Significance
Riboswitches are a broadly distributed means of regulation of gene expression in bacteria that solely rely on RNA. Seven distinct families of riboswitches bind S-adenosylmethionine (SAM) as their effector, regulating genes involved in sulfur metabolism across a broad spectrum of bacterial species. Further, SAM riboswitches regulate expression of genes essential for survival and/or virulence in medically important pathogens, suggesting they might be important targets for the development of new antimicrobial agents. Our studies reveal the atomic-resolution structure of a unique peripheral architecture that supports a SAM-binding core shared among three families that make up the “SAM clan” and how this subdomain facilitates both ligand binding and gene regulation.
Keywords: RNA structure, X-ray crystallography, chemical probing, isothermal titration calorimetry, gene regulation
Abstract
In bacteria, sulfur metabolism is regulated in part by seven known families of riboswitches that bind S-adenosyl-l-methionine (SAM). Direct binding of SAM to these mRNA regulatory elements governs a downstream secondary structural switch that communicates with the transcriptional and/or translational expression machinery. The most widely distributed SAM-binding riboswitches belong to the SAM clan, comprising three families that share a common SAM-binding core but differ radically in their peripheral architecture. Although the structure of the SAM-I member of this clan has been extensively studied, how the alternative peripheral architecture of the other families supports the common SAM-binding core remains unknown. We have therefore solved the X-ray structure of a member of the SAM-I/IV family containing the alternative “PK-2” subdomain shared with the SAM-IV family. This structure reveals that this subdomain forms extensive interactions with the helix housing the SAM-binding pocket, including a highly unusual mode of helix packing in which two helices pack in a perpendicular fashion. Biochemical and genetic analysis of this RNA reveals that SAM binding induces many of these interactions, including stabilization of a pseudoknot that is part of the regulatory switch. Despite strong structural similarity between the cores of SAM-I and SAM-I/IV members, a phylogenetic analysis of sequences does not indicate that they derive from a common ancestor.
Riboswitches are noncoding RNA elements generally found in the leader of bacterial mRNAs that regulate expression via direct binding of a specific cellular metabolite (reviewed in refs. 1 and 2). To date, at least 25 different families of riboswitches have been identified and validated, binding a diverse set of effectors, including nucleobases and nucleosides, amino acids, protein cofactors, metal ions, and second messengers (1). Effector binding promotes formation of a downstream regulatory structure that generally directs transcription and/or translation of the message. As the repertoire of known riboswitches continues to expand, relationships among the families are emerging. One of the best-characterized examples is the purine family of riboswitches, which bind three distinct effector molecules: guanine, adenine, and 2′-deoxyguanosine (3). These RNAs are highly similar at all structural levels, with only two nucleotides in the binding pocket required to alter binding selectivity, indicating that these three distinct subfamilies likely diverged from a common ancestor (4, 5). Conversely, the two known families of riboswitches that bind cyclic diguanylate or pre-Q1 are structurally distinct and recognize the effector in different fashions, pointing to independent evolutionary origins (3).
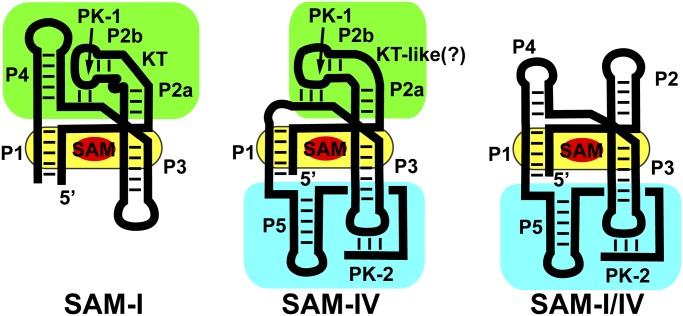
Intermediate between these two extreme cases are the three families of riboswitches making up the “SAM clan” [SAM-I (RF00162), SAM-IV (RF00642), and SAM-I/IV (RF01725)], whose members share a common binding core but have widely divergent peripheral architectures (6–9). Structures of the SAM-I family revealed that S-adenosylmethionine is recognized by features within and surrounding a central four-way junction (yellow, Fig. 1) (10, 11). Nucleotides crucial for effector binding, along with the secondary structural features in which they are embedded, are nearly invariant within the clan (7). However, the three families of the SAM clan significantly differ in the peripheral architectural features surrounding the ligand-binding core (7). Within the SAM-I family, the central ligand-binding core is organized by the “PK-1” peripheral subdomain, defined by a pseudoknot (PK-1) between L2 and J3/4 (green, Fig. 1) (10, 11). PK-1 is facilitated by an essential kink-turn module in P2 that redirects the terminal loop back toward the core. These peripheral tertiary interactions serve to preorganize the core for SAM recognition (12). In the SAM-IV family, this subdomain differs by a non-kink-turn motif in P2 that presumably introduces a similar bend in the helix, along with the absence of the P4 helix (7). Because loss of P4 destabilizes the core (13), a second peripheral subdomain is observed, called “PK-2,” comprising a new hairpin following P1 (P5) and a 3′-tail that base pairs with L3 to form a second pseudoknot (PK-2; Fig. 1). Before this work, the structure of the PK-2 subdomain and its relationship to the SAM-binding core was, to the authors' knowledge, unknown. The SAM-I/IV family lacks the PK-1 subdomain, only having the PK-2 subdomain of the SAM-IV family (cyan, Fig. 1) (8).
Fig. 1.
Cartoon of the secondary structure of the three families of the SAM clan of riboswitches. The phylogenetically conserved SAM-binding core shared by all members of the clan is highlighted in yellow, and the two types of peripheral subdomains, the P4/PK-1/P2 (called the PK-1 subdomain) and P5/PK-2 (called the PK-2 subdomain), are highlighted in green and cyan, respectively.
The two distinct peripheral subdomains of the SAM clan have radically different relationships to the regulatory secondary structural switch controlling expression of the mRNA. In the SAM-I family, the first (P1) helix of the aptamer domain competes with an alternative secondary structure in the expression platform (14). For SAM-dependent transcriptional termination by these riboswitches, an alternative antiterminator helix can form at the expense of P1, enabling RNA polymerase to synthesize the