Significance
Ion-translocating, light-activated membrane proteins known as rhodopsins are found in all three domains of life. Proton-pumping rhodopsins, such as proteorhodopsin, are known to be broadly distributed in marine bacteria. The first known sodium-pumping rhodopsin was recently described in marine flavobacterium. We report the discovery and characterization of a unique type of light-activated ion-translocating rhodopsin that translocates chloride ions into the cell and is evolutionarily distinct from the other known rhodopsin chloride pump, halorhodopsin, found in haloarchaea. Our data show that rhodopsins with different ion specificities have evolved independently in marine bacteria, with individual strains containing as many as three functionally different rhodopsins.
Keywords: evolution, ecology, photoproteins, photoheterotroph, retinal
Abstract
Light-activated, ion-pumping rhodopsins are broadly distributed among many different bacteria and archaea inhabiting the photic zone of aquatic environments. Bacterial proton- or sodium-translocating rhodopsins can convert light energy into a chemiosmotic force that can be converted into cellular biochemical energy, and thus represent a widespread alternative form of photoheterotrophy. Here we report that the genome of the marine flavobacterium Nonlabens marinus S1-08T encodes three different types of rhodopsins: Nonlabens marinus rhodopsin 1 (NM-R1), Nonlabens marinus rhodopsin 2 (NM-R2), and Nonlabens marinus rhodopsin 3 (NM-R3). Our functional analysis demonstrated that NM-R1 and NM-R2 are light-driven outward-translocating H+ and Na+ pumps, respectively. Functional analyses further revealed that the light-activated NM-R3 rhodopsin pumps Cl− ions into the cell, representing the first chloride-pumping rhodopsin uncovered in a marine bacterium. Phylogenetic analysis revealed that NM-R3 belongs to a distinct phylogenetic lineage quite distant from archaeal inward Cl−-pumping rhodopsins like halorhodopsin, suggesting that different types of chloride-pumping rhodopsins have evolved independently within marine bacterial lineages. Taken together, our data suggest that similar to haloarchaea, a considerable variety of rhodopsin types with different ion specificities have evolved in marine bacteria, with individual marine strains containing as many as three functionally different rhodopsins.
Rhodopsins are a class of membrane proteins spanning seven transmembrane domains that contain retinal as the light-absorbing chromophore (1). All bacterial and archaeal rhodopsins are classified as type I rhodopsins that have various functions, including light-driven outward translocating H+ pumps [bacteriorhodopsin (BR), proteorhodopsin (PR), and xanthorhodopsin-like rhodopsin (XLR)], light-driven inward translocating Cl− pumps [halorhodopsin (HR)], and light-activated signal transducers (sensory rhodopsin I and II) (2–7). Microbial rhodopsins such as BR and HR were originally discovered in extremely halophilic archaea in the 1970s (2, 3). For some time, noneukaryotic rhodopsins were thought to be restricted to hypersaline archaea, until a new type of proton-pumping rhodopsin, proteorhodopsin (PR), was discovered in marine bacteria (6). Subsequent culture-independent surveys indicated that rhodopsin genes are taxonomically and geographically broadly distributed among marine bacteria and archaea and occur in a high proportion of surface-dwelling oceanic microbial species (8). Furthermore, recent whole genome analyses coupled with biochemical and physiological analyses have revealed that some marine bacteria also have a newly discovered rhodopsin functionality, an outward Na+-pumping rhodopsin (NaR) (9). These studies indicate that light utilization by microbial rhodopsins is a favorable survival strategy that has been broadly distributed via both vertical and horizontal transmission events, and has resulted in considerable diversification of rhodopsin's functional properties and taxon distributions.
Rhodopsins have a number of potential physiological roles related to the physiology, growth strategies, and energy budgets of diverse microbial taxa (10). Outward cation- or inward anion-pumping rhodopsins can convert light energy into a proton motive force that directly provides biochemical energy for cells via proton translocating ATPases (3, 10, 11). In addition to ATP production, the transmembrane ion gradient created by H+ or Na+ pumping (BR, PR, and NaR) can be used to drive flagellar rotation (12) or active ion transport via secondary transport systems. The inward Cl− translocation by HR also may be used for the maintenance of osmotic balance in saturated brines (13). Previous studies have shown that H+-pumping rhodopsins, such as BR and PR, have proton acceptor and donor amino acids that protonate the retinal Schiff base. These H+ acceptors and donors are carboxylic amino acid residues that include residues D85 and D96 in BR and D85 and E96 in PR (BR numbering), respectively (14–16). In contrast, these carboxylic residues are replaced by neutral amino acids in HR and NaR (T85 and A96 in HR; N85 and Q96 in NaR). Inoue et al. (9) suggested that NaR, carboxylic residue D89 (BR numbering) has a significant role in Na+ pumping, because a D89N mutant of NaR lost Na+-pumping activity. For BR, PR, and HR, neutral amino acid residues are found at the same positions (T89 in BR, T89 in PR, and S89 in HR). These data suggest that the amino acids located in 85, 89, and 96 (BR numbering) play critical roles in determining the specificity of ion-pumping activity.
