Significance
In higher organisms, DNA is bound to proteins and tightly packed within the nucleus, leaving only certain regions accessible for gene expression. As the enzyme RNA polymerase II (RNAPII) travels along the DNA template synthesizing RNA, it must contend with forces generated by various obstacles, and some of these forces are known to produce transient pausing and even transcriptional arrest. How do organisms deal with such problems? Using optical-trapping technology, we performed a single-molecule study of RNAPII interactions with transcription factors TFIIS and TFIIF, which are involved in modulating and regulating transcriptional elongation. By applying controlled loads to RNAPII and combinations of these factors, we learned about the mechanisms by which pausing and arrest are overcome.
Keywords: Pol II, optical tweezers, optical trap
Abstract
Recent evidence suggests that transcript elongation by RNA polymerase II (RNAPII) is regulated by mechanical cues affecting the entry into, and exit from, transcriptionally inactive states, including pausing and arrest. We present a single-molecule optical-trapping study of the interactions of RNAPII with transcription elongation factors TFIIS and TFIIF, which affect these processes. By monitoring the response of elongation complexes containing RNAPII and combinations of TFIIF and TFIIS to controlled mechanical loads, we find that both transcription factors are independently capable of restoring arrested RNAPII to productive elongation. TFIIS, in addition to its established role in promoting transcript cleavage, is found to relieve arrest by a second, cleavage-independent mechanism. TFIIF synergistically enhances some, but not all, of the activities of TFIIS. These studies also uncovered unexpected insights into the mechanisms underlying transient pauses. The direct visualization of pauses at near-base-pair resolution, together with the load dependence of the pause-entry phase, suggests that two distinct mechanisms may be at play: backtracking under forces that hinder transcription and a backtrack-independent activity under assisting loads. The measured pause lifetime distributions are inconsistent with prevailing views of backtracking as a purely diffusive process, suggesting instead that the extent of backtracking may be modulated by mechanisms intrinsic to RNAPII. Pauses triggered by inosine triphosphate misincorporation led to backtracking, even under assisting loads, and their lifetimes were reduced by TFIIS, particularly when aided by TFIIF. Overall, these experiments provide additional insights into how obstacles to transcription may be overcome by the concerted actions of multiple accessory factors.
The expression of most genes is carefully regulated at the level of transcription. As a consequence, RNA polymerase II (RNAPII)—the enzyme responsible for mRNA synthesis in eukaryotic organisms—is at the nexus of an exquisite network of regulatory pathways, many of which are controlled by transcription factors. The control pathways associated with RNAPII recruitment to, and initiation at, promoter sites have been studied extensively (1), but it has become increasingly clear that significant regulatory activity also occurs during postinitiation steps and, in particular, at the level of transcript elongation (2).
Productive transcript elongation (3–5) is characterized by periods of unidirectional motion by RNAPII along the DNA template, adding one nucleotide at a time to the growing RNA transcript. Elongation—both in vitro in highly purified systems (6, 7) and in vivo (8, 9)—is frequently interrupted by transcriptional pauses, at least some fraction of which are associated with enzyme backtracking, a process by which RNAPII reverses its normal direction of motion and moves upstream on the template (6, 7). Entry into backtracked states appears to confer a high degree of force sensitivity to elongating RNAPII molecules (7) that likely governs their response to physical obstacles encountered in vivo, including nucleosomes and other DNA-binding proteins (8, 10), signals in the underlying template sequence, and multiple RNAPII molecules transcribing the same gene simultaneously (11). Long-lifetime pauses can supply regulatory mechanisms that poise genes for rapid induction (9), control the expression of alternatively spliced protein isoforms (12, 13), couple transcription to other RNA-processing events, and implement epigenetic control mechanisms by the interactions of RNAPII with modified histones (2) and methylated DNA (13).
Paused elongation complexes are targeted by transcription factors, including TFIIS and TFIIF. These factors are believed to work in concert to enhance the efficiency of productive elongation (14–17). TFIIS is known to enhance the intrinsic endonucleolytic cleavage activity of RNAPII, leaving a fresh 3′ end of the RNA aligned with the active site, which allows transcription to resume after any backtracking and facilitates multiple attempts to overcome transcriptional barriers (7, 10, 18, 19). Transcript cleavage in response to nucleotide misincorporation may also play a role in maintaining fidelity (20, 21), although evidence for the relevance of this activity in vivo is conflicting (22, 23). Transcription factor TFIIF, in addition to its essential function as an initiation factor, is thought to continue interacting at least transiently with RNAPII throughout the elongation phase. In vitro, TFIIF has been shown to contribute to the efficiency of the very earliest stages of elongation (24–26) and to stimulate overall transcription rates (15, 27, 28), at least in part by suppressing pauses. In vivo, TFIIF appears to predominantly associate with RNAPII in promoter-proximal regions (29, 30), although there is evidence for interactions in the late stages of elongation as well (31). Structural characterization suggests that TFIIF binds to the periphery of RNAPII, where it may interact with DNA entering the main channel (32–34) and stimulate the opening of the downstream end of the transcription bubble (35) without apparent access to the active site. Consequently, TFIIF is thought to exert its effects indirectly, by inducing as-yet-uncharacterized conformational changes in RNAPII.
