Significance
Despite being one of the oldest groups of land plants, the majority of living ferns resulted from a relatively recent diversification following the rise of angiosperms. To exploit fully the new habitats created by angiosperm-dominated ecosystems, ferns had to evolve novel adaptive strategies to cope with the low-light conditions exerted by the angiosperm canopy. Neochrome, an unconventional photoreceptor that allows ferns to “see the light” better, was likely part of the solution. Surprisingly, we discovered that fern neochrome was derived from a bryophyte lineage via horizontal gene transfer (HGT). This finding not only provides the first evidence that a plant-to-plant HGT can have a profound evolutionary impact but also has implications for the evolution of photosensory systems in plants.
Keywords: phototropism, chloroplast movement
Abstract
Ferns are well known for their shade-dwelling habits. Their ability to thrive under low-light conditions has been linked to the evolution of a novel chimeric photoreceptor—neochrome—that fuses red-sensing phytochrome and blue-sensing phototropin modules into a single gene, thereby optimizing phototropic responses. Despite being implicated in facilitating the diversification of modern ferns, the origin of neochrome has remained a mystery. We present evidence for neochrome in hornworts (a bryophyte lineage) and demonstrate that ferns acquired neochrome from hornworts via horizontal gene transfer (HGT). Fern neochromes are nested within hornwort neochromes in our large-scale phylogenetic reconstructions of phototropin and phytochrome gene families. Divergence date estimates further support the HGT hypothesis, with fern and hornwort neochromes diverging 179 Mya, long after the split between the two plant lineages (at least 400 Mya). By analyzing the draft genome of the hornwort Anthoceros punctatus, we also discovered a previously unidentified phototropin gene that likely represents the ancestral lineage of the neochrome phototropin module. Thus, a neochrome originating in hornworts was transferred horizontally to ferns, where it may have played a significant role in the diversification of modern ferns.
Plant growth and development are modulated by photoreceptor systems that provide information about the surrounding environment. Major peaks in the action spectra of these informational photoreceptors lie either in the UV-blue (e.g., cryptochromes and phototropins) or red/far-red (phytochromes) light regions (1). The chimeric photoreceptor neochrome is a remarkable exception. It fuses red-sensing phytochrome and blue-sensing phototropin modules into a single molecule (Fig. 1A) that mediates phototropic responses (1–4). Neochromes have a restricted occurrence in the plant tree of life and are hypothesized to have had two independent origins (5)—one in the green alga Mougeotia scalaris and another in ferns. The possession of neochrome may be evolutionarily advantageous, as evidenced by the greatly enhanced phototropic responses in ferns with neochrome (3, 4) and by its phylogenetic distribution within the fern lineage. The early-diverging fern orders Osmundales and Schizaeales do not possess neochrome (3). It has been reported only in Cyatheales (6) and Polypodiales (3, 6), lineages that mostly diversified following the Cretaceous/Tertiary establishment of low-light, angiosperm-dominated forest canopies (7, 8). As a result, it has been suggested that the evolution of neochrome was a key innovation that conferred a phototropic advantage on ferns growing under low-light conditions, facilitating their modern diversification in the “shadow of angiosperms” (3, 7, 8). Although potentially significant from an evolutionary standpoint, the origin of fern neochrome has remained a mystery.
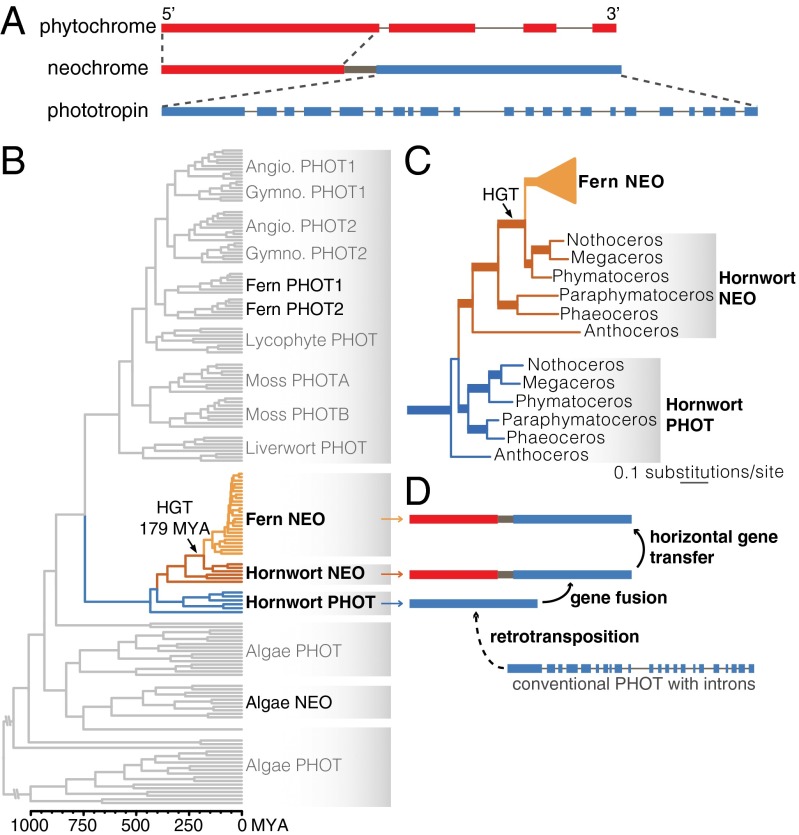
Fig. 1.
