Significance
DNA, the blueprint of the cell, is constantly damaged by chemicals and radiation. Because DNA damage may cause cell death or mutations that may lead to diseases such as cancer, cells are armed with an arsenal of several distinct mechanisms for repairing the many types of DNA damage that occur. DNA repair capacity (DRC) varies among individuals, and reduced DRC is associated with disease risk; however, the available DRC assays are labor intensive and measure only one pathway at a time. Herein, we present powerful new assays that measure DRC in multiple pathways in a single assay. We use the assays to measure interindividual DRC differences and inhibition of DNA repair, and to uncover unexpected error-prone transcriptional bypass of a thymine dimer.
Keywords: personalized medicine, disease prevention, flow cytometry, high-throughput sequencing
Abstract
The capacity to repair different types of DNA damage varies among individuals, making them more or less susceptible to the detrimental health consequences of damage exposures. Current methods for measuring DNA repair capacity (DRC) are relatively labor intensive, often indirect, and usually limited to a single repair pathway. Here, we describe a fluorescence-based multiplex flow-cytometric host cell reactivation assay (FM-HCR) that measures the ability of human cells to repair plasmid reporters, each bearing a different type of DNA damage or different doses of the same type of DNA damage. FM-HCR simultaneously measures repair capacity in any four of the following pathways: nucleotide excision repair, mismatch repair, base excision repair, nonhomologous end joining, homologous recombination, and methylguanine methyltransferase. We show that FM-HCR can measure interindividual DRC differences in a panel of 24 cell lines derived from genetically diverse, apparently healthy individuals, and we show that FM-HCR may be used to identify inhibitors or enhancers of DRC. We further develop a next-generation sequencing-based HCR assay (HCR-Seq) that detects rare transcriptional mutagenesis events due to lesion bypass by RNA polymerase, providing an added dimension to DRC measurements. FM-HCR and HCR-Seq provide powerful tools for exploring relationships among global DRC, disease susceptibility, and optimal treatment.
DNA is under constant assault from damaging agents that produce a vast array of lesions. Left unrepaired, these lesions have the potential to alter cellular function and compromise the health of the organism, leading to degenerative diseases, cancer, and premature aging (1–5). Consequently, interindividual variations in DNA repair capacity (DRC) are thought to contribute to the fact that people have different susceptibilities to these diseases (6–10). Furthermore, the efficacy of cancer chemotherapy with DNA-damaging agents depends on the DRC of the targeted cells (11). Thus, DRC measurements potentially might be used to personalize both treatment and prevention of disease.
We define DRC as the basal ability of cells to eliminate DNA damage from the genome; however, it should be noted that some DRC assays, such as mutagen sensitivity assays, also may reflect changes in gene expression and the activation of non-DNA repair pathways upon treatment of cells with DNA-damaging agents. A wide variety of methods are used to estimate DRC. Many studies focus on indirect assessments of DRC through transcriptional profiling, proteomics, and single-nucleotide polymorphism (SNP) screens (12–16). However, SNPs in DNA repair genes are not informative if the relevant gene is not expressed. Likewise, gene expression data are not informative for cases in which the gene product is inactive or cannot be assembled into a functional complex. More direct measurements of DRC in vitro using cell lysates have overcome some of this complexity by integrating these factors into a single readout (17–20); however, these methods require separate assays for measurements in more than one repair pathway, and may not be representative of DRC in cells. Measuring the consequences of DNA repair in intact cells by monitoring sister chromatid exchanges, chromosome aberrations, or DNA strand breaks by comet assays also integrates complexity into a single readout, but it requires labor-intensive analyses that make it refractory to implementation in a clinical setting (21–23). Although recently developed high-throughput comet assays provide an excellent alternative, they are limited to DNA damage that leads to, or can be converted to, DNA strand breaks (24).
Host cell reactivation (HCR) assays report the ability of cells to repair DNA damage that blocks transcription of a transiently transfected reporter gene (8, 9, 25–28). Repair of the transcription-blocking lesion reactivates reporter gene expression, thus providing a quantitative readout for DRC. However, HCR assays generally cannot report repair of DNA lesions that do not block the progression of the RNA polymerase, and current methods for measuring DRC are limited further by the need for separate experiments to measure repair capacity in more than one pathway, or at more than one dose of DNA damage. Herein, we introduce a new high-throughput, fluorescence-based multiplex HCR assay (FM-HCR) for measuring DRC in living cells that overcomes these limitations. We first present a multicolor fluorescence assay that simultaneously measures DNA repair at multiple doses of DNA damage. We then demonstrate simultaneous DRC measurements for up to four repair pathways in human and rodent cells, using reporters for the repair of both transcription-blocking lesions and lesions that are bypassed by RNA polymerase. To demonstrate potential applications of FM-HCR to population studies, we measure interindividual DRC differences in five pathways in a panel of 24 lymphoblastoid cell lines derived from apparently healthy individuals. We also show that FM-HCR may be used to identify agents that inhibit or enhance DRC. Finally, to increase throughput further and to establish a single generalized detection method for the repair of any lesion that alters transcription of a reporter gene, we added to the HCR reporter protein paradigm, a reporter transcript assay that leverages the extraordinary power of next-generation sequencing (HCR-Seq). We use HCR-Seq to measure 20 independent reporter signals in a single assay and detect error-prone transcriptional bypass at a bulky DNA lesion in human cells. FM-HCR and HCR-Seq provide rapid, high-throughput methods of assessing DRC in multiple pathways and represent a major improvement over standard methods currently used in basic, clinical, and epidemiological research addressing the relationship among DNA damage, DNA repair, and disease susceptibility.
Results
Validation of FM-HCR.
FM-HCR was used to assay DRC in 55 cell lines (Table 1). Expression levels of five fluorescent reporter proteins were quantitated simultaneously using flow cytometry (Fig. S1). Use of 96-well electroporation plates reduced the time required for transfection to less than an hour per plate, and a BD High Throughput Sampler permitted data acquisition in less than 10 min active time.
Table 1.
Fifty-five cell lines used for this study
| Cell line | Cell type | Repair deficiency | Repair defect |
| GM01630 (i) | Fibroblast | XPA | NER, severe |
| GM01953 | Lymphoblastoid | WT | None |
| GM02246 | Lymphoblastoid | XPC | NER, moderate |
| GM02249 | Lymphoblastoid | XPC | NER, mild |
| GM02253 | Lymphoblastoid | XPD | NER, severe |
| GM02344 (i) | Lymphoblastoid | XPA | NER, severe |
| GM02345 | Lymphoblastoid | XPA | NER, severe |
| GM03657 (ii) | Lymphoblastoid | WT | None |
| GM03658 (ii) | Fibroblast | WT | None |
| GM07752 (iii) | Lymphoblastoid | WT | None |
| GM07753 (iii) | Fibroblast | WT | None |
| GM14878 (iv) | Lymphoblastoid | XPC | NER, very mild |
| GM14879 (iv) | Fibroblast | XPC | NER, very mild |
| GM21071 (v) | Fibroblast | XPB | NER, severe |
| GM21148 (v) | Lymphoblastoid | XPB | NER, severe |
| GM21677 (vi) | Lymphoblastoid | WT | None |
| GM21833 (vii) | Lymphoblastoid | WT | None |
| GM23249 (vi) | Fibroblast | WT | None |
| GM23251 (vii) | Fibroblast | WT | None |
| TK6 | Lymphoblastoid | MGMT | DR of O6-MeG |
| MT1 | Lymphoblastoid | MGMT, MSH6 | MMR and DR of O6-MeG |
| TK6+MGMT | Lymphoblastoid | WT | None |
| HCT116 | Colorectal carcinoma | MLH1 | MMR |
| HCT116+3 | Colorectal carcinoma | WT | None |
| M059J | Glioblastoma | DNA PKcs | NHEJ |
| M059K | Glioblastoma | WT | None |
| WT MEFS | MEFs | WT | None |
| Ogg1−/− MEFS | MEFs | Ogg1 | BER of 8-oxoG |
| V79 (hamster) | Fibroblasts | WT | None |
| VC8 (hamster) | Fibroblasts | BRCA2 | HR |
| xrs6 (hamster) | CHO | Ku80 | NHEJ |
| GM15029 (1) | Lymphoblastoid | WT | None |
| GM15036 (2) | Lymphoblastoid | WT | None |
| GM15215 (3) | Lymphoblastoid | WT | None |
| GM15223 (4) | Lymphoblastoid | WT | None |
| GM15245 (5) | Lymphoblastoid | WT | None |
| GM15224 (6) | Lymphoblastoid | WT | None |
| GM15236 (7) | Lymphoblastoid | WT | None |
| GM15510 (8) | Lymphoblastoid | WT | None |
| GM15213 (9) | Lymphoblastoid | WT | None |
| GM15221 (10) | Lymphoblastoid | WT | None |
| GM15227 (11) | Lymphoblastoid | WT | None |
| GM15385 (12) | Lymphoblastoid | WT | None |
| GM15590 (13) | Lymphoblastoid | WT | None |
| GM15038 (14) | Lymphoblastoid | WT | None |
| GM15056 (15) | Lymphoblastoid | WT | None |
| GM15072 (16) | Lymphoblastoid | WT | None |
| GM15144 (17) | Lymphoblastoid | WT | None |
| GM15216 (18) | Lymphoblastoid | WT | None |
| GM15226 (19) | Lymphoblastoid | WT | None |
| GM15242 (20) | Lymphoblastoid | WT | None |
| GM15268 (21) | Lymphoblastoid | WT | None |
| GM15324 (22) | Lymphoblastoid | WT | None |
| GM15386 (23) | Lymphoblastoid | WT | None |
| GM15061 (24) | Lymphoblastoid | WT | None |
To facilitate comparison of data, the seven individuals from whom both lymphoblastoid and fibroblast cultures were derived have been assigned indexes i through vii. The numbers in parentheses (1–24) correspond to labels in Fig. 4A; five-digit GM identifiers were assigned by the Coriell Cell Repository.
