Significance
The degradative dNTP triphosphohydrolase activity of the sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) enzyme helps maintain optimal dNTP balances for DNA replication and also serves as an HIV-1 restriction factor in resting CD4+ target cells of HIV by depleting dNTP substrates of reverse transcriptase. This study shows that full activation of SAMHD1 involves ordered binding of GTP and substrate dNTPs to activator and substrate sites on the enzyme, leading to ordered assembly of the tetramer active form. After the enzyme is activated, it no longer communicates with free activator nucleotides, which contributes to efficient depletion of dNTP pools in resting T cells.
Keywords: dNTP induced oligomerization, enzyme catalysis, innate immunity
Abstract
The HIV-1 restriction factor sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) is a tetrameric protein that catalyzes the hydrolysis of all dNTPs to the deoxynucleoside and tripolyphosphate, which effectively depletes the dNTP substrates of HIV reverse transcriptase. Here, we establish that SAMHD1 is activated by GTP binding to guanine-specific activator sites (A1) as well as coactivation by substrate dNTP binding to a distinct set of nonspecific activator sites (A2). Combined activation by GTP and dNTPs results in a long-lived tetrameric form of SAMHD1 that persists for hours, even after activating nucleotides are withdrawn from the solution. These results reveal an ordered model for assembly of SAMHD1 tetramer from its inactive monomer and dimer forms, where GTP binding to the A1 sites generates dimer and dNTP binding to the A2 and catalytic sites generates active tetramer. Thus, cellular regulation of active SAMHD1 is not determined by GTP alone but instead, the levels of all dNTPs and the generation of a persistent tetramer that is not in equilibrium with free activators. The significance of the long-lived activated state is that SAMHD1 can remain active long after dNTP pools have been reduced to a level that would lead to inactivation. This property would be important in resting CD4+ T cells, where dNTP pools are reduced to nanomolar levels to restrict infection by HIV-1.
The steady-state composition and concentration of deoxynucleotide triphosphate pools in mammalian cells are highly regulated because of the mutagenic consequences of dNTP imbalances in dividing cells (1, 2) as well as the important antiviral effects of dNTP pool depletion in quiescent cells (3, 4). In all cell types, the ultimate pool balance is determined by dNTP-dependent regulatory pathways that affect the activities of enzymes involved in both synthesis and degradation of dNTPs (5–7). The most important highly up-regulated synthetic enzyme during S phase of dividing cells is the R1/R2 isoform of ribonucleotide triphosphate reductase, which ensures that dNTP precursors are plentiful for DNA synthesis (8). However, in quiescent cells of the immune system (resting CD4+ T cells, macrophages, and dendritic cells), where dNTP pools are ∼10-fold lower than dividing cells, the ultimate pool levels are likely determined by a balance between the activities of the R1/p53R2 isoform of ribonucleotide triphosphate reductase and the degradative dNTP triphosphohydrolase sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) (9). The highly dynamic nature of dNTP pools demands finely tuned mechanisms for feedback regulation of these enzymes by dNTPs as well as coarse regulatory mechanisms (posttranslational modifications, transcriptional regulation, and proteasomal targeting) that serve to turn these activities on and off at appropriate stages of the cell cycle and in specific cell types (10, 11).
dNTP triphosphohydrolase enzymes, such as SAMHD1, are conserved from bacteria to humans and carry out the unusual conversion of dNTPs to the nucleoside (dN) and tripolyphosphate (12, 13). The tripolyphosphate is likely degraded to 3Pi through the action of plentiful cellular pyrophosphatases or tripolyphosphatases (14). Presumably, degradation to the level of nucleoside rather than deoxynucleoside diphosphate (dNDP) or deoxynucleoside monophosphate (dNMP) is to make the process energetically or kinetically costly to reverse, with the additional possibility that the neutral nucleoside will be irreversibly transported out of the cell (9). The unusual triphosphohydrolase activity of SAMHD1 in quiescent immune cells has received significant attention, because HIV-1 and HSV-1 are severely restricted in their ability to infect quiescent cells that have severely depressed dNTP pools, which has been directly linked to SAMHD1 enzymatic activity (15, 16). Mutations in the SAMHD1 gene have also been linked to Aicardi–Goutières syndrome, a rare genetic autoimmune encephalopathy with a chronic inflammatory pathology that resembles congenital viral infections (17). As would be expected, resting CD4+ T cells from Aicardi–Goutières syndrome patients are permissive to HIV-1 infection (18).
A key question with SAMHD1 is how its activity is tuned in response to dNTP pool levels. Early reports established that the dNTPase activity of SAMHD1 required the presence of dGTP as an activator when dATP, dTTP, or dCTP was used as a substrate (15, 17). Subsequently, it was reported that the dGTP activator/substrate can be replaced by GTP, which serves as an activator, but not a substrate (19). Two recent structures of SAMHD1 in complex with dNTPs show a tetrameric quaternary structure with dGTP molecules bound to the (A1A2)4 activator sites and one dNTP bound to each of four catalytic sites (20). Comparison with an earlier dimeric structure of the free enzyme suggests that activator and/or substrate binding drive the enzyme into the tetrameric form (15, 21).
The importance of this enzyme in innate immunity (15), autoimmunity (17), and control of the transition from G1 to S phase in the cell cycle (5) makes an understanding of its regulatory and catalytic mechanism of interest. In addition, such mechanistic work can also provide useful insights to guide the design of mechanism-based activators and inhibitors of SAMHD1 that could have research or therapeutic uses. In this study, we have characterized the coordinated effects of activator and substrate binding on oligomerization and activation of the enzyme and discovered a long-lived activated state of SAMHD1 that is not in equilibrium with free activator nucleotides. We propose that GTP combined with any dNTP serve as the preferred activators for SAMHD1 in the cell and that the long-lived activated state is key for efficient dNTP depletion.
Results
Activation of dUTP Hydrolysis by GTP.
The complex dGTP/GTP-dependent activation mechanism for dNTP hydrolysis and the possibility that SAMHD1 might exist in multiple active oligomeric states present a challenging system for designing kinetic experiments that reveal useful insights into function and mechanism. Because activation by dGTP is complicated by the fact that it is also a substrate, we began by investigating activation by GTP, which does not compete for substrate binding to the catalytic site.
The activation of dUTP hydrolysis by GTP was studied using 3H-labeled dUTP (Fig. 1 A and B). dUTP was selected for these studies, because it is present at high concentrations in resting immune cells that express SAMHD1 and it reacts similarly as all other dNTPs (Fig. S1). Variable concentrations of dUTP in the presence of five fixed GTP concentrations (0.01–5 mM) were incubated with SAMHD1 (0.5 μM monomer). The concentration dependence of the velocities showed no evidence of sigmoidicity, and accordingly, the data were globally fit to Eq. 1 (Fig. 1C). The 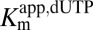 values decreased as the GTP activator was increased, giving a limiting
values decreased as the GTP activator was increased, giving a limiting  at saturating GTP activator with
at saturating GTP activator with 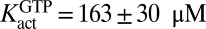 and
and  . Normalization of
. Normalization of 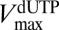 to monomer concentration is justified, because plots of velocity against SAMHD1 monomer concentration were linear in the range of 0.2–10 μM when the GTP activator concentration was ≥50 μM. (Fig. S2).
to monomer concentration is justified, because plots of velocity against SAMHD1 monomer concentration were linear in the range of 0.2–10 μM when the GTP activator concentration was ≥50 μM. (Fig. S2).
