Significance
Indeterminate growth of shoots continually produces new tissues from the dividing apical meristem. In contrast, determinate growth of the floral meristem produces flowers of a particular size and form by specification of floral organs and termination of stem-cell divisions in the meristem. To achieve this specification, floral organs do not form meristems in organ axils, although leaves can form axillary meristems and thus have reiterative developmental potential. In this study, we found that the classic floral homeotic gene APETALA1 directly regulates homeostasis of the plant hormones (i.e., cytokinins) to inhibit the formation of sepal axil stem-cell niches. A deeper understanding of axil lateral meristem activity provides crucial information for enhancing yield by engineering crops that produce more elaborated racemes.
Keywords: axillary meristem, indeterminacy
Abstract
In angiosperms, after the floral transition, the inflorescence meristem produces floral meristems (FMs). Determinate growth of FMs produces flowers of a particular size and form. This determinate growth requires specification of floral organs and termination of stem-cell divisions. Establishment of the FM and specification of outer whorl organs (sepals and petals) requires the floral homeotic gene APETALA1 (AP1). To determine FM identity, AP1 also prevents the formation of flowers in the axils of sepals. The mechanisms underlying AP1 function in the floral transition and in floral organ patterning have been studied extensively, but how AP1 terminates sepal axil stem-cell activities to suppress axillary secondary flower formation remains unclear. Here we show that AP1 regulates cytokinin levels by directly suppressing the cytokinin biosynthetic gene LONELY GUY1 and activating the cytokinin degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3. Restoring the expression of these genes to wild-type levels in AP1-expressing cells or suppressing cytokinin signaling inhibits indeterminate inflorescence meristem activity caused by ap1 mutation. We conclude that suppression of cytokinin biosynthesis and activation of cytokinin degradation mediates AP1 function in establishing determinate FM. A deeper understanding of axil-lateral meristem activity provides crucial information for enhancing yield by engineering crops that produce more elaborated racemes.
Formation of flowers marks the beginning of the reproductive stage, a critical process in the angiosperm life cycle (1, 2). After the floral transition, the shoot apical meristem (SAM) transforms into an inflorescence meristem (IM), which produces floral meristems (FMs). FMs may share similar origins with the axillary meristems (AMs) produced during vegetative growth (3). Similar to the SAM, AMs maintain indeterminate developmental potential; in contrast, the FM undergoes determinate growth to form a flower with particular numbers of floral organs and specific size. This transition to determinate growth requires FM identity genes, which terminate the production of stem cells. In the center of the FM, AGAMOUS (AG) terminates the FM stem-cell niche by repressing WUSCHEL (WUS) expression (4, 5). In addition, the reiterative developmental potential of leaves is suppressed in floral organs so that no meristem forms at lateral organ axils. APETALA1 (AP1) mediates the suppression of the AM-like stem-cell niche in the sepals, the first-whorl floral organs (6–8). Because indeterminate growth within the inflorescence results in branching and produces diverse inflorescence architectures (9), a better understanding of meristem activity of flowers will improve our understanding of how human selection produced elaborated racemes to enhance crop yield (10).
FM formation requires AP1, LEAFY, and CAULIFLOWER (CAL) (8, 11–15), which activate floral organ identity genes. After FM formation, AP1 functions as a class A floral organ identity gene to specify sepals and petals, the outer two whorls of floral organs (6, 7). AP1 also prevents the formation of secondary flowers in the axils of sepals to maintain determinate growth of flowers (6, 7). Mutations in AP1 result in ectopic formation of secondary flowers in the axils of sepals (Fig. 1 A–C). Reiteration of this pattern, so that tertiary flowers form in sepal axils of secondary flowers, results in indeterminate growth of the FM. AP1 encodes a MADS-domain (MCM1, AGAMOUS, DEFICIENS, and SERUM RESPONSE FACTOR) transcription factor initially expressed throughout FMs and later restricted to sepals and petals (7). The molecular networks underlying AP1’s function in floral transition and floral-organ patterning have been studied extensively (1, 2), but how AP1 inhibits indeterminate growth in sepal axils remains to be unraveled. AP1 down-regulates three flowering-time MADS-domain genes, SHORT VEGETATIVE PHASE (SVP), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and AGAMOUS-LIKE 24 (AGL24), to inhibit secondary flower formation partially in sepal axils (16). In addition, these three transcription factors also act in floral patterning and regulating the floral transition (17, 18). How AP1 and additional transcription factors inhibit the stem-cell fate of sepal axil cells remains an open question.
Fig. 1.