Here, we present a single-molecule optical-trapping study of the roles of TFIIS and TFIIF in transcript elongation by RNAPII. Individual elongation complexes containing RNAPII, alone or in conjunction with TFIIF and/or TFIIS, were monitored in real time at near-base-pair resolution as controlled mechanical loads were applied between the DNA template and the complexes, biasing their motions to favor forward elongation (assisting load) or pausing and arrest pathways (hindering load).
Results
TFIIF and TFIIS Reactivate Arrested RNAPII by Three Distinct Mechanisms.
To probe the effects of the factors on RNAPII under conditions favoring pausing and arrest, we measured transcription against increasing hindering loads (Fig. 1A), stepping up the force after every ∼200 base pairs (bp) transcribed (Fig. 1B). Both isolated RNAPII molecules and stable RNAPII⋅TFIIF complexes (36, 37) were probed; TFIIS was added as indicated. Transcriptional arrest, operationally defined as an event in which a previously transcribing enzyme stalled and no significant net forward translocation occurred over a period of ∼300 s, was consistently observed at hindering loads in the −5 to −20 pN range (SI Appendix, Fig. S1). The high-resolution data reveal the entry process into the arrested state in previously unattainable detail. As seen in Fig. 1B and quantified in SI Appendix, Fig. S2, entry into this state typically corresponded to a single, large backtracking event (∼5–15 bp) that distinguished it from transient pausing, which, by contrast, displayed a more limited degree of backtracking. In the absence of added factors, arrested molecules of RNAPII remained transcriptionally inactive, even after the force was lowered (after ∼300 s) to −2 pN, a value that is readily overcome by elongating RNAPII complexes.
Fig. 1.

TFIIS and TFIIF reactivate arrested RNAPII by distinct mechanisms. (A) Experimental geometry for the optical-trapping assay. (B) A representative single-molecule record of RNAPII elongation (displacement converted to bp; left vertical axis) against opposing load (trace is color-coded according to the force). Force was increased stepwise every ∼200 bp (gray trace; right vertical axis). A large backtrack (blue arrow) led to force-induced arrest; after ∼300 s, force was lowered to −2 pN to test for transcription restart. (C) Representative elongation records, color-coded as in B, with transcription factors as follows. (i) RNAPII + 1 µM WT TFIIS. (ii) RNAPII⋅TFIIF. (iii) RNAPII⋅TFIIF + 1 µM WT TFIIS. In backtrack-and-rescue events (red and green double arrows), RNAPII recovered from a large backtrack and resumed elongation under high load. In low-force restart events (black arrows), elongation only resumed once the force was lowered. (D) Probability of low-force restart. (E) Average number of backtrack-and-rescue events per molecule for the factor combinations indicated. Error bars are SE.