Fig. 1.
Activation of dUTP hydrolysis by GTP. SAMHD1 (0.5 μM) was incubated with increasing concentrations of 3H-dUTP in the presence of five GTP activator concentrations (0.01–5 mM). (A) Time courses for dUTP hydrolysis using the C18-RP TLC plate assay. (B) Linear initial rates for formation of the 3H-dU product. (C) Initial velocities were found to follow a hyperbolic dependence on dUTP concentration with no evidence of sigmoidicity. Curves are from global nonlinear least squares best fit to all of the data using Eq. 1. (D) Secondary plot of  against 1/[GTP]. (E) GTP concentration dependence of the dUTP hydrolysis rates at increasing [dUTP]. (F) Secondary plot of
against 1/[GTP]. (E) GTP concentration dependence of the dUTP hydrolysis rates at increasing [dUTP]. (F) Secondary plot of 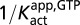 against [dUTP].
against [dUTP].
A secondary plot of 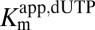 against 1/[GTP] activator concentration clearly shows that the catalytic site has undetectable affinity for dUTP in the absence of GTP (Fig. 1D) and that GTP acts to lower the
against 1/[GTP] activator concentration clearly shows that the catalytic site has undetectable affinity for dUTP in the absence of GTP (Fig. 1D) and that GTP acts to lower the 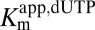 without changing
without changing 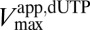 (Fig. S3A). Therefore, GTP is a Km-type essential activator, and high dUTP substrate concentrations always pull the equilibrium to the active enzyme-activator-substrate (EAS) complex, even at low activator concentrations. The cellular implication of this mechanism is that, as long as the activator concentration exceeds that of the enzyme, dNTP binding can, in principle, pull the system all of the way to the activated EAS state.
(Fig. S3A). Therefore, GTP is a Km-type essential activator, and high dUTP substrate concentrations always pull the equilibrium to the active enzyme-activator-substrate (EAS) complex, even at low activator concentrations. The cellular implication of this mechanism is that, as long as the activator concentration exceeds that of the enzyme, dNTP binding can, in principle, pull the system all of the way to the activated EAS state.
To confirm the ordered essential activation mechanism, we also performed a kinetic analysis, where dUTP hydrolysis was measured at fixed dUTP concentrations (0.2–5 mM) and the GTP activator concentration was varied in the range of 0.01–5 mM (Fig. 1E). As expected, the secondary replot of  against [dUTP] was linear, with the y intercept providing the true
against [dUTP] was linear, with the y intercept providing the true 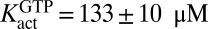 (Fig. 1F). The important implication from this result is that GTP binds to activator site(s) of SAMHD1 monomers or dimers in the absence of substrate. The secondary plot
(Fig. 1F). The important implication from this result is that GTP binds to activator site(s) of SAMHD1 monomers or dimers in the absence of substrate. The secondary plot 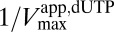 against 1/[dUTP] also showed the expected linear response from an ordered essential activation mechanism (Fig. S3B), and order-of-addition experiments showed that these kinetic properties of SAMHD1 are invariant (Fig. S4). We note that oligomerization is enzyme concentration-dependent, and therefore, the Kact parameters for activators of SAMHD1 are apparent values that hold for the dilute concentrations of monomer used in these experiments.
against 1/[dUTP] also showed the expected linear response from an ordered essential activation mechanism (Fig. S3B), and order-of-addition experiments showed that these kinetic properties of SAMHD1 are invariant (Fig. S4). We note that oligomerization is enzyme concentration-dependent, and therefore, the Kact parameters for activators of SAMHD1 are apparent values that hold for the dilute concentrations of monomer used in these experiments.
To summarize, SAMHD1 shows classical ordered binding of activator and substrate. Despite the complex nature of SAMHD1 activation, these data indicate that binding of the dUTP substrate to the four catalytic sites as well as GTP activator binding to the guanine-specific A1 activator sites occur independently without any detectable cooperativity. Accordingly, in terms of kinetic models, it is sufficient to consider only a phenomenological EAS complex. We defer to Discussion the likely mechanistic basis for the apparent absence of cooperativity in this reaction. In addition, these data in isolation do not provide information as to whether the A2 activator site is occupied by GTP or alternatively, dUTP.
Activation and Inhibition by dGTP and 2′-Deoxyguanosine-5′-[α-thio] Triphosphate Lithium Salt.
Unlike GTP, dGTP can activate its own hydrolysis and is known to serve as a transactivator for hydrolysis of other substrate dNTPs at low concentrations (17). However, it should also act as an inhibitor of dNTP hydrolysis at higher concentrations (20, 22). To explore these properties of dGTP, we determined the concentration dependence of the initial rates of dGTP hydrolysis at an SAMHD1 concentration of 0.2 μM and variable concentrations of dGTP in the range of 5 μM to 5 mM (Fig. 2A, black circles). Although a self-activating substrate is expected to show a sigmoidal concentration dependence to the initial velocities (23), these data did not require the use of the Hill equation (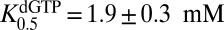 and
and 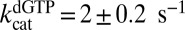 ). We discuss later how the unusual activation mechanism of SAMHD1 can mask expected cooperativity (Discussion).
). We discuss later how the unusual activation mechanism of SAMHD1 can mask expected cooperativity (Discussion).
Fig. 2.
