequences in both ends, the transcription originating from the cartridge may produce longer transcripts functioning as antisense RNA to interfere with the expression of the contiguous genes ORF206a and nifVZT. Second, the constitutive overexpression of cnfR driven by the T5 promoter may cause some side effects on the cells. Irrespective of the reasons for the partial complementation, the appearance of nitrogenase activity under nitrate-replete conditions indicates that all of the genes required for the production of active nitrogenase are expressed (exemplified by nifHDK; Fig. 3C, Inset) even under conditions in which these nif genes are repressed in WT cells, a finding that strongly supports the assignment of CnfR as the master regulator.
The nitrogenase activity of NK4CS under the nitrate-replete conditions was much lower (∼20%) than that of cells under nitrate-depleted conditions (Fig. 4C, lanes e and f), which was consistent with the difference between transcript levels of nifHDK under nitrate-replete and depleted conditions (Fig. 4C, Inset, lanes 3 and 4). Despite the same levels of the cnfR transcript, the lower transcriptional levels of nifHDK under nitrate-replete conditions imply two possibilities. One is that the activity of CnfR is regulated at the posttranscriptional level in response to the presence of nitrate or some other combined nitrogen form. The other is that other factors are also involved in the negative regulation of nif genes in response to the cellular nitrogen status.
Based on the results and features of the CnfR amino acid sequence, we hypothesize the following regulatory mechanism of the nif genes by CnfR (Fig. 5): the transcription of cnfR is induced in response to nitrogen starvation (Fig. 4A). CnfR is maintained as an inactive form under aerobic conditions. Once the cellular oxygen level becomes low enough to drive nitrogenase, CnfR is converted to an active form to activate the transcription of the target nif genes to produce active nitrogenase. Two [4Fe-4S] clusters in the N-terminal domain may contribute in sensing the cellular oxygen level. This hypothesis is in accordance with the requirement of two conditions, nitrogen starvation and micro-oxic conditions, for the expression of nitrogen fixation. Further studies of the mechanism by which cnfR is induced in response to nitrogen starvation and of biochemical features of CnfR are required to support this hypothesis.
The gene arrangement of subcluster nifB-fdxN-nifSUHDK is conserved in heterocystous cyanobacteria such as Anabaena 7120 and Anabaena variabilis ATCC 29413. Ungerer et al. (36) reported that the high expression of nifH1 is supported by two promoters: a strong one is located upstream of nifB1, and the other weak one is located in the coding region of nifU1 in A. variabilis. In addition, they pointed out that a secondary structure of 5′ UTR just upstream of nifH1 might contribute to stabilize the processed nifH1 transcript. We previously identified a monocistronic nifH transcript in diazotrophically grown L. boryana cells by Northern blot analysis (25). Short inverted repeats forming similar secondary structures in the 5′ and 3′ UTRs of nifH of L. boryana (25) might stabilize the nifH transcript contributing higher levels of the monocistronic nifH transcript than those of other nif genes similar to A. variabilis. In Anabaena 7120, Suzuki et al. (37) reported that AnCrpA (Anabaena cAMP receptor protein) is involved in a positive regulation of the nif and cox genes, which was supported by microarray data and specific binding of AnCrpA to the 5′ upstream regions of nifB, hesA, and coxB in the presence of cAMP. In the draft genome of L. boryana strain dg5, there is a probable ortholog of AnCrpA (76% identity). Thus, the nif gene cluster of L. boryana may be also regulated by some other mechanisms common to heterocystous strains such as CrpA in addition to the CnfR system.
Nitrogen fixation in L. boryana is directly regulated by the action of CnfR in transcription of nif genes and indirectly by the action of ChlR in the Chl supply under micro-oxic conditions. ChlR contributes not only to maintain the photosynthesis activity but also to enhance respiratory activity by the inducible cytochrome oxidase encoded by coxB2A2C2 for protecting nitrogenase from oxygen damage. Nitrogen fixation is separated from oxygenic photosynthesis in a temporal manner in nonheterocystous cyanobacteria (9, 10). The two regulatory proteins ChlR and CnfR would play critical roles in maintaining circadian outputs in response to the cellular oxygen levels to separate these incompatible processes. How ChlR and CnfR sense the cellular oxygen level is of particular interest for future studies of cyanobacterial nitrogen fixation.
Materials and Methods
The L. boryana (formerly P. boryanum) strain dg5 and mutants were cultivated on BG-11 medium. For nitrogen deprivation treatment, BG-110 medium with no combined nitrogen sources was used. For growth under micro-oxic conditions, agar plates were incubated in an anaerobic jar (BBL GasPak anaerobic systems; BD Biosciences) with a sachet for producing anoxic conditions (Gas Generating Kit Anaerobic System; Oxoid) (28) under light intensity of 40 µmolphoton⋅m–2⋅s–1 for 6 h to 4 d. Preparation of genomic DNA, gene walking, construction of plasmids, preparation of RNA, RT-PCR, and pigment analysis are described in SI Materials and Methods (Tables S3 and S4). For the induction of nitrogenase, the cells were transferred onto BG-110 agar plates and were incubated in an anaerobic jar in the light for 6–12 h. Nitrogenase activity was assayed by acetylene reduction, as described in SI Materials and Methods. The nucleotide sequences of the 50-kb nif gene cluster, the chlAII-ho2-hemN gene cluster, and rpoA have been deposited in GenBank under the accession numbers of AB808482, AB808629, and AB808630, respectively.