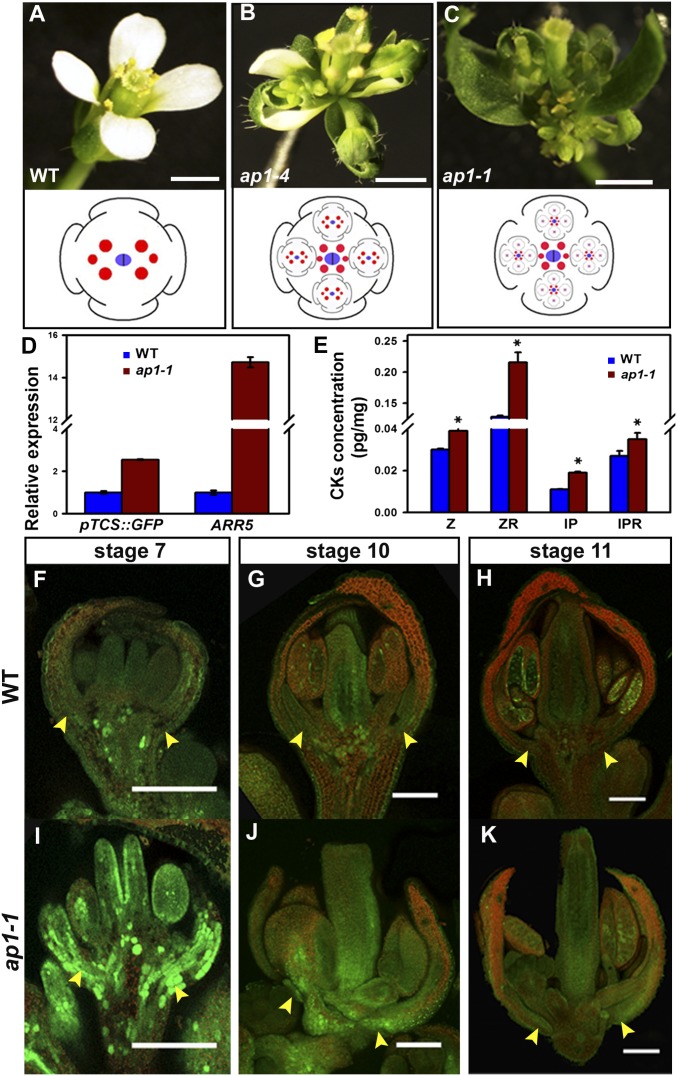
AP1 inhibits cytokinin signaling and reduces cytokinin levels in the FM. (A–C) Flower phenotype of Ler (A), ap1-4 (B), and ap1-1 (C) showing sepal axil secondary flowers in ap1 mutants. (D) Relative expression of pTCS::GFP-ER and ARR5 in wild-type and ap1-1 inflorescences. Transcript levels were measured by qRT-PCR of three independently collected samples. Results were normalized against the expression of TUB6. Error bars indicate the SD of three biological experiments, each run in triplicate. (E) Mass spectrometric measurements of cytokinins (CKs) in wild-type and ap1-1 inflorescences. IP, isopentenyladenine; IPR, isopentenyladenine riboside; Z, zeatin riboside 5′-monophosphate; ZR, zeatin riboside. Mean values of four replicates are shown. Error bars indicate the SD of three biological experiments, each run in triplicate. *P < 0.01 between wild-type and ap1-1 inflorescences. (F–K) pTCS::GFP-ER (green) in longitudinal sections of developing flowers at stage 7 (F and I), stage 10 (G and J), and stage 11 (H and K) of wild-type (D–F) and ap1-1 (G–I) plants. Autofluorescence is shown in red. Arrowheads indicate sepal axils. (Scale bars: A–C, 1 mm; D–I, 100 μm).
The cytokinin plant hormones play pivotal roles in many aspects of plant development, such as promoting shoot development. Shoot meristem marker SHOOT MERISTEMLESS (STM) and related KNOTTED1-like homeobox transcription factors (19, 20) activate cytokinin biosynthesis. In addition, SAM functions may involve positive feedback between cytokinin and the stem-cell regulator WUS (21, 22).
In this study, we show that AP1 inhibits the establishment of a stem-cell niche in sepal axils by suppressing cytokinin biosynthesis and by activating cytokinin degradation. First, we show enhanced cytokinin signaling and cytokinin levels in ap1 mutants. In addition, elevated cytokinin levels in flowers phenocopy the ap1 axillary secondary flower phenotype, whereas cytokinin signaling mutants partially rescue this phenotype in the ap1-1 mutant. We demonstrate that AP1 directly inhibits the expression of the cytokinin biosynthetic gene LONELY GUY1 (LOG1) and directly promotes expression of the cytokinin degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3). Restoring the expression of these genes to wild-type levels partially suppressed the indeterminate stem-cell activity in sepal axils caused by a loss of AP1 function.
Results
AP1 Suppresses Cytokinin Signaling and Levels in FMs.
Sepals in the outermost floral whorl are thought to be modified leaves (23) and the ground state of floral organs. However, unlike leaves, sepals lack meristematic cells in their axils, a difference that requires AP1. We hypothesized that AM and FM initiation requires cytokinin signaling and that AP1 might suppress cytokinin signaling to inhibit meristematic activity in sepal axils. To test this notion, we used quantitative RT-PCR (qRT-PCR) to measure the mRNA levels of ARABIDOPSIS RESPONSE REGULATOR 5 (ARR5), which encodes a classical cytokinin-inducible type-A ARR (24). We found that ARR5 mRNA levels strongly increased in ap1-1 mutant flowers as compared with wild-type flowers (Fig. 1D). To confirm the enhanced cytokinin signaling, we introduced pTCS::GFP-ER, a synthetic reporter for downstream activation of the cytokinin signaling pathway (25), into the ap1-1 background (Fig. 1 F–K). By qRT-PCR we found elevated GFP transcript levels in ap1-1 floral tissues as compared with wild-type pTCS::GFP-ER homozygous siblings (Fig. 1D).