We report here that the genome sequence of an orange-pigmented marine flavobacterium, Nonlabens marinus strain S1-08T, encodes three different rhodopsins (NM-R1, NM-R2, and NM-R3). Phylogenetic analyses revealed that NM-R1 and NM-R2 are affiliated with the PR clade (D85, T89, and E96; DTE motif) and the NaR clade (NDQ motif), respectively. The sequence of NM-R3 was associated with a unique cluster consisting of rhodopsins containing an NTQ motif, different from that of functionally characterized rhodopsins. Subsequent in vitro functional analyses of the heterologously expressed rhodopsins revealed different ion specificities for each. In addition, culture experiments using low carbon-containing seawater media were conducted to determine the effect of light on growth of S1-08T.
Results
N. marinus Strain S1-08T Has Three Different Rhodopsins.
Genome sequence analysis revealed that N. marinus S1-08T has a single 2.9-Mbp chromosome (one scaffold consisting of two contigs), encoding 2,790 genes (SI Appendix, Fig. S1). Three predicted rhodopsin genes (N. marinus rhodopsins: NM-R1, NM-R2, and NM-R3) are encoded in different regions in the S1-08T genome. With respect to the ion transport-relevant amino acid sites, rhodopsin NM-R1 contains residues D85, T89, and E96 (BR numbering, DTE motif), a typical feature of proton-pumping rhodopsins. In the same protein region, NM-R2 and NM-R3 contain residues N85, D89, and Q96 (NDQ motif) and N85, T89, and Q96 (NTQ motif), respectively. Residue K216, the site of Schiff base linkage to the retinal chromophore, is conserved in all three rhodopsins. The genome of N. marinus S1-08T contains all of the genes required for retinal biosynthesis, including crtE (geranylgeranyl pyrophosphate synthase), crtI (phytoenedehydrogenase), crtB (phytoene synthase), and crtY (lycopene cyclase). The blh gene encoding 15,15′-β-carotene dioxygenase (17) is located adjacent to the NM-R1 opsin gene.
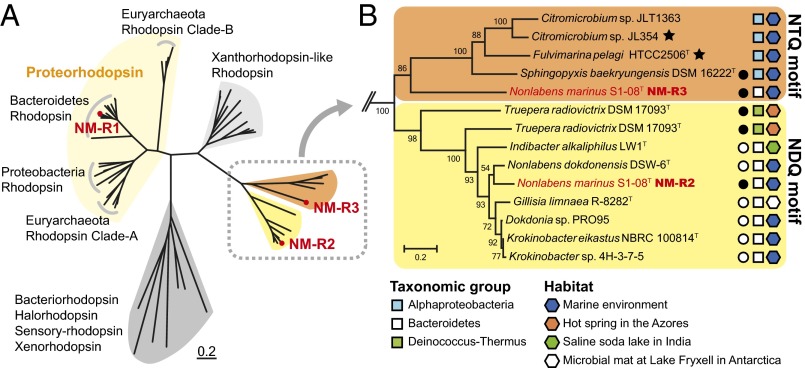
The phylogenetic relationships among the three N. marinus S1-08T rhodopsins suggest that NM-R1 is affiliated with the PR clade (DTE motif) and NM-R2 is affiliated with the NaR clade (NDQ motif). Thus, the NM-R1 and NM-R2 opsins might function similarly as their close homologs, proton and sodium ion pumps, respectively. However, because NM-R3 is affiliated with a functionally uncharacterized clade of NTQ motif rhodopsins (Fig. 1A), it was not possible to infer its ion specificity on the basis of sequence motifs and phylogenetic position alone.
Fig. 1.
Phylogenetic relationships among microbial rhodopsins. (A) Unrooted phylogenetic tree of microbial rhodopsin amino acid sequences. The three rhodopsin genes (NM-R1, NM-R2, and NM-R3) encoded in the S1-08T genome are indicated by red circles. Orange and yellow indicate the NTQ motif and the NDQ motif rhodopsin clade, respectively. (B) Detailed phylogenetic relationships of NTQ (orange) and NDQ (yellow) motif rhodopsin clades. NM-R2 and NM-R3 are indicated by the red characters. Stars indicate AAP bacteria. Closed and open circles indicate strains containing three opsins and two opsins, respectively. Taxonomic groups and the habitat of origin for each are indicated by the different symbols.
Comparative analyses provided further insight into the taxonomic distributions and phylogenetic relationships of other bacterial rhodopsins containing the NTQ or NDQ motifs (Fig. 1B). With the exception of S1-08T, all other sequenced genomes containing rhodopsin sequences encoding the NTQ motif are classified as alphaproteobacteria, including two aerobic anoxygenic phototrophic (AAP) bacteria (18, 19). In contrast, NDQ motif rhodopsin genes were found in the genomes of five strains in Bacteroidetes and one strain of the phylum Deinococcus-Thermus. NDQ motif rhodopsin encoding has been found in bacteria inhabiting ecologically diverse environments, including Truepera radiovictrix DSM 17093T, isolated from a hot spring (20); Indibacter alkaliphilus LW1T, isolated from a saline soda lake in India; and Gillisia limnaea R-8282T, isolated from a microbial mat at Lake Fryxell, Antarctica (21, 22). In contrast, the NTQ-carrying strains whose genomes have been sequenced to date are found only in marine environments (23, 24).
