Significance
The glutamyl-tRNA reductase (GluTR)-catalyzed reduction of glutamyl-tRNA is the rate-limiting and a pivotal regulation step in the tetrapyrrole biosynthetic pathway. In chloroplast-containing photosynthetic organisms, GluTR binding protein (GluBP) is a newly identified spatial regulator that allocates GluTR for synthesis of different tetrapyrrole products. We find that GluBP stimulates GluTR catalytic efficiency. The structure of the GluTR–GluBP complex shows that GluBP binding promotes GluTR to a hydride-transferring state, the second step of the glutamyl-tRNA reduction, revealing structural details for the catalytic process. These findings clarify a series of arguments regarding the activation and regulation of GluTR. The GluBP structure also suggests that GluBP may have a novel role in heme metabolism.
Abstract
Tetrapyrrole biosynthesis in plants, algae, and most bacteria starts from the NADPH-dependent reduction of glutamyl-tRNA by glutamyl-tRNA reductase (GluTR). The GluTR-catalyzed reaction is the rate-limiting step, and GluTR is the target of multiple posttranslational regulations, such as heme feedback inhibition, for the tetrapyrrole biosynthetic pathway. A recently identified GluTR regulator, GluTR binding protein (GluBP), has been shown to spatially organize tetrapyrrole synthesis by distributing GluTR into different suborganellar locations. Here we report the complex structure of GluTR–GluBP from Arabidopsis thaliana. The dimeric GluBP binds symmetrically to the catalytic domains of the V-shaped GluTR dimer via its C-terminal domain. A substantial conformational change of the GluTR NADPH-binding domain is observed, confirming the postulated rotation of the NADPH-binding domain for hydride transfer from NADPH to the substrate. Arg146, “guarding the door” for metabolic channeling, adopts alternative conformations, which may represent steps involved in substrate recognition and product release. A coupled enzyme assay shows that GluBP stimulates GluTR catalytic efficiency with an approximate threefold increase of the 5-aminolevulinic acid formation rate. In addition, the GluTR activity can be inhibited by heme in a concentration-dependent way regardless of the presence of GluBP. A structural alignment indicates that GluBP belongs to a heme-binding family involved in heme metabolism. We propose a catalytic mechanism model for GluTR, through which photosynthetic organisms can achieve precise regulation of tetrapyrrole biosynthesis.
Tetrapyrroles, including chlorophyll, heme, siroheme, and phytochromobilin, are essential in virtually all living organisms and play key roles in a variety of biological processes such as photosynthesis and respiration (1). 5-Aminolevulinic acid (ALA) is the universal precursor for tetrapyrrole biosynthesis. In plants, algae, and most bacteria, ALA is synthesized by a two-step reaction starting from tRNA-bound glutamate (2). First, the initial substrate glutamyl-tRNA is reduced to glutamate-1-semialdehyde (GSA) by glutamyl-tRNA reductase (GluTR). Then the unstable metabolic intermediate GSA is isomerized to ALA by GSA-2,1-aminomutase (GSAM). The NADPH-dependent reduction of glutamyl-tRNA catalyzed by GluTR is rate-limiting for ALA formation, and GluTR is the hub of a multilayered regulation for all of the tetrapyrrole biosynthetic pathway in photosynthetic organisms at the transcriptional and posttranslational levels (3, 4). Fine tuning of GluTR activity in chloroplasts is achieved by a combination of three known factors: (i) the feedback inhibitor heme (5), (ii) the negative regulator of chlorophyll biosynthesis FLU (6, 7), and (iii) the spatial organizer GluTR binding protein (GluBP) (8). GluBP was previously identified as proton gradient regulation 7 (PGR7) because its truncated mutant is defective in photosynthetic electron transport (9). This pgr7 phenotype presumably results from a deficiency in heme for the cytochrome b6f complex in the photosynthetic electron transport chain (8). It has been suggested that GluBP ensures heme biosynthesis during dark periods by preventing GluTR from binding to FLU, and thus distinguishes chlorophyll and heme-synthesizing pathways (10). The fact that GluBP is found in all chloroplast-containing organisms indicates that GluBP is a common regulator of GluTR (8). Despite the insight gained from these studies, the molecular basis underlying GluTR regulation by GluBP remains to be explored.
The X-ray structure of the dimeric GluTR from the deep-sea archaeon Methanopyrus kandleri (MkGluTR) has been obtained (11). Its open V-shape provides a complementary surface for the association of a GSAM dimer (12). The proposed GluTR–GSAM complex enables an intermolecular channel for GSA transfer to the active site of GSAM without exposure to the aqueous environment (11). The structure of MkGluTR has substantiated a thioester-mediated catalytic mechanism for GluTR (13, 14), but steps including hydride transfer from NADPH to the thioester intermediate and product GSA release are not clear (15, 16). Furthermore, MkGluTR is an atypical GluTR because M. kandleri is a hyperthermophile and its genome does not contain the genes to convert uroporphyrinogen III into heme (17). The lack of structural information for GluTR from photosynthetic organisms has hampered elucidation of the mechanisms underlying GluTR catalysis and posttranslational regulation as mentioned earlier.