Subsequent to this force-induced arrest, two characteristic activities were observed that were factor-dependent (Fig. 1 C–E). In “backtrack-and-rescue” events, RNAPII recovered from a large backtrack—one that would otherwise lead to arrest in the absence of factors—and resumed forward elongation under continued (high) hindering loads (red and green arrows). In “low-force restart” events, elongation resumed only after the force had been dropped (black arrows). The following evidence suggests that these activities arise from distinct reactivation mechanisms. Low-force restart was found to be strictly factor-dependent (Fig. 1D) and stimulated by both TFIIF and WT TFIIS, with additive effects exerted by TFIIF and WT TFIIS. Interestingly, low-force restart could be stimulated, and at similar levels, both by WT TFIIS and a cleavage-defective mutant, TFIIS Δ2–146 E291H (henceforth called TFIISΔNmut) (38, 39). These findings imply that the restart mechanism is independent of any TFIIS-catalyzed transcript cleavage. We note that cleavage-independent reactivation would require significant forward translocation, to reverse any backtracking and restore the transcript 3′ end to the active site, and that such a motion is strongly disfavored by hindering loads. Consistent with the foregoing, TFIISΔNmut failed to trigger any discernible recovery activity under high hindering loads (SI Appendix, Fig. S3). Together, the evidence shows that either TFIIS or TFIIF can promote low-force restart independently and that neither of the two mechanisms involved requires transcript cleavage. Backtrack-and-rescue events (Fig. 1E), by contrast, were observed at low baseline levels for RNAPII alone, and these were strongly stimulated by WT TFIIS, but not by TFIIF or TFIISΔNmut. These data suggest that backtrack-and-rescue strictly requires cleavage promoted by TFIIS. We note that cleavage-mediated transcript rescue bypasses the need for forward translocation to reverse any prior backtracking, hence remaining a possibility even under high hindering loads. Backtrack-and-rescue therefore constitutes a third reactivation mechanism, distinct from the other two, which both facilitate low-force restart. TFIIF, when present, synergistically enhanced the backtrack-and-rescue activity of WT TFIIS, but not that of the fully cleavage-competent, N-terminal truncation mutant TFIIS Δ2–146 (henceforth called TFIISΔN) in which domain I was deleted (38), suggesting that domain I, the role of which is only poorly understood, is likely required for this effect. A defined interaction between TFIIF and TFIIS domain I has recently been established by cross-linking experiments (34).
The most dramatic effects on RNAPII elongation under high loads were observed when TFIIS and TFIIF were present simultaneously (Fig. 1 C–E and SI Appendix, Figs. S4 and S5). This synergy is compatible with an enhanced recruitment, or stabilization, of WT TFIIS in the elongation complex by its interaction with TFIIF. Under such conditions, a fraction of molecules was observed to cycle between bursts of forward transcription and periods of processive backtracking, often extending over hundreds of seconds and without any appreciable (net) forward translocation. This behavior is broadly consistent with a previous report of the action of TFIISΔN on elongating RNAPII (7). There, TFIISΔN produced an increase in the average force required to halt RNAPII (the stall force), together with a bimodal stall force distribution, which was interpreted in terms of both TFIIS-bound and -unbound subpopulations. In our work, by contrast, none of the transcription factor combinations altered the RNAPII