To visualize the cytokinin response at high resolution, we examined the pTCS::GFP-ER reporter in early-stage flowers. Secondary FMs initiate in sepal axils between flower stages 6 and 10 (Fig. S1). We observed a stronger GFP signal in ap1-1 flowers than in wild-type sibling flowers when the pTCS::GFP-ER transgene was homozygous in both plants and when the same imaging setup was used (Fig. 1 F–K). We also observed elevated GFP signal in floral organs other than sepals and petals in ap1-1 plants, even when AP1 was no longer expressed in inner whorls. This signal may reflect possible indirect effects of AP1 on cytokinin levels. The difference in GFP signal was most dramatic at stage 7 (Fig. 1 F and I) and became reduced in older flowers (Fig. 1 G and J). However, the difference in GFP signals was still visible by stage 11 (Fig. 1 H and K). Although elevated GFP signal is ubiquitous in all floral organs and pedicels in ap1-1 flowers, sepals had more enhanced signals, especially in the proximal part (Fig. 1 I–K) including the sepal axil where secondary flowers initiate. This finding is consistent with the expression of AP1 in the outer whirls after floral bud formation (7).
We directly measured cytokinins to test the possibility that the ap1 mutation causes elevated cytokinin levels. We quantified endogenous levels of zeatin riboside 5′-monophosphate, zeatin riboside, isopentenyladenine, and isopentenyladenine riboside in young inflorescence. All measured cytokinins increased in ap1-1 mutants, but the most significant increase was observed for zeatin riboside and isopentenyladenine (Fig. 1E). These results might suggest that AP1 reduces cytokinin levels and cytokinin responses.
AP1 Suppresses Axil Stem Cells Through Cytokinin Activity.
We further tested the role of cytokinin in the initiation of sepal AMs. To this end, we treated inflorescences with the cytokinin analog benzylaminopurine. As previously reported (26), application of cytokinin phenocopies the sepal axil secondary flower phenotype of ap1 (Fig. 2 A–D). Without cytokinin, no secondary flowers form after mock treatment (Fig. 2A). Consistent with previous reports, cytokinin treatment also resulted in enlarged FMs (22, 26).
Fig. 2.
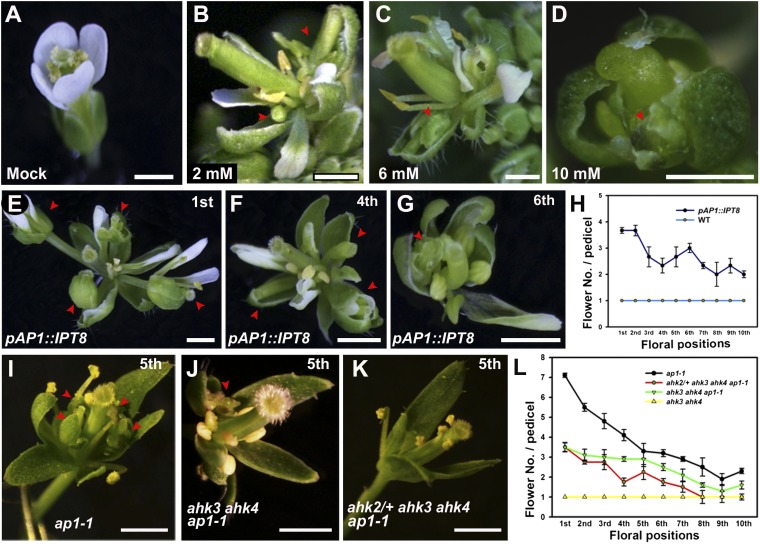
The initiation of ectopic secondary flowers involves cytokinin signaling. (A–D) Flower phenotype after mock (A), 2 mM benzylaminopurine (BAP) (B), 6 mM BAP (C), and 10 mM BAP (D) treatment. Ectopic secondary flowers (arrowheads) are observed after BAP treatment. (E–H) Flower phenotype of pAP1::IPT8 transgenic plants. Ectopic secondary flowers (arrowheads) observed in the first (E), fourth (F), and sixth (G) flowers are shown. (H) Mean number of flowers per pedicel in pAP1::IPT8 and in Ler plants at different floral positions at the main inflorescence are shown; floral position 1 refers to the basal flower. Error bars indicate SD. (I–L) Flower phenotype of ap1-1 (I), ahk3 ahk4 ap1-1 (J), and ahk2/+ ahk3 ahk4 ap1-1 (K). The fifth flower is shown for each genotype. (L) Mean number of flowers per pedicel at different floral positions at the main inflorescence. Error bars indicate SD. (Scale bars: 1 mm.)