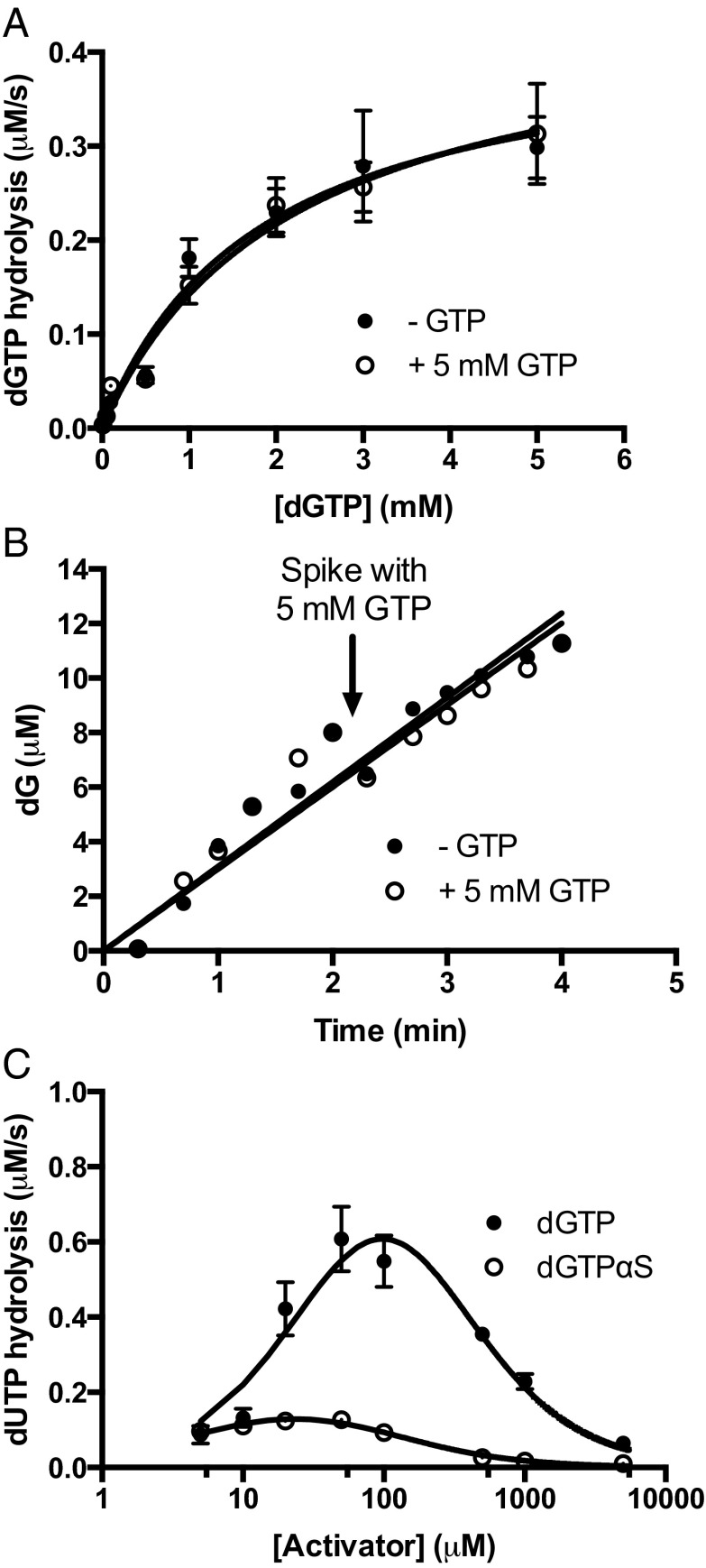
Self-activation, transactivation, and inhibition by dGTP. (A) dGTP self-activation of dGTP hydrolysis and transactivation of dGTP hydrolysis by 5 mM GTP. The datasets were normalized to total active sites and are indistinguishable. (B) Addition of 5 mM GTP during steady-state hydrolysis of 25 μM dGTP by 0.5 μM SAMHD1 provides no additional activation. (C) Activation and inhibition of dUTP hydrolysis by dGTP and dGTPαS. Curves were generated using the simulation program Dynafit to obtain the activation and inhibition constants for dGTP (expressions 5, 6, 7, and 8).
We found that GTP was unable to stimulate the hydrolysis of dGTP beyond the self-activation provided by dGTP alone. Addition of a saturating 5 mM fixed concentration of GTP to reactions where dGTP was the variable substrate resulted in a simple hyperbolic velocity profile that was indistinguishable from dGTP alone (Fig. 2A, white circles). The steady-state kinetic parameters for dGTP hydrolysis derived from these data are very similar to the parameters observed above in the GTP activation of the dUTP reaction and the dGTP self-activation data (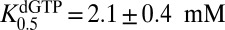 and
and  ). Additionally, we discovered that the steady-state turnover of SAMHD1 with a low concentration of dGTP (25 μM) could not be stimulated by spiking with 5 mM GTP activator (Fig. 2B). This intriguing finding suggested that GTP cannot access its A1 activator site during steady-state turnover of the activated enzyme form. The basis for this intriguing property is explored in the experiments below.
). Additionally, we discovered that the steady-state turnover of SAMHD1 with a low concentration of dGTP (25 μM) could not be stimulated by spiking with 5 mM GTP activator (Fig. 2B). This intriguing finding suggested that GTP cannot access its A1 activator site during steady-state turnover of the activated enzyme form. The basis for this intriguing property is explored in the experiments below.
We further explored whether dGTP could activate the hydrolysis of dUTP at low concentrations and then inhibit the reaction of dUTP by competitive binding to the catalytic site as the concentration of dGTP was increased. These outcomes were born out in steady-state rate measurements (Fig. 2C), where the dUTP concentration was held constant at 1 mM and dGTP was varied in the concentration range from 5 μM to 5 mM. These data were simulated by fixing the 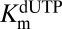 at the value obtained with the activator GTP, while allowing
at the value obtained with the activator GTP, while allowing 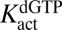 ,
, 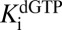 , and
, and 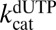 to float (expressions 5, 6, 7, and 8). If
to float (expressions 5, 6, 7, and 8). If 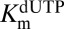 was allowed to float, similar results were obtained, but the fitting errors increased. The constants obtained for dGTP were
was allowed to float, similar results were obtained, but the fitting errors increased. The constants obtained for dGTP were 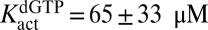 ,
, 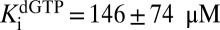 , and
, and 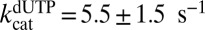 . Thus, GTP and dGTP have similar activation constants for dUTP hydrolysis, but dGTP results in less activation, because it is also a strong competitive inhibitor
. Thus, GTP and dGTP have similar activation constants for dUTP hydrolysis, but dGTP results in less activation, because it is also a strong competitive inhibitor  .
.
The crystal structure of SAMHD1 bound to 2′-deoxyguanosine-5′-[α-thio] triphosphate lithium salt (dGTPαS) shows that the R stereoisomer of this analog occupies all activator sites and catalytic sites of the tetramer, suggesting that it should serve as a stable dGTP analog for mechanistic studies (20). We found that racemic dGTPαS was a very slow substrate in the absence and presence of transactivation by GTP (t1/2 = 24 h) (Fig. S5), which is consistent with the crystal structure showing that dGTPαS can bind to the A1 and A2 activator sites as well as the catalytic sites (20). Supporting the suggestion that dGTPαS can self-activate, it also served as a weak transactivator for dUTP hydrolysis at low concentrations (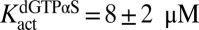 and
and 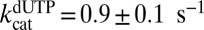 ) and an inhibitor at higher concentrations
) and an inhibitor at higher concentrations 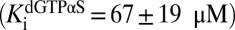 (Fig. 2C). These properties of dGTPαS, which may underestimate its potency by a factor of two because of the use of a racemic mixture, make it useful for comparing the discrete effect of a nonsubstrate activator (GTP) with that of activators that are known to bind to all activator and substrate sites of SAMHD1 (dGTP and dGTPαS) (20, 22).
(Fig. 2C). These properties of dGTPαS, which may underestimate its potency by a factor of two because of the use of a racemic mixture, make it useful for comparing the discrete effect of a nonsubstrate activator (GTP) with that of activators that are known to bind to all activator and substrate sites of SAMHD1 (dGTP and dGTPαS) (20, 22).
Long-Lived Activated States of SAMHD1.