The genome size of N. marinus S1-08T is the smallest among the known NDQ motif-containing rhodopsins, and approximately 1 Mbp smaller than that of its closest relative, Nonlabens dokdonensis DSW-6T (16S rRNA sequence similarity, 95.02%) (SI Appendix, Table S1). Although most NDQ motif (NaR) opsin-containing bacterial strains also contained opsins with the DTE motif (PR), the NTQ motif rhodopsin was found only in the strain S1-08T genome.
To further examine the evolutionary history of the various rhodopsin genes, we compared genomic flanking regions around each type (SI Appendix, Fig. S2) among the available genome sequences. Interestingly, nearly all strains containing rhodopsins with either NTQ or NDQ motifs (except for JLT1363, JL354, and HTCC2506T) encoded more than one rhodopsin gene in their genomes; for example, all of the NaR-containing flavobacterial strains also contained a PR gene with a blh gene adjacent to it. With respect to NTQ motif alphaproteobacterial rhodopsins, blh genes were found adjacent to NTQ motif rhodopsin in HTCC2506T and DSM 16222T, but no blh genes were detected in strains JL354 and JLT1363. A protein of unknown function domain 2254 (DUF2254) was found adjacent to NTQ motif rhodopsin in the genomes of S1-08T, JL354, and JLT1363. Taken together, these findings indicate that the gene sequences surrounding each rhodopsin (except for the blh gene) generally are not highly conserved. Given the available data, whether S1-08T acquired the NTQ motif rhodopsin gene by horizontal acquisition remains unclear.
To further examine rhodopsin versus retinal biosynthesis pathway evolutionary relationships, we compared blh and crtY gene phylogenies among rhodopsin-containing strains (SI Appendix, Figs. S3 and S4). These phylogenetic trees suggest that NTQ motif rhodopsin- or xenorhodopsin (XeR)-containing proteobacteria harbor blh and crtY genes that form a distinct clade compared with other PR-harboring proteobacteria. In contrast, blh and crtY genes form a coherent lineage among Bacteroidetes. Similar trends were observed in phylogenies based on crtI and crtB genes previously reported by Klassen (25). These data suggest that retinal biosynthesis pathways in NTQ motif rhodopsin or XeR-containing versus solely PR-containing proteobacteria have different evolutionary pathways that are distinct from the monophyletic carotenoid/retinal biosynthetic pathway in the Bacteroidetes.
Functional Analyses of NM-R1, NM-R2, and NM-R3.
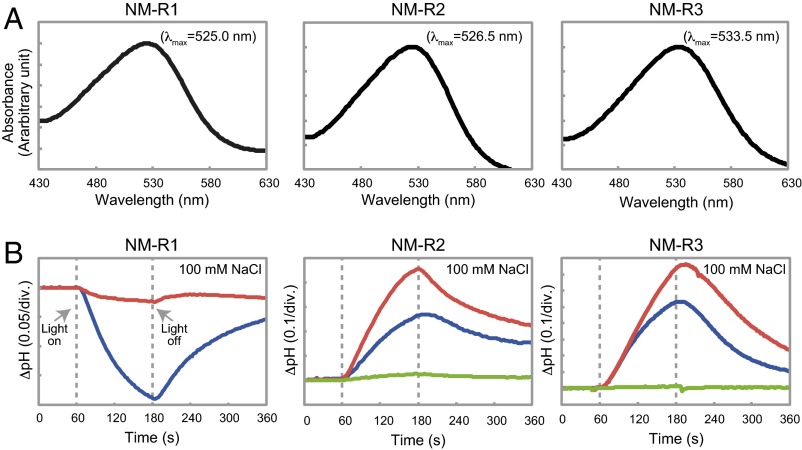
For characterization of the functional properties of NM-R1, NM-R2, and NM-R3, each opsin gene sequence was chemically synthesized and overexpressed in Escherichia coli. The three N. marinus rhodopsins have similar absorption spectra (Fig. 2A), similar to those of other known green-light absorbing rhodopsins (6, 9). Light-induced pH changes in E. coli cell suspensions expressing NM-R1, NM-R2, or NM-R3 were determined (Fig. 2B). In NM-R1, a light-induced pH drop was observed, and in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), this pH change was abolished. These data indicate that NM-R1 is a light-driven H+ pump, consistent with the phylogenetic position of this rhodopsin in the PR clade. In contrast, light-induced pH increases were seen in NM-R2– and NM-R3–expressing cell suspensions, which was accelerated with CCCP but abolished with CCCP and tetraphenylphosphonium bromide (TPP+) (Fig. 2B). These findings demonstrate that passive inward H+ translocation occurs in response to transmembrane electrical potential, and suggest that NM-R2 and NM-R3 pump ions other than H+ (9, 26).