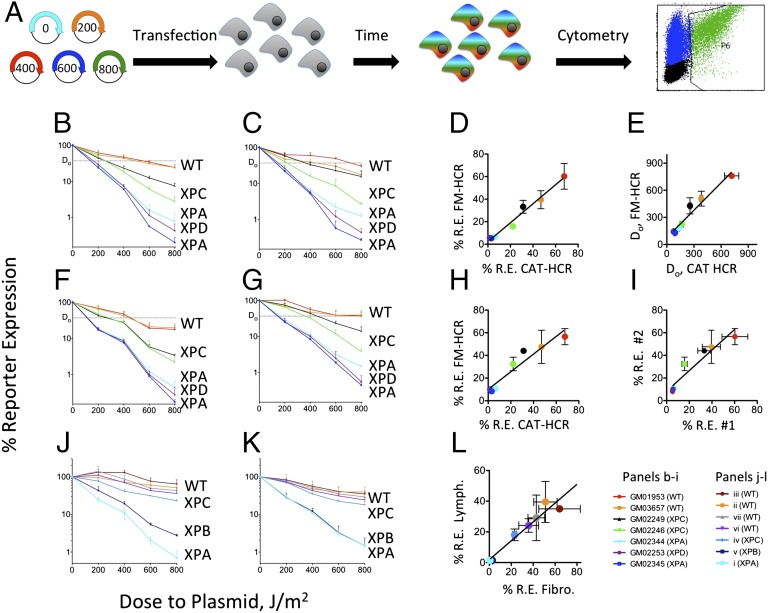
In vitro treatment of plasmids with UV-C light resulted in a dose-dependent reduction in reporter expression. When each of the five fluorescent reporter plasmids was treated with a unique dose of UV-C (plasmid combination 1 in Table 2) and subsequently cotransfected into cells, a dose–response curve was generated from a single experiment that required only two transfections (Fig. 1A). Dose–response curves spanning up to three decades of percent reporter expression (%R.E.) were obtained for seven lymphoblastoid cell lines (Fig. 1 B and C), chosen because they were characterized for their capacity to repair UV-irradiated plasmid DNA by another, much more laborious method more than 20 y ago (8). Differences in DRC were most pronounced at the highest dose to plasmid (800 J/m2), with %R.E. values varying over a range of about 100-fold among the cell lines. As expected, the highest DRC was observed for lymphoblastoid cell lines derived from apparently healthy individuals, referred to as wild type (WT) (Table 1). Moderately reduced DRC was observed for two xeroderma pigmentosum group C (XPC) cell lines, and severe defects were evident for xeroderma pigmentosum group A (XPA) and xeroderma pigmentosum group D (XPD) cell lines. Between 18 and 40 h, %R.E. increased for most cell lines (Fig. 1 B and C), consistent with time-dependent repair of transcription blocking lesions.
Table 2.
Combinations of reporter plasmids and types of DNA damage used in each experiment
| Combination | TagBFP | AmCyan | EGFP | mOrange | mPlum |
| 1 | 600 J/m2 | No lesion | 800 J/m2 | 200 J/m2 | 400 J/m2 |
| 2 | No lesion | 200 J/m2 | 400 J/m2 | 600 J/m2 | 800 J/m2 |
| 3 | No lesion | — | 800 J/m2 | G:G* | O6-MeG |
| 4 | DSB | No lesion | 800 J/m2 | G:G* | O6-MeG |
| 5 | 800 J/m2 | No lesion | A:C† | 8-oxoG | O6-MeG |
| 6 | DSB | — | DSB | — | No lesion |
| 7 | No lesion | 200 J/m2 | T<>T‡ | 400 J/m2 | 800 J/m2 |
G:G mismatch.
A:C mismatch.
Site-specific thymine dimer.
Fig. 1.
Measurements of DRC by FM-HCR. (A) Assay work flow. DNA lesions are introduced into fluorescent reporter plasmids in vitro. Numbers labeling the plasmids represent the dose (in joules per square meter) of UV radiation. Following treatment, plasmids were combined and cotransfected into cells. After 18 or 40 h incubation, cells were assayed for fluorescence by flow cytometry. Comparison of fluorescence signals with those from cells transfected with undamaged plasmids yields a dose–response curve (experimental data for GM02344 with plasmid combination 1 in Table 2) (B) Dose–response curves for seven cell lines 18 h after transfection with plasmid combination 1 (Table 2) (C) Dose–response curves for the cells in B at 40 h. (D) Comparison of %R.E. as measured by FM-HCR at 400 J/m2 is plotted against %CAT as measured by conventional HCR for the same cell lines at 300 J/m2. (E) Do values calculated from FM-HCR data plotted against those reported in the literature (8). Error bars represent the SD calculated from biological triplicates. (F) Dose–response curves for seven cell lines 18 h after transfection with plasmid combination 2 (Table 2). (G) Dose–response curves at 40 h. (H) Comparison of %R.E. as measured by FM-HCR at 400 J/m2 with plasmid combination 2 is plotted against %CAT as measured by conventional HCR for the same cell lines at 300 J/m2. (I) Comparison of FM-HCR data for plasmids treated at 400 J/m2 in experiments 1 and 2. (J) Dose–response curves generated by FM-HCR for lymphoblastoid cell lines 40 h after transfection with plasmid combination 2 (Table 2). (K) Corresponding dose–response curves for primary skin fibroblasts from the same seven individuals. (L) Correlation between %R.E. from plasmids irradiated at 800 J/m2 in the lymphoblastoid and fibroblast cells isolated from the same individuals. Each color in J–L corresponds to one of the individuals (i–vii) in Table 1. Error bars represent the SD calculated from biological triplicates. See also Fig. S1.
The FM-HCR data presented in Fig. 1C reproduce those from the previous study that also monitored transcription inhibition on UV-damaged plasmids 40 h after transfection (8). In that study, chloramphenicol acetyl transferase (CAT) levels in cell-free extracts were used as the reporter. Two complementary methods were used to compare our data with the historical data. First, the percent CAT expression (%CAT) reported at a single dose of UV irradiation (300 J/m2 in ref. 8) was highly correlated (R2 = 0.92, P = 0.0006) with %R.E. at a single dose (400 J/m2) in the present study (Fig. 1D). The relative repair capacity of multiple cell lines also can be compared by calculating the parameter Do, corresponding to the dose at which HCR falls below 37% R.E (29). The Do values calculated from our experimental data also were highly correlated with the historical Do values (R2 = 0.92, P < 0.0001) (Fig. 1E).