In addition to exogenous cytokinin treatment, we elevated in vivo cytokinin levels in flowers by expressing the Arabidopsis adenosine phosphate-isopentenyltransferase 8 (IPT8), which encodes a rate-limiting enzyme in cytokinin biosynthesis (27), from the AP1 promoter. Consistent with exogenous cytokinin treatment results, we found wild-type Ler plants carrying the pAP1::IPT8 transgene phenocopied the sepal axil secondary flower phenotype of ap1 mutants (Fig. 2 E–H and Fig. S2 A–C). Although we did not observe defects in flower initiation, as previously reported (28), we found that pAP1::IPT8 lines with strong transgene expression lacked petals (Fig. S2 A–D), suggesting the involvement of cytokinins in petal development as well.
If sepal axil flower formation requires the cytokinin response, disruption of the cytokinin-signaling pathway may rescue the ap1 mutant phenotype, at least partially. To test this prediction, we introduced cytokinin receptor mutations into the ap1-1 background. According to the current model for cytokinin signal transduction, three Arabidopsis histidine kinase receptors (AHK2, AHK3, and AHK4/CRE1/WOL) perceive cytokinin (29). We generated ahk3-7 cre1-2 ap1-1 and ahk2-5/+ ahk3-7 cre1-2 ap1-1 mutants (quadruple homozygous mutants fail to form flowers) and found that the secondary flower phenotype was alleviated in the mutants (Fig. 2 I–L and Fig. S2 E–G). The frequency of the secondary flower phenotype was reduced to the wild-type level in stage 8 and younger flowers in ahk2-5/+ ahk3-7 cre1-2 ap1-1 mutants (Fig. 2L). Double mutants of ap1-1 with one ahk exhibited weaker phenotypic rescue, suggesting that the AHK receptors have redundant roles. Finally, the defect in sepal initiation, but not the defect in petal initiation, was slightly reduced by ahk mutations (Fig. S2 E–H).
Coordinated Regulation of Cytokinin Homeostasis Genes by AP1.
Our recent cell-specific genome expression analysis found low expression of several cytokinin biosynthesis genes in AP1-expressing cells and high expression of a few cytokinin degradation enzyme genes (30). A reexamination of the genome-wide binding data for AP1 (31) identified the cytokinin-activating enzyme gene LOG1 and the cytokinin dehydrogenase gene CKX3 as being involved in cytokinin degradation, as putative direct targets of AP1, although these two genes were not included in the original shortlist with higher statistical cutoff (31). Using qRT-PCR, we quantified the RNA levels of LOG1 and CKX3 in young inflorescences and found that ap1-1 mutant flowers have higher LOG1 expression and lower CKX3 expression than seen in wild-type plants (Fig. 3A).
Fig. 3.
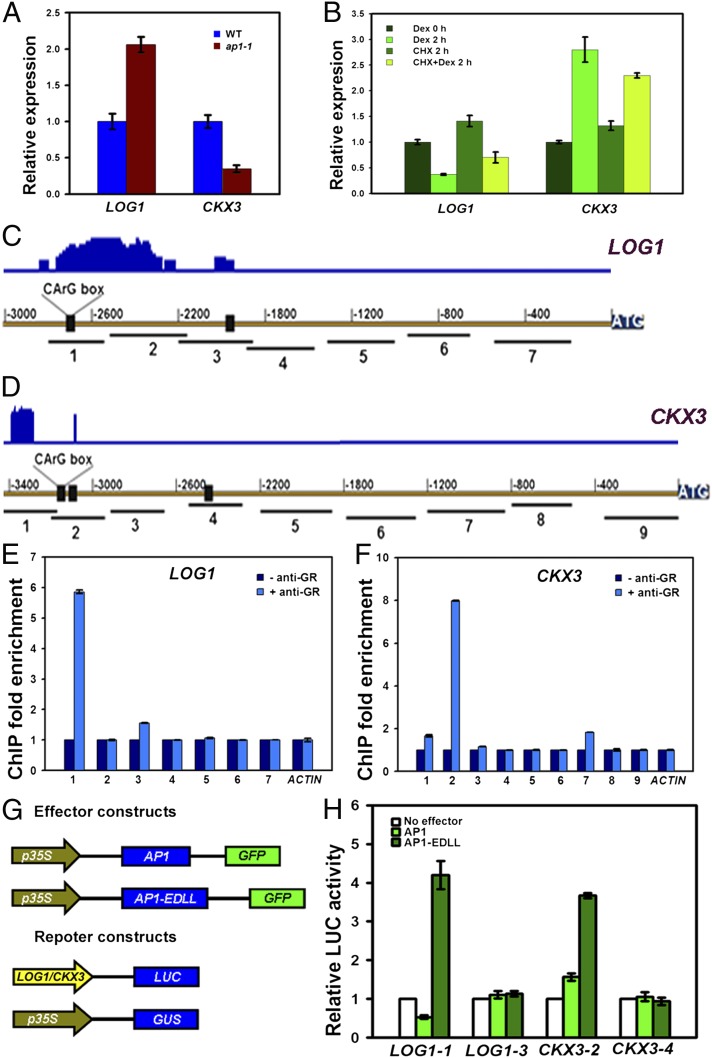
AP1 regulates LOG1 and CKX3 expression via binding to a conserved promoter motif. (A) Real-time RT-PCR analysis of LOG1 and CKX3 in Ler and in ap1-1 inflorescences. (B) Real-time RT-PCR analysis of LOG1 and CKX3 using the p35S::AP1-GR ap1-1 cal-1 inflorescences before and after Dex treatment or simultaneous Dex and CHX treatment for 2 h. The vertical axis indicates relative mRNA amount compared with the amount before Dex treatment. Error bars indicate SD. (C) (Lower) Schematic of the LOG1 genomic region. Black boxes indicate the sites containing the consensus binding sequence (CArG box), ATG denotes the translation start site. (Upper) Reported binding profiles of AP1 by ChIP-seq. (31) for the same region. Seven PCR fragments were designed for ChIP analysis. (D) Schematic of the CKX3 genomic region (Lower) and reported AP1 binding profiles (Upper) (31). Nine PCR fragments were designed for ChIP analysis. (E) ChIP enrichment test by PCR shows binding of AP1-GR to the region