Fig. 2.
Absorption spectra and light-induced pH changes of NM-R1, NM-R2, and NM-R3 proteins. (A) Absorption spectra of purified NM-R1, NM-R2, and NM-R3 proteins. (B) Light-induced pH changes of E. coli cell suspensions expressing NM-R1, NM-R2, or NM-R3 in 100 mM NaCl (blue line). The pH changes after addition of CCCP (red line) or CCCP + TPP+ (green line) are indicated as well.
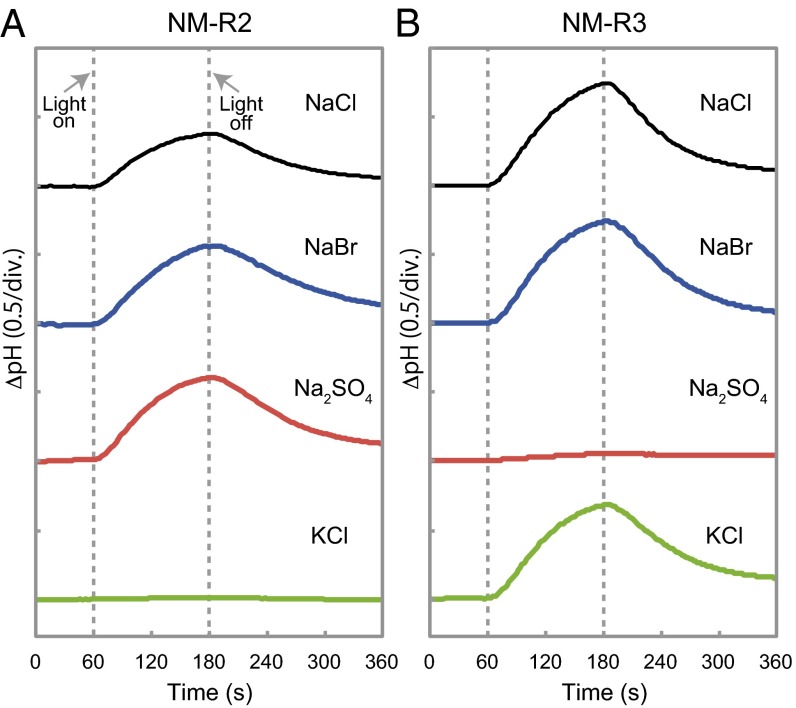
To determine the ion specificities of NM-R2 and NM-R3, we performed similar measurements in various salt solutions, as described previously (9). In the case of NM-R2, the same light-induced pH increases as observed in NaCl solutions were also observed in NaBr and Na2SO4 solutions, but not in KCl solutions (Fig. 3A). This result indicates that NM-R2 is a light-driven outward Na+ pump, like NaR in Krokinobacter eikastus NBRC 100814T (9). Although the NaR of NBRC 100814T was shown to pump H+ outward in the absence of Na+ ions, the NM-R2 NaR was not able to pump H+ in a KCl solution. The third rhodopsin of N. marinus, NM-R3, showed similar light-induced pH increases in NaCl and NaBr as for NM-R2. These pH changes were also observed in KCl and NaI solutions, with a slight pH increase in an NaNO3 solution as well; however, no light-induced pH changes were observed in Na2SO4 solutions (Fig. 3B and SI Appendix, Fig. S5). These halide-dependent pumping behaviors are very similar to those of HR in haloarchaea, and suggest that NM-R3 is a light-driven inward Cl− pump.
Fig. 3.
Light-induced pH changes of E. coli cell suspensions expressing NM-R2 or NM-R3 in different salt solutions (100 mM). (A) Effect of light on NM-R2 in different salt solutions. (B) Effect of light on NM-R3 in different salt solutions.
We also measured light-induced pH changes in different concentrations of NaCl (SI Appendix, Fig. S6). In NM-R2, similar light-induced pH increases were observed in both 100 mM and 200 mM NaCl. In contrast, NM-R3 showed elevated pH increases in the presence of 200 mM NaCl compared with 100 mM NaCl. Previous studies of HR and BR in haloarchaea (26) have shown that varying extracellular salt concentration affect the activity of inward, but not outward, ion-pumping rhodopsins, suggesting that NM-R3 is an inward Cl− pump and NM-R2 is an outward Na+ pump. Taken together, these findings support the conclusion that NM-R2 is an outward Na+ pump and that NM-R3 is an inward Cl− pump (dubbed ClR).
Light-Enhanced Growth in N. marinus S1-08T at Low Nutrient Concentrations.