In this study, we describe the structure of Arabidopsis thaliana GluTR in complex with GluBP. The conformation observed for GluTR resembles an active state during the NADPH-dependent reduction. By using a coupled enzyme assay with GSAM, we show that GluBP stimulates GluTR activity. Based on the biochemical and structural information, we ascribe a new role to GluBP in the fine tuning of GluTR activity, and suggest a model for the GluTR catalytic mechanism that may provide a basis for GluTR regulation in all chloroplast-containing photosynthetic organisms.
Results
Overall Structure of the GluTR–GluBP Complex.
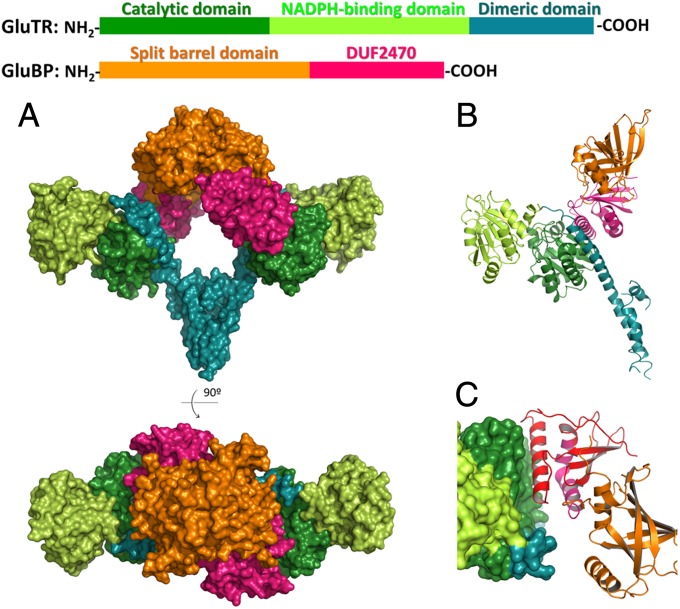
The recombinant mature GluTR and GluBP were expressed and purified separately and then mixed for complex formation. The stable 2:2 complexes were isolated by size-exclusion chromatography (SEC) and crystallized. Despite the sequence similarity between Arabidopsis GluTR and MkGluTR (Fig. S1), molecular replacement was not successful using MkGluTR as the search model. The structure of the GluTR–GluBP complex was determined by the selenomethionyl multiwavelength anomalous dispersion method and refined to 2.8-Å resolution (Fig. 1A and 1B and Table S1). Surprisingly, GluBP binds directly to the cleft of the V-shaped GluTR dimer, in a manner similar to that which has been proposed for the GluTR–GSAM complex (11, 15, 16, 18, 19). This is contrary to the assumption that GluBP does not interfere with the GluTR–GSAM interaction, and therefore GluBP attaches to GluTR’s NADPH-binding domain (10) or dimerization domain (20).
Fig. 1.
The GluTR–GluBP complex. (A) Surface representation of the overall structure, colored by domains. (B) A monomeric half of the GluTR–GluBP complex in ribbon representation showing secondary structure elements. (C) The GluTR–GluBP interface. GluTR is shown in surface representation and GluBP is in ribbon representation. The C-terminal 84-resiude sequence corresponding to the truncated pgr7 mutant is colored in red.
The overall architecture of the GluTR dimer upon GluBP binding does not substantially change in comparison with the MkGluTR structure (11), which has an rmsd value of 3.15 Å. This large rmsd value is mainly a result of the rotation of the NADPH-binding domains at both ends of the V shape (Fig. S2). This rotation can be largely accounted for by a twist of an extended spinal α-helix, which spans from the joint to the end of the V shape and spatially connects the three domains of GluTR along each arm: the C-terminal dimerization domain, the N-terminal catalytic domain, and the NADPH-binding domain. The twist of this spinal helix confirms the hypothesized tilting of the NADPH-binding domain to allow hydride transfer from NADPH to the thioester intermediate (11, 15).