The origin of fern neochrome. (A) Neochrome is a chimeric photoreceptor in which the N terminus consists of a phytochrome sensory module fused to an almost complete phototropin sequence at the C terminus. Thick and thin lines represent exons and introns, respectively (length not to scale). (B) Dated phylogeny of phototropin and neochrome, showing neochrome HGT from hornworts to ferns (details are given in SI Appendix, Fig. S5). The blue, brown, and yellow branches represent hornwort phototropin, hornwort neochrome, and fern neochrome, respectively. (C) Portion of the phototropin phylogeny showing relationships of fern neochrome (Fern NEO), hornwort phototropin (Hornwort PHOT), and hornwort neochrome (Hornwort NEO), with highly supported branches thickened (details are shown in SI Appendix, Figs. S1 and S2). (D) A schematic depicting the origin of fern neochrome involving retrotransposition of a phototropin gene (and hence the loss of introns), its fusion with a phytochrome, and HGT from hornworts to ferns.
In this study we investigated the origin of neochrome by searching for homologous sequences in 434 transcriptomes, and 40 whole or draft genomes of plants and algae (SI Appendix, Table S1) and surprisingly discovered neochrome homologs from hornworts (Fig. 1B and SI Appendix, Figs. S1–S3). Analyses of the hornwort draft genome (Anthoceros punctatus) suggest that neochrome originated in hornworts, independent from the green algae. Large-scale phylogenetic analyses and divergence time estimates further demonstrate that ferns acquired neochrome from hornworts via horizontal gene transfer (HGT).
Results and Discussion
Algal Neochrome.
The only published algal neochrome is from a single species, Mougeotia scalaris (5). We identified homologs of neochrome in the transcriptomes of all 10 sampled members of the Zygnemataceae (9), including Mougeotia, Mesotaenium, Cylindrocystis, and Zygnemopsis, but in no other algal transcriptomes surveyed (SI Appendix, Table S1 and Fig. S1).
Neochrome in Hornworts.
Among land plants, we documented the occurrence of neochrome in 25 additional fern species (SI Appendix, Figs. S1–S3). Surprisingly, we also discovered neochrome in hornworts, a small bryophyte lineage that diverged early in the history of land plants. The exact branching order among the three bryophyte lineages (hornworts, mosses, and liverworts) is not resolved with certainty; previous analyses have suggested that hornworts are sister to vascular plants (lycophytes, ferns, and seed plants) (10), but this relationship was challenged recently, and the monophyly of all bryophytes was proposed (11). We confirmed the presence of neochrome in hornworts through PCR and cloning, and isolated neochrome sequences from the genera Nothoceros, Megaceros, Phymatoceros, Phaeoceros, Paraphymatoceros, and Anthoceros, representing four of the five hornwort orders (namely, Dendrocerotales, Phymatocerotales, Notothyladales, and Anthocerotales). We were unable to obtain adequate material of the monotypic hornwort Leiosporoceros to test for the presence of neochrome in Leiosporocerotales. To confirm that our hornwort neochrome sequence data were indeed derived from the hornwort nuclear genome and not from contaminant algae or ferns, we performed genome-walking in Nothoceros aenigmaticus to obtain flanking genomic sequences. Downstream of neochrome we found a pseudogene for imidazoleglycerol-phosphate dehydratase (IGPD) and, because its sequence is most closely related to other hornwort IGPD genes (SI Appendix, Fig. S4), we are confident that neochrome is present in the hornwort genome.
Neochrome HGT from Hornworts to Ferns.
The phylogenetic distribution of neochrome in land plants (present only in hornworts and ferns) could be explained by (i) an ancient origin along the branch that unites hornworts and tracheophytes, followed by losses from lycophytes and seed plants, (ii) independent origins in ferns and hornworts, or (iii) one or more instances of HGT between hornworts and ferns. To distinguish among these three possible scenarios, we compiled comprehensive sequence alignments of phototropin and phytochrome from across all land plants and algae, which included the corresponding domains from hornwort and fern neochromes, and evaluated the resultant gene phylogenies. Maximum likelihood and Bayesian estimates of phototropin and phytochrome phylogenies revealed that fern neochromes are embedded within hornwort neochromes with very strong branch support (Fig. 1 B and C and SI Appendix, Figs. S1–S3). This nested relationship indicates that neochrome was transferred horizontally from hornworts to ferns, along the stem lineage leading to Phymatoceros + Nothoceros + Megaceros (Fig. 1C, arrow, and SI Appendix, Figs. S1–S3). The alternative possibilities, suggesting either an ancient vertical transfer of neochrome (i.e., fern and hornwort neochromes were reciprocally monophyletic) or an independent origin of neochrome (i.e., fern neochromes were monophyletic with either fern phototropins or phytochromes) were both rejected (P < 10−30) and were never observed in the Bayesian posterior tree samples.
We used estimates of divergence time to assess our HGT hypothesis further, reasoning that in a case of HGT the split between hornwort and fern neochrome should be younger than the split between the hornwort and fern lineages themselves. By integrating fossil calibrations (SI Appendix, Table S2) with a Bayesian relaxed molecular clock analysis, we estimated the divergence date between hornwort and fern neochrome to be ∼179 Mya with a 95% highest posterior density interval of 133 and 229 Mya (Fig. 1B and SI Appendix, Fig. S5). This date is far more recent than published divergence estimates between ferns and hornworts (at least 400 Mya) (12) but is congruent with the date estimates for the stem branch leading to Phymatoceros + Nothoceros + Megaceros (85–244 Mya) (13). The disparity in divergence times rejects the hypothesis invoking multiple neochrome origins or losses and reinforces the HGT scenario.