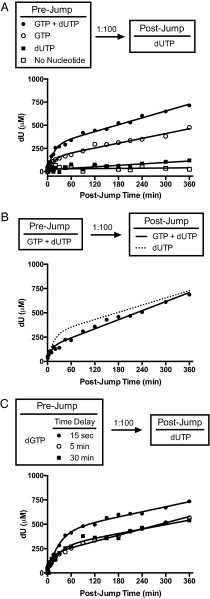
The observation that a high concentration of GTP was not able to further stimulate SAMHD1 dGTPase activity at low dGTP concentrations (Fig. 2B) required us to consider that the activated form of the enzyme was not in equilibrium with free activator during turnover. To test this hypothesis, we performed a series of dilution-jump experiments, where a high concentration of SAMHD1 (10 μM) was preincubated with high concentrations of GTP activator and/or dUTP substrate and then rapidly diluted 100-fold into a solution that contained millimolar concentrations of the dUTP substrate but no activator (Fig. 3A). This experiment probes the kinetic stability of activated enzyme form(s) that are generated in the prejump conditions.
Fig. 3.
Dilution-jump kinetic experiments for dUTP hydrolysis. High concentrations of SAMHD1 and various nucleotide combinations were incubated as indicated in pre- and postjump reactions. (A) SAMHD1 was incubated with GTP (0.5 mM) and dUTP (5 mM), GTP alone (0.5 mM), dUTP alone (5 mM), or no nucleotide before dilution. (B) Inclusion of 0.5 mM GTP in the postjump reaction did not enhance the postjump kinetics. For comparison, the dashed line shows the curve from the dUTP-only postjump condition in A. (C) Time dependence of prejump reactions containing only dGTP (5 mM).
When GTP and dUTP were present in the prejump solution, a rapid initial burst rate of dUTP hydrolysis was observed in the postjump reaction (vi = 0.28 ± 0.02 μM/s) that was sixfold greater than a standard steady-state reaction of fully activated SAMHD1 under equivalent conditions (5 mM GTP, 1 mM dUTP, 100 nM SAMHD1 monomer) (Fig. 3A). This burst phase corresponds to over 2,500 turnovers of enzyme active sites. We attribute the postjump burst–decay rate constant to the slow disappearance of the highly active form of SAMHD1 generated in the prejump solution (kinact = 0.0011 ± 0.0001 s−1; t1/2 = 10 ± 1 min). At the conclusion of the burst–decay period, a second lower-activity form persisted and produced a linear rate of dUTP hydrolysis for at least 6 h (vss = 0.02 ± 0.002 μM/s). This rate is comparable with the calculated steady-state rate of fully activated SAMHD1 under these conditions, indicating that this long-lived form is the steady-state active form of SAMHD1. Thus, using prejump conditions containing both dUTP substrate and GTP activator, two distinct forms of SAMHD1 can be detected in the postjump reaction. These forms differ with respect to their half-lives for decay (∼10 min and greater than 6 h) as well as their catalytic activities.
The postjump results were very dependent on the composition of the prejump solution. When GTP alone was present in the prejump, the same two kinetic phases were observed as with the combination [GTP + dUTP]. However, when the prejump contained only SAMHD1 or enzyme and dUTP alone, the burst was abolished, and exceedingly slow linear rates were observed in the postjump reaction with dUTP (Fig. 3A, black and white squares). With only GTP in the prejump, the postjump burst amplitude was reduced by 50%, but the burst–decay rate constant and steady-state rate (normalized for active enzyme) were essentially unchanged (kinact = 0.0010 ± 0.0003 s−1, vss = 0.02 ± 0.001 μM/s). Consistent with the surprising finding that GTP could not stimulate steady-state turnover using low concentrations of dGTP (Fig. 2B), addition of GTP to the postjump solution containing 1 mM dUTP did not enhance the burst properties or steady-state linear rate obtained from a prejump solution that contained both GTP and dUTP (Fig. 3B). This finding establishes that the GTP-activated form of SAMHD1 behaves like the dGTP-activated enzyme and is not in communication with free activators during steady-state turnover.
When the substrate/self-activator dGTP was present in the prejump, the postjump behavior was identical to the combination [dUTP + GTP] (Fig. 3C). We could establish a requirement for dGTP binding to the A2 or catalytic sites by inserting a prejump delay before dilution, thereby consuming any free dGTP that was not protected by being bound to the enzyme. As the prejump delay was increased, the postjump burst amplitude decreased until it reached a plateau level of 50% of maximal. This result is identical to that observed above when only GTP activator was present in the prejump (Fig. 3A). These results are consistent with a model where GTP and dGTP behave identically when interacting with the guanine-specific A1 site, but binding of dGTP or dUTP to the A2 and catalytic sites is required to achieve full activation.
We conclude from the above dilution-jump results that activator and substrate must be present in the prejump solution to fully generate the two active enzyme species that are detected in the postjump phase. Maximal activation is provided by (i) the combination GTP (A1 site only) and dUTP (A2 and catalytic sites) or (ii) dGTP alone (all sites). Lesser activation is observed with GTP alone, which can only occupy the A1 site. Additional evidence for this interpretation is found below and in Discussion. Importantly, after activation occurs, SAMHD1 is no longer in equilibrium with free activator while it performs steady-state turnover. We report additional salient kinetic properties of the dilution-jump reactions in Fig. S6.
(d)NTP Induced Changes in Oligomeric State of SAMHD1.
We hypothesized that the slowly decaying active species present in the dilution-jump experiments might reflect dissociation of the active tetramer or alternatively, dissociation of one or more activating nucleotides. To elucidate the oligomeric species that were present during the different phases of the postjump period, we performed an analogous procedure, except that the samples taken from the postjump reactions at various times were mixed with 50 mM glutaraldehyde (Fig. 4). The standard dilution-jump cross-linking experiment involves preincubation of SAMHD1 (10 μM) with various combinations of GTP activator (0.5 mM), dUTP (1 mM), dGTP (1 mM), and dGTPαS (5 mM) for 10 s before rapid 100-fold dilution into a postjump solution containing 1 mM dUTP substrate.
Fig. 4.
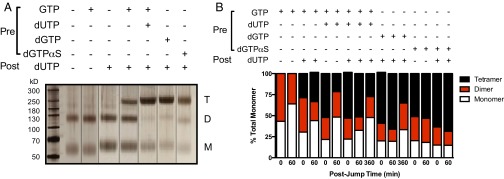
Dilution-jump cross-linking experiments for elucidating the oligomeric states of SAMHD1. (A) Denaturing polyacrylamide gel separation of the cross-linked species. SAMHD1 (10 μM) was incubated under the indicated prejump conditions before dilution into postjump solutions, and immediate chemical cross-linking with 50 mM glutaraldehyde was performed. Prejump (GTP = 0.5 mM; dUTP, dGTP, and dGTPαS = 5 mM) and postjump (dUTP = 1 mM) nucleotide concentrations were evaluated for their effect on oligomerization. Visualization was by silver staining, and monomer, dimer, and tetramer bands were imaged and quantified using Quantity-One. The figure is a composite of multiple gels with the order of lanes designed to clarify presentation of the data. The positions of monomer (M), dimer (D), and tetramer (T) are indicated. (B) Time dependence of dimer and tetramer stabilities under various pre- and postjump conditions. Reactions were performed as in A, and glutaraldehyde was added at the indicated times. The concentrations of pre- and postjump nucleotides were the same as stated in A.