near the number 1 fragment. More controls are shown in Fig. S4C. (F) ChIP enrichment test by PCR shows the binding of AP1-GR to the region near the number 2 fragment. More controls are shown in Fig. S4D. Error bars indicate SD. (G and H) Transcriptional activity assays in Arabidopsis protoplasts. EDLL is a transcriptional activation domain; p35S::GUS is the internal control. (H) Relative LUC reporter gene expression. The LOG1-1 and LOG1-3 promoter regions (indicated as in C) and the CKX3-2 and CKX3-4 promoter regions (indicated as in D) were assayed. Data are mean ± SD. Error bars are derived from three independent biological experiments, each run in triplicate.
To determine whether AP1 directly elicits cytokinin homeostasis, we monitored the effects on LOG1 and CKX3 expression after activation of AP1 function. To this end, we used a line in which an AP1-glucocorticoid receptor (GR) fusion protein is expressed from the constitutive 35S promoter in ap1-1 cal-1 double-mutant plants. Nuclear translocation of the AP1-GR fusion protein can be triggered specifically through the steroid hormone dexamethasone (Dex) (Fig. S3). AP1 activation in p35S::AP1-GR ap1-1 cal-1 plants rescues the ap1 phenotype (31). We measured the effect of AP1 activation in p35S::AP1-GR ap1-1 cal-1 plants on the expression of LOG1 and CKX3 by qRT-PCR. AP1 activation resulted in a rapid reduction of LOG1 mRNA levels and a rapid elevation of CKX3 mRNA levels within 2 h of AP1 induction in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Fig. 3B). Because CHX was shown previously to be effective in this system (31), our results strongly suggest that suppression of LOG1 and induction of CKX3 do not require de novo protein synthesis and that these genes are likely direct targets of AP1.
AP1 Regulates LOG1 and CKX3 Expression via Binding to a Conserved Promoter Motif.
We performed further ChIP assays to examine whether AP1 directly controls LOG1 and CKX3 expression. We scanned the LOG1 and CKX3 genomic sequence for CC(A/T)6GG (CArG) motifs, the canonical binding site for MADS-domain proteins (32). We designed primers near identified motifs and other regions to measure DNA enrichment (Fig. 3 C and D). AP1-GR associated with the region near the LOG1-1 fragment containing a CArG motif, but only after Dex treatment (Fig. 3E and Fig. S4A). Similarly, AP1-GR associated with the region near the CKX3-2 fragment containing two CArG motifs only after Dex treatment (Fig. 3F and Fig. S4B). The CArG motifs closer to the start codon in the LOG1 and CKX3 genomic regions did not associate with AP1-GR. ChIP-PCR results generally were consistent with a recent, large-scale ChIP-seq analysis (31).
A transient transfection assay in protoplasts further confirmed that AP1 and AP1-EDLL (Fig. 3G), a fusion protein between AP1 and the EDLL transcriptional activation domain (33), bound to the upstream promoter fragments containing the upstream CArG motif but not to the ones closer to the start codon (Fig. 3H). Although the AP1-EDLL fusion protein activated the expression of the luciferase (LUC) reporter gene driven by both the LOG1 and CKX3 promoter regions, native AP1 repressed the expression of the LOG1 promoter driving LUC gene but activated the expression of the CKX3 promoter driving LUC gene. If the transcriptional regulation activity is conferred by additional proteins, their association with AP1 should be bridged by the sequences containing the CArG motif used in this assay. The selective binding of AP1 to different CArG motifs also highlighted the critical roles of flanking sequences on protein–DNA interactions.
Flower-Specific Regulation of LOG1 and CKX3 Expression by AP1.