The origin of land plant neochrome within the hornwort lineage is supported by its relationship to hornwort phototropin. The single hornwort phototropin gene in the Anthoceros punctatus draft genome completely lacks introns (Fig. 1D) and thus closely resembles the C-terminal end of both fern and hornwort neochromes. We found this intron-free phototropin in all hornworts examined by using PCR on genomic DNA (SI Appendix, Fig. S2 and Table S3). All other phototropins characterized to date, including those of ferns, contain more than 20 introns. We explored whether this gene might be a partial neochrome masquerading as a phototropin by using inverse PCR to obtain the 5′ upstream genomic region in N. aenigmaticus. Multiple stop codons were encountered upstream of the Nothoceros phototropin gene, and there was no indication of nearby phytochrome domains. These data suggest that hornworts might not have a canonical phototropin gene. Instead, hornwort phototropins are most closely related to fern and hornwort neochromes (Fig. 1 B and C and SI Appendix, Fig. S2), implying that they likely represent the ancestral, retrotransposed phototropin lineage that gave rise to neochrome through fusion with the phytochrome module (Fig. 1D). On the other hand, in the phytochrome phylogeny, hornwort phytochromes are not resolved as sister to hornwort and fern neochromes (SI Appendix, Fig. S3), although there is no branch support for this nonmonophyletic relationship. The phytochrome progenitor of neochrome therefore remains unclear.
Recurrent Fern-to-Fern HGT.
We detected an extraordinary incongruence between our fern neochrome gene tree and the published phylogeny of ferns (SI Appendix, Fig. S6) (14). By examining the entire Bayesian posterior tree sample, we found that none of the trees resolved neochromes from the same fern family as being monophyletic. This conflicting pattern is not observed in other fern phylogenies based on nuclear genes (15) and is not seen in the hornwort neochrome tree (Fig. 1C), which perfectly mirrors the published phylogeny of hornworts (13). Here we investigate and discuss the possible causes of the incongruent gene tree/species tree in ferns.
Incomplete sampling of extant neochrome homologs is not likely to be the explanation, because neochrome has been shown by Southern blotting to be a single-copy gene in Adiantum capillus-veneris (2). This result was corroborated by the cloning efforts that produced most of our sequence data (SI Appendix, Table S3). Except for Deparia spp., in which two divergent sequences were found (SI Appendix, Fig. S6, arrowheads), we were able to isolate only a single neochrome from each fern species.
Next, we investigated whether an aberrant nucleotide substitution process may have misled the phylogenetic reconstruction. For example, pervasive positive selection or variation in guanine/cytosine (GC) content can obscure true phylogenetic signal (16–18), thereby causing a gene tree to be incongruent with the species tree. Using codon models for tree inference potentially can accommodate complex selection profiles by allowing different nonsynonymous/synonymous substitution rate ratios to fall into distinct classes (19). However, we found that incorporating codon models did not improve the incongruence between the gene tree and species tree; the resultant tree largely matches that from the nucleotide substitution model, with comparable branch support values (SI Appendix, Figs. S6 and S7). Similarly, inferences based on first + second-codon positions or only on third-codon positions also yielded topologies discordant with the species tree (SI Appendix, Fig. S7).
We then used a random effects branch-site model to infer the dynamics of positive selection across the neochrome tree (20). Only five fern branches were identified as having experienced significant episodic positive selection (SI Appendix, Fig. S7D), and the proportion of positively selected codon sites along each of these five branches is very low (<3%). These results suggest that positive selection operated on very few codons over a limited number of branches. Similarly, a sliding window analysis of GC content found none of the fern sequences to be deviant in base composition (SI Appendix, Fig. S7E). Taken together, the nucleotide substitution processes among fern neochromes appear to be unexceptional and are not likely to explain the incongruence between the gene tree and species tree.
We therefore hypothesized that the incongruent tree could be the result of (i) multiple fern-to-fern HGT events, (ii) an elevated gene turnover rate that may have been selected for after HGT (21, 22), or (iii) a combination of both factors. We have some evidence suggesting recurring fern-to-fern HGT might have been involved. For example, we discovered neochrome genes from two early-diverging fern orders [Gleicheniales (Dipteris conjugata) and Cyatheales (Alsophila podophylla and Plagiogyria spp.)] that likely were derived from secondary HGT events (SI Appendix, Fig. S6, arrows). These neochromes are not phylogenetically resolved, as would be predicted based on published fern species relationships (14), but instead are nested among Polypodiales (SI Appendix, Fig. S6). Furthermore, the split between these and other fern neochromes (81 Mya, 95% highest posterior density interval: 59–106 Mya; SI Appendix, Fig. S5) occurred long after the estimated organismal divergence dates for Gleicheniales (276 Mya) and Cyatheales (223 Mya) (8), a pattern that may be explained best by fern-to-fern HGT.