stall force significantly (SI Appendix, Fig. S1). The difference may be attributable to the higher stall force observed here for RNAPII alone: −9.8 ± 0.6 pN, compared with −7.4 ± 2.0 pN reported in ref. 7. However, the average stall forces for RNAPII with WT TFIIS (−10.1 ± 0.6 pN) and RNAPII with TFIISΔN (−10.2 ± 0.5 pN) measured here agree well with the weighted-average stall force (−10.5 ± 2 pN) of the putative TFIIS-bound and -unbound populations reported in ref. 7. In our hands, therefore, the transcription factors did not enhance the force-production capability of RNAPII. Instead, the factors served to suppress arrests and to retain the enzyme in a transcriptionally competent state for extended periods. Maintaining this state may be important for overcoming barriers in vivo, including sequence-specific DNA-binding proteins, which transiently dissociate, or nucleosomes, which transiently unravel in a manner that permits transcription (40).
RNAPII Elongation and Pausing Under Assisting and Hindering Loads.
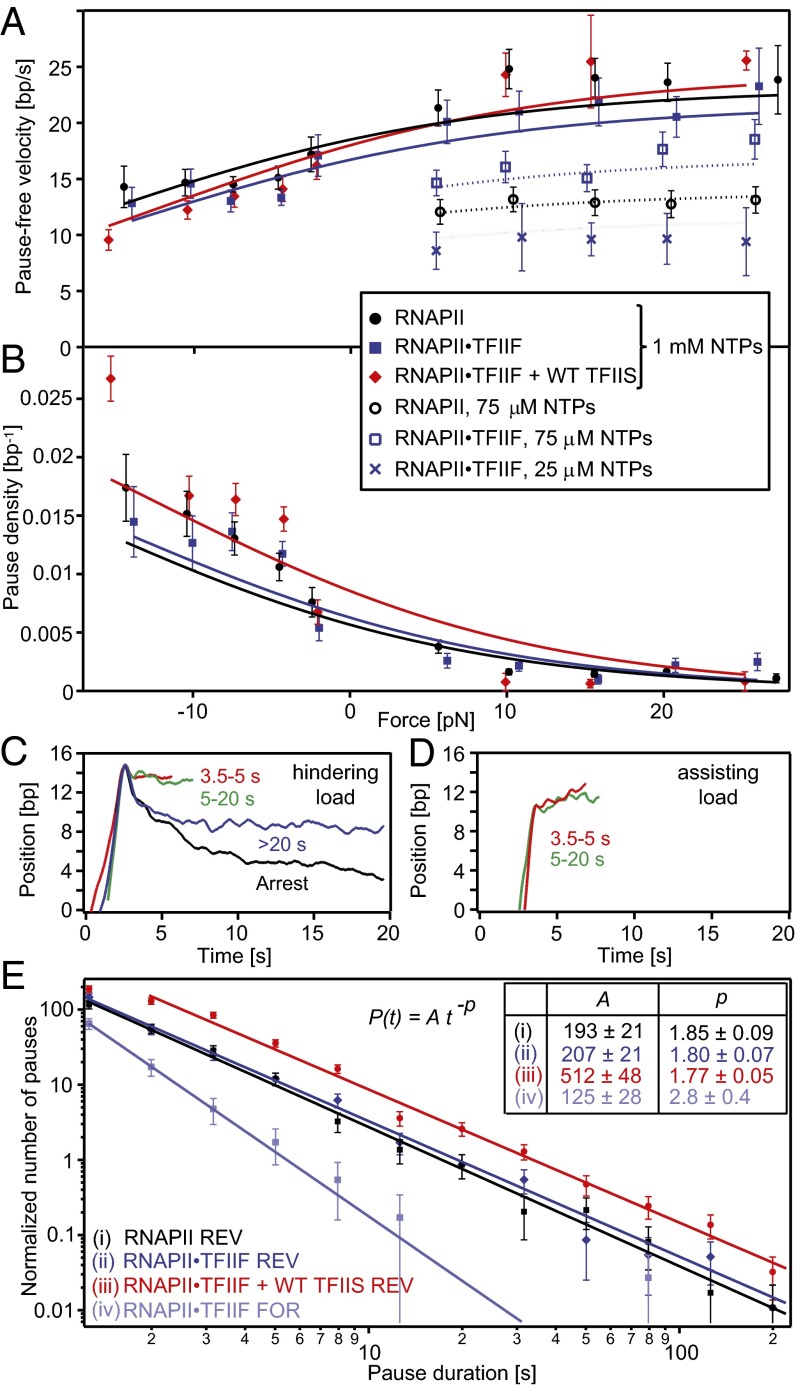
We examined the effects of TFIIS and TFIIF on pause production and elongation rate under controlled loads (Fig. 2). Over a range of forces that we estimate to be comparable to those experienced in vivo (+5 to −5 pN), the (pause-free) velocity dropped from 23 bp/s under high assisting loads to 14 bp/s under high hindering loads. Concomitantly, the pause density rose sharply, from a nearly force-independent level of ∼1.5 kb−1 under assisting load to ∼15 kb−1 under hindering load. The experimental data (Fig. 2) could be fit by a two-state Boltzmann expression, which relates the enzyme translocation register (i.e., the position of the RNAPII active site relative to the template nucleotide of DNA) to the force, and a Michaelis–Menten expression, which relates the enzyme velocity to the nucleoside triphosphate (NTP) concentration (SI Appendix). Fixing the shift in the translocation register to 1 bp, corresponding to a shuttling motion between the pre- and posttranslocated states of RNAPII (41), gave reasonable fits to the data (Fig. 2 A and B). Slightly better fits could be obtained by allowing this parameter to assume the larger value of 1.16 nm (∼3.4 bp) (SI Appendix, Fig. S6), which would imply larger-scale enzyme motions. SI Appendix, Fig. S7 shows a possible pathway for RNAPII elongation and pausing that is consistent with our data.
Fig. 2.
Influence of TFIIS and TFIIF on transcription speeds and pausing. (A) The pause-free elongation velocity at saturating (filled symbols) and subsaturating (open symbols and crosses) NTP concentrations. (B) Pause density (pauses per bp transcribed) under assisting (+) and hindering (−) loads. (C and D) Averaged position records during pause entry for RNAPII and RNAPII⋅TFIIF. (C) Under hindering loads, short (<20 s) pauses exhibited small backtracks (∼1–2 bp); long pauses (≥20 s) and arrests were backtracked by ∼6 and ∼10 bp on average, respectively. (D) Pauses under assisting loads were not backtracked and exhibited slow forward creep. (E) Distributions of pause lifetimes under assisting (FOR) and hindering (REV) loads were approximated by distinct power laws (fit parameters for amplitude, A, and exponent, p, with SE). The distribution was normalized as described in Materials and Methods. Numbers of pauses scored were as follows: RNAPII⋅TFIIF FOR, 248; RNAPII⋅TFIIF REV, 278; RNAPII REV, 266; RNAPII⋅TFIIF + TFIIS REV, 638. Fits were restricted to ranges of duration with six or more pauses per bin as follows: RNAPII⋅TFIIF FOR, 1–3.5 s; RNAPII⋅TFIIF REV, 1–35 s; RNAPII REV, 1–25 s; RNAPII⋅TFIIF + TFIIS REV, 1–55 s. Error bars are SE.