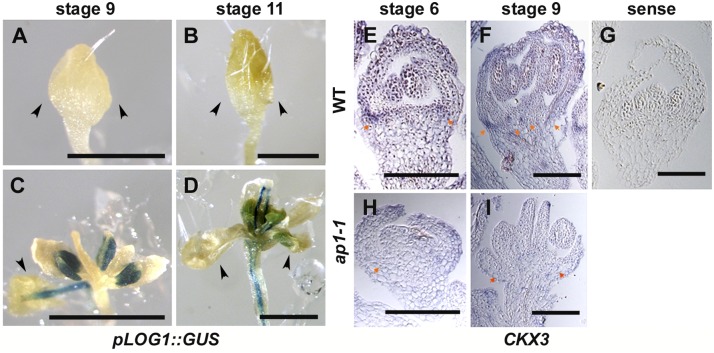
To determine precisely the effect of AP1 on LOG1 expression, we crossed a pLOG1::GUS reporter (34) into ap1-1. We detected no GUS expression in flowers of the wild-type siblings until stage 12 (Fig. 4 A and B). In contrast, we detected GUS expression in ap1-1 flowers between stages 7 and 11, with strong expression in sepals and stamens (Fig. 4 C and D). Consistent with this finding, secondary flowers form in sepal axils of ap1-1 flowers between stages 6 and 10 (Fig. S1). The enhanced GUS expression in stamens, where AP1 is not expressed, in early-stage ap1-1 flowers may indicate an indirect regulation of LOG1 by AP1. Interestingly, GUS expression in stamens after stage 12 was detected in wild-type flowers but not in ap1-1 flowers, suggesting the existence of complicated indirect regulation. Nevertheless, this later suppression of LOG1 expression in ap1-1 does not overlap spatiotemporally with secondary flower development. Similarly, we compared the expression of CKX3 in ap1-1 and Ler flowers by in situ hybridization (Fig. 4 E–I). CKX3 is expressed in floral organs throughout stages 6–9 in wild-type flowers, and the highest expression was detected at floral organ axils (Fig. 4 E and F). CKX3 expression generally is reduced in ap1-1 flowers, particularly in sepal axils, where secondary flowers could form (Fig. 4 H and I).
Fig. 4.
Ectopic expression of LOG1 and CKX3 in ap1-1. (A–D) Representative GUS staining patterns of wild-type (A and B) and ap1-1 (C and D) flower buds containing pLOG1::GUS at stage 9 (A and C) and stage 11 (B and D). Arrowheads indicate sepals. (E–I) Patterns of CKX3 transcript accumulation in wild-type (E–G) and ap1-1 (H and I) flowers. Longitudinal sections through stage 6 (E–G) and stage 9 (H and I) flowers were hybridized with CKX3 antisense (E, F, H, and I) and sense (G) probes, respectively. In Ler wild-type plants, CKX3 transcript accumulation was observed in sepal axils (arrows in E and F). This transcript accumulation was not obvious in ap1-1 sepal axils (arrows in H and I). (Scale bars: A–D, 500 μm; E–I, 100 μm.)
Restoring Normal Levels of CKX3 and LOG1 Expression Suppresses the ap1 Axil Flower Phenotype.
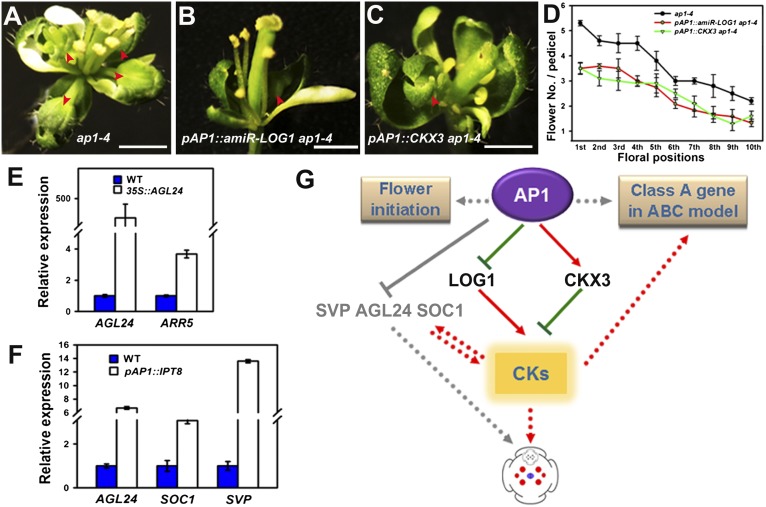
To test whether the activation of LOG1 expression and the suppression of CKX3 expression in ap1 mutant plants are relevant to the ectopic formation of sepal axil secondary flowers, we expressed an artificial microRNA that specifically targets LOG1 mRNA (amiR-LOG1) but not paralogous LOG genes expressed in flowers (Fig. S5A). We also expressed CKX3 cDNA under the control of the AP1 promoter. In transgenic ap1-4 mutant lines containing either pAP1::amiR-LOG1 or pAP1::CKX3, we found partial rescue of the sepal axil secondary flower phenotype (Fig. 5 A–C). For pAP1::amiR-LOG1 ap1-4, we observed a clear positive correlation between remaining LOG1 expression levels and the frequency of secondary flowers (Fig. S5B). Moreover, we observed phenotypic rescue in flowers at all positions (Fig. 5D). These observations suggest that the effect of AP1 on secondary flower formation can be short-circuited by direct manipulation of AP1 targets in flowers initiated at different ages.