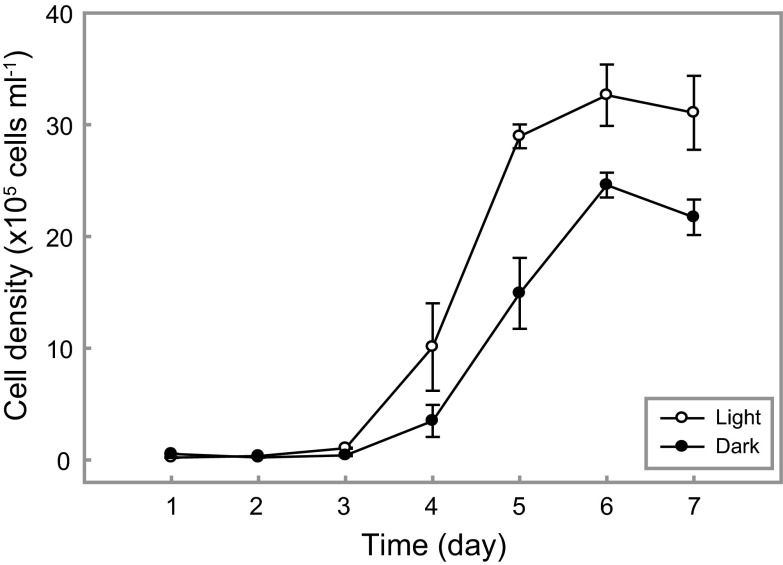
To evaluate the effect of light on the growth of S1-08T, we examined cultures in light and dark conditions when grown in a basal seawater media amended with carbon, nitrogen, and phosphorus (0.14 mM carbon). Differences in cell densities under light or dark conditions was observed after 4 d of incubation (Fig. 4). So far, light-stimulated growth of PR-containing bacteria has been observed only in Dokdonia sp. MED134 in low-carbon artificial seawater (27) and Psychroflexus torquis ATCC 700755T in marine broth with added salt (28). Similar to previous studies with MED134 (27), in the present study, growth yields for N. marinus S1-08T were considerably enhanced at low carbon concentrations in light (Fig. 4). Other PR-containing strains, as well as the NaR-containing strain PRO95, have not been shown to exhibit growth stimulation in light versus dark conditions, but the reasons for this are unclear (23, 29, 30). The data reported here indicate that light stimulation of growth is not unique to MED134, and likely is more widespread among rhodopsin-containing flavobacteria.
Fig. 4.
Light-stimulated growth in N. marinus S1-08T. S1-08T was grown in sterile ASW containing 0.14 mM carbon at 22 °C. Error bars denote SD for triplicate cultures.
Discussion
Our genomic analysis of marine flavobacterial isolate S1-08T revealed that this strain has multiple rhodopsins, reminiscent of the many multiple rhodopsin-containing haloarchaea found in hypersaline habitats. Functional analyses of each N. marinus rhodopsin revealed a typical outward H+-pumping rhodopsin (PR), an outward Na+-pumping rhodopsin (NaR), and a new type of inward Cl− pumping rhodopsin (ClR). The N. marinus S1-08T genome encodes all three different ion-pumping rhodopsin types in one cell. The function of ClR appears to be similar to that of the haloarchaeal inward Cl−-pumping rhodopsin (HR) that has been considered uniquely adapted to hypersaline environments (13). Importantly, the crucial amino acid residues for Cl− ion pumping in HR (TSA motif) were not conserved in ClR, which instead contains an NTQ motif in this region (85, 89, and 96 in BR numbering; Table 1). Phylogenetic analysis revealed that the lineages of ClR and HR are quite distinct from one another. These results strongly suggest that these two different light-induced inward Cl−-pumping rhodopsins, one found in archaea (HR) and the other found in bacteria (ClR), have evolved independently.
Table 1.
Amino acid sequences in each type of rhodopsin from residues D85–D96 in BR (PR) numbering
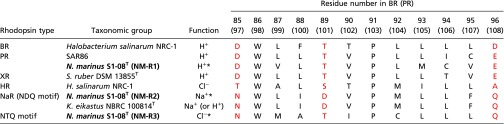 |
The positions corresponding to D85, T89, and D96 in BR are colored red. Bold characters indicate three rhodopsins of S1-08T.
Indicates the functions of rhodopsins investigated in this study.
We found ClR genes in the genomes of four other marine isolates as well (Fig. 1). Like N. marinus S1-08T, the alphaproteobacterium strain DSM 16222T encodes more than one rhodopsin, including one with an NTQ motif. Other AAP proteobacteria (HTCC2506T and JL354), as well as the proteobacterium JLT1363 (17), which has been reported to have lost most of its phototrophic genes (19), encode only one opsin, a putative ClR, in their genomes. The interplay between bacteriochlorophyll-based photosystems with different opsins of various ion specificities is not known at present, but available data suggest that at least CIRs may be functionally compatible with the bacteriochlorophyll-based photosystems of AAP bacteria.
Comparative analyses showed that the genomic regions flanking the ClR are not highly conserved. Furthermore, the NM-R3 (ClR) gene was not found in predicted horizontal gene transfer (HGT) island regions in the genome (SI Appendix, Fig. S1). Orthologous gene comparisons between S1-08T and other NaR-containing flavobacteria suggest that the upstream region of ClR gene has not been particularly well conserved (SI Appendix, Fig. S7, dashed line). Thus, the data presented here provide no clear evidence as to whether HGT acquisition or loss underlies the patterns of distribution and diversification of ClRs reported here.