Samples subjected to this procedure and cross-linked immediately after dilution are shown in Fig. 4A after separation of monomer, dimer, and tetramer using denaturing PAGE and visualization with silver staining. The salient findings from these data are (i) SAMHD1 in the absence of guanine nucleotides exists as an inactive monomer–dimer equilibrium, even in the presence of 1 mM dUTP in the postjump, (ii) highly efficient tetramer formation requires dGTP alone, [GTP + dUTP], or dGTPαS in the prejump solution, and (iii) GTP alone in the prejump induces about 25% of the amount of tetramer as the combined presence of [GTP + dUTP] as long as dUTP is present in the postjump. These effects of nucleotides on the formation of tetramer exactly parallel their effects on activity in the initial times of the postjump kinetic assay.
We performed a series of cross-linking studies as a function of postjump time to show that the kinetic phases in the kinetic dilution-jump experiments were not correlated with dissociation of the tetrameric state of SAMHD1. The relative amounts of each oligomeric species at each postjump time were quantified by imaging the silver-stained gels as summarized in Fig. 4B. In all conditions where tetramer was formed in the prejump step (dGTP, GTP, dGTPαS, and [GTP + dUTP]), it was found to persist intact during the entire postjump burst–decay time (60 min). Thus, the initial burst–decay period cannot be attributed to dissociation of the tetramer. Moreover, tetramer was still present in significant amounts 6 h after the dilution jump when dUTP + GTP or dGTP was present in the prejump (Fig. 4B). Thus, the observed changes in activity must result from other more subtle alterations in the enzyme structure that are not detected by the cross-linking method.
We were curious if the highly active form of SAMHD1 present in the activator-free postjump solution retained tightly bound activator nucleotides. To address this question, we performed reactions similar to the prejump kinetics, where SAMHD1 was mixed with [γ-32P]GTP and [5-3H]dUTP and then subjected to a spin column gel filtration chromatography step to separate free and bound (d)NTPs (Fig. S7A). To evaluate the time course for release of the bound nucleotides that were retained after the first column step, a second spin column step was performed on the flow-through fraction from the first column. The simultaneous use of [γ-32P]GTP and [5-3H]dUTP and dual-channel scintillation counting allowed the precise ratio of both nucleotides to be determined. Furthermore, determination of the amount of tetramer present after each column step allowed for estimation of the stoichiometry of GTP and dUTP binding to each monomer subunit of the tetramer (Materials and Methods and Fig. S7B). After the first column, an average of 1.3 and 0.87 equivalents of [5-3H]dUTP and [γ-32P]GTP was bound per monomer of the tetramer form, consistent with the A1 and A2 sites being occupied by GTP and dUTP, respectively. Because the catalytic site has a high Km, we assume that dUTP is rapidly released from this site during the spin column step, leaving only the A2 site occupied. The time course for release of the bound GTP and dUTP was largely complete in the 2-min delay time before running the second spin column, from which we estimate a half-life for activator release of less than 2 min (Fig. S7B). We conclude that GTP and dUTP activators are bound in 1:1 stoichiometry with each monomer subunit of the tetramer and that their release greatly precedes that of tetramer dissociation.
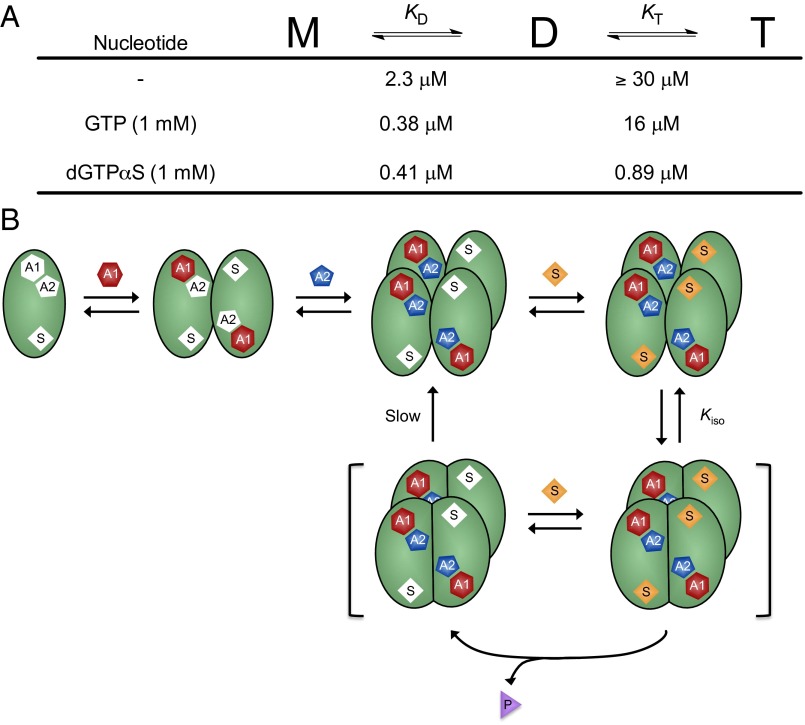
We used sedimentation velocity ultracentrifugation to complete the characterization of the linkage between the oligomeric equilibria of SAMHD1 and binding of the nucleotides GTP and dGTPαS. The raw scan data were converted to a c(S) distribution and then fitted to a monomer–dimer–tetramer model as described in Materials and Methods. SAMHD1 in the concentration range of 2–8 μM total monomer largely existed in a monomer–dimer equilibrium in the absence of nucleotides (Fig. 5A, red and Fig. S8). The monomer form dominated at the lower concentrations, but at 8 μM total monomer, the monomer–dimer equilibrium slightly favored dimer [KD = 2.3 μM, 95% confidence interval (95% CI) = 2.2–2.4]. Even at the highest monomer concentration tested, less than 4% of total monomer was in the tetramer form in the absence of nucleotide (KT ≥ 30 μM). Addition of 1 mM GTP pushed the monomer–dimer equilibrium significantly to dimer (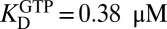 , 95% CI = 0.33–0.42) but induced a much smaller increase in tetramer (
, 95% CI = 0.33–0.42) but induced a much smaller increase in tetramer (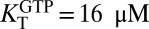 , 95% CI = 15–17) (Fig. 5A). In contrast, the predominant effect of 1 mM dGTPαS was to shift the dimer–tetramer equilibrium to favor tetramer (
, 95% CI = 15–17) (Fig. 5A). In contrast, the predominant effect of 1 mM dGTPαS was to shift the dimer–tetramer equilibrium to favor tetramer (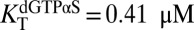 , 95% CI = 0.38–0.43;
, 95% CI = 0.38–0.43; 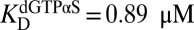 , 95% CI = 0.77–1.0) (Fig. 5B). These effects of GTP and dGTPαS on the oligomeric equilibria recapitulate their effects on the dilution-jump kinetics and cross-linking and support the model where GTP induces dimerization but occupation of the A2 and substrate sites is required for tetramerization.
, 95% CI = 0.77–1.0) (Fig. 5B). These effects of GTP and dGTPαS on the oligomeric equilibria recapitulate their effects on the dilution-jump kinetics and cross-linking and support the model where GTP induces dimerization but occupation of the A2 and substrate sites is required for tetramerization.
Fig. 5.