Neither the addition of TFIIS nor TFIIF exerted strong effects on the elongation velocity at saturating NTP concentrations (Fig. 2), suggesting that these factors do not influence any rate-limiting steps that may be associated with, say, phosphodiester bond formation or pyrophosphate release. At subsaturating NTP concentrations, however, TFIIF exerted a mild stimulatory effect on NTP binding, which could be seen most clearly in the 75 μM data of Fig. 2A (KD = 29 ± 5 μM for RNAPII⋅TFIIF vs. 72 ± 9 μM for RNAPII), possibly by facilitating NTP entry into the enzyme active site.
RNAPII Exhibits both Backtracked and Backtrack-Independent Pause Mechanisms.
Together, the load dependence of the pause density and the averaged template position during pause entry (Fig. 2 B–D) suggest that elongation is associated with more than one pause mechanism. Under assisting loads, records of the average template position displayed no statistically significant evidence of enzyme backtracking beyond the ∼1-bp noise level. This observation, together with the independence of a baseline pause density on external load in the assisting-load regime, is fully consistent with a backtracking-independent mechanism for pausing. Under hindering loads, by contrast, records of the average template position showed significant backtracking (∼2–10 bp), and the pause density displayed a strong force dependence. Moreover, the extent of backtracking at pause entry correlated with the pause duration: Short pauses (<20 s) were associated with limited backtracking (∼1–2 bp), whereas long pauses (>20 s) were entered, on average, through longer backtracks (∼3–8 bp). Transcriptional arrests, in which no recovery to the elongation phase was scored, arose from significantly greater backtracks (approximately ≥10 bp). These results contrast strikingly with bacterial RNAP from Escherichia coli, where the most common transcriptional pauses, appearing at a density of 1 per 100 bp, are not backtracked and occur at a rate that is nearly independent of the load (42).
Normalized distributions of the pause lifetimes for RNAPII, obtained in the presence and absence of transcription factors, were approximately linear in log–log plots (Fig. 2E and SI Appendix, Fig. S8), implying that pause durations can be approximated by a simple power law, P(t) ∼ t−p, where P(t) is the associated probability, t is the lifetime, and p is a power-law exponent. Fits to our data yielded estimates for this exponent ranging from 1.8 to 2.8. A theory that assumes that backtracking is the only mechanism responsible for pausing, and which models backtracking as a diffusive process involving a random walk over the array of backtracked bases on the template, predicts that the asymptotic behavior of the distribution at long times should decay no faster than t−3/2, namely, with p = 1.5 (7, 43). Pause lifetimes determined under assisting loads yielded a significantly steeper power-law exponent, p ∼ 2.8 ± 0.4, and likely reflect an additional mechanism for pausing that is not purely diffusive. We also note that lifetime distributions can be fit to other functional forms at comparable levels of statistical significance, including sums of multiple exponentials (SI Appendix, Fig. S8). The latter functions correspond to the distributions generated by conventional biochemical kinetic models—i.e., by discrete states connected by reaction rates (6, 42). Distinguishing theoretical models for pausing solely on the basis of the shapes of their lifetime distributions is problematic. In particular, the models that have been proposed differ mainly in their asymptotic behavior at long times, where the experimental statistics are poor. Moreover, in this regime, long-lifetime pauses become difficult to distinguish from transcriptional arrests, which can be generated by altogether different mechanisms.
TFIIS Facilitates Entry into, but Not Exit from, Backtracked Pauses.
In the presence of WT TFIIS, the density of backtracked pauses was enhanced under hindering loads (Fig. 2B), i.e., TFIIS facilitated entry into backtracked states. Pauses under assisting loads, by contrast, were not strongly influenced. The lifetimes under hindering loads (Fig. 2E) showed that TFIIS enhanced the density of pauses of >2 s, but apparently not that of shorter pauses. TFIIS did not significantly alter the power-law exponent for pause lifetimes, however. This finding seemingly runs contrary to the expectation that TFIIS-induced cleavage of backtracked RNA might contribute to a specific reduction of longer pauses. The records of averaged position at pause entry (SI Appendix, Fig. S9) indicate that TFIIS limited the extent and velocity of backtracking for long pauses (>20 s) and arrests, a finding that is consistent with a predicted steric clash between backtracked RNA and TFIIS in the RNAPII funnel (38, 44).