Our hypothesis of potentially recurrent HGT events within ferns is not unprecedented. In angiosperms, rampant HGTs have been documented for the mitochondrial cox1 homing intron. This intron is believed to have experienced one initial “seed transfer” from fungi that was followed by at least 80 incidents of plant-to-plant HGT among 833 diverse angiosperm species (23–25). Perhaps neochrome is similarly associated with mobile elements that may have facilitated its movement across species boundaries.
Evolutionary and Physiological Implications of Neochrome in Hornworts.
Our discovery of neochrome in hornworts is an important step toward understanding the evolution of photosensory systems in plants. In the moss Physcomitrella patens, both red and blue light can elicit directional chloroplast movements, and these movements are mediated by molecular interactions between physically separate phytochrome and phototropin proteins (26). The hornwort neochrome represents a strikingly different strategy for integrating these two photosensory systems, combining them into a single, chimeric gene. Light-induced directional chloroplast movement has not yet been observed in hornworts, probably because their epidermal cells usually contain only one chloroplast that occupies most of the cellular space. However, nearly 50 y ago, Burr (27) documented an unusual chloroplast photoresponse in Megaceros hornworts; she discovered that the large chloroplasts “contract” to form compact shapes under strong light. Although we confirmed this phenomenon in hornworts (SI Appendix, Fig. S8), future studies are needed to examine if neochrome is responsible for the contraction response and to explore other possible physiological roles.
Evolutionary Significance of Plant-to-Plant HGT.
This study pinpoints the origin of land plant neochrome within the hornwort lineage and demonstrates that neochrome was transferred horizontally from hornworts to ferns. The life history of ferns may help explain their hypothesized susceptibility to HGT. Most land plants share a common sexual life cycle that alternates between a diploid sporophyte and a haploid gametophyte; only in ferns and lycophytes are the sporophytic and gametophytic phases both free-living and fully independent. Seed plants insulate their gametophytes from outside interactions with relatively impervious cell walls in microgametophytes and by embedding megagametophytes within protective sporophyte tissues. In contrast, almost all fern gametophytes are not enclosed and grow in direct, intimate contact with other fern and bryophyte gametophytes (including those of hornworts). These characteristics may facilitate the entrance of foreign genetic elements into fern germ lines (i.e., via the gamete-producing structures, the antheridia and archegonia, that are exposed to the environment) (28).
To date, most documented examples of plant-to-plant HGT involve mitochondrial DNA and/or parasite–host transfers (29–36); only a handful of cases include functional nuclear genes (34, 37, 38), and even fewer have possible adaptive implications (39). Consequently, plant-to-plant HGT generally has been overlooked as a potentially significant factor in plant evolution. Given that neochrome may have played a major role in promoting the diversification of ferns under the Cretaceous/Tertiary angiosperm canopy (3, 7, 8), our study has important implications for the macroevolutionary significance of plant-to-plant HGT.
Materials and Methods
Mining Transcriptomes and Whole-Genome Sequences for Homologs of Neochrome, Phototropin, and Phytochrome.
All but one of the 434 transcriptomes used were generated by the One Thousand Plants Project (1KP; www.onekp.com); these transcriptomes were derived from a diverse selection of brown algae, red algae, green algae, bryophytes, lycophytes, ferns, and seed plants (SI Appendix, Table S1). Details on RNA extraction, sequencing, and assembly for 1KP can be found in Johnson et al. (40). Additionally, a whole-plant normalized Illumina transcriptome library was constructed and sequenced for Pteridium aquilinum using pooled RNA from six sporophyte tissues (young sporeling leaf, rhizome tip, fiddlehead, mature sterile pinnae, and pinnae with developing and mature sporangia). The Pteridium transcriptome was assembled using default parameters in the Trinity RNA-seq pipeline version r2012-01–25p1 (41). The sequencing reads were deposited in National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under experiment SRX423244.
For the 1KP transcriptomes, we used both Short Oligonucleotide Assembly Package (SOAP) de novo and SOAP de novo trans assemblies. For each assembly, a BLAST database was constructed using the BLAST+ package (42). Neochrome, phototropin, and phytochrome sequences were queried separately (by tBLASTn for 1KP and by BLASTn for Pteridium assemblies), and the significant hits to transcriptome scaffolds were extracted (e-value threshold of <10−5). For each scaffold, the best ORF was identified, the sequence was translated into amino acids, and then BLASTp queried against the NCBI nonredundant protein database (nr). The scaffolds were discarded if they did not match neochrome, phototropin, or phytochrome homologs in the nr database with an e-value threshold of <0.001. For 1KP transcriptomes, the filtered scaffolds from SOAP de novo and SOAP de novo trans assemblies were merged using CAP3 (43). We carried out the above procedures using our Python pipeline BlueDevil (see http://dx.doi.org/10.5061/dryad.fn2rg). We also searched for and obtained photoreceptor homologs from 39 plant and algae whole-genome sequences through Phytozome (44) and the Amborella Genome Database (www.amborella.org) (45).
Assembling and Mining an Anthoceros punctatus Draft Genome for Homologs of Neochrome, Phototropin, and Phytochrome.