Sedimentation velocity experiments to evaluate the effects of GTP and dGTPαS on the oligomeric state of SAMHD1. (A) SAMHD1 alone (8 μM), in the presence of 1 mM GTP or (B) SAMHD1 alone (8 μM), and in the presence of 1 mM dGTPαS were dialyzed and analyzed by sedimentation velocity centrifugation. Raw scan data were fitted to a c(S) distribution as described in Materials and Methods.
Discussion
Nucleotide-Dependent Ordered Assembly of SAMHD1.
These diverse measurements allow for the construction of a minimal model for nucleotide-dependent activation of SAMHD1 (Fig. 6). In the absence of nucleotides, SAMHD1 is trapped in an equilibrium between monomer and dimer forms and is unable to generate active tetramer (Fig. 6A). Binding of GTP to the A1 site (but not the A2 or catalytic sites; see below) strongly shifts the equilibrium to dimer, with little effect on the tetramerization equilibrium (Fig. 6A). The structural basis for the increased dimer affinity apparently results from docking of the guanine base of GTP into the highly specific guanine binding site on one monomer and the formation of electrostatic interactions between its triphosphate group and the second monomer of the dimer (Fig. S9 A and B) (22). Hence, GTP serves as a nucleotide tether to bring together monomers. Because GTP concentrations exceed that of dGTP by about 1,000-fold in the cell (3) and there is no large difference in the activation constants for these nucleotides, the implication of these findings is that GTP occupies the guanine-specific A1 site under physiological conditions (not dGTP). An additional advantage of using GTP as the primary activator is that GTP levels are not depleted by SAMHD1, allowing the activated tetramer to persist (see below).
Fig. 6.
Nucleotide-dependent oligomeric equilibria of SAMHD1 and mechanism of long-lived activation. (A) Summary of the effects of GTP and dGTPαS on the oligomeric equilibria. The values for KD and KT are for the indicated concentration of nucleotide. (B) Model for ordered essential activation and oligomerization by GTP activators and dNTP coactivator/substrates involving the formation of a long-lived activated tetramer. The E(A1A2)4S4 complex is depicted as isomerizing (Kiso) to a long-lived tetramer (brackets) that performs steady-state turnover. This form only slowly reconverts to the loosely associated tetramer form that is in equilibrium with monomer, dimer, and free nucleotides. The model also indicates that the activator sites of the long-lived tetramer do not communicate with free (d)GTPs during steady-state turnover of substrate dNTPs. Although not depicted, the actively cycling tetramer does not require the presence of bound activator nucleotides (in the text).
Based on the limited ability of GTP alone to induce tetramerization and the results from structural modeling (Fig. S9C), we propose that the 2′-hydroxyl group of GTP sterically prevents binding of GTP to both the A1 and A2 activator sites. Thus, as suggested above, GTP is restricted to the A1 activator site, and the A2 site must be occupied by a dNTP coactivator to efficiently produce tetramer (i.e., dUTP in the studies here). The contention that any dNTP can bind to the A2 site is supported by a crystal structure of SAMHD1 with dGTP bound to the specific A1 site and dATP bound to the A2 site (22), structural modeling showing that the A2 site nicely accommodates dUTP (Fig. S9 D and E), binding of one equivalent of dUTP to each monomer only in the presence of GTP (Fig. S7B), and dilution-jump cross-linking showing that, when dUTP is added in addition to the A1 site activator GTP, tetramer is efficiently formed (Fig. 4).
To understand the combined effect of activator and substrate on oligomerization, we used the relatively inert dGTPαS analog (20). Binding of dGTPαS shifted the equilibrium dramatically to tetramer compared with GTP alone, an effect that resulted from the same increase in dimer affinity observed with GTP and additional enhancement of the tetramerization constant (Figs. 5 and 6). This result is consistent with the crystal structure showing that dGTPαS can occupy the A1, A2, and catalytic sites and that binding of dGTPαS to the A2 site forms bridging interactions within the tetramer (dimer–dimer) interface involving its base and triphosphate groups (Fig. S10A) (20).
Ordered Pathway for Activation.
The combined activation and oligomerization data using GTP alone, [GTP + dUTP], dGTP, and dGTPαS define an ordered pathway for nucleotide activation of SAMHD1. The first event is occupation of the A1 site by GTP, which promotes dimerization, followed by occupancy of the A2 site by a coactivator dNTP, which is thermodynamically coupled with binding of substrate dNTP to the catalytic site. The combined effect of dNTP binding to the A2 and catalytic sites pulls the entire set of equilibria to active tetramer. This effect is the structural and temporal basis for the ordered essential activation mechanism. The dilution-jump and direct binding measurements of GTP and dUTP indicate that, after activating nucleotides induce tetramer formation, it persists without additional requirement for bound activator nucleotides. This result leads to the unanticipated conclusion that activating nucleotides serve as small molecule chaperones during tetramer formation.
Quite surprisingly, none of the experimental data for activation and catalysis revealed any evidence for cooperativity. This nonintuitive and perplexing finding was brought into focus using kinetic simulations that included the step of forming a long-lived activated state, such as that depicted in Fig. 6B. These simulations revealed that cooperativity would only be apparent in the presteady-state regime when such a mechanism operates. Because initial velocities are assumed to be instantaneous, the hysteresis in the system would not be observed, and simple steady-state initial velocity equations would still hold.
Long-Lived SAMHD1.
An ordered essential activation mechanism involving GTP activation and a long-lived tetramer has several important implications for the cellular activity of SAMHD1. Because all dNTPs are substrates for SAMHD1, the driving force for formation of the active Michaelis complex would be provided by the concentration of the entire dNTP pool. The observation that an active tetrameric form of SAMHD1 is stable for over 6 h in the absence of activating nucleotides is relevant to the condition where dNTP pools have been largely depleted by SAMHD1. If SAMHD1 immediately dissociated to inactive dimer and monomer states under low dNTP conditions and if dGTP were the sole A1 site activator, the pool levels would not readily reach the nanomolar regime observed in resting macrophages or CD4+ T cells. Such low dNTP pool levels have been implicated in the HIV-1 restriction activity of SAMHD1 in these cell types, because reverse transcriptase is much less efficient when dNTP substrates are scarce (24, 25).
An enigmatic feature of SAMHD1 activity in resting immune cells is the presence of elevated dUTP levels in an otherwise depleted dNTP pool background (26, 27). Given our findings that dUTP is an excellent substrate, there must be undiscovered regulatory mechanisms that allow dUTP levels to persist or accumulate after the other dNTPs have been depleted. Because dUTP incorporation into HIV-1 cDNA has been implicated as a related antiviral defense mechanism (28, 29), additional understanding of the mechanisms that regulate nucleotide pools during the transition from active to resting states of cells will likely be relevant to viral infectivity and persistence in these cell types.
Materials and Methods
General Reagents.
dNTPs were obtained from Roche, [5-3H]dUTP and [8-3H]dGTP were from Moravek Biochemicals, dGTPαS was from ChemCyte, and C18-reversed phase TLC plates were purchased from Macherey-Nagel. Guanosine-5′-triphosphate, 2′-deoxyguanosine (dG), and tripolyphosphate were from Sigma-Aldrich.
SAMHD1 Enzyme Cloning and Overexpression.