Phylogenetic analyses of retinal biosynthesis pathway genes among different NaR and ClR flavobacterial strains suggest that they represent distinct lineages that differ from their homologs found in PR-containing alphaproteobacteria and gammaproteobacteria. The retinal biosynthetic pathway appears to be evolutionarily conserved within rhodopsin-bearing Bacteroidetes. Like other flavobacteria, the ClR- and NaR-containing flavobacterial strains have other carotenoid pigment-producing genes in addition to crtB, crtI, and crtY, unlike PR-containing proteobacteria, which contain a minimal gene set for producing retinal (25). For example, some ClR-containing strains (i.e., S1-08T, HTCC2506T, and DSM 16222T) also encode a β-carotene hydroxylase gene (crtZ) that converts β-carotene to zeaxanthin. In this context, the extremely halophilic bacterium Salinibacter ruber has recently been shown to use different carotenoid types as accessory pigments in the xanthorhodopsin photocycle (7). Thus, it seems possible that in the rhodopsin-containing Bacteroidetes, alternative carotenoid pigments could be involved in photocycle, function, and the evolution of different opsin types, such as NaR and ClR.
Environmental metagenomics and whole genome sequencing of bacterial isolates have shown that light-induced outward H+ pumping rhodopsins, such as BR, PR, and XLR, are abundant and widely distributed in surface-dwelling aquatic microbes (31–34). Given their ability to capture energy from sunlight, it is apparent that the light-driven H+-pumping activity of these rhodopsins is favorable for survival in the photic zone of aquatic environments. Light-induced inward Cl−-pumping rhodopsins like HR were once thought to be restricted to extreme halophiles. Here we have shown that light-induced Cl−-pumping rhodopsins appear also to have evolved independently within bacteria. Furthermore, in addition to a chloride pumping rhodopsin, some marine isolates also contain a proton and a sodium pumping rhodopsin as well.
Given that Cl− and Na+ are the two most predominant dissolved ionic components of seawater, it is not surprising that light-induced inward or outward translocation of these ions has evolved among marine bacteria. Notably, all three rhodopsins in N. marinus S1-08T have very similar absorption spectra, with a maximum around 525 nm. This finding implies that all three rhodopsins can function efficiently and simultaneously in the same light field, each contributing to the transmembrane potential using different ion gradients. Whether each rhodopsin is actually expressed simultaneously or, alternatively, serves a different physiological role (e.g., during different growth stages of the cell) remains to be determined.
Taken together, our findings suggest that marine bacterial chloride-pumping rhodopsins have evolved independently from those found in hypersaline habitats. Similar to the situation in haloarchaea, multiple different rhodopsin types, each with different ion specificities, can be found within a single strain of marine bacteria, particularly within the Bacteroidetes. Given that marine ClR and NaR, like BR and PR, can generate a transmembrane potential, the light-mediated energy generation by these rhodopsins represents a unique mode of energy acquisition for marine bacteria. Future laboratory and field studies of ClR- and NaR-containing marine bacteria are required to further illuminate the physiological and ecological significance of these functionally versatile bacterial rhodopsins.
Materials and Methods
Strain Information of N. marinus Strain S1-08T.
Strain S1-08T was isolated from a surface seawater sample collected from the northwestern Pacific Ocean (30°11′ N, 145°05′ E) by the R/V Mirai (Japan Agency for Marine-Earth Science and Technology) on February 9, 2010 (MR10-01 cruise). S1-08 is type strain of N. marinus. Details of culture conditions and phenotypic traits have been described previously (35).
Genome Sequencing and Annotation.
The genome sequence of S1-08T was determined using Roche 454 FLX titanium sequencer (8-kb mate-paired library; 44,326,991-bp sequences; 15-fold coverage) and Illumina MiSeq (Nextera paired-end library; 489,343,336-bp sequences; 168-fold coverage) sequencing platforms. Assembly was performed using Newbler version 2.8. Gaps between adjacent contigs were closed by sequencing PCR amplicons from genomic DNA. The final assembly comprised two contigs in one scaffold chromosome without plasmids. Annotation of coding DNA sequence (CDS) and calculation of the genomic guanine-cytosine (GC) content were performed with the RAST (Rapid Annotation using Subsystem Technology; http://rast.nmpdr.org/) server using default parameters (36). The genome sequence of N. marinus S1-08T was deposited in GenBank (accession no. AP014548).
Genome Analysis.
Circular genome maps were visualized using Circos (37). Genomic island and membrane proteins were predicted using IslandViewer (www.pathogenomics.sfu.ca/islandviewer/query.php) (38) and SOSUI engine version 1.10 (39), respectively. GC skew and GC content were calculated in each 100-bp window. For syntenic analysis, DomClust (40) was adopted to detect orthologous genes.
Phylogenetic Analyses.
The amino acid sequences were aligned using MUSCLE (41), and the evolutionary distances were calculated with the JTT matrix-based method (42). Phylogenetic trees were constructed using the neighbor-joining method, with bootstrap values based on 1,000 replications (43, 44). All analyses were performed using MEGA5 software (45).