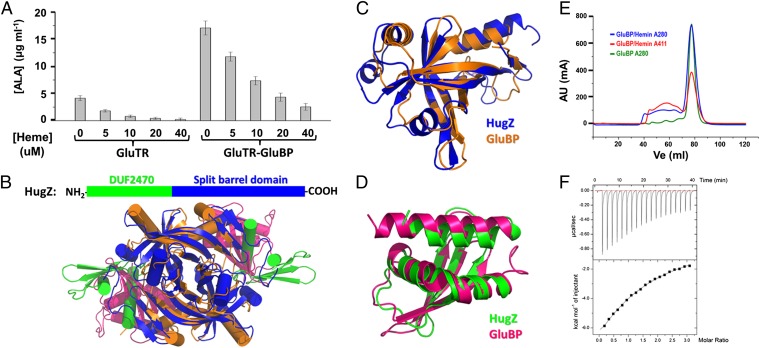
Consistent with previous theoretical prediction (9), GluBP is composed of two domains: an N-terminal flavin mononucleotide (FMN)-binding split barrel domain, and a C-terminal domain of unknown function 2470 (DUF2470) according to the Pfam database (21). The FMN-binding split barrel domain is responsible for dimerization of GluBP, and the DUF2470 domain confers interaction with GluTR. The GluTR–GluBP interface is exclusively between the catalytic domain of GluTR and the last α-helix of the DUF2470 domain (Fig. 1C). This explains the loss of GluTR-binding ability of the defective pgr7 mutant, whose C-terminal 84 residues are truncated (8, 9).
Hydride Transfer from NADPH to Glutamate.
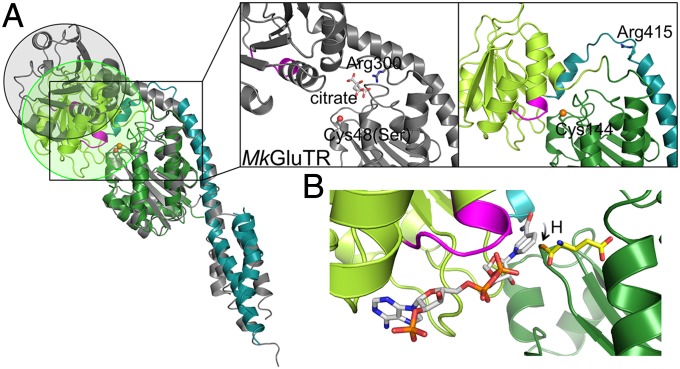
It is known that GluTR employs hydride transfer from NADPH to the thioester-bound glutamate to produce GSA (2, 13). However, in the MkGluTR structure, the NADPH-binding domain is ∼21 Å distant from the active-site cysteine (Cys48 in MkGluTR) (11). To transfer hydride from NADPH to the thioester intermediate, the NADPH-binding domain needs to swing toward the catalytic domain, and thus the MkGluTR structure is described as a “preactive” state (11). Such a proposed conformational change is observed in the structure of the GluTR-GluBP complex: the twist of the spinal α-helix brings the NADPH-binding domain to the vicinity of the active site (Fig. 2A). Interestingly, the twisted region of the spinal α-helix includes a conserved arginine (Arg415 in Arabidopsis GluTR) that has been predicted to recognize the tRNA moiety of the substrate glutamyl-tRNA (11, 15). In the symmetric MkGluTR structure, the equivalent arginine (Arg300) is located near an opening between the catalytic domain and the NADPH-binding domain, and is bound to a citrate, which is deemed to partly mimic the tRNA acceptor arm. Here, Arg415 is in a flexible loop that is inaccessible from the substrate pocket, and the opening that can accommodate the tRNA acceptor arm as observed in the MkGluTR structure is closed (Fig. 2A). Thus, the GluTR–GluBP complex structure may offer a snapshot of the NADPH-dependent reduction step, which follows formation of the thioester intermediate.
Fig. 2.
Rotation of the NADPH-binding domain. (A) Structure comparison of MkGluTR (gray) and the GluBP-bound GluTR (colored as in Fig. 1). The NADPH-recognition motif (GXGXXG) is in magenta. The nucleophile sulfur is in sphere and the conserved arginine on the spinal helix is in stick. (B) Model of NADPH (white) bound to GluTR. A thioester-bound glutamate (yellow) is illustrated.
Because we could not obtain the cocrystal of GluTR–GluBP with NADPH, we generated a NADPH-binding model of GluTR (Fig. 2B) by using the homologous structure of a NADP-binding domain (22). The close contact between the nicotinamide ring of NADPH and the nucleophile Cys144 allows the transfer of hydride from NADPH to the thioester-bound glutamate, supporting the suggestion that the GluTR conformation in the complex represents a hydride transfer state.
Tunnel for Product GSA Release.