SAMHD1 full-length (1,881 bp) construct was PCR-amplified using the forward primer (5′-GTAACTCATATGCAGCGAGCCGATTCC-3′ and the reverse primer 5′-GCACCTGGATCCCTACATTGGGTCATCTTT-3′) from the cloned cDNA vector from the Invitrogen Ultimate ORF collection (clone IOH55544). The SAMHD1 gene was ligated into a pET19b-His10-PreScission protease site (PPS; Novagen) plasmid (Nde1 and BamH1), and the sequence was confirmed by sequencing both DNA strands. The full-length construct was expressed as an N-terminal His10-PPS fusion. Escherichia coli BL21-DE3 cells (Novagen) were transformed with the pET19b-His10-PPS plasmid and grown in LB medium at 37 °C. After an OD600 = 0.5 was achieved, the temperature was reduced to 16 °C on ice. Expression was induced by addition of 0.25 mM isopropylthio-β-galactoside. After expression for 24 h at 16 °C, cells were harvested by centrifugation (4,000 × g) and frozen at −80 °C. Cells were resuspended in lysis buffer containing 50 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10% (vol/vol) glycerol, and one protease inhibitor mixture tablet (Sigma). Cells were lysed by the addition of 0.5 mg/mL hen egg white lysozyme. The resulting cell lysate was clarified by centrifugation (30,000 × g), filtered with a 0.2-μm syringe top filter, and loaded onto 10 mL Ni-nitrilotriacetic acid resin (Qiagen) that had been equilibrated in buffer A [50 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% (vol/vol) glycerol] at 4 °C. Unbound material was removed with 3 column volumes buffer A supplemented with 50 mM imidazole, and bound SAMHD1 protein was eluted using a linear gradient of buffer A containing 0–100% (vol/vol) 500 mM imidazole. Fractions containing SAMHD1 were pooled and incubated with PPS for 1 h at 4 °C to remove N-terminal His10 tag. Crude protein was diluted 10-fold into buffer A and loaded onto an SP-Sepharose cation-exchange column (GE Healthcare). Bound protein was eluted with a 0–100% (vol/vol) linear gradient of buffer A containing 500 mM NaCl. Fractions containing protein were analyzed by SDS/PAGE and judged to be ∼99% pure. Purified proteins were buffer-exchanged into storage buffer [50 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% (vol/vol) glycerol] by chromatography using sephacryl 200HR gel filtration resin (Sigma-Aldrich). Protein concentrations were determined by absorbance measurements at 280 nm using the calculated (Protparam tool; ExPASy) molar extinction coefficients for full-length human SAMHD1 monomer (ε = 76,500 M−1 cm−1). Protein yields were typically 20 mg/L bacterial culture. Purified proteins were stored at −80 °C in small portions. Experiments were initiated by thawing a single (20 μL) aliquot, stored at −20 °C, and used over the course of 3 d before being discarded.
Steady-State Kinetic Measurements.
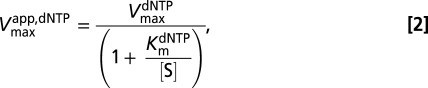
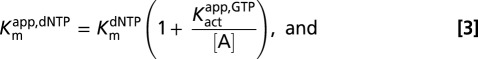
Standard reaction conditions for steady-state kinetic measurements were 0.01–5 mM GTP (activator), 50 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, and 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride in a 12 μL total reaction volume at 22 °C. Concentrations of the [5-3H]dUTP or [8-3H]dGTP substrates were varied in the range of 0.01–5 mM, and standard reactions were initiated by the addition of SAMHD1. Two-microliter samples were removed at indicated times and quenched by spotting onto a C18-reversed phase TLC plate. The TLC plate was developed in 50 mM KH2PO4 (pH 4.0) to separate substrate [dGTP (Rf = 0.80) or dUTP (Rf = 0.97)] from products [dG (Rf = 0.20) or dU (Rf = 0.54)]. Plates were exposed to a tritium-sensitive screen for 5 h and then scanned on a Typhoon phosphoimager (GE Healthcare), and the counts present in the substrate and product were quantified using the program Quantity One (Bio-Rad). The amount of product formed at each time point was calculated from the ratio (cpm dN product)/(cpm dNTP substrate + cpm dN product)(initial [dNTP]). Initial rates of product formation were obtained from the slopes of linear plots of [dN] vs. time at reaction extents of less than 20%. The reaction rates (micromolar per second) were plotted vs. dNTP substrate concentration at various concentrations of GTP activator as described. For GTP activation with dUTP as the substrate, the kinetic parameters were determined by fitting to an ordered essential activation mechanism (Eq. 1), where [S] is dUTP, [A] is GTP, Kmapp,dUTP is the apparent Michaelis constant at a given concentration of GTP activator, Kactapp,GTP is the activation constant for GTP at a given [S], and Vmaxapp,dUTP is the apparent maximal velocity for dUTP hydrolysis at a given activator concentration:
 |
 |
 |
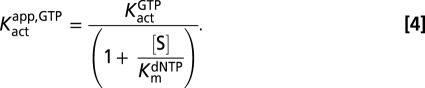 |
Mechanism of Activation and Inhibition by dGTP.
dGTP is both a cis-activator and a substrate, and sigmoidal plots of velocity vs. [dGTP] would be expected. However, sigmoidicity was not observed, and the steady-state velocity data in the presence and absence of the transactivator GTP were fitted to a simple hyperbolic expression (Eq. 1). To investigate the activation and inhibition of dUTP hydrolysis by dGTP, we used a fixed concentration of the dUTP substrate (1 mM) and varying concentrations of dGTP in the range from 5 μM to 5 mM. These data were fitted to the mechanism in expressions 5, 6, 7, and 8 using numerical integration methods and nonlinear least squares optimization to the data (30):
dGTPαS: Inhibition, Activation, and Reactivity.
Inhibition by dGTPαS was evaluated under reaction conditions containing SAMHD1 (0.5 μM) and fixed dUTP (1 mM) substrate while varying the concentrations of dGTPαS (5 μM to 5 mM). Because dGTP behaves both as a cis-activator and a substrate, we presumed that dGTPαS would behave similarly. Therefore, the data were fitted by simulation to the mechanism in expressions 5, 6, 7, and 8 to obtain  and
and  .
.
Dilution-Jump Kinetic Measurements.
In the standard assay, SAMHD1 (10 μM) was incubated for 10 s with varying concentrations of GTP, dUTP, and/or dGTP before a 100-fold dilution into standard reaction buffer containing [5-3H]dUTP (1 mM). Time points were then quenched on a TLC plate, and product formation was quantified as described above. Plots of [dU] against time were fitted to the equation [dU] = A[1 − exp(−kinactt)] + νt, where A is the burst amplitude (micromolar), kinact is the rate constant for the burst decay (seconds−1), and ν is the linear steady-state rate (micromolar per second). Variations of the standard assay were performed as indicated above.
Dilution-Jump Glutaraldehyde Cross-Linking.