To generate a draft genome for Anthoceros punctatus, genomic DNA was sheared into ∼400-bp fragments and sequenced using Illumina HiSeq2000, giving a total of 25 million 90-bp paired-end reads (about 20× genome coverage). The reads were subjected to two cycles of read error correction using the ALLPATHS-LG FindError program (46) before being assembled using Velvet (47). Assemblies were generated for a range of kmer values (k = 21, 31, 41, 51, and 61) and then were combined. The redundant scaffolds were removed using Usearch (48), and overlapping contigs were subject to additional assembly using CAP3 to produce a draft genome assembly. The final assembly contains 29,582 contigs with a total combined assembly length of 99.5 Mb and the N50 contig length of 4,955 bp. The contig length ranges from 919 bp to 76.5 kb, with 1,643 contigs over 10 kb. The median and mean assembled contig coverage is 18.1× and 44×, respectively. The raw reads were deposited in NCBI under SRA096687.
This assembly was searched for homologs of neochrome, phototropin, and phytochrome using tBLASTn. Although phototropin and phytochrome genes were readily identified, no contig was found containing a putative neochrome sequence. To search for any A. punctatus neochrome gene that perhaps failed to be assembled, all sequencing reads were searched against a library of neochrome protein sequences using BLASTx. Reads obtaining an e-value of ≤10−10 were isolated and assembled using Velvet with liberal assembly parameters (-cov_cutoff 1 -min_pair_count 1 -edgeFractionCutoff 0.1 -scaffolding yes -min_contig_lgth 90) at five different values for kmer length (21, 31, 41, 51, and 61). The resulting assemblies were combined, redundant contigs were discarded using Usearch, and overlapping contigs were merged using CAP3. All sequencing reads then were mapped to these seed contigs using Bowtie2 (49) with the very-sensitive-local option. Paired-end reads where at least one read mapped to the seed contigs were selected. All the selected reads then were reassembled as above. This mapping and assembly process was repeated until no further reads could be identified and contigs could no longer be extended. The final assembly contained a single contig comprising a 797-bp fragment of neochrome. This fragment then was extended to include almost the entire ORF using a combination of PCR (see below) and additional read mapping and assembly.
Cloning of Neochrome, Phototropin, and Phytochrome.
To verify empirically the presence of the hornwort photoreceptor genes found in the transcriptomes and to obtain intron/exon information, we cloned the genes from genomic DNA from five hornwort species (SI Appendix, Table S3). In addition, neochrome sequences were obtained from 25 fern species by PCR and cloning (SI Appendix, Table S3). Genomic DNA was extracted using the Qiagen DNAeasy Plant Mini Kit (Qiagen). The gene fragments were amplified using Phusion DNA polymerase (New England Biolabs) or Denville Choice Taq (Denville). The primers and detailed PCR conditions are summarized in SI Appendix, Tables S3 and S4. The amplified products were cloned into Promega pGEM-T (Promega) and sequenced.
Genome-Walking In Hornwort Phototropin and Neochrome.
To rule out the possibility that the phototropin gene found in hornworts might be a partial neochrome, we used inverse PCR (50) to obtain the flanking genomic region. Genomic DNA of N. aenigmaticus was digested by apoI (New England Biolabs) and self-ligated using T4 DNA ligase (New England Biolabs). Then nested PCRs were conducted on the circularized DNA. The amplicons were cloned using Promega pGEM-T and sequenced. To search for the genes flanking neochrome in N. aenigmaticus, we used the Clontech GenomeWalker kit (Clontech) and followed the manufacturer’s manual. The resulting PCR amplicons were cloned and sequenced. In total, we obtained 3,291-bp and 4,578-bp regions up- and down-stream, respectively, of neochrome. The primers for the above PCR reactions are listed in SI Appendix, Table S4.
Sequence Alignment for Neochrome, Phototropin, and Phytochrome.
We built two large alignments for phototropin and phytochrome, with each alignment including the corresponding domains from hornwort and fern neochrome. The phototropin dataset contains 163 sequences from 106 species, and the phytochrome dataset includes 139 sequences from 76 species. To reduce ambiguities in sequence alignment, we included only the conserved domains (i.e., LOV1, LOV2, and STK for phototropins; PAS, GAF, PHY, PAS repeats, HisKA, and HATPase for phytochromes). The domain boundaries were identified by querying each scaffold against the NCBI Conserved Domain Database (51). Each domain was aligned separately (based on the amino acid sequences) using Muscle (52) and then was concatenated. We developed a Python script, DomainDivider (see http://dx.doi.org/10.5061/dryad.fn2rg), to automate these processes. We also generated a separate alignment for hornwort and fern neochromes. This alignment was based on entire neochrome sequences rather than on domains. All alignments were inspected manually, and ambiguously aligned regions were excluded before phylogenetic analyses. The phototropin, phytochrome, and neochrome alignments contain 1,716, 2,802, and 4,002 bp, respectively. The GenBank accession numbers are listed in SI Appendix, Figs. S1–S3.
Phylogenetic Analyses of Phototropin and Neochrome.