Fig. 5.
(A–D) Rescue of the ap1-4 sepal axil flower phenotype by local manipulation of CKX3 and LOG1 expression. Representative flowers are shown for ap1-4 (A), pAP1::amiR-LOG1 ap1-4 (B), and pAP1::CKX3 ap1-4 (C). (Scale bars: 1 mm.) The mean number of flowers per pedicel at different floral positions at the main inflorescence is shown in D. Error bars indicate SD. (E) Relative expression of AGL24 and ARR5 in Col-0 wild-type and p35S::ALG24 inflorescences. (F) Relative expression of AGL24, SOC1 and SVP in Ler wild-type and pAP1::IPT8 inflorescences. Transcript levels were measured by qRT-PCR of three independently collected samples. Results were normalized against the expression of TUB6. Error bars indicate SD and are derived from three independent biological experiments, each performed in triplicate. (G) Model depicting AP1 suppression of sepal axil stem-cell activity through the inhibition of cytokinin biosynthesis (LOG1) and activation of cytokinin degradation (CKX3) to reduce active cytokinin levels in sepals. Elevated cytokinin levels activate sepal axil meristem activity to induce secondary flower formation. Cytokinin signaling also interacts positively and reciprocally with flowering-time MADS-domain genes.
Reciprocal Interactions Between Cytokinin Signaling and Flowering-Time Genes.
Finally, we tested if cytokinin interacted with the flowering-time genes SVP, SOC1, and AGL24, whose overexpression promotes secondary flower formation (16). To this end, we used qRT-PCR to measure the mRNA levels of cytokinin-inducible ARR5 in a p35S::AGL24 line and to measure the mRNA levels of these three flowering-time genes in an intermediate pAP1::IPT8 line. We found that ARR5 mRNA levels strongly increased in p35S::AGL24 inflorescence (Fig. 5E) and that SVP, SOC1, and AGL24 mRNA levels strongly increased in pAP1::IPT8 inflorescence (Fig. 5F). There results suggest that cytokinin signaling and these flowering-time genes activate each other reciprocally.
Discussion
Flowers, the reproductive structures of angiosperms, develop from FMs after the floral transition to reproductive growth. The FM can be considered a type of shoot meristem and shares many similarity with AMs (3), which also derive from the SAM. Nevertheless, the FM has a distinct feature: determinate growth and production of a defined number of floral organs by limited meristem activity. In the center of a floral bud, AG mediates the determination of the FM, and ag mutants exhibit indeterminate FM growth, producing double flowers (4, 5). In the outer whorls of a floral bud, AP1 suppresses indeterminate AM or IM activity, and ap1 mutants have secondary flowers in sepal axils (6–8). Repetition of this pattern leads to the production of tertiary flowers in secondary flower sepal axils, and so forth (Fig. 1C). AG suppresses WUS expression and terminates meristem activity in the center of the floral bud (4, 5). Although three flowering-time genes encoding the MADS-box transcription factor have been identified as direct AP1 targets regulating determinate FM identity and floral patterning (16–18), it remains unknown why AP1 and these additional transcription factor genes inhibit meristem activity in sepal axils.
Our findings have revealed a genetic pathway inhibiting FM formation in sepal axils that, to our knowledge, has not been known previously. A parallel pathway also functions in the control of AM activity commonly found in leaf axils in higher plants (Fig. 5G). Our combined results indicate that AP1 acts upstream of cytokinin, affecting cytokinin biosynthesis and degradation to suppress meristem activity in sepal axils (Fig. 5G). Formation of AM, a potentially related developmental process (3), also may require the cytokinin response. We found that AP1 directly suppresses LOG1 expression and activates CKX3 expression to orchestrate cytokinin levels. Consistent with this finding, a recent large-scale analysis of AP1 binding targets found both activation and suppression of direct target gene expression (31). AP1 may recruit other transcription factors, chromatin regulators, or other cofactors to regulate target gene activation or suppression differentially. Our results indicate that AP1 binding and transcriptional regulation are controlled not only by the core CArG motif but also by flanking sequences (Fig. 3 E–H). Activation of cytokinin signaling in the entire FM, as reported by pTCS::GFP-ER, was observed in ap1-1 (Fig. 1 F–K). Enhanced TCS signals outside sepals and petals, where AP1 expresses, may suggest a non–cell-autonomous indirect effect of AP1 on cytokinin signaling. Enhanced LOG1 expression outside the AP1-expressing domain, such as in stamens (Fig. 4 A–D) and the termination of LOG1 expression after stage 12 in mature ap1-1 flowers but not in wild-type flowers support the existence of additional indirect AP1 regulation of LOG1 expression. In addition, the non–cell-autonomous effect of AP1 on cytokinin levels may result, at least partially, from cytokinin translocation within the FM.