The standard dilution-jump procedure was performed, except that samples from the diluted reactions were taken at various times and mixed with 50 mM glutaraldehyde. Samples were incubated at 22 °C for 15 min, quenched by the addition of 1 M Tris (pH 7.5), and loaded on to a 4–12% Bis⋅Tris denaturing polyacrylamide gel. Protein samples were visualized by silver staining. Briefly, gels were fixed overnight in a 50 mL solution containing 50% (vol/vol) methanol, 10% (vol/vol) acetic acid, and 0.02% paraformaldehyde. Gels were washed three times for 20 min in 20% (vol/vol) ethanol followed by a 5-min incubation in sensitizing solution containing 0.02% sodium thiosulfate. Sensitizing solution was removed by washing two times with 50 mL ddH2O and immediately transferred to an equivalent volume of staining solution containing 0.2% silver nitrate and 0.03% paraformaldehyde. Finally, gels were developed for 2–5 min in a 6% (wt/vol) sodium carbonate and 0.02% paraformaldehyde (50 mL) solution. Developer was quenched by direct addition of an equivalent volume of 50% (vol/vol) methanol and 10% (vol/vol) acetic acid. Gels were imaged immediately using an Epson V750 flat-bed scanner. Quantitation of monomer, dimer, and tetramer populations was performed using Quantity-One (Bio-Rad) software.
Binding Stoichiometry and Kinetic Release of Activator Nucleotides.
The binding of nucleotides was measured by mixing SAMHD1 (10 μM) with [γ-32P]GTP (0.5 mM) and [5-3H]dUTP (1 mM) in standard reaction buffer. This solution was immediately applied to a buffer-equilibrated Micro Bio-Spin P-30 gel filtration spin column (Bio-Rad), and the protein was eluted according to the manufacturer’s instructions. After 2 min, the eluted protein was filtered through a second Micro Bio-Spin P-30 column. After each spin step, the protein eluate was cross-linked with glutaraldehyde to quantify the percentage of tetrameric SAMHD1 as described above. The remaining solution present after the first and second columns (50–60 μL) was added to 10 mL scintillation fluid, and 32P and 3H decays were counted using appropriate channel windows on a Beckman LS-6500 scintillation counter. Control reactions without enzyme were run to determine the background contribution from unretained nucleotides (<1%), and the recovery of SAMHD1 from the column was 96%. After correcting for the background and the protein recovery, the stoichiometry of bound dUTP and GTP per monomer subunit of tetrameric SAMHD1 was calculated from the specific activity of the labeled nucleotide solutions and the fraction tetramer determined from imaging the band intensities in the cross-linked protein gel (Fig. S7B).
Analytical Ultracentrifugation.
Sedimentation velocity experiments were performed on a Beckman XL-I analytical ultracentrifuge equipped with an An-Ti 60 rotor and interference optics. All runs were performed at 50,000 rpm and 20 °C, with scans taken every 30 s for 9 h. SAMHD1 protein samples were dialyzed overnight at 22 °C against a buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 5 mM MgCl2, 0.5 mM DTT, and 5% (vol/vol) glycerol. After loading the cells, the rotor was allowed to thermally equilibrate for 3 h before the start of the run. SEDNTERP was used to determine partial specific volume, density, and viscosity of the protein and buffer (31). These parameters were then used in SEDFIT to fit the scan data to a c(S) distribution (32). To determine the dissociation constants for the monomer–dimer (KD) and dimer–tetramer equilibria (KT), scans at different concentrations were globally fit using SEDANAL by assigning an arbitrary slow dissociation rate constant of 10−5 s−1. The 95% CIs were determined from the integrated F-statistics module in SEDANAL (33).
Computational Modeling.
Structural models of SAMHD1 bound to dUTP/GTP and GTP/GTP were made by modification of the crystal structure containing tetrameric SAMHD1 bound to dGTP in both activator sites (Protein Data Bank ID code 4MZ7). The nucleobase and sugar moieties were built using Discovery Studio (Accelrys) and then positioned into the allosteric pockets in the same planes and configurations as the original ligands. No additional refinement of the dUTP structure was required. However, the GTP/GTP ribose sugars showed steric conflict and were subjected to 1,000 cycles of steepest descent energy minimization using the CHARMM27 molecular dynamics force field within HyperChem7 (HyperCube, Inc.) in an unsuccessful attempt to alleviate the steric clash. Visualization of the resultant structures was done using PyMOL.
Supplementary Material
Acknowledgments
We thank Dr. Heng Zhu for the cloning vector containing the human SAMHD1 gene. We thank Dr. Katherine Tripp and the Center for Molecular Biophysics at The Johns Hopkins University for assistance with the sedimentation velocity experiments. This work was supported by National Institutes of Health Grants T32GM080189, T32GM008763, T32GM008403, and GM056834 (to J.T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.F.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401706111/-/DCSupplemental.
References
- 1.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O’Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci USA. 2013;110(32):12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992;267(6):3589–3596. [PubMed] [Google Scholar]
- 3.Gavegnano C, Kennedy EM, Kim B, Schinazi RF. The impact of macrophage nucleotide pools on HIV-1 reverse transcription, viral replication, and the development of novel antiviral agents. Mol Biol Int. 2012;2012(5270):625983. doi: 10.1155/2012/625983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franzolin E, et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci USA. 2013;110(35):14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 7.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41(2):462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 8.Engström Y, et al. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260(16):9114–9116. [PubMed] [Google Scholar]
- 9.Rampazzo C, et al. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat Res. 2010;703(1):2–10. doi: 10.1016/j.mrgentox.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Cribier A, Descours B, Valadão ALC, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3(4):1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol Lett. 2000;74(3):221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 12.Vorontsov II, et al. Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J Biol Chem. 2011;286(38):33158–33166. doi: 10.1074/jbc.M111.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornberg SR, Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J Biol Chem. 1958;233(1):159–162. [PubMed] [Google Scholar]
- 14.Kohn G, et al. High inorganic triphosphatase activities in bacteria and mammalian cells: Identification of the enzymes involved. PLoS ONE. 2012;7(9):e43879. doi: 10.1371/journal.pone.0043879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 16.Kim ET, White TE, Brandariz-Núñez A, Diaz-Griffero F, Weitzman MD. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J Virol. 2013;87(23):12949–12956. doi: 10.1128/JVI.02291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286(51):43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldauf H-M, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18(11):1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amie SM, Bambara RA, Kim B. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J Biol Chem. 2013;288(35):25001–25006. doi: 10.1074/jbc.C113.493619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji X, et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol. 2013;20(11):1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem. 2013;288(15):10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu C, et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 23.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley; 1993. pp. 227–272. [Google Scholar]
- 24.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279(49):51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien WA, et al. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68(2):1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy EM, et al. Abundant non-canonical dUTP found in primary human macrophages drives its frequent incorporation by HIV-1 reverse transcriptase. J Biol Chem. 2011;286(28):25047–25055. doi: 10.1074/jbc.M111.234047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aquaro S, et al. Macrophages and HIV infection: Therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 2002;55(2):209–225. doi: 10.1016/s0166-3542(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 28.Weil AF, et al. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc Natl Acad Sci USA. 2013;110(6):E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priet S, Sire J, Quérat G. Uracils as a cellular weapon against viruses and mechanisms of viral escape. Curr HIV Res. 2006;4(1):31–42. doi: 10.2174/157016206775197673. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237(2):260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 31.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54(5):328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 32.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: Curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108(1-3):231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 33.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton JC, editors. Cambridge, United Kingdom: Royal Society of Chemistry; 1991. pp. 90–125. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.