Supplementary Material
Acknowledgments
We thank Kenshiro Oshima, Yuu Hirose, and Masahira Hattori for the genome sequence of L. boryana strain dg5; Haruki Yamamoto for technical help in using anaerobic techniques and gas chromatography; Rina Aoki for pigment analysis of the chlR mutant; Yuto Hiraide and Kazuma Uesaka for draft genome analysis of L. boryana; and Tsuyoshi Matsumoto and Kazuyuki Tatsumi for arrangements of anaerobic chamber and growth cabinet. We also thank Kaori Ohki, Tatsuo Omata, and Kazuki Terauchi for valuable discussion. This work was supported by the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research 23370020 and for Specially Promoted Research 23000007) and the Japan Science and Technology Agency (Advanced Low Carbon Technology Research and Development Program and Precursory Research for Embryonic Science and Technology).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB808482, AB808629, and AB808630).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323570111/-/DCSupplemental.
References
- 1.Smith BE, Richards RL, Newton WE. Catalysts for Nitrogen Fixation: Nitrogenases, Relevant Chemical Models and Commercial Processes. Dordrecht, Netherlands: Kluwer Academic Publishers; 2004. [Google Scholar]
- 2.Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eady RR. Isolation and characterization of various nitrogenase. Methods Enzymol. Vol 69. San Diego: Academic; 1980. pp. 753–778. [Google Scholar]
- 4.Stal LJ, Zehr JP. Cyanobacterial nitrogen fixation in the ocean: Diversity, regulation, and ecology. In: Herrero A, Flores E, editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Norfolk, UK: Caister Academic Press; 2008. pp. 423–446. [Google Scholar]
- 5.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2(4):a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiura K, Itoh S. Single-cell confocal spectrometry of a filamentous cyanobacterium Nostoc at room and cryogenic temperature. Diversity and differentiation of pigment systems in 311 cells. Plant Cell Physiol. 2012;53(8):1492–1506. doi: 10.1093/pcp/pcs093. [DOI] [PubMed] [Google Scholar]
- 7.Awai K, Wolk CP. Identification of the glycosyl transferase required for synthesis of the principal glycolipid characteristic of heterocysts of Anabaena sp. strain PCC 7120. FEMS Microbiol Lett. 2007;266(1):98–102. doi: 10.1111/j.1574-6968.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 8.Valladares A, Herrero A, Pils D, Schmetterer G, Flores E. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol. 2003;47(5):1239–1249. doi: 10.1046/j.1365-2958.2003.03372.x. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui A, et al. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photosynthetically. Nature. 1986;323(6090):720–722. [Google Scholar]
- 10.Grobbelaar N, Huang TC, Lin HY, Chow TJ. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol Lett. 1986;37(2):173–177. [Google Scholar]
- 11.Taniuchi Y, Yoshikawa S, Maeda S, Omata T, Ohki K. Diazotrophy under continuous light in a marine unicellular diazotrophic cyanobacterium, Gloeothece sp. 68DGA. Microbiology. 2008;154(Pt 7):1859–1865. doi: 10.1099/mic.0.2008/018689-0. [DOI] [PubMed] [Google Scholar]
- 12.Bandyopadhyay A, et al. Novel metabolic attributes of the genus cyanothece, comprising a group of unicellular nitrogen-fixing Cyanothece. MBio. 2011;2(5):e00214–11. doi: 10.1128/mBio.00214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capone DG, Zehr JP, Paerl HW, Bergmen B, Carpenter EJ. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276(5316):1221–1229. [Google Scholar]
- 14.Stewart WD, Lex M. Nitrogenase activity in the blue-green alga Plectonema boryanum strain 594. Arch Mikrobiol. 1970;73(3):250–260. doi: 10.1007/BF00410626. [DOI] [PubMed] [Google Scholar]
- 15.Bandyopadhyay A, Elvitigala T, Liberton M, Pakrasi HB. Variations in the rhythms of respiration and nitrogen fixation in members of the unicellular diazotrophic cyanobacterial genus Cyanothece. Plant Physiol. 2013;161(3):1334–1346. doi: 10.1104/pp.112.208231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steunou AS, et al. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J. 2008;2(4):364–378. doi: 10.1038/ismej.2007.117. [DOI] [PubMed] [Google Scholar]
- 17.Finzi-Hart JA, et al. Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci USA. 2009;106(15):6345–6350. doi: 10.1073/pnas.0810547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohki K, Taniuchi Y. Detection of nitrogenase in individual cells of a natural population of Trichodesmium using immnocytochemical methods for fluorescence cells. J Ocenanography. 2009;65(3):427–432. [Google Scholar]