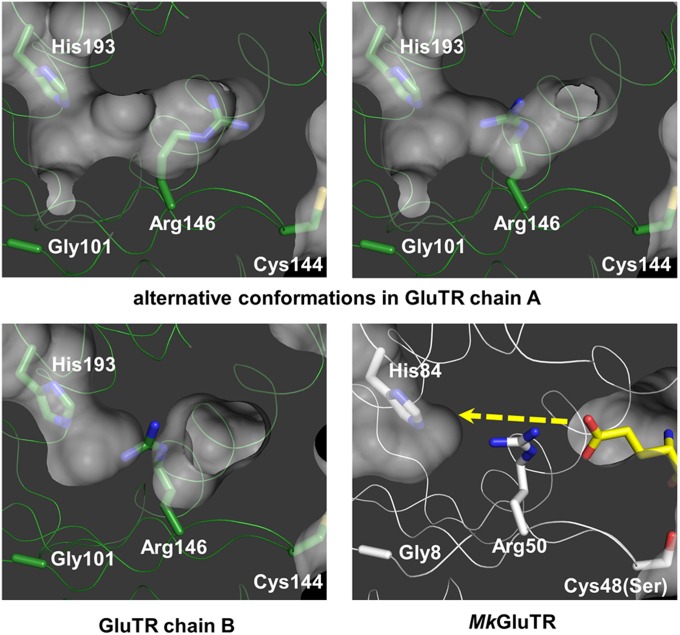
Because the isomerization of GSA to ALA requires an asymmetric allosteric activation of the GSAM dimer (12, 23), we examined whether asymmetry also occurs in GluTR for GSA formation. We compared the two monomeric halves (denoted chain A and chain B) of the GluTR dimer and found a critical difference. Arg146 of chain A has two conformations, which enable the opening of the GSA channel to the “exit” His193 (Fig. 3 and Fig. S3). In chain B, Arg146 is of similar conformation to Arg50 in MkGluTR, which is one of the determinants for substrate glutamate recognition (11). The alternative conformations observed here demonstrate a dual function of this conserved arginine for substrate recognition and product release. It is of note that a pocket is formed by Gly101 on β1, which provides a comfortable space for GSA (Fig. 3). This glycine is also strictly conserved, suggesting that the tunnel observed here may be a common feature for GluTR. The tunnel observed for GSA release in the GluTR–GluBP complex verifies the proposed “back door” for GSA channeling to GSAM, thus providing evidence for a theoretical model developed earlier (11, 14–16, 18).
Fig. 3.
The tunnel for GSA channeling. The yellow dashed line indicates the proposed pathway of GSA. Gly101, Cys144, Arg146, and His193 (corresponding to Gly8, Cys48Ser, Arg50, and His84 in MkGluTR) are shown as sticks and labeled. The glutamate moiety of glutamycin in the MkGluTR structure is shown in yellow stick representation.
GluBP Stimulates GluTR Activity and Regulates GSA Release.
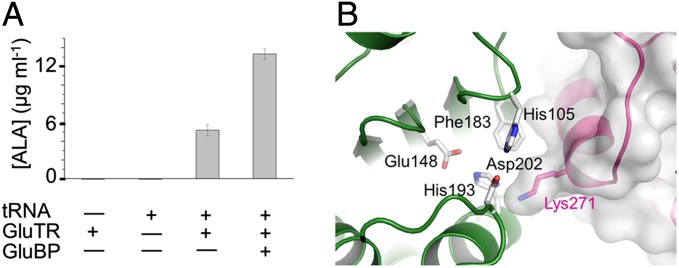
The active conformation of GluTR observed in the GluTR–GluBP complex suggests a stimulatory role of GluBP in GluTR regulation. To test this, we measured the effect of GluBP on GluTR activity by determining ALA formation by using a coupled enzyme assay containing GluTR and GSAM as described previously (24–26). Addition of GluBP to the assay mixture increases ALA formation approximately threefold vs. the level in the absence of GluBP (Fig. 4A). This is reminiscent of GSAM, which stimulates GluTR activity by ∼2.5-fold in a NADPH oxidation assay (26). Notably, the aminomutase activity is dispensable because the inactive and WT forms of GSAM have equal stimulatory effects on GluTR (26). Our results suggest that, like GSAM, GluBP binds and promotes the transition of GluTR to the hydride-transferring active state, thus accelerating the formation of GSA. Considering the similar effects of GSAM and GluBP on GluTR, the transition of GluTR from preactive to active state is most likely stimulated by GluTR–GSAM interaction or by GluTR–GluBP interaction.
Fig. 4.
Effect of GluBP on GluTR activity. (A) GluBP significantly increases ALA synthesis. Data are presented as the mean ± SEM of six independent experiments. (B) Residues in the GluTR–GluBP interface incorporating the exit pocket of GSA, shown in stick representation and labeled.
The GluTR-GluBP interface incorporates the exit pocket of GSA (Fig. 4B). The side chain of GluBP Lys271 stretches to a concaved surface of the GluTR catalytic domain, which is just outside the tunnel where the conserved His193 is located [corresponding to His84 in MkGluTR (11, 15)]. Lys271 largely shields the GSA exit tunnel, implying a regulatory role for GluBP in GSA release.
GluBP Belongs to a Heme-Binding Protein Family.