Effects of TFIIS and TFIIF Probed by ITP Misincorporation.
Conditions favoring nucleotide misincorporation can serve as a testbed for studying factor interactions. We examined the effect of 0.4 mM inosine triphosphate (ITP), a nucleotide analog that partially mimics GTP. Following ITP misincorporation, the rate of next-nucleotide addition slows substantially, leading to pausing and backtracking (6, 41). Misincorporation-induced pausing facilitates cleavage of the backtracked RNA fragment, either through the slow, intrinsic endonucleolytic capability of RNAPII or, when present, by faster TFIIS-induced cleavage. Removal of the RNA fragment containing the misincorporated base constitutes a mechanism for error correction. Fig. 3 shows the effects of TFIIS and TFIIF on elongation under assisting loads, which minimize the background of misincorporation-independent pauses. (Experiments conducted under hindering loads confirm the same conclusions; SI Appendix, Fig. S10.) In agreement with earlier studies (6, 41), ITP increased the density and duration of pauses while leaving the pause-free velocity nearly unchanged [Fig. 3A; the minor reduction in the pause-free elongation velocity is likely due to an increase in pauses that are too short (<1 s) to be detected in our assays]. When WT TFIIS was added to RNAPII⋅TFIIF complexes in the presence of ITP, pause density and mean duration were restored to levels observed without ITP, whereas the addition of TFIIS in the absence of TFIIF led to only a slight reduction in the mean duration.
Fig. 3.
Effects of TFIIS and TFIIF under assisting load and conditions favoring misincorporation, induced by ITP. (A) ITP (0.4 mM; other NTPs at 1 mM) increased both the average density and duration of pauses but left pause-free velocities nearly unchanged. In the presence of ITP, TFIIS restored both mean pause density and duration for RNAPII⋅TFIIF to normal levels, while reducing the pause duration for RNAPII (alone) only slightly. (B) ITP did not significantly affect the power-law exponent in fits to pause lifetime distributions (shown for RNAPII⋅TFIIF); TFIIS restored the original pause distribution. The number of pauses was normalized as in Fig. 2E. Numbers of pauses scored were as follows: RNAPII⋅TFIIF FOR, 248 (same data as Fig. 2E; shown for comparison); RNAPII⋅TFIIF FOR + ITP, 243; RNAPII⋅TFIIF FOR + ITP + WT TFIIS, 54. Fit domains were as follows: RNAPII⋅TFIIF FOR, 1–3.5 s; RNAPII⋅TFIIF FOR + ITP, 1–8.5 s; RNAPII⋅TFIIF FOR + ITP + WT TFIIS, 1–3.5 s; Error bars are SE.
These results are notable in two ways. First, they support a model whereby WT TFIIS selectively recognizes and suppresses ITP-induced pauses while leaving other types of pause largely unaffected. Averaged records of position at pause entry acquired under assisting loads (SI Appendix, Fig. S11), while at the limit of our spatial resolution, suggest that RNAPII may backtrack by ∼1 bp after pausing due to ITP misincorporation, but not in the absence of ITP. Second, the efficient removal by TFIIS of even the shortest observable ITP-induced pauses clearly requires access by this factor to the enzyme active site on a short timescale (∼1 s or less). The rapid response of backtracked RNAPII to TFIIS produced by misincorporation events stands in stark contrast to the apparent inability of this same factor to rescue backtracked pauses induced by hindering loads on similarly fast timescales, irrespective of the presence of TFIIF (Fig. 2E; see also Fig. 1C and SI Appendix, Fig. S5).
Discussion
Long-lifetime pauses for RNAPII, as well as transcriptional arrests, are countered by specific responses, mediated both independently and synergistically by TFIIS and TFIIF. Their molecular mechanisms do not appear to confer the capacity to override any mechanical regulatory signals, but, rather, serve to render transcription more persistent overall by preserving RNAPII in a transcriptionally competent state during extended periods of pausing, from which elongation can eventually resume once mechanical barriers are reduced or removed.
Two distinct pause mechanisms for RNAPII, one backtracked and the other backtrack-independent, are revealed by this work. This dichotomy has been a source of some controversy (7, 43). TFIIS stimulates entry into backtracked pauses, an effect that has been observed in past biochemical studies of metazoan TFIIS (15) and of a cleavage-incompetent yeast TFIIS mutant (45). Backtrack-independent pauses, by contrast, are not strongly modulated—and perhaps even reduced slightly—by TFIIS.