In addition to its long association with SAM activities, recent studies have suggested connections between cytokinin and branching meristems. The rice LOG gene is specifically expressed in primary panicle branch meristems, and panicle branching is dramatically reduced in log mutant alleles (35). On the other hand, a recent study reported promoted FM activities in a ckx3 ckx5 mutant line but did not report the secondary flower phenotype (36). The difference in phenotypes observed in the ckx3 ckx5 mutant and the pAP1::IPT8 transgenic lines may suggest that a high level of active cytokinin is required for secondary flower formation and that CKX genes may be suppressed by AP1. Indeed, we did not observe secondary flowers in pAP1::IPT8 transgenic lines with a low level of transgene expression. Alternatively, the difference in phenotype may reflect the importance of tissue-specific hormone action in development, because CKXs have broad expression and pleotropic effects on development (37) that may interfere with the formation of secondary flowers.
AP1 acts as a master regulator of flower development. Extensive study over the past two decades has revealed how AP1 controls the onset of flower development and how AP1 specifies sepal and petal identities as an A function gene (31). Our findings uncover a previously unidentified regulatory mechanism of AP1 and link AP1 function directly to the regulation of hormone homeostasis in establishing determinate growth in the outer whorl of flowers.
Materials and Methods
Plants were grown in the greenhouse on soil at 22 °C under long-day conditions (16 h light/8 h dark). Cytokinin treatment was performed as described (22). Standard genetic and molecular biology techniques were used for crossing and for the construction of plasmids and reporter transgenes. RT-PCR, quantitative real-time PCR, and ChIP were performed as previously described (31). Primers are given in Tables S1–S4. Confocal imaging was performed using a Nikon C2 confocal microscope with a 40× objective. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Chu, X. Sun, and C. Yan at the National Center for Plant Gene Research for cytokinin measurement; H.-S. Guo, B. Müller, H. Sakakibara, H. Yu, J. Zuo, and the Arabidopsis Biological Research Center for seeds and plasmids; Y. Wang for advice on genetic analysis and imaging; J. L. Riechmann and F. Wellmer for sharing ChIP protocols; and Y. Xu and K. Chong for help with protoplast assay. We also thank F. Wellmer, J. Zuo, and two anonymous reviewers for their constructive comments. This work was supported by National Basic Research Program of China (973 Program) Grant 2014CB943500, National Natural Science Foundation of China Grants 31222033 and 31171159, Strategic Priority Research Program of the Chinese Academy of Science (CAS) Grant XDA08020105, and the Hundred Talents Program of the CAS.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online atwww.pnas.org/lookup/suppl/doi:10.1073/pnas.1318532111/-/DCSupplemental.
References
- 1.Krizek BA, Fletcher JC. Molecular mechanisms of flower development: An armchair guide. Nat Rev Genet. 2005;6(9):688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- 2.Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends Genet. 2010;26(12):519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Dev Biol. 2000;218(2):341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- 4.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105(6):805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 5.Lohmann JU, et al. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105(6):793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 6.Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell. 1990;2(8):741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360(6401):273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 8.Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119(3):721–743. [Google Scholar]
- 9.Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316(5830):1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Li J. Molecular basis of plant architecture. Annu Rev Plant Biol. 2008;59:253–279. doi: 10.1146/annurev.arplant.59.032607.092902. [DOI] [PubMed] [Google Scholar]
- 11.Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377(6549):522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- 12.Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267(5197):522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 13.Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377(6549):495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- 14.Weigel D, Meyerowitz EM. Activation of floral homeotic genes in Arabidopsis. Science. 1993;261(5129):1723–1726. doi: 10.1126/science.261.5129.1723. [DOI] [PubMed] [Google Scholar]
- 15.Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127(4):725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 16.Liu C, et al. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development. 2007;134(10):1901–1910. doi: 10.1242/dev.003103. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013;24(6):612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Xi W, Shen L, Tan C, Yu H. Regulation of floral patterning by flowering time genes. Dev Cell. 2009;16(5):711–722. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Jasinski S, et al. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol. 2005;15(17):1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15(17):1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 21.Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438(7071):1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 22.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106(38):16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124(4):1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453(7198):1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venglat SP, Sawhney VK. Benzylaminopurine induces phenocopies of floral meristem and organ identity mutants in wild-type Arabidopsis plants. Planta. 1996;198(3):480–487. doi: 10.1007/BF00620066. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, et al. The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol. 2003;131(1):167–176. doi: 10.1104/pp.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 30.Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328(5974):85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 32.de Folter S, Angenent GC. trans meets cis in MADS science. Trends Plant Sci. 2006;11(5):224–231. doi: 10.1016/j.tplants.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Tiwari SB, et al. The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012;70(5):855–865. doi: 10.1111/j.1365-313X.2012.04935.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuroha T, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21(10):3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 36.Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23(1):69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner T, Schmülling T. Cytokinin action in plant development. Curr Opin Plant Biol. 2009;12(5):527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.