GluBP was proposed to be involved in heme-mediated metabolism (8). To examine the effect of heme on GluBP activity, we measured ALA formation in the presence of GluBP and heme by using the coupled enzyme assay described earlier. The results show that to reach a similar repression level of ALA formation, the heme concentration needs to be approximately fourfold higher in the presence than in the absence of GluBP (Fig. 5A). Interestingly, a Dali search (27) reveals that GluBP is structurally homologous to a newly discovered heme-binding protein, heme utilization gene Z (HugZ) (28). Both GluBP and HugZ are composed of an FMN-binding split barrel domain and a DUF2470 domain, except that the two domains are rearranged (Fig. 5B). The N- and C-terminal domains of GluBP can be superimposed on the C- and N-terminal domains of HugZ, with rmsd values of 2.2 Å (for 115 aligned Cα) and 1.7 Å (for 77 aligned Cα; Fig. 5 C and D). These findings raise the possibility that GluBP can bind heme. To test this, we first examined the interactions between GluBP and heme by SEC. A comigration was observed during SEC (Fig. 5E), indicating binding of GluBP to heme. Then, we used isothermal titration calorimetry (ITC) to determine their stoichiometry. The titration curve reveals a 1:1 ratio of heme to GluBP (Fig. 5F). From these results, we conclude that GluBP is a heme-binding protein.
Fig. 5.
GluBP belongs to a heme-binding protein family. (A) Heme represses ALA synthesis in a concentration-dependent manner. Data are presented as the mean ± SEM of six independent experiments. (B) Ribbon representation showing the overall structures of GluBP and HugZ. (C) Superposition of the FMN-binding split barrel domains. (D) Superposition of the DUF2470 domains. (E) Elution profiles of GluBP in the absence (green line) and presence (blue line) of hemin. After incubation with a twofold excess of hemin, GluBP sample was subjected to Superdex 200 column. The red line corresponds to absorbance at 411 nm. (F) ITC titration of GluBP with hemin. The fitted parameters for a single site binding model are as follows: n = 1.08 ± 0.14, Kd = 0.20 ± 0.02 mM, ΔH = −21.6 ± 3.5 kcal/mol, and ΔS = −56.7 cal/mol/K.
Discussion
Here we report the crystal structure of the Arabidopsis GluTR–GluBP complex and provide new insights into the activation and regulation of GluTR. A comparison with the archaeon MkGluTR structure substantiates the longstanding hypothesis that rotation of the NADPH-binding domain is required for the NADPH-dependent reduction step. The alternative conformations observed for Arg146 also support the predicted GSA channel, whose gating requires a rotation of the side chain of this arginine. The alternative conformations may be viewed as snapshots along the channeling process. Interestingly, each GluTR monomer within the GluTR–GluBP complex is not exactly twofold symmetric, as only chain A has two conformations for Arg146. Considering the asymmetric allosteric activation mechanism of the GSAM dimer (12, 23), the asymmetry observed for GluTR allows the possibility of a concerted process involving GSA release from GluTR and subsequent capture by GSAM.
Unexpectedly, GluBP binds to the catalytic domain of GluTR at the site predicted to form an intermolecular channel for GSA transfer. A coupled enzyme assay with GSAM by measuring ALA formation demonstrates that GluBP stimulates GluTR. This stimulatory effect of GluBP on GluTR for ALA synthesis could explain that, in the Arabidopsis pgr7 mutant, both chlorophyll and heme contents are reduced (8). However, the different reduction degree of heme and chlorophyll in the pgr7 mutant indicates the existence of separate regulation mechanisms for different tetrapyrrole production pathways (10).
GluBP is structurally homologous to the heme utilization protein, HugZ, which is essential for heme acquisition by bacteria (29). According to Pfam classification (21), the N-terminal domain of GluBP and the C-terminal domain of HugZ belong to the PNP oxidase-like family, a member of the FMN-binding split-barrel superfamily. However, we could not detect binding of flavin to GluBP, which is consistent with previous studies (8). For the DUF2470 domain shared by GluBP and HugZ, two functions can be assigned. In GluBP, it contributes exclusively to GluTR recognition, and, in HugZ, it partially shields the heme-binding pocket (27). ITC experiments indicate a 1:1 binding ratio of heme to GluBP, confirming that GluBP is a heme-binding protein.