- 19.Bandyopadhyay A, Stöckel J, Min H, Sherman LA, Pakrasi HB. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat Commun. 2010;1:139. doi: 10.1038/ncomms1139. [DOI] [PubMed] [Google Scholar]
- 20.Stöckel J, et al. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA. 2008;105(16):6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aryal UK, et al. Dynamic proteome analysis of Cyanothece sp. ATCC 51142 under constant light. J Proteome Res. 2012;11(2):609–619. doi: 10.1021/pr200959x. [DOI] [PubMed] [Google Scholar]
- 22.Weare NM, Benemann JR. Nitrogenase activity and photosynthesis in Plectonema boryanum. J Bacteriol. 1974;119(1):258–265. doi: 10.1128/jb.119.1.258-265.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagatani HH, Haselkorn R. Molybdenum independence of nitrogenase component synthesis in the non-heterocystous cyanobacterium Plectonema. J Bacteriol. 1978;134(2):597–605. doi: 10.1128/jb.134.2.597-605.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misra HS, Tuli R. Differential expression of photosynthesis and nitrogen fixation genes in the cyanobacterium Plectonema boryanum. Plant Physiol. 2000;122(3):731–736. doi: 10.1104/pp.122.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita Y, Takahashi Y, Shonai F, Ogura Y, Matsubara H. Cloning, nucleotide sequences and differential expression of the nifH and nifH-like (frxC) genes from the filamentous nitrogen-fixating cyanobacterium Plectonema borynaum. Plant Cell Physiol. 1991;32(7):1093–1106. [Google Scholar]
- 26.Fujita Y, Takahashi Y, Chuganji M, Matsubara H. The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1992;33(1):81–92. [Google Scholar]
- 27.Liang J, Scappino L, Haselkorn R. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc Natl Acad Sci USA. 1992;89(12):5655–5659. doi: 10.1073/pnas.89.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki R, Takeda T, Omata T, Ihara K, Fujita Y. MarR-type transcriptional regulator ChlR activates expression of tetrapyrrole biosynthesis genes in response to low-oxygen conditions in cyanobacteria. J Biol Chem. 2012;287(16):13500–13507. doi: 10.1074/jbc.M112.346205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih PM, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110(3):1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamizaki K, Mizoguchi T, Goto T, Tamiaki H, Fujita Y. Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 2008;283(5):2684–2692. doi: 10.1074/jbc.M708954200. [DOI] [PubMed] [Google Scholar]
- 31.Aoki R, Goto T, Fujita Y. A heme oxygenase isoform is essential for aerobic growth in the cyanobacterium Synechocystis sp. PCC 6803: Modes of differential operation of two isoforms/enzymes to adapt to low oxygen environments in cyanobacteria. Plant Cell Physiol. 2011;52(10):1744–1756. doi: 10.1093/pcp/pcr108. [DOI] [PubMed] [Google Scholar]
- 32.Goto T, Aoki R, Minamizaki K, Fujita Y. Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2010;51(4):650–663. doi: 10.1093/pcp/pcq023. [DOI] [PubMed] [Google Scholar]
- 33.Aoki R, Hiraide Y, Yamakawa H, Fujita Y. A novel “oxygen-induced” greening process in a cyanobacterial mutant lacking the transcriptional activator ChlR involved in low-oxygen adaptation of tetrapyrrole biosynthesis. J Biol Chem. 2014;289(3):1841–1851. doi: 10.1074/jbc.M113.495358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min H, Sherman LA. Genetic transformation and mutagenesis via single-stranded DNA in the unicellular, diazotrophic cyanobacteria of the genus Cyanothece. Appl Environ Microbiol. 2010;76(22):7641–7645. doi: 10.1128/AEM.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones KM, Buikema WJ, Haselkorn R. Heterocyst-specific expression of patB, a gene required for nitrogen fixation in Anabaena sp. strain PCC 7120. J Bacteriol. 2003;185(7):2306–2314. doi: 10.1128/JB.185.7.2306-2314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ungerer JL, Pratte BS, Thiel T. RNA processing of nitrogenase transcripts in the cyanobacterium Anabaena variabilis. J Bacteriol. 2010;192(13):3311–3320. doi: 10.1128/JB.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T, Yoshimura H, Ehira S, Ikeuchi M, Ohmori M. AnCrpA, a cAMP receptor protein, regulates nif-related gene expression in the cyanobacterium Anabaena sp. strain PCC 7120 grown with nitrate. FEBS Lett. 2007;581(1):21–28. doi: 10.1016/j.febslet.2006.11.070. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto H, Kurumiya S, Ohashi R, Fujita Y. Oxygen sensitivity of a nitrogenase-like protochlorophyllide reductase from the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol. 2009;50(9):1663–1673. doi: 10.1093/pcp/pcp111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.