Phototropin and neochrome phylogenies were inferred based on their nucleotide alignments. We used PartitionFinder (53) to identify the optimal data partition schemes and nucleotide substitution models under the Akaike Information Criterion. Based on this analysis, each codon position was treated as a distinct partition. For phototropin, the first, second, and third positions were assigned GTR+Γ+I substitution models; for neochrome, GTR+Γ+I, GTR+Γ+I, GTR+I models were applied to each codon position respectively. We used Garli (54) to obtain the maximum likelihood tree under the aforementioned models, with genthreshfortopoterm set to 1,000,000 and eight independent runs. Multiparametric bootstrapping was done using RAxML (55) with 1,000 replicates. For the neochrome alignment, we also carried out the same maximum likelihood analyses on the first + second-codon positions and on the third-codon positions separately. We used MrBayes (56) to conduct Bayesian tree inference under the same models, with two independent Markov chain Monte Carlo (MCMC) runs, four chains each, and trees sampled every 1,000 generations. Substitution parameters were unlinked, and the rate prior was set to vary among partitions. The MrBayes output was inspected using Tracer (57) to ensure proper convergence and mixing (effective sample sizes all >200), and 25% of the total generations were discarded as burn-in before making the 50% majority consensus tree. Because the stationary, homogeneous assumptions of GTR might be violated in cases associated with HGT and deep divergence (58), we also used a nonstationary, heterogeneous nucleotide substitution model implemented in nhPhyML (59) to infer the phototropin tree. The analysis was carried out with 10 discrete categories of GC equilibrium frequencies, and the required starting tree was the best tree from the Garli analysis. To conduct bootstrapping in nhPhyML, we created a Python wrapper, and for each replicate, RAxML was used to input the starting tree. In addition to the nucleotide substitution model, we also used codon models to infer phylogenies, which were carried out in CodonPhyML (19) under a maximum likelihood framework. We used the Goldman-Yang (60) model with four categories of nonsynonymous/synonymous substitution rate ratios drawn from the discrete gamma distribution, and codon frequencies were estimated from the data under the F3 × 4 model (19). The tree topology search was done using the nearest neighbor interchange approach, and branch support was estimated using the SH-like aLRT (61, 62) method.
Phylogenetic Analyses of Phytochrome.
For the phytochrome phylogeny, we used the protein alignment following the analytical strategy of Mathews et al. (63). Using ProtTest (64), JTT + F was found to be the best empirical substitution model under the Akaike Information Criterion. For the maximum likelihood analyses, we used Garli to search for the maximum likelihood tree, with genthreshfortopoterm set to 1,000,000 and eight independent runs, and RAxML to conduct the multiparametric bootstrapping with 1,000 replicates. For Bayesian tree inference, we used MrBayes with two independent MCMC runs, four chains each, and trees sampled every 1,000 generations. After 25% of the total generations were removed, the 50% majority consensus tree was calculated. Codon-based tree inference also was carried out as described above.
Topology Test.
We used the Swofford–Olsen–Waddell–Hillis test (65) to compare the inferred HGT tree topology (i.e., fern neochromes embedded within hornworts) against the alternative topologies suggestive of vertical inheritance or independent origin, using the program sowhat (66) with RAxML and Seq-Gen (67). For testing the vertical inheritance topology, topological constraints forcing fern and hornwort neochromes to be reciprocally monophyletic were used; for independent origin, constraints were placed to have all fern genes to be monophyletic (i.e., monophyly either as neochrome + phototropin or neochrome + phytochrome). To calculate the posterior probability of the vertical transfer and independent origin topologies, we filtered the posterior tree samples from MrBayes and calculated the frequency of trees given the monophyly constraints. The filtering was done by PAUP* (68). We applied this same approach to examine the posterior distribution of fern neochrome gene trees. We searched for topologies that exhibited better congruence with the published species relationships (compared with the inferred gene tree). The constraint for tree filtering required that neochromes from the same fern family be monophyletic.
Phylogenetic Analysis of the IGPD Gene.
As a result of genome-walking in N. aenigmaticus, we discovered an IGPD pseudogene downstream from neochrome. To place this pseudogene in phylogenetic context, we resolved an IGPD phylogeny for land plants. A subset of the transcriptomes and whole-genome sequences was mined for IGPD homologs (SI Appendix, Fig. S4) using BlueDevil, and an alignment of IGPD (624 bp in length) was constructed manually. We partitioned the data by codon position, with each partition given a GTR+Γ+I model as suggested by PartitionFinder under the Akaike Information Criterion. Maximum likelihood analyses were carried out in RAxML with 100 random starting trees, and multiparametric bootstrapping was done with 1,000 replicates.
Divergence Time Estimation of the Phototropin Gene Family.
We used BEAST (69) to infer simultaneously the divergence times and phylogeny of the phototropin gene family. As recommended by PartitionFinder, the phototropin dataset was partitioned by codon position, each with the GTR+Γ+I substitution model. A total of 15 calibration priors were used (see SI Appendix, Table S2 for details) (8, 13, 70–77), and a birth–death speciation prior was used as the tree prior. We used the uncorrelated relaxed-clock model with rates drawn from a lognormal distribution. A starting tree was first estimated by r8s (78) and was provided to BEAST to initiate the run. Two independent MCMC runs were carried out, and the output was inspected in Tracer to ensure convergence and mixing (effective sample sizes all >200). The trees from the two runs were combined in LogCombiner (69) with a 25% burn-in and were summarized in TreeAnnotator (69). It should be noted that the stationary, homogeneous GTR model used here could be violated, especially in the case of HGT, and might affect the divergence estimates. However, there is no nonstationary, heterogeneous model that is currently implemented in divergence time analyses, and our results should be revisited in the future when more sophisticated methods are available.
Inferring Episodic Selection and GC Content Variation in Neochrome Evolution.
To investigate whether fern neochromes had experienced pervasive episodic positive selection, we used the unrestricted, random effects branch-site model (20) implemented in the HyPhy package (20, 79). Branches with episodic positive selection were identified by the sequential likelihood ratio test (20). The neochrome alignment and the best maximum likelihood tree were used as the input data. The analyses were carried out on the Datamonkey server (79, 80). A GC content sliding window was constructed using a custom Python script; each window is 400 bp in size, and the window slides every 50 bp.