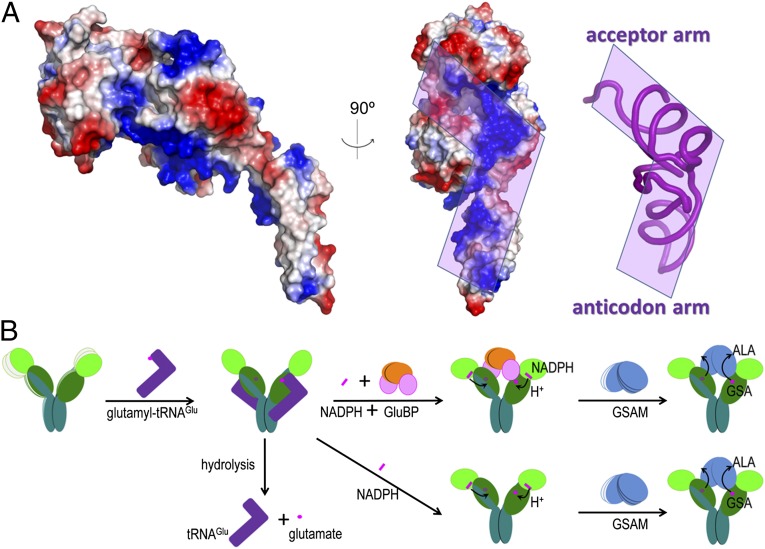
A continuous stretch of positively charged surface on the GluTR catalytic domain may define a region for tRNA binding (Fig. 6A). The L-shaped tRNA can be placed on the GluTR surface with its anticodon positioned near the dimerization domain, in a manner comparable to glutamyl-tRNA synthetase (30). The opening between the catalytic domain and the NADPH-binding domain is required for accommodating the acceptor arm, and thus recognizing the integrity of tRNA (31). Compared with the MkGluTR structure, the structure of GluTR complexed with GluBP may exist in a state following tRNA release.
Fig. 6.
Glutamyl-tRNAGlu binding site and a proposed working model of GluTR. (A) The electrostatic surface potential representation of the GluTR monomer. The region predicted to interact with tRNA phosphate backbone is indicated in purple. (B) Model of GluTR catalysis and its regulation by GluBP. The presence of GluBP favors the NADPH-dependent reduction of the thioester-bound glutamate and thus increases the GSA formation rate. In the following step, GSAM captures GSA through metabolic channeling and isomerizes the unstable GSA to ALA.
Based on the structure of the GluTR-GluBP complex and the new findings on GluBP function, a model for the GluTR catalytic mechanism is presented that may provide a basis for GluTR regulation (Fig. 6B). In this model, the stimulatory role of GluBP is achieved by driving a conformational change of the preactive GluTR, which is depicted in the MkGluTR structure. As predicted previously (11), this conformational change mainly occurs in the extended α-helix connecting the three domains of GluTR. A possible consequence of GluBP binding is the stabilization of the V-shape, whose two arms, when in the absence of GluBP or GSAM, are in wobbling motion with respect to each other. This is in accordance with the role of the citrate anion that stabilizes the MkGluTR structure, where this anion is predicted to partly mimic the acceptor stem of the substrate glutamyl-tRNA (11). Likewise, during the NADPH-dependent reduction step, which follows formation of the thioester intermediate and release of tRNA, GSAM as the counterpart is needed to maintain the otherwise wobbling V-shaped conformation. The biochemical role of GluBP is to shift the conformational equilibrium toward the active state, providing a strategy to regulate the GluTR-GSAM enzyme relay system.
In summary, we report the crystal structure of Arabidopsis GluTR in complex with its stimulator protein GluBP at 2.8-Å resolution. This structure represents an active state and provides novel insights into the catalysis and regulation of GluTR.
Materials and Methods
Protein Expression and Purification.
The genes of Arabidopsis GluTR (At1g58290) and GluBP (At3g21200) lacking the plastid-targeting sequences were cloned into pMAL-c5X (New England Biolabs) and pET-28a(+) (Novagen) vectors, respectively. Escherichia coli cells expressing GluTR were sonicated in a solution containing 20 mM Na-Hepes, pH 8.1, 1M NaCl, and 5 mM DTT (solution A). The cleared lysate was applied to a maltose binding protein (MBP) affinity column. The MBP fusion GluTR was eluted by solution A supplemented with 40 mM maltose, and the MBP tag was then cleaved by tobacco etch virus protease. Protein aggregates and the MBP tag were removed by SEC by using a HiLoad 16/60 Superdex 200 column (GE Healthcare). The fractions corresponding to GluTR dimer were pooled and concentrated, then shock-frozen in liquid nitrogen and stored at −80 °C for further crystallization or activity assay. E. coli cells expressing GluBP were sonicated in a solution containing 20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 20 mM imidazole (solution B). The cleared lysate was loaded onto a Ni-NTA column (Qiagen). The recombinant GluBP protein was eluted by solution B supplemented with 200 mM imidazole. The GluTR–GluBP complex was obtained by mixing the purified GluTR and GluBP at a ratio of 1:1.2. Then, excess GluBP was removed by SEC by using a HiLoad 16/60 Superdex 200 column (GE Healthcare). The GluTR–GTRBP complex was pooled and concentrated to 5 mg/mL with a 100-kDa-cutoff centrifugal filter (Millipore). SDS/PAGE analysis confirmed the homogeneity of the purified complex. The selenomethionine derivatives of GluTR and GluBP were expressed in E. coli B834 (DE3) cells and purified as described earlier.