Gene Preparations and Protein Expression of NM-R1, NM-R2, and NM-R3.
The codon-optimized genes with NdeI and XhoI restriction enzyme sites were chemically synthesized by Operon Biotechnology. These fragments were inserted into NdeI and XhoI sites of pET21a vectors (Novagene); consequently, the plasmids encoded hexahistidines at the C terminus. These vectors were transformed into E. coli C41(DE3) (Lucigen). All rhodopsins were overexpressed in cells grown at 37 °C in 200 mL of 2xYT medium supplemented with ampicillin (final concentration, 100 μg/mL) and induced at an OD600 of 0.4–0.6 with 0.2 mM isopropyl β-d-1-thiogalactopyranoside and 10 μM all-trans-retinal.
Functional Analyses of NM-R1, NM-R2, and NM-R3.
The rhodopsin-expressing cells were collected by centrifugation (3000 × g for 5 min), washed three times, and then resuspended in the solvent for measurement. Then 4 mL of cell suspension was placed in darkness and illuminated using a tungsten halogen lump (LS2; Hansatech) through a glass filter (46-217; Edmund Optics) for 2 min. Measurements were repeated under the same condition after the addition of CCCP (final concentration, 30 μM) and further addition of TPP+ (final concentration, 20 mM). The light-induced pH changes were measured with a Horiba F-72 pH meter. All measurements were performed at 4 °C. For pumping activity analyses at different NaCl concentrations, rhodopsin-expressed cells were maintained in 100 mM NaCl at 4 °C overnight. These measurements were obtained within a few minutes after washing with the solvent.
Protein Purification and Spectral Analysis of NM-R1, NM-R2, and NM-R3.
The expressed rhodopsin proteins with six histidines at the C terminus were purified using a His GraviTrap affinity column (GE Healthcare) according to the modified manufacturer’s protocol (adding 1.0% and 0.1% n-dodecyl β-d-maltoside binding and elution buffer, respectively). Absorption spectra of purified rhodopsins were measured with a Beckman Coulter DU 800 UV-visible spectrophotometer.
Culture Experiments in Low Carbon Concentration.
S1-08T was grown in 500 mL of artificial seawater (ASW; 35 practical salinity units, prepared from sea salts; Sigma-Aldrich) containing a low concentration of dissolved organic carbon (0.14 mM carbon), NH4Cl (225 μM), and Na2HPO4·H2O (44.7 μM) according to the methodology of Kimura et al. (46). Acid-washed borosilicate glass culture bottles (1 L; VWR) were used in this experiment. The growth medium was filtered through a 0.2-μM pore size bottle-top vacuum filter system (Nalgene) and autoclaved. A colony grown on a marine agar plate (Difco) streaked from frozen stock was inoculated in a full-strength broth medium (containing 0.5% peptone and 0.1% yeast extract in ASW). The starter culture was covered with aluminum foil (Reynolds) and grown in the dark at 22 °C until near the middle of the exponential phase. Cell density of the starter culture was determined by the direct cell counting method. The starter culture was stored at 4 °C for several days, and used as the inoculum in subsequent growth experiments. Experimental cultures were inoculated to a starting concentration of 1,000 cells/mL and grown in either the light (∼150 mmol of photons m−2 s−1) or the dark at 22 °C.
Bacterial cell density was determined by epifluorescent direct counts on a preblackened isopore membrane filter (pore size 0.22 μM; Millipore). Bacterial cells on the filter were stained with SYBR Green I (1:100 dilution; Molecular Probes) for 15 min and counted under an epifluorescence microscope (Axioskop 2; Zeiss). All culture experiments were performed in triplicate.
Supplementary Material
Acknowledgments
We thank the officers and crew of R/V Mirai for assistance and support in sample collection, Professor Hideki Kandori and Dr. Inoue Keiichi for their technical support in functional analyses of microbial rhodopsins, and Haruka Ozaki and Tsukasa Fukunaga for their advice on genome analysis. This work was supported in part by Japan Society for the Promotion of Science, Kakenhi (Grants 24681003 and 23710231); the Canon Foundation; the Japan Society for the Promotion of Science Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation (Grant G2401); a Grant-in-Aid for Scientific Research on Innovative Area, “Genome Science,” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 221S0002); the Japan Science and Technology Agency; the Gordon and Betty Moore Foundation GBMF 492.01 (to E.F.D.); and the National Science Foundation (Grant EF0424599, to E.F.D.). This work is a contribution of the Center for Microbial Oceanography: Research and Education.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper has been deposited in the GenBank database (accession no. AP014548, BAO54106, BAO54132, BAO54171, BAO54172, and BAO55276).
See Commentary on page 6538.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403051111/-/DCSupplemental.