The observed distributions for pause lifetimes, and the modulatory effects found here for TFIIF and TFIIS, do not appear to fully support the notion that enzyme backtracking follows a random walk, but may instead be modulated by mechanisms intrinsic to RNAPII. Candidate mechanisms have been identified in recent structural work (44). A “gating tyrosine” residue was found to contact the 3′ end of backtracked RNA, facilitating a 1-bp motion upstream, but restricting any further backtracking. When backtracked RNA manages to bypass this tyrosine, however, a more substantially backtracked state (∼8-bp) could be stabilized, in principle, by additional interactions between the backtracked RNA and amino acid residues that form a “backtrack site” in the RNAPII pore and funnel. These residues are highly conserved among eukaryotes but have no clear homologs in bacterial RNAP. In the presence of TFIIS, structural work (44) suggests that for long backtracks, a steric clash between RNA and TFIIS occurs inside the funnel, in which TFIIS weakens the grip of RNAPII on backtracked RNA and displaces the RNA, through competitive binding to the backtrack site. This activity does not require transcript cleavage and therefore provides a mechanism for transcriptional restart at low forces, as observed here. Additional cleavage-independent effects of TFIIS have been reported, in which TFIIS was identified as a component of preinitiation complexes (PICs) (1), stimulating PIC assembly in a cleavage-independent manner in vitro (46) and in vivo (47, 48).
Strong stimulatory effects of metazoan TFIIF on overall rates of RNA synthesis have been reported (27, 49, 50) and attributed to positive effects on catalytic rates and the suppression of pausing. No such reports exist for yeast TFIIF effects on elongation, however. Here, we find that yeast TFIIF, by contrast, does not affect the catalytic rate of RNAPII or its behavior on pause entry and exit. Instead, its effects seem limited to facilitating transcriptional restart from long-distance backtracks. The differences between metazoan and yeast TFIIF may reflect functional differences, or they may arise from the fact that in vitro assays using mammalian RNAPII (27, 49), unlike yeast RNAPII (41), have not yet been able to achieve fast elongation rates that are comparable to those measured in vivo. Here, yeast TFIIF was found to exert a synergistic effect on TFIIS activity. We speculate that this interaction may be attributable to binding between TFIIF and TFIIS domain I, which recruits and stabilizes TFIIS in the elongation complex. In this context, we note that TFIIF specifically enhances the cleavage-dependent activities of TFIIS (backtrack-and-rescue and the removal of misincorporation pauses), whereas the cleavage-independent activity of TFIIS (low-force restart) remains unaffected.
Materials and Methods
A detailed description of materials and methods is given in SI Appendix, SI Materials and Methods.
Single-Molecule Optical-Trapping Assay.
Biotinylated RNAPII or preformed RNAPII·TFIIF complexes from Saccharomyces cerevisiae (SI Appendix) were initiated on a DNA:RNA scaffold as described (41). Individual RNAPII or RNAPII⋅TFIIF molecules were tethered to 0.60-μm diameter polystyrene beads by biotin–avidin linkages. To apply controlled loads assisting (or hindering) forward elongation, upstream (or downstream) DNA was attached to a second, 0.82-μm diameter bead, and the beads were captured by separate optical traps (5). Transcription was started by introducing 1 mM NTP unless otherwise indicated; 1 μM recombinant TFIIS was added where indicated.
Data Collection and Analysis.
Records of position data were acquired and analyzed by using custom software as described (41). Pause-free elongation velocities for single RNAPII molecules were determined by fitting experimental velocity distributions to a sum of two Gaussians (one centered around zero and corresponding to pausing; the other to active elongation) as described (42). Pauses were identified and scored as described (6). Records of elongation were boxcar-filtered at 0.5 s for display. For display purposes, small vertical artifacts (∼10 bp) associated with rapid template reequilibration when the force was stepped up or down were removed from the records in Fig. 1 and SI Appendix, Figs. S3 and S5. Transcriptional arrests were scored as events in which a previously transcribing enzyme stalled and no significant forward translocation occurred over a period of ≥300 s. Backtrack-and-rescue events were scored as events in which backtracking by ≥5 nm was recorded, followed by a transcriptional restart extending for ≥10 nm. Only events at forces |F| ≥ 5 pN were included for analysis. Low-force restart events were scored when, consequent to arrest (as defined above), the applied force was dropped to ∼2 pN, and a subsequent restart of transcription was observed within 100 s. Averaged position records were generated by aligning a series of individual traces with respect to the pause start time (event scored automatically; record alignments were adjusted manually in cases where the algorithm failed to identify the start). Pauses scored for forces |F| <5 pN, where thermal noise compromises the data quality, were not included in averaged position records. Pause distributions (Figs. 2 and 3) were normalized as follows: Histogram bin widths were allocated in increments of equal log(pause duration) to account for the rarity of longer pauses. The numbers of pauses per unit time were then computed for each bin and plotted.