Crystallization, Data Collection, and Structure Determination.
Initial crystallization screening was performed by the vapor-diffusion method in a sitting drop consisting of 1 µL protein solution mixed with 1 µL well solution at 289 K. Diffraction-quality crystals of the complex were grown against a reservoir solution consisting of 22.5% (wt/vol) PEG 3,350, 0.3 M ammonium citrate tribasic, pH 7.0. The crystals were transferred step by step into drops of the mother liquid supplemented with 5%, 10%, 20% (vol/vol) glycerol before being flash-frozen in liquid nitrogen. All X-ray diffraction data sets were collected at beam line BL17U of the Shanghai Synchrotron Radiation Facility at a wavelength of 0.9793 Å at 100 K, integrated, and scaled by using DENZO and SCALEPACK as implemented in HKL2000 (32). Selenium positions of GluBP in the complex were determined by using the 4.2-Å selenomethionine derivative data by the program AutoSol in PHENIX suite (33), and 22 selenium sites were found in two GluBP molecules per asymmetric unit. These 22 selenium sites were applied to the 3.1-Å selenomethionine derivative data, and additional 34 selenium sites were found for the GluTR dimer. The identified selenium sites were then refined, and the initial phases were generated using the program AutoBuild in PHENIX suite. Additional missing residues in the auto-built model were added according to the 2Fo−Fc and Fo−Fc electron density maps by manually model building using COOT (34) and the model was refined with PHENIX. The overall quality of final structure was assessed by PROCHECK (35), with 94.7%, 4.8%, and 0.5% of the amino acids in the most favored, additionally allowed, and disallowed regions of the Ramachandran plot, respectively. Data collection and structure refinement statistics are summarized in Table S1. The protein structure figures were prepared by using the program PyMOL (www.pymol.org).
GluTR Activity Assay.
GluTR activity was measured in a reconstitution assay with purified recombinant E. coli glutamyl-tRNA synthetase and GSAM (24–26). The 250-µL assay mixture contained 50 µg glutamyl-tRNA synthetase, 156 µg GluTR, 195 µg GSAM, 1 mM NADPH, 200 µM glutamate, and 360 µg E. coli total tRNA, in a buffer consisting of 50 mM K-Hepes, pH 7.9, 1 mM DTT, and 25 mM MgCl2. The ALA synthetic reactions were performed by incubating the mixture at 25 °C for 3 h and stopped by adding 30 µL 7% (wt/vol) perchloric acid and cooling on ice. Reaction mixture without GluTR or tRNA for each condition used served as background controls. The protein precipitates were removed by centrifugation, and the supernatant containing ALA was analyzed by colorimetric analysis (25, 26). The light absorption was measured at 553 nm, at which the maximum absorption of the Ehrlich salt of ALA-pyrrole occurs. For assay with GluBP, the mixture was prepared as described earlier except that the appropriate amount of GluBP was added to replace the same volume of buffer.
Heme Preparation.
Hemin (ferriprotoporphyrin IX chloride; Sigma-Aldrich) was dissolved in 0.1 N NaOH and neutralized to pH 7.5 using 1 M Tris⋅HCl. The solution was diluted with appropriate buffers to desired concentrations. Fresh heme solution was prepared to ensure no aggregation for each assay.
ITC.
Calorimetric measurements were carried out on a MicroCal iTC200 system (GE Healthcare) (36). Twenty injections of 2 μL of 2 mM hemin were made into a 100 µM GluBP solution in the cell. The heat of dilution of hemin upon injection into the buffer solution (20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl) was subtracted from the experimental titration data. Baseline correction and integration of the calorimeter signals were performed by using the MicroCal Origin software provided by the manufacturer.
Supplementary Material
Acknowledgments
We acknowledge assistance by the staff at the Shanghai Synchrotron Radiation Facility of China and thank Mingzhu Wang [Institute of Biophysics of the Chinese Academy of Sciences (IBP-CAS)] for experimental support and Sarah Perrett (IBP-CAS) for critically reading the manuscript. This research was supported by National Natural Science Foundation of China Grant 31370759, Ministry of Science and Technology of China Grant 2011CBA00901, and the Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4N7R).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400166111/-/DCSupplemental.