Supplementary Material
Acknowledgments
We thank Y.-L. Qiu, C.-W. Li, J. Nelson, B. Shaw, J. Duff, D. Long, and L. Forrest for helping us obtain hornwort materials; L. Huiet for laboratory assistance; P. G. Wolf, C. W. dePamphilis, and P. E. Ralph for help in collecting the Pteridium aquilinum transcriptome data; J. Leebens-Mack, S. Joya, S. Ellis, and others for contributing plant tissues for 1KP transcriptome sequencing; D. L. Swofford and S. Wu for help with the phylogenetic analyses; and J. D. Palmer, M. Chen, J. Meireles, two anonymous reviewers, members of the K.M.P. laboratory, and members of the Duke Systematics Discussion Group for insightful comments on a draft manuscript. This work was supported by research grants from the Society of Systematic Biologists, Sigma Xi, American Society of Plant Taxonomists, and Duke University (to F.-W.L.), National Science Foundation Doctoral Dissertation Improvement Grant DEB-1407158 (to F.-W.L. and K.M.P.), DEB-1145614 (to K.M.P.), and a European Research Council Advanced Investigator Award (to J.A.L.). The 1000 Plants (1KP) initiative, led by G.K.-S.W., is funded by the Alberta Ministry of Enterprise and Advanced Education, Alberta Innovates Technology Futures, Innovates Centre of Research Excellence, Musea Ventures, and BGI-Shenzhen. This article is part of a doctoral dissertation in Biology at Duke University by F.-W.L., supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequences reported here are deposited in the GenBank database (accession nos. KJ128382, KJ128383, KJ128384, and KJ194997–KJ195254). The 1KP transcriptomes can be accessed at www.bioinfodata.org/Blast4OneKP. The sequencing reads for Anthoceros punctatus draft genome and Pteridium aquilinum transcriptome are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under SRA096687 and SRX423244, respectively. The assembled P. aquilinum transcriptome is deposited in NCBI Transcriptome Shotgun Assembly under GASP00000000. Alignments and tree files are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.fn2rg.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319929111/-/DCSupplemental.
References
- 1.Möglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 2.Nozue K, et al. A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc Natl Acad Sci USA. 1998;95(26):15826–15830. doi: 10.1073/pnas.95.26.15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai H, et al. Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature. 2003;421(6920):287–290. doi: 10.1038/nature01310. [DOI] [PubMed] [Google Scholar]
- 4.Kanegae T, Hayashida E, Kuramoto C, Wada M. A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proc Natl Acad Sci USA. 2006;103(47):17997–18001. doi: 10.1073/pnas.0603569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc Natl Acad Sci USA. 2005;102(38):13705–13709. doi: 10.1073/pnas.0504734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Qi X, Sen L, Su Y, Wang T. Cloning and sequence analysis of red/blue light chimeric photoreceptor genes from three fern species (Coniogramme intermedia var. glabra, Plagiogyria distinctissima and Pronephrium lakhimpurnense) Am Fern J. 2010;100(1):1–15. [Google Scholar]
- 7.Schneider H, et al. Ferns diversified in the shadow of angiosperms. Nature. 2004;428(6982):553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- 8.Schuettpelz E, Pryer KM. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci USA. 2009;106(27):11200–11205. doi: 10.1073/pnas.0811136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gontcharov AA, Melkonian M. Molecular phylogeny and revision of the genus Netrium (Zygnematophyceae, Streptophyta): Nucleotaenium gen. nov. J Phycol. 2010;46(2):346–362. [Google Scholar]
- 10.Qiu Y-L, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103(42):15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox CJ, Li B, Foster PG, Embley TM, Civán P. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst Biol. 2014;63(2):272–279. doi: 10.1093/sysbio/syt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges SB, Kumar S. The Timetree of Life. Oxford, UK: Oxford Univ Press; 2009. [Google Scholar]
- 13.Villarreal JC, Renner SS. Hornwort pyrenoids, carbon-concentrating structures, evolved and were lost at least five times during the last 100 million years. Proc Natl Acad Sci USA. 2012;109(46):18873–18878. doi: 10.1073/pnas.1213498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuettpelz E, Pryer KM. Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon. 2007;56(4):1037–1050. [Google Scholar]
- 15.Rothfels CJ, et al. Transcriptome-mining for single-copy nuclear markers in ferns. PLoS ONE. 2013;8(10):e76957. doi: 10.1371/journal.pone.0076957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanderson MJ, Shaffer HB. Troubleshooting molecular phylogenetic analyses. Annu Rev Ecol Syst. 2002;33:49–72. [Google Scholar]
- 17.Kapralov MV, Filatov DA. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol Biol. 2007;7:73. doi: 10.1186/1471-2148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nabholz B, Künstner A, Wang R, Jarvis ED, Ellegren H. Dynamic evolution of base composition: Causes and consequences in avian phylogenomics. Mol Biol Evol. 2011;28(8):2197–2210. doi: 10.1093/molbev/msr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil M, Zanetti MS, Zoller S, Anisimova M. CodonPhyML: Fast maximum likelihood phylogeny estimation under codon substitution models. Mol Biol Evol. 2013;30(6):1270–1280. doi: 10.1093/molbev/mst034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosakovsky Pond SL, et al. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 2011;28(11):3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lind PA, Tobin C, Berg OG, Kurland CG, Andersson DI. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol Microbiol. 2010;75(5):1078–1089. doi: 10.1111/j.1365-2958.2009.07030.x. [DOI] [PubMed] [Google Scholar]