References
- 1.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 2.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233(39):149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 3.Matsuno-Yagi A, Mukohata Y. Two possible roles of bacteriorhodopsin: A comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun. 1977;78(1):237–243. doi: 10.1016/0006-291x(77)91245-1. [DOI] [PubMed] [Google Scholar]
- 4.Spudich EN, Spudich JL. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogomolni RA, Spudich JL. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79(20):6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béjà O, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289(5486):1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 7.Balashov SP, et al. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309(5743):2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: An array of physiological roles? Nat Rev Microbiol. 2008;6(6):488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 9.Inoue K, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 10.Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc Natl Acad Sci USA. 2007;104(13):5590–5595. doi: 10.1073/pnas.0611470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshizawa S, Kawanabe A, Ito H, Kandori H, Kogure K. Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ Microbiol. 2012;14(5):1240–1248. doi: 10.1111/j.1462-2920.2012.02702.x. [DOI] [PubMed] [Google Scholar]
- 12.Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA. 2007;104(7):2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oesterhelt D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr Opin Struct Biol. 1998;8(4):489–500. doi: 10.1016/s0959-440x(98)80128-0. [DOI] [PubMed] [Google Scholar]
- 14.Otto H, et al. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc Natl Acad Sci USA. 1989;86(23):9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braiman MS, et al. Vibrational spectroscopy of bacteriorhodopsin mutants: Light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- 16.Dioumaev AK, Wang JM, Bálint Z, Váró G, Lanyi JK. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry. 2003;42(21):6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 17.Sabehi G, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3(8):e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang I, et al. Genome sequence of Fulvimarina pelagi HTCC2506T, a Mn(II)-oxidizing alphaproteobacterium possessing an aerobic anoxygenic photosynthetic gene cluster and Xanthorhodopsin. J Bacteriol. 2010;192(18):4798–4799. doi: 10.1128/JB.00761-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q, et al. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS ONE. 2012;7(4):e35790. doi: 10.1371/journal.pone.0035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova N, et al. Complete genome sequence of Truepera radiovictrix type strain (RQ-24) Stand Genomic Sci. 2011;4(1):91–99. doi: 10.4056/sigs.1563919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anil Kumar P, Srinivas TNR, Madhu S, Manorama R, Shivaji S. Indibacter alkaliphilus gen. nov., sp. nov., an alkaliphilic bacterium isolated from a haloalkaline lake. Int J Syst Evol Microbiol. 2010;60(Pt 4):721–726. doi: 10.1099/ijs.0.014076. [DOI] [PubMed] [Google Scholar]
- 22.Van Trappen S, Vandecandelaere I, Mergaert JS, Swings J. Gillisia limnaea gen. nov., sp. nov., a new member of the family Flavobacteriaceae isolated from a microbial mat in Lake Fryxell, Antarctica. Int J Syst Evol Microbiol. 2004;54(Pt 2):445–448. doi: 10.1099/ijs.0.02922-0. [DOI] [PubMed] [Google Scholar]
- 23.Riedel T, et al. Genomics and physiology of a marine flavobacterium encoding a proteorhodopsin and a xanthorhodopsin-like protein. PLoS ONE. 2013;8(3):e57487. doi: 10.1371/journal.pone.0057487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon SK, et al. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol Evol. 2013;5(1):187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klassen JL. Phylogenetic and evolutionary patterns in microbial carotenoid biosynthesis are revealed by comparative genomics. PLoS ONE. 2010;5(6):e11257. doi: 10.1371/journal.pone.0011257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257(17):10306–10313. [PubMed] [Google Scholar]
- 27.Gómez-Consarnau L, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445(7124):210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 28.Feng S, Powell SM, Wilson R, Bowman JP. Light-stimulated growth of proteorhodopsin-bearing sea-ice psychrophile Psychroflexus torquis is salinity dependent. ISME J. 2013;7(11):2206–2213. doi: 10.1038/ismej.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giovannoni SJ, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438(7064):82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- 30.Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. The SAR92 clade: An abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol. 2007;73(7):2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 32.Vollmers J, et al. Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS ONE. 2013;8(5):e63422. doi: 10.1371/journal.pone.0063422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLong EF, Béjà O. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 2010;8(4):e1000359. doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodaker I, Suzuki MT, Oren A, Béjà O. Dead Sea rhodopsins revisited. Environ Microbiol Rep. 2012;4(6):617–621. doi: 10.1111/j.1758-2229.2012.00377.x. [DOI] [PubMed] [Google Scholar]
- 35.Park S, et al. Nonlabens marinus [corrected] sp. nov., a novel member of the Flavobacteriaceae isolated from the Pacific Ocean. Antonie van Leeuwenhoek. 2012;102(4):669–676. doi: 10.1007/s10482-012-9765-4. [DOI] [PubMed] [Google Scholar]
- 36.Aziz RK, et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langille MGI, Brinkman FSL. IslandViewer: An integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25(5):664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14(4):378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 40.Uchiyama I. Hierarchical clustering algorithm for comprehensive orthologous domain classification in multiple genomes. Nucleic Acids Res. 2006;34(2):647–658. doi: 10.1093/nar/gkj448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence-limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura H, Young CR, Martinez A, Delong EF. Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 2011;5(10):1641–1651. doi: 10.1038/ismej.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.