References
- 1.Mochizuki N, et al. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15(9):488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Jahn D, Verkamp E, Söll D. Glutamyl-transfer RNA: A precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci. 1992;17(6):215–218. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 4.Czarnecki O, Grimm B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot. 2012;63(4):1675–1687. doi: 10.1093/jxb/err437. [DOI] [PubMed] [Google Scholar]
- 5.Beale SI. Enzymes of chlorophyll biosynthesis. Photosynth Res. 1999;60:43–73. [Google Scholar]
- 6.Meskauskiene R, et al. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98(22):12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goslings D, et al. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40(6):957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 8.Czarnecki O, et al. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell. 2011;23(12):4476–4491. doi: 10.1105/tpc.111.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung HS, et al. Arabidopsis thaliana PGR7 encodes a conserved chloroplast protein that is necessary for efficient photosynthetic electron transport. PLoS ONE. 2010;5(7):e11688. doi: 10.1371/journal.pone.0011688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czarnecki O, Grimm B. New insights in the topology of the biosynthesis of 5-aminolevulinic acid. Plant Signal Behav. 2013;8(2):e23124. doi: 10.4161/psb.23124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser J, et al. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 2001;20(23):6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennig M, Grimm B, Contestabile R, John RA, Jansonius JN. Crystal structure of glutamate-1-semialdehyde aminomutase: An α2-dimeric vitamin B6-dependent enzyme with asymmetry in structure and active site reactivity. Proc Natl Acad Sci USA. 1997;94(10):4866–4871. doi: 10.1073/pnas.94.10.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser J, Lorenz S, Hubschwerlen C, Rompf A, Jahn D. Methanopyrus kandleri glutamyl-tRNA reductase. J Biol Chem. 1999;274(43):30679–30685. doi: 10.1074/jbc.274.43.30679. [DOI] [PubMed] [Google Scholar]
- 14.Schauer S, et al. Escherichia coli glutamyl-tRNA reductase. Trapping the thioester intermediate. J Biol Chem. 2002;277(50):48657–48663. doi: 10.1074/jbc.M206924200. [DOI] [PubMed] [Google Scholar]
- 15.Schubert WD, Moser J, Schauer S, Heinz DW, Jahn D. Structure and function of glutamyl-tRNA reductase, the first enzyme of tetrapyrrole biosynthesis in plants and prokaryotes. Photosynth Res. 2002;74(2):205–215. doi: 10.1023/A:1020963711861. [DOI] [PubMed] [Google Scholar]
- 16.Lüer C, et al. Glutamate recognition and hydride transfer by Escherichia coli glutamyl-tRNA reductase. FEBS J. 2007;274(17):4609–4614. doi: 10.1111/j.1742-4658.2007.05989.x. [DOI] [PubMed] [Google Scholar]
- 17.Slesarev AI, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci USA. 2002;99(7):4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lüer C, et al. Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J Biol Chem. 2005;280(19):18568–18572. doi: 10.1074/jbc.M500440200. [DOI] [PubMed] [Google Scholar]
- 19.Schulze JO, Schubert WD, Moser J, Jahn D, Heinz DW. Evolutionary relationship between initial enzymes of tetrapyrrole biosynthesis. J Mol Biol. 2006;358(5):1212–1220. doi: 10.1016/j.jmb.2006.02.064. [DOI] [PubMed] [Google Scholar]
- 20.Richter AS, Grimm B. Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway in plants. Front Plant Sci. 2013;4:371. doi: 10.3389/fpls.2013.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker PJ, Sawa Y, Shibata H, Sedelnikova SE, Rice DW. Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase. Nat Struct Biol. 1998;5(7):561–567. doi: 10.1038/817. [DOI] [PubMed] [Google Scholar]
- 23.Stetefeld J, Jenny M, Burkhard P. Intersubunit signaling in glutamate-1-semialdehyde-aminomutase. Proc Natl Acad Sci USA. 2006;103(37):13688–13693. doi: 10.1073/pnas.0600306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vothknecht UC, Kannangara CG, von Wettstein D. Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc Natl Acad Sci USA. 1996;93(17):9287–9291. doi: 10.1073/pnas.93.17.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava A, Beale SI. Glutamyl-tRNA reductase of Chlorobium vibrioforme is a dissociable homodimer that contains one tightly bound heme per subunit. J Bacteriol. 2005;187(13):4444–4450. doi: 10.1128/JB.187.13.4444-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogaj LA, Beale SI. Physical and kinetic interactions between glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase of Chlamydomonas reinhardtii. J Biol Chem. 2005;280(26):24301–24307. doi: 10.1074/jbc.M502483200. [DOI] [PubMed] [Google Scholar]
- 27.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, et al. Crystal structure of HugZ, a novel heme oxygenase from Helicobacter pylori. J Biol Chem. 2011;286(2):1537–1544. doi: 10.1074/jbc.M110.172007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilks A, Burkhard KA. Heme and virulence: How bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24(3):511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 30.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat Struct Biol. 2001;8(3):203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 31.Randau L, et al. tRNA recognition by glutamyl-tRNA reductase. J Biol Chem. 2004;279(33):34931–34937. doi: 10.1074/jbc.M401529200. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59(pt 3):228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26(2):283–291. [Google Scholar]
- 36.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179(1):131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.