To confirm that the dose–response curves in Fig. 1 B and C could be obtained independently of the choice of fluorescent reporters, the experiment was repeated with the plasmids shuffled with regard to which plasmid received a particular UV dose (plasmid combination 2 in Table 2). The resulting dose–response curves obtained at 18 and 40 h are presented in Fig. 1 F and G, respectively. Once again, the FM-HCR data collected at 40 h reproduce the historical data (8) (Fig. 1H). Likewise, the FM-HCR data collected with plasmid combination 2 reproduce those obtained using plasmid combination 1 (Fig. 1I).
FM-HCR assays also were carried out on seven primary untransformed skin fibroblast cell lines and compared with Epstein–Barr virus-transformed lymphoblastoid cell lines derived from the same individuals (represented as individuals i–vii in Table 1); cells were from four apparently healthy individuals and three xeroderma pigmentosum patients. A similar pattern of dose–response curves was obtained for both fibroblasts and lymphoblastoid cells (Fig. 1 J and K, respectively). Overall, absolute nucleotide excision repair (NER) capacity measured in fibroblasts appeared to be somewhat higher than that in the lymphoblastoid cell lines; however, a comparison of DRC measured at 800 J/m2 indicated that NER phenotype is strongly correlated (R2 = 0.94, P = 0.0003) between the two cell types (Fig. 1L).
Development of FM-HCR Assays for DNA Mismatch Repair and Direct Reversal of O6-Methylguanine.
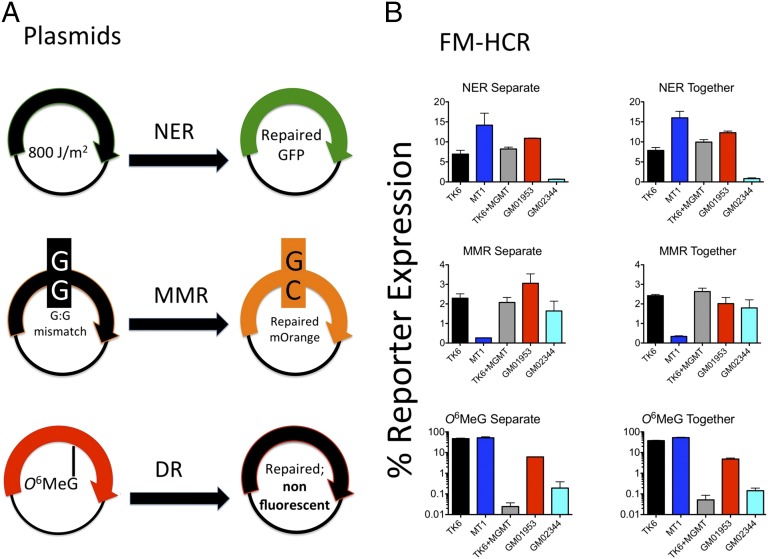
Fluorescent plasmid reporters for direct reversal of O6-methylguanine (O6-MeG) and mismatch repair (MMR) capacity were developed (Fig. S2). For MMR assays, a G:G mismatch-containing plasmid was constructed; it previously was shown that G:G mismatches are repaired inefficiently in extracts from MMR-deficient cells (30), and we confirmed this observation in MMR-deficient HCT116 cells and MMR-proficient HCT116+3 cells that were complemented with human chromosome 3 (Fig. S2D), as well as MMR-deficient MT1 cells that lack MSH6, and MMR-proficient TK6 cells (Fig. 2). The MMR reporter expresses a nonfluorescent protein unless a repair event restores a cytosine in the transcribed strand at the site of the mismatch, leading to WT orange fluorescent protein. Because the plasmid lacks a strand discrimination signal, the theoretical upper limit of reporter expression (relative to a similarly constructed WT homoduplex control) is 50%. For direct reversal of O6-MeG, a plasmid that encodes a nonfluorescent protein in the absence of the lesion was prepared. Introduction of a site-specific O6-MeG lesion into the transcribed strand causes transcription errors (31), producing transcripts encoding the WT mPlum fluorescent protein. Thus, cells deficient for methylguanine methyltransferase (MGMT)-mediated O6-MeG repair express relatively high levels of the mPlum reporter, whereas cells expressing MGMT remove the source of transcription errors, thus reducing reporter expression.
Fig. 2.
Simultaneous measurements of DRC in three pathways. (A) Plasmids used in the multipathway FM-HCR (also see plasmid combination 3 in Table 2). (B) DRC for several cell lines obtained by assaying each pathway in separate transfection experiments (Left) or by assaying all three pathways at once in a single experiment with cotransfected reporter plasmids (Right). Error bars represent the SD calculated from biological triplicates. See also Fig. S2.
Simultaneous Measurement of DRC in Three Pathways with FM-HCR.
DRC for three pathways, namely NER, MMR, and the direct reversal of O6-MeG (MGMT) was measured in five lymphoblastoid cell lines (Fig. 2). DRC for each pathway was first measured in separate experiments. The severe NER defect for the XPA-deficient GM02344 cell line was reproduced, whereas relatively high NER capacity was confirmed for the four other cell lines with no known NER defect (GM01953, MT1, TK6, and TK6+MGMT). The lymphoblastoid cell line MT1, known to be deficient for MMR (32), expressed the MMR reporter at a level up to 10-fold lower than that of the other four cell lines that have no known MMR defects. As expected, the O6-MeG direct-reversal reporter was highly expressed in MT1 and TK6, owing to the absence of MGMT. Expression of the same reporter was reduced nearly 1,000-fold in TK6+MGMT cells expressing a high level of MGMT, and was reduced ∼8-fold and 250-fold in GM01953 and GM02344, respectively, both of which express active MGMT, but at different levels (33).
One of our goals is to increase the throughput of DRC assays by measuring the activity of multiple DNA repair pathways in a single assay. To test whether our DRC reporters could be combined in a single experiment without affecting the accuracy of the assay, we cotransfected three reporter plasmids, each targeting a different pathway, along with an internal transfection control (plasmid combination 3 in Table 2). This yielded DRC profiles for NER, MMR, and MGMT nearly identical to those obtained when the reporters were transfected individually in separate experiments (Fig. 2B).
Simultaneous Measurement of Four DNA Repair Pathways, Including BER and NHEJ.
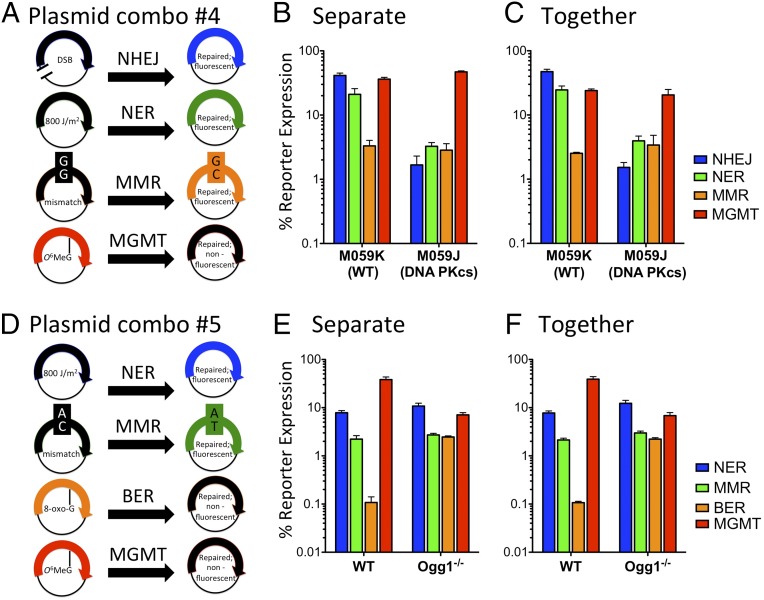
We next sought to measure DRC in four pathways including base excision repair (BER) or double-strand break (DSB) repair (Fig. 3). To add a fourth pathway to the FM-HCR in Fig. 2, a blue fluorescent protein (BFP) reporter for repair of a double-strand break by the nonhomologous end joining (NHEJ) pathway was developed (Fig. 3A and Fig. S3). This assay was validated by using the M059J and M059K cell lines, which were derived from a single glioblastoma (34). The M059J cell line is deficient for the DNA-dependent protein kinase catalytic subunit (DNA PKcs) required for NHEJ (35). As expected, M059J cells expressed ∼40-fold lower levels of the NHEJ reporter relative to the WT M059K cells when the reporter was transfected separately from other reporters (Fig. 3B). To test whether DRC could be measured in four pathways simultaneously, we cotransfected the NHEJ reporter with the reporters described above for NER, MMR, and MGMT (plasmid combination 4 in Table 2). As with the three-pathway measurements described above, the cotransfected reporters yielded DNA repair profiles nearly identical to those of the separately transfected reporters (Fig. 3 B and C). MMR and MGMT capacity was similar in the two cell lines; however, NER capacity was reduced in M059J by approximately sevenfold relative to M059K. It was observed previously that the XPC and ERCC2 genes are overexpressed in M059J vs. M059K cells, and that the M059J cells are slightly more sensitive than M059K cells to UV irradiation (36). Inefficient NER in the presence of excess XPC protein also has been noted in vitro (37).
Fig. 3.
Simultaneous measurements of DRC in four pathways. (A) Plasmids used in the FM-HCR for NHEJ, NER, MMR, and MGMT (also see plasmid combination 4 in Table 2). Note that the undamaged plasmid (AmCyan) included to control for transfection efficiency is not shown. (B) DRC for several cell lines obtained by assaying each pathway in a separate transfection experiment. (C) DRC measured simultaneously following cotransfection of the reporter plasmids. (D) Plasmids used in the FM-HCR for NER, MMR, BER, and MGMT (also see plasmid combination 5 in Table 2). (E) DRC for several cell lines obtained by assaying each pathway in a separate transfection experiment. (F) DRC measured simultaneously following cotransfection of the reporter plasmids. Error bars represent the SD calculated from biological triplicates. See also Fig. S3.
To demonstrate the versatility of FM-HCR further, we performed an assay including reporters for BER, NER, MMR, and MGMT (Fig. 3D). An mOrange fluorescent reporter for base excision repair of 8-oxoguanine (8-oxoG) was developed that produces WT mOrange transcripts when RNA polymerase incorporates adenine opposite a site-specific 8-oxoG lesion. Deficient 8-oxoG repair is expected to result in a higher reporter expression. In keeping with this expectation, mouse embryonic fibroblasts (MEFs) deficient for 8-oxoG DNA glycosylase (Ogg1) expressed an ∼20-fold higher level of mOrange than WT MEFs when the reporter was transfected separately from the other reporters (Fig. 3E). When the 8-oxoG reporter was cotransfected with three other reporters for NER, MMR, and MGMT (plasmid combination 5 in Table 2), we once again observed DRC profiles nearly identical to those obtained when the reporters were transfected separately (Fig. 3 E and F). MMR and NER capacity were similar in the two cell lines, but MGMT reporter expression was approximately fivefold higher in the WT MEFs. Because the WT MEFs were derived from C57BL/6J mice, the differences in MGMT repair capacity might be a result of the mixed (C57BL/6J and 129S) background of the Ogg1−/− mouse from which the MEFs were derived (38).
Analysis of DRC for Five Pathways in a Panel of 24 Cell Lines Derived from Apparently Healthy Individuals.
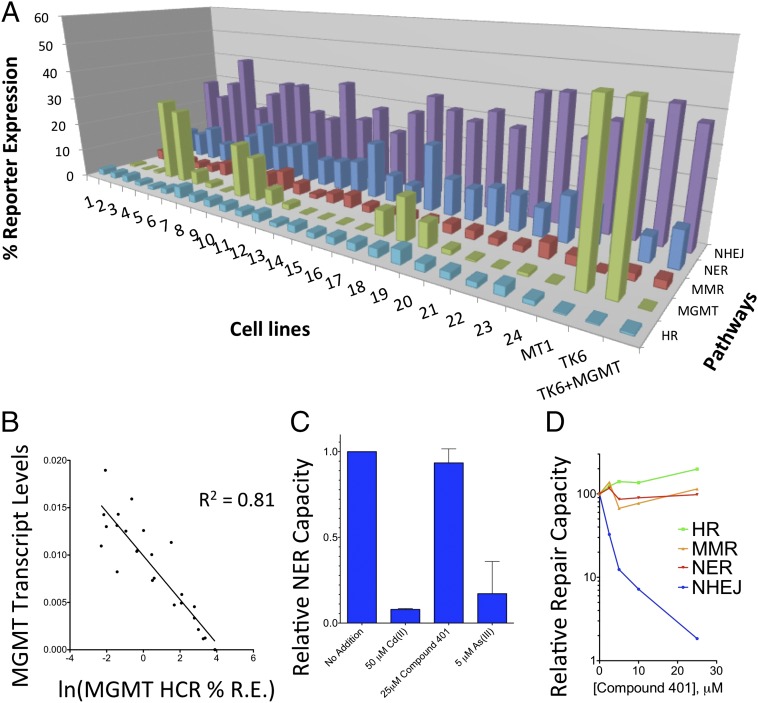
A previously described assay for repair of a double-strand break by homologous recombination (HR) was incorporated into FM-HCR experiments and was validated using cell lines with known defects in double-strand break repair (Fig. S4) (39). Assays for NER, MMR, MGMT, HR, and NHEJ capacity (see plasmid combinations 3 and 6) were carried out on a panel of three control cell lines (TK6, MT1, and TK6+MGMT) and 24 human lymphoblastoid cell lines derived from apparently healthy individuals of diverse ancestry (40). Each of the 24 cell lines exhibited a unique DNA repair profile (Fig. 4A), and a range of DRC was observed across the 24 cell lines for each pathway (MGMT, 285-fold; MMR, 4.4-fold; HR, 3.7-fold; NER, 3.2-fold; and NHEJ, 2.1-fold). To further validate the FM-HCR assay for MGMT, transcript levels measured by TaqMan quantitative PCR (qPCR) for the cell lines in Fig. 4A were compared with fluorescent reporter expression. A nonlinear relationship was observed between MGMT FM-HCR %R.E. and transcript levels (not shown); however, log-transformed FM-HCR data correlated extremely well (R2 = 0.81) with MGMT transcript levels (Fig. 4B).
Fig. 4.
Applications of FM-HCR to assessment interindividual DRC differences and identification of DNA repair inhibitors and enhancers. (A) FM-HCR analysis of repair capacity in five pathways for 27 cell lines. Cells were transfected with plasmid combination 3 or 6. (B) Correlation between MGMT transcript levels measured by TaqMan qPCR and %R.E. (log transformed) from the MGMT HCR. (C) FM-HCR measurements of NER inhibition. (D) FM-HCR measurements of compound 401 inhibition of DRC in four pathways. For all experiments, cells were assayed by flow cytometry 18 h after transfection. See also Fig. S4.
Application of FM-HCR to Assays for DRC Inhibition.
To further demonstrate the potential applications of FM-HCR, the assay was used to detect inhibition of DRC by metals and a small molecule. Cadmium and arsenic previously were shown to inhibit NER (41, 42). FM-HCR confirmed NER inhibition in the presence of low-micromolar concentrations of the two metals (Fig. 4C), whereas no effect on NER was detected in the presence of compound 401, known to inhibit a critical NHEJ factor, namely DNA PKcs (43). Moreover, compound 401 was shown to inhibit NHEJ in a dose-dependent manner (Fig. 4D), whereas MMR, NER, and HR were unaffected.
Taken together, the FM-HCR data demonstrate a versatile method for measuring, in a single assay, the repair of multiple DNA damage substrates, with either different doses or different types of damage. Although measuring four DNA repair pathways simultaneously represents a significant advance, the degree of multiplexing possible for the FM-HCR assay depends on the number of fluorescent reporters that can be measured simultaneously. We therefore developed an additional assay that does not require the detection of fluorescent proteins.
Deep Sequencing Analysis of Cells Transfected with Reporter Plasmids.
To increase the potential number of reporters that can be detected in a single assay, we developed HCR-Seq, a method of distinguishing and quantifying multiple full-length reporter transcripts using next-generation sequencing. Two cell lines exhibiting a large difference in their NER capacity (GM02344 and GM01953) were selected for a direct comparison of DRC measured by HCR-Seq vs. FM-HCR (Fig. 5). Each cell line was transfected with plasmid combination 7 (Table 2), and at 18 h, cells were analyzed by both HCR-Seq and FM-HCR. Plasmid combination 7 included a modified GFP reporter containing a single site-specific cyclobutane pyrimidine dimer (CPD). This reporter was included to allow a focused analysis of possible transcriptional errors induced by a bulky DNA lesion.
Fig. 5.
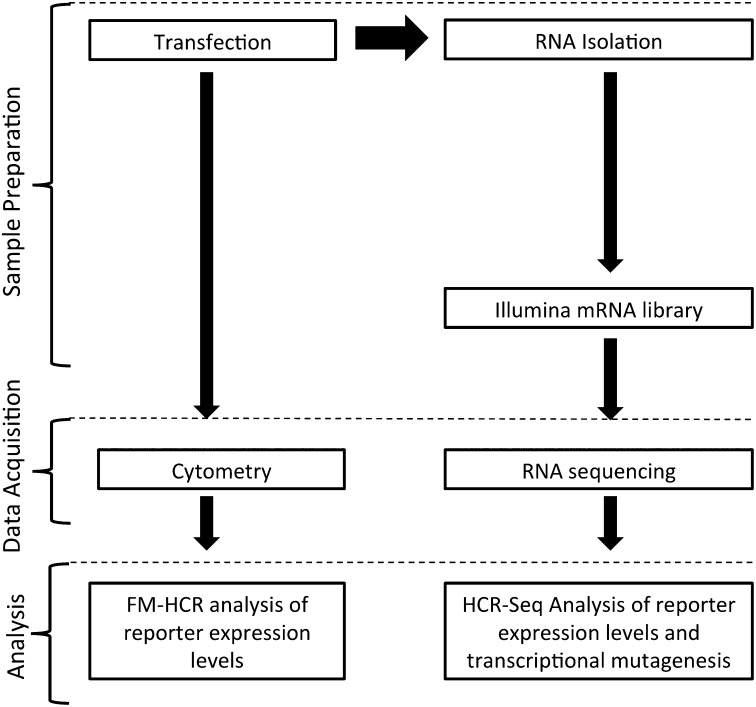
Workflow for experiments comparing two methods of analyzing reporter expression. Following transfection, an aliquot of cells is analyzed by flow cytometry. From the remaining cells, RNA is isolated and an aliquot is subjected to Illumina sample preparation and sequencing. FM-HCR analysis of fluorescent reporter expression is compared with HCR-Seq analysis of reporter transcript expression, measured as RPKM. See also Tables S1–S5.
For HCR-Seq analysis, total RNA was isolated and subjected to standard Illumina mRNA-Seq sample preparation and analysis (44). A total of 358,281,302 reads were generated for two replicates of four multiplexed samples, with each replicate analyzed in a separate HiSeq lane (Table S1). Between 30 million and 50 million reads were assigned to each original sample. Of these, 315,574,792 reads (88%) mapped properly to genes annotated for the human genome plus the five reporter sequences. In each sequencing lane, all five reporter transcripts were detected for each of the four samples; each sequencing lane simultaneously measured expression levels for 20 reporters. Alignment statistics, the criteria used to define proper alignment, and reasons for excluding the remaining 12% of reads from subsequent analysis are detailed in Tables S2–S5.
DNA Repair and Transcriptional Mutagenesis Detected by RNA Sequencing.
Relative transcript levels in WT (GM01953) vs. XPA (GM02344) cell lines were determined for both host genes and plasmid reporter genes. Plasmid reporters were found to be among the most highly expressed genes (Fig. S5A). As expected, reporter expression from UV-treated plasmids was reduced in a dose-dependent manner (Fig. 6A), and the reduced expression for the XPA cell line (GM02344) was far greater than that for WT cells (GM01953). More importantly, reporter transcript expression mirrored closely the dose–response curves obtained from the same transfected cells using FM-HCR (Fig. 6B). Reporter expression from plasmids containing a single site-specific CPD in the transcribed strand likewise was reduced relative to that from undamaged plasmids (Fig. S5B).
Fig. 6.
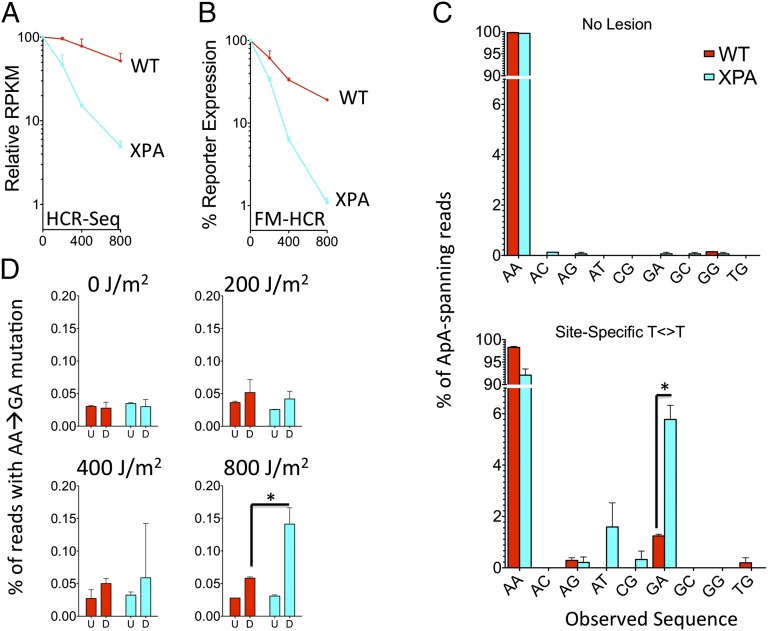
mRNA-Seq analysis of reporter expression. (A) Dose–response curves for reporter expression from randomly UV-damaged plasmids generated from mRNA-Seq analysis. (B) Dose–response curves for the same transfected cells generated from flow-cytometric (FM-HCR) analysis. (C) Sequence variants detected in transcripts at the position corresponding to the site-specific thymine dimer in the absence (Upper) or presence of the lesion (Lower). Frequencies are reported for the expected sequence (AA) as well as all variants that were observed in at least one sample. (D) Frequencies of AA→GA mutations in transcripts expressed from randomly damaged plasmids as a function of dose (combination 7 in Table 2). The undamaged case (0 J/m2) refers to the frequency of mutations as measured in transcripts expressed from the BFP transfection control. D, data for cells transfected with reporters irradiated as indicated in Table 2; U, HCR-Seq data from cells in which all the reporter plasmids were undamaged; *, differences deemed to be statistically significant (P < 0.05) by a t test. See also Fig. S5.
With respect to global gene expression in the transfected cells, fewer than 10 host transcripts showed a greater than twofold change in expression in the presence of UV-treated plasmids vs. undamaged plasmids (Table S5). Among these, only three (SMN1, RPL21, and RN5-8S1) were observed in both cell lines, but in no case was a change in the same direction observed for both replicates. Thus, no significant transcriptional response to the presence of DNA damage in plasmids was evident under our experimental conditions. However, consistent with the FM-HCR data (Fig. 2) suggesting higher MGMT activity in GM02344 cells compared with GM01953 cells, the mRNA-Seq data indicated an approximately threefold higher expression of the MGMT transcript in GM02344 vs. GM01953. Furthermore, XPA transcripts were expressed at lower levels in GM02344 vs. GM01953, and they were spliced correctly only rarely in GM02344 (Fig. S5C). These data reproduce previously reported splicing errors in GM02344 due to a homozygous 555G > C mutation in the XPA gene (45). Finally, to assess the potential for DNA contamination in RNA-Seq samples, the density of reads aligning to intergenic regions (which are not expected to be represented in transcripts) was compared with the density of reads aligning to exons, and the ratio of exonic/intergenic reads was found to be greater than 1,000, indicating an RNA purity >99.9%.
Sequence-level analysis of mRNA-Seq data revealed base substitutions in reporter transcripts at the position corresponding to the site-specific CPD; this was true for both cell lines (Fig. 6C). The most frequently observed base change, an A→G mutation at the 5′ adenine in the ApA sequence opposite the CPD (hereafter AA→GA), was detected at a frequency of 1.3% in cells with no known repair defect (GM01953) and 5.8% in NER-deficient GM02344 cells. In transcripts expressed from the undamaged plasmid, the frequency of the AA→GA mutations at this position was less than 0.2%. A potential experimental concern is that trace-contaminating plasmid DNA might be amplified during Illumina sample preparation, thus giving rise to DNA fragments with base substitutions due to error-prone CPD bypass by DNA polymerase. However, nearly identical frequencies for AA→GA mutations were found by using a second sample preparation method that excludes the possibility of contaminating lesion-containing plasmid (Fig. S5D). It therefore appears that human RNA polymerase can bypass a thymine dimer in vivo, albeit in an error-prone manner.
AA→GA mutations also were induced in a dose-dependent manner in transcripts expressed from UV-irradiated reporter plasmids containing thymine dimers that were not site specific (Fig. 6D). As expected for randomly induced DNA damage, the absolute frequency of the base substitution was much lower than that observed for transcripts expressed from the reporter with the site-specific thymine dimer. Once again, base changes occurred at a higher frequency in NER-deficient vs. WT cells. These data provide additional evidence for error-prone transcriptional bypass of thymine dimers.
Discussion
We have established new methods that enable rapid, high-throughput measurements of DRC for DNA lesions that either block RNA polymerase II-mediated transcription or alter the sequence of reporter transcripts. These methods do not require the generation of cell lysates or the use of in vitro assays and can measure in vivo DRC at multiple doses or in multiple repair pathways simultaneously. As a result, these assays outperform current methods of measuring DRC (20, 27) and have the potential to be used to personalize the prevention and treatment of cancer and other diseases caused by inefficient repair of DNA damage.
We have demonstrated an application of the FM-HCR to the question of whether NER capacity in human lymphoblastoid cells is representative of repair capacity in other tissues. Lymphoblastoid cells provide a convenient source of cells for use in human variability studies; however, the extent to which they represent a faithful surrogate for other cells in primary tissues has been called into question (46–48). The present data indicate a strong correlation between NER capacity in primary human skin fibroblasts and transformed B-lymphoblastoid cells from the same individuals (Fig. 1L). The strong correlation further illustrates that the assay can be carried out reproducibly in primary or transformed cells from multiple tissues.
To our knowledge, our use of HCR to measure combinations of NER, MMR, BER, NHEJ, HR, or MGMT capacity simultaneously is the first example of a quantitative assay capable of measuring repair of DNA damage by multiple distinct pathways in parallel. One of the strengths of FM-HCR is that it yields a single readout (fluorescence) in place of multiple unique outputs from very different experimental procedures that were used previously to characterize the same repair pathways in the cell lines for which data are presented (Figs. 2 and 3) (8, 32, 33, 35, 38, 49). The fluorescent reporters for direct reversal of O6-MeG and BER of 8-oxoG illustrate the use of transcriptional mutagenesis to measure the repair of DNA lesions that are bypassed in an error-prone manner by RNA polymerase. This paradigm, in which the presence of a DNA lesion changes the expressed reporter sequence to one that encodes a functional protein, holds promise as a general method of measuring DRC, because a wide variety of toxic and mutagenic DNA lesions are known to induce transcriptional errors (50, 51).
We have presented several applications of FM-HCR to demonstrate the broad utility of the assay. The flow-cytometric fluorescence-based FM-HCR method accurately reproduces data collected previously for a set of cell lines known to differ in NER capacity (8). A screen of a much larger set of cell lines derived from apparently healthy individuals illustrates the potential to measure DRC efficiently in multiple pathways in large sample sets (Fig. 4A) and to identify agents that inhibit or enhance DRC (Fig. 4 C and D). Screens for DRC inhibitors or enhancers are expected to identify some agents for which the mechanism of action is unknown; indeed, uncertainty remains as to the precise mechanisms by which arsenic and cadmium exposure lead to reduced DRC (41, 52). In view of this uncertainty, the strength of FM-HCR lies in the ability to measure changes in DRC as an important functional endpoint.
By using multiple fluorescent reporters, a 96-well format, and automated flow-cytometric sample processing, FM-HCR is rapid and less labor intensive than the standard CAT-based HCR assay. For example, the total active laboratory time required to perform the analysis to generate the triplicate data in Fig. 1 B and C is ∼12 h, or 1–2 h per cell line, using flow cytometers equipped with a high-throughput sampler to enable automated data acquisition. In addition, experimental error is reduced by cotransfection of reporters, allowing normalization of expression from a damaged plasmid to that of an undamaged control plasmid included in every transfection. Through these technical improvements, FM-HCR removes a major barrier to epidemiological studies of DRC that include large populations and multiple DNA repair pathways. Furthermore, because standard oncology laboratories are equipped with flow cytometers, the assay also has the potential to be useful in a clinical setting.
The use of next-generation sequencing to essentially count reporter transcripts (HCR-Seq), rather than measuring their fluorescent translation products, presents an opportunity to increase throughput vastly and overcomes important limitations on assay throughput and versatility that otherwise are imposed by the need to detect fluorescent reporter proteins. We have validated the HCR-Seq approach by showing that HCR of UV-irradiated plasmids analyzed by mRNA-Seq yields a pattern of dose–response curves similar to those obtained previously by using a CAT-based HCR assay (8), as well as those obtained in the current study by FM-HCR analysis (Fig. 1). Because next-generation sequencing may be used to quantitate the expression levels of thousands of transcripts simultaneously, our assay has the potential to measure expression of dozens of reporters for multiple individuals in a single experiment; this would make characterization of global DRC in large populations both efficient and affordable (SI Materials and Methods, Supplementary Note).
HCR-Seq constitutes a paradigm shift in the quantitation of DRC because of the ability to measure the repair of any lesion that either inhibits transcription or induces transcriptional mutagenesis. Base misincorporation opposite DNA lesions that are bypassed by DNA polymerase during replication has been studied extensively for many lesions (53). Misincorporation during transcription by RNA polymerase has been documented for a growing number of lesions and often mirrors that of DNA polymerase during replication (54). As a result, most mutagenic lesions may be expected to have a transcriptional mutagenic signature. The HCR-Seq strategy therefore should be useful in DRC measurements for nearly any pathway. The data in Fig. 6 also illustrate the power of this unbiased approach to detect rare events that are specific to transcription of damaged DNA.
The two major applications to human health that we foresee for these assays relate to personalized prevention and treatment of cancer. The available published data indicate that DRC is an important factor both in cancer susceptibility and in the efficacy of cancer treatment with DNA-damaging agents, and that plasmid-based HCR assays can be applied readily to primary human tissue samples, including stimulated peripheral blood mononuclear cells (8–11, 20, 27). FM-HCR and HCR-Seq now open the door to a comprehensive analysis of DRC as a biomarker for disease susceptibility. For personalized disease prevention, FM-HCR might be applied to human blood cells to identify individuals who may have a higher risk of disease. In terms of personalized treatment, the assays might be used to measure DRC in blood cells to predict patient tolerance for a particular cancer therapy (55), or to measure DRC in cancer cells to predict the efficacy of treatment with DNA-damaging chemotherapeutic agents in a manner analogous to using MGMT promoter methylation to predict the response of cancers to alkylating chemotherapy agents, such as temozolomide (56). Indeed, the data in Fig. 4 show that FM-HCR data reproduce the results of a standard TaqMan qPCR assay for MGMT gene expression in lymphoblastoid cell lines. The functional FM-HCR and HCR-Seq assays might be expected to outperform promoter methylation assays because (i) they provide a direct, quantitative readout of repair activity rather than an indirect estimate of DNA repair gene expression, and (ii) they provide data for repair capacity in additional pathways, such as MMR, which also contributes importantly to alkylation sensitivity (32). Finally, the ability of FM-HCR to identify agents that either inhibit or enhance DRC in human cells (Fig. 4 C and D) opens the door to screens for novel compounds that might be used either to potentiate the effects of DNA damage-based anticancer agents or to mitigate the deleterious effects of environmental exposure to DNA-damaging agents (57–59).
In addition to the possible clinical applications described above, HCR-Seq has the potential to reveal new biological phenomena in the basic research setting. The mRNA-Seq data presented here provide evidence that transcriptional errors result when human RNA polymerase II bypasses a CPD. Because the plasmids are not replicated in the cell, and transcript sequence changes were observed at an elevated rate in repair-deficient cells, these changes likely reflect transcriptional mutagenesis events due to unrepaired DNA lesions in the transcribed DNA strand. Although it was reported previously that in vivo bypass of a CPD by RNA polymerase may result in rare deletions, and that bypass of a bulky 8,5′-cyclo-2′-deoxyadenosine lesion may result in both deletions and base substitutions (60), our observation of frequent base misincorporation opposite a CPD by RNA polymerase II appears to be without precedent. A recent in vitro analysis indicated that transcriptional CPD bypass followed a so-called A-rule, resulting in error-free bypass (61); although base misincorporation was observed, subsequent extension of transcripts beyond the misincorporated base was strongly inhibited. The present data provide in vivo evidence of error-prone transcriptional bypass of bulky DNA lesions in human cells followed by completion and polyadenylation of the transcript. A lower limit (about 6%) for the frequency of bypass events resulting in an AA→GA mutation can be estimated from the data in Fig. 6C. Because it is expected that reporter plasmids that already have been repaired will be transcribed at a higher rate, and because error-free bypass (according to an A-rule) cannot be distinguished from transcripts arising from repaired plasmid, the rate of bypass likely is higher than 6%.
Conclusions
FM-HCR and HCR-Seq represent powerful new tools for high-throughput measurements of human DRC and provide a rapid functional characterization that complements existing, indirect measures of DRC. FM-HCR permits the simultaneous measurement of repair for up to four different doses of DNA damage, or types of DNA damage, in a single assay. HCR-Seq has the potential to measure thousands of reporter sequences in a single assay, with bar codes providing unique identifiers for the type or dose of DNA damage as well as for the individual whose cells are being analyzed. Both methods expand the scope of lesions whose repair can be measured to include those that do not block transcription, and as additional substrates are developed, we anticipate that our assays will permit measurements of DRC in all the major DNA repair pathways in a single assay. Our assays hold an advantage over in vitro assays because the transcription-based reporters limit the readout to DNA that has been repaired in vivo in chromatinized DNA, thus increasing the likelihood of recapitulating physiological DNA repair phenotypes. The assays have the potential to reduce the cost and labor required for DRC measurements to a level compatible with large-scale epidemiological studies and clinical diagnostic/prognostic applications. The data presented herein also illustrate the utility of the assays as a research tool that can reveal mechanisms of DNA repair and damage tolerance and that may provide a means of screening chemical libraries for inhibitors or enhancers of DRC.
Materials and Methods
DNA Repair Reporter Plasmids.
Detailed methodology for the construction of reporter plasmids and the methods used to transfect plasmids into cells may be found in Supporting Information (Figs. S2–S4). Briefly, plasmids for expression of the fluorescent proteins AmCyan, EGFP, mOrange, and mPlum were purchased from Clontech, and that for tagBFP was purchased from Axxora. Reporter genes were subcloned into the pmaxCloning Vector (Lonza). NER reporters were prepared by irradiating plasmids with UV-C light. The resulting DNA damage prevents fluorescent reporter expression by blocking transcription; repair by NER restores reporter expression. The NHEJ reporter comprised a linearized fluorescent reporter; because double-strand breaks constitute an absolute block to transcription, NHEJ-dependent recircularization of the plasmid is required to restore reporter expression. MMR reporters consisted of heteroduplex DNA engineered such that the transcribed strand encoded a nonfluorescent protein. Repair of a single, site-specific mismatch restores the WT sequence to the transcribed strand and results in fluorescent reporter expression. Reporters for repair of 8-oxoG or O6-MeG were engineered such that transcriptional mutagenesis in the presence of the DNA lesion led to expression of WT fluorescent reporter protein. Because repair removes the source of transcriptional mutagenesis, repair of these plasmids is inversely proportional to the measured fluorescence.
Flow Cytometry.
Cells suspended in culture media were analyzed for fluorescence on a BD LSR II cytometer running FACSDiva software. Cell debris, doublets, and aggregates were excluded based on their side-scatter and forward-scatter properties. TO-PRO–3 was added to cells 5–10 min before analysis and was used to exclude dead cells from the analysis. The following fluorophores and their corresponding detectors (in parentheses) were used: tagBFP (Pacific Blue), AmCyan (AmCyan), EGFP (FITC), mOrange (phycoerythrin; PE), mPlum (PE-Cy5-5), and TO-PRO–3 (allophycocyanin; APC). The linear range for the corresponding photomultiplier tubes was determined using BD Rainbow fluorescent beads and unlabeled polystyrene beads based on the signal-to-noise ratio, % coefficient of variation, and M1/M2 parameters as previously described (62). Compensation was set by using single-color controls. Regions corresponding to cells positive for each of the five fluorescent proteins were established by using single-color dropout controls. For reporters that required compensation in more than one detector channel, fluorescence in the reporter channel was plotted separately against each of the channels requiring compensation. Using these plots, both single controls and the dropout control (in which the reporter of interest was excluded from the transfection) were used to establish regions corresponding to positive cells (Fig. S1A). Equations used to calculate fluorescent reporter expression are detailed in Supporting Information.
mRNA-Seq.
Total RNA was isolated by using standard procedures detailed in Supporting Information. Total RNA samples were submitted to the Massachusetts Institute of Technology BioMicroCenter for preparation and sequencing. Briefly, total RNA was poly-A purified, fragmented, and converted to cDNA by using the Illumina TruSeq protocol. Library construction from cDNA was performed using the Beckman Coulter SPRIworks system. During library amplification, a unique bar code was introduced for each of the eight samples corresponding to the four transfections performed in duplicate (Table S1) and from which total RNA was generated. Four samples from each replicate were clustered on a separate sequencing lane and run on an Illumina HiSeq 2000 instrument. Image analysis, base calling, and sequence alignment to a synthetic genome consisting of the human genome and the five fluorescent reporter genes were performed using the Illumina Pipeline. Aberrant expression of the XPA gene in GM02344 cells provided an internal confirmation of the identity of the cell lines; reduced expression and an expected lack of regular splicing-junction reads spanning intron 4 of the XPA gene from GM02344 was observed (Fig. S5C), confirming a previously reported missplice in XPA transcripts due to the homozygous 555G > C mutation (45).
To ensure that trace DNA contamination of the RNA-Seq samples did not contribute significantly to the observed frequency of base substitutions in transcripts expressed from reporter plasmids (Fig. 6), a second complementary sample preparation was performed and analyzed by Illumina sequencing. Details of the experimental procedures are available in Supporting Information; briefly, mRNA isolated from cells transfected with reporter plasmids was treated with DNase and reverse transcribed to generate a cDNA library. PCR amplification of reporter cDNA was not detected when mRNA that was not reverse transcribed was used as a template (Fig. S5D), confirming cDNA as the template for PCR amplification, and hence ruling out significant plasmid contamination. Amplicons were fragmented and submitted for standard Illumina sample preparation.
Next-Generation Sequencing Data Analysis.
Illumina sequencing data were analyzed using the Tuxedo software suite. Mapped reads were aligned to the hg19 human genome assembly and the five reporter gene sequences by using TopHat version 2.0.6, and junction reads were determined. Additional details of all analyses, including input parameters, are available in Tables S2–S5. Cufflinks version 2.0.2 was run to quantify reads in terms of reads per kilobase of transcript per million mapped reads (RPKM) (63). Samtools mpileup (version 0.1.16 r963:234) was used to aggregate reads at all positions in the alignment file. By using the pileup file as input, single-nucleotide variants, as well as insertions and deletions (indels) present in the mRNA-Seq data, were identified using the software package VarScan v2.3.4 (64). All positions meeting a minimum read depth of 8 were considered further; however, no minimum variant frequency threshold was set to detect rare variants and to establish the sequencing error rate. Custom Python scripts were used to generate a list of all deletions spanning an ApA sequence. The frequencies for base substitutions at each ApA sequence in the reporter transcripts also were determined.
Statistics.
Statistics were performed with the GraphPad Prism 5.0 software package. The correlations between datasets in Figs. 1 and 4 were assessed by using a linear regression model that reports R2 for the goodness of fit and a P value for the slope of the line being significantly different from zero. The P values in Fig. 6 were calculated from a two-tailed unpaired t test. Error bars in Figs. 1–5 report the SD of at least three biological replicates.
Supplementary Material
Acknowledgments
We thank Dr. Jennifer Calvo for establishing the immortalized MEF cell lines, Prof. Penny Jeggo for the V79 and xrs6 cell lines, and Prof. Ryan Jensen for the VC8 cell line. We acknowledge funding support from National Institutes of Health Grant DP1-ES022576. L.D.S. is an American Cancer Society Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.W.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401182111/-/DCSupplemental.
References
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286(5446):1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12(2):104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralhan R, Kaur J, Kreienberg R, Wiesmüller L. Links between DNA double strand break repair and breast cancer: Accumulating evidence from both familial and nonfamilial cases. Cancer Lett. 2007;248(1):1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DM, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711(1-2):100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 1991;51(21):5786–5793. [PubMed] [Google Scholar]
- 9.Decordier I, Loock KV, Kirsch-Volders M. Phenotyping for DNA repair capacity. Mutat Res. 2010;705(2):107–129. doi: 10.1016/j.mrrev.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Jalal S, Earley JN, Turchi JJ. DNA repair: From genome maintenance to biomarker and therapeutic target. Clin Cancer Res. 2011;17(22):6973–6984. doi: 10.1158/1078-0432.CCR-11-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkaria JN, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14(10):2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452(7187):564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 14.Ellis NC. Obtaining and Using Genetic Information. New York: Springer; 2003. [Google Scholar]
- 15.Li CY, et al. Polymorphisms of nucleotide excision repair genes predict melanoma survival. J Invest Dermatol. 2013;133(7):1813–1821. doi: 10.1038/jid.2012.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warmoes M, et al. Proteomics of mouse BRCA1-deficient mammary tumors identifies DNA repair proteins with potential diagnostic and prognostic value in human breast cancer. Mol Cell Proteomics. 2012;11(7):013334. doi: 10.1074/mcp.M111.013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redaelli A, Magrassi R, Bonassi S, Abbondandolo A, Frosina G. AP endonuclease activity in humans: Development of a simple assay and analysis of ten normal individuals. Teratog Carcinog Mutagen. 1998;18(1):17–26. [PubMed] [Google Scholar]
- 18.Geng H, et al. In vitro studies of DNA mismatch repair proteins. Anal Biochem. 2011;413(2):179–184. doi: 10.1016/j.ab.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgiadis P, Polychronaki N, Kyrtopoulos SA. Progress in high-throughput assays of MGMT and APE1 activities in cell extracts. Mutat Res. 2012;736(1-2):25–32. doi: 10.1016/j.mrfmmm.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Leitner-Dagan Y, et al. N-methylpurine DNA glycosylase and OGG1 DNA repair activities: Opposite associations with lung cancer risk. J Natl Cancer Inst. 2012;104(22):1765–1769. doi: 10.1093/jnci/djs445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans RG, Norman A. Radiation stimulated incorporation of thymidine into the DNA of human lymphocytes. Nature. 1968;217(5127):455–456. doi: 10.1038/217455a0. [DOI] [PubMed] [Google Scholar]
- 22.Perry P, Evans HJ. Cytological detection of mutagen carcinogen exposure by sister chromatid exchange. Nature. 1975;258(5531):121–125. doi: 10.1038/258121a0. [DOI] [PubMed] [Google Scholar]
- 23.Parshad R, Sanford KK, Jones GM. Chromatid damage after G2 phase X-irradiation of cells from cancer-prone individuals implicates deficiency in DNA repair. Proc Natl Acad Sci USA. 1983;80(18):5612–5616. doi: 10.1073/pnas.80.18.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci USA. 2010;107(22):10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao YW, et al. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509(1-2):165–174. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 26.Ramos JM, et al. DNA repair and breast carcinoma susceptibility in women. Cancer. 2004;100(7):1352–1357. doi: 10.1002/cncr.20135. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Wang L-E, Wei Q. DNA repair phenotype and cancer susceptibility—a mini review. Int J Cancer. 2009;124(5):999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez P, et al. A modified host-cell reactivation assay to quantify DNA repair capacity in cryopreserved peripheral lymphocytes. DNA Repair (Amst) 2011;10(6):603–610. doi: 10.1016/j.dnarep.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Jagger J. Ultraviolet inactivation of biological systems. In: Wang SY, editor. Photochemistry and Photobiology of Nucleic Acids. Vol 2. New York: Academic; 1976. pp. 147–186. [Google Scholar]
- 30.Parsons R, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 31.Burns JA, Dreij K, Cartularo L, Scicchitano DA. O6-methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 2010;38(22):8178–8187. doi: 10.1093/nar/gkq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kat A, et al. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90(14):6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhukovskaya N, Rydberg B, Karran P. Inactive O6-methylguanine-DNA methyl transferase in human cells. Nucleic Acids Res. 1992;20(22):6081–6090. doi: 10.1093/nar/20.22.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allalunis-Turner MJ, Barron GM, Day RS, 3rd, Dobler KD, Mirzayans R. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat Res. 1993;134(3):349–354. [PubMed] [Google Scholar]
- 35.Lees-Miller SP, et al. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267(5201):1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 36.Galloway AM, Allalunis-Turner J. cDNA expression array analysis of DNA repair genes in human glioma cells that lack or express DNA-PK. Radiat Res. 2000;154(6):609–615. doi: 10.1667/0033-7587(2000)154[0609:ceaaod]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271(14):8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 38.Klungland A, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96(23):13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiziltepe T, et al. Delineation of the chemical pathways underlying nitric oxide-induced homologous recombination in mammalian cells. Chem Biol. 2005;12(3):357–369. doi: 10.1016/j.chembiol.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8(12):1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 41.Hartwig A. Mechanisms in cadmium-induced carcinogenicity: Recent insights. Biometals. 2010;23(5):951–960. doi: 10.1007/s10534-010-9330-4. [DOI] [PubMed] [Google Scholar]
- 42.Hartwig A, et al. Interaction of arsenic(III) with nucleotide excision repair in UV-irradiated human fibroblasts. Carcinogenesis. 1997;18(2):399–405. doi: 10.1093/carcin/18.2.399. [DOI] [PubMed] [Google Scholar]
- 43.Griffin RJ, et al. Selective benzopyranone and pyrimido 2,1-alpha isoquinolin-4-one inhibitors of DNA-dependent protein kinase: Synthesis, structure-activity studies, and radiosensitization of a human tumor cell line in vitro. J Med Chem. 2005;48(2):569–585. doi: 10.1021/jm049526a. [DOI] [PubMed] [Google Scholar]
- 44.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 45.Satokata I, Tanaka K, Yuba S, Okada Y. Identification of splicing mutations of the last nucleotides of exons, a nonsense mutation, and a missense mutation of the XPAC gene as causes of group A xeroderma pigmentosum. Mutat Res. 1992;273(2):203–212. doi: 10.1016/0921-8777(92)90081-d. [DOI] [PubMed] [Google Scholar]
- 46.Choy E, et al. Genetic analysis of human traits in vitro: Drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4(11):e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis AR, Kohane IS. Expression differences by continent of origin point to the immortalization process. Hum Mol Genet. 2009;18(20):3864–3875. doi: 10.1093/hmg/ddp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stark AL, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10(6):505–512. doi: 10.1038/tpj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci USA. 1999;96(19):10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brégeon D, Peignon PA, Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5(7):e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bregeon D, Doetsch PW. Assays for transcriptional mutagenesis in active genes. DNA Repair, Pt B, Methods in Enzymology. Vol 409. San Diego: Academic; 2006. pp. 345–357. [DOI] [PubMed] [Google Scholar]
- 52.Bhattacharjee P, Banerjee M, Giri AK. Role of genomic instability in arsenic-induced carcinogenicity. A review. Environ Int. 2013;53:29–40. doi: 10.1016/j.envint.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Shrivastav N, Li DY, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31(1):59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brégeon D, Doetsch PW. Transcriptional mutagenesis: Causes and involvement in tumour development. Nat Rev Cancer. 2011;11(3):218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alapetite C, Thirion P, de la Rochefordière A, Cosset JM, Moustacchi E. Analysis by alkaline comet assay of cancer patients with severe reactions to radiotherapy: Defective rejoining of radioinduced DNA strand breaks in lymphocytes of breast cancer patients. Int J Cancer. 1999;83(1):83–90. doi: 10.1002/(sici)1097-0215(19990924)83:1<83::aid-ijc16>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 56.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 57.Srinivasan A, Gold B. Small-molecule inhibitors of DNA damage-repair pathways: An approach to overcome tumor resistance to alkylating anticancer drugs. Future Med Chem. 2012;4(9):1093–1111. doi: 10.4155/fmc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim K, et al. High throughput screening of small molecule libraries for modifiers of radiation responses. Int J Radiat Biol. 2011;87(8):839–845. doi: 10.3109/09553002.2011.560994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zellefrow CD, et al. Identification of druggable targets for radiation mitigation using a small interfering RNA screening assay. Radiat Res. 2012;178(3):150–159. doi: 10.1667/rr2810.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8(4):388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walmacq C, et al. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46(1):18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P, Roederer M. Quality assurance for polychromatic flow cytometry. Nat Protoc. 2006;1(3):1522–1530. doi: 10.1038/nprot.2006.250. [DOI] [PubMed] [Google Scholar]
- 63.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.