- 22.Näsvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338(6105):384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA. 1998;95(24):14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol. 2008;25(8):1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Puerta MV, et al. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended co-conversion of flanking exons. BMC Evol Biol. 2011;11:277. doi: 10.1186/1471-2148-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaedicke K, Lichtenthäler AL, Meyberg R, Zeidler M, Hughes J. A phytochrome-phototropin light signaling complex at the plasma membrane. Proc Natl Acad Sci USA. 2012;109(30):12231–12236. doi: 10.1073/pnas.1120203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burr FA. 1968. Chloroplast Structure and Division in Megaceros Species. PhD Dissertation (Univ of California, Berkeley)
- 28.Huang J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays. 2013;35(10):868–875. doi: 10.1002/bies.201300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 30.Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101(51):17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science. 2004;305(5684):676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- 32.Davis CC, Anderson WR, Wurdack KJ. Gene transfer from a parasitic flowering plant to a fern. Proc Biol Sci. 2005;272(1578):2237–2242. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida S, Maruyama S, Nozaki H, Shirasu K. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science. 2010;328(5982):1128. doi: 10.1126/science.1187145. [DOI] [PubMed] [Google Scholar]
- 34.Renner SS, Bellot S. 2012. in Genomics of Chloroplasts and Mitochondria (Springer Netherlands, Dordrecht), Vol 35, Advances in Photosynthesis and Respiration, pp 223–235.
- 35.Xi Z, et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9(2):e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 37.Xi Z, et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol Biol. 2013;13:48. doi: 10.1186/1471-2148-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christin P-A, et al. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Curr Biol. 2012;22(5):445–449. doi: 10.1016/j.cub.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 40.Johnson MTJ, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE. 2012;7(11):e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342(6165):1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 46.Maccallum I, et al. ALLPATHS 2: Small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10(10):R103. doi: 10.1186/gb-2009-10-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 54.Zwickl DJ. 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. PhD dissertation (Univ of Texas at Austin, Austin)
- 55.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 56.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambaut A, Drummond AJ. 2009. Tracer v1.5. Available at http://tree.bio.ed.ac.uk/software/tracer/. Accessed June 2013.
- 58.Verbyla KL, Yap VB, Pahwa A, Shao Y, Huttley GA. The embedding problem for markov models of nucleotide substitution. PLoS ONE. 2013;8(7):e69187. doi: 10.1371/journal.pone.0069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boussau B, Gouy M. Efficient likelihood computations with nonreversible models of evolution. Syst Biol. 2006;55(5):756–768. doi: 10.1080/10635150600975218. [DOI] [PubMed] [Google Scholar]
- 60.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11(5):725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 61.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 62.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 63.Mathews S, Clements MD, Beilstein MA. A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants. Philos Trans R Soc Lond B Biol Sci. 2010;365(1539):383–395. doi: 10.1098/rstb.2009.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 65.Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49(4):652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 66.Church SH, Ryan JF, Dunn CW. 2014. Sowhat. Available from GitHub repository https://github.com/josephryan/sowhat. Accessed January 2014.
- 67.Rambaut A, Grassly NC. Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13(3):235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 68.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0a131. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 69.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clarke JT, Warnock RCM, Donoghue PCJ. Establishing a time-scale for plant evolution. New Phytol. 2011;192(1):266–301. doi: 10.1111/j.1469-8137.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- 71.Hubers M, Kerp H. Oldest known mosses discovered in Mississippian (late Visean) strata of Germany. Geol. 2012;40(8):755–758. [Google Scholar]
- 72.Guo C-Q, et al. Riccardiothallus devonicus gen. et sp. nov., the earliest simple thalloid liverwort from the Lower Devonian of Yunnan, China. Rev Palaeobot Palynol. 2012;176-177(C):35–40. [Google Scholar]
- 73.Prasad V, Strömberg CAE, Alimohammadian H, Sahni A. Dinosaur coprolites and the early evolution of grasses and grazers. Science. 2005;310(5751):1177–1180. doi: 10.1126/science.1118806. [DOI] [PubMed] [Google Scholar]
- 74.Kotyk ME, Basinger JF, Gensel PG, de Freitas TA. Morphologically complex plant macrofossils from the Late Silurian of Arctic Canada. Am J Bot. 2002;89(6):1004–1013. doi: 10.3732/ajb.89.6.1004. [DOI] [PubMed] [Google Scholar]
- 75.Skog JE, Banks HP. Ibyka amphikoma, gen. et sp. n., a new protoarticulate precursor from the late Middle Devonian of New York State. Am J Bot. 1973;60(4):366–380. [Google Scholar]
- 76.Galtier J, Wang S-J, Li C-S, Hilton J. A new genus of filicalean fern from the Lower Permian of China. Bot J Linn Soc. 2001;137(4):429–442. [Google Scholar]
- 77.Trivett ML. Growth architecture, structure, and relationships of Cordaixylon iowensis nov. comb. (Cordaitales) Int J Plant Sci. 1992;153(2):273–287. [Google Scholar]
- 78.Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19(2):301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 79.Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 80.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26(19):2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.