Note.
Subsequent to the online publication of a preliminary report of this work (51), and as the present work was being prepared for publication, ref. 52 was published on a similar topic.
Supplementary Material
Acknowledgments
We thank Jing Zhou for contributions to the initial stages of this work and Kyle Eagen for his help with producing yeast constructs. This research was supported by National Institutes of Health Grants GM57035 (to S.M.B.), GM97260 (to C.D.K.), GM36659, and AI21144 (to R.D.K.); Welch Foundation Grant A-1763 (to C.D.K.); a Kazato Research Foundation grant (to K.M.); and Damon Runyon Postdoctoral Fellowship DRG-2059-10 (to V.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405181111/-/DCSupplemental.
References
- 1.Sikorski TW, Buratowski S. The basal initiation machinery: Beyond the general transcription factors. Curr Opin Cell Biol. 2009;21(3):344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44(2-3):117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Schweikhard V, Block SM. Single-molecule studies of RNAPII elongation. Biochim Biophys Acta. 2013;1829(1):29–38. doi: 10.1016/j.bbagrm.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438(7067):460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426(6967):684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446(7137):820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 8.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18(1):97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 11.Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35(2):191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Close P, et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484(7394):386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9(12):1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Yan H, Burton ZF. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J Biol Chem. 2003;278(50):50101–50111. doi: 10.1074/jbc.M307590200. [DOI] [PubMed] [Google Scholar]
- 16.Luse DS, Spangler LC, Újvári A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286(8):6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem. 2001;276(25):23109–23114. doi: 10.1074/jbc.M101445200. [DOI] [PubMed] [Google Scholar]
- 18.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325(5940):626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16(6):955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 20.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93(4):627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 21.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93(24):13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama H, Ito T, Nakanishi T, Sekimizu K. Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells. 2007;12(5):547–559. doi: 10.1111/j.1365-2443.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- 23.Nesser NK, Peterson DO, Hawley DK. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci USA. 2006;103(9):3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Q, Moreland RJ, Conaway JW, Conaway RC. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem. 1999;274(50):35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 25.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282(30):21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 26.Újvári A, Pal M, Luse DS. The functions of TFIIF during initiation and transcript elongation are differentially affected by phosphorylation by casein kinase 2. J Biol Chem. 2011;286(26):23160–23167. doi: 10.1074/jbc.M110.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276(45):42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 28.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267(19):13647–13655. [PubMed] [Google Scholar]
- 29.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17(10):1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 30.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol. 2002;22(20):6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cojocaru M, et al. Genomic location of the human RNA polymerase II general machinery: Evidence for a role of TFIIF and Rpb7 at both early and late stages of transcription. Biochem J. 2008;409(1):139–147. doi: 10.1042/BJ20070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen ZA, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29(4):717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29(4):706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami K, et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science. 2013;342(6159):1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18(4):461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Murakami K, et al. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J Biol Chem. 2013;288(9):6325–6332. doi: 10.1074/jbc.M112.433623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami K, et al. Tfb6, a previously unidentified subunit of the general transcription factor TFIIH, facilitates dissociation of Ssl2 helicase after transcription initiation. Proc Natl Acad Sci USA. 2012;109(13):4816–4821. doi: 10.1073/pnas.1201448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30(5):547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 41.Larson MH, et al. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci USA. 2012;109(17):6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115(4):437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 43.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96(6):2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 45.Imashimizu M, et al. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425(4):697–712. doi: 10.1016/j.jmb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim B, et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci USA. 2007;104(41):16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA. 2007;104(41):16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prather DM, Larschan E, Winston F. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(7):2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nedialkov YA, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278(20):18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- 50.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 51.Schweikhard V, et al. TFIIF and TFIIS enhance the mechanical persistence of transcript elongation by RNA polymerase II. Biophys J. 2014;106(2 Suppl 1):486 (abstr). [Google Scholar]
- 52.Ishibashi T, et al. Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci USA. 2014;111(9):3419–3424. doi: 10.1073/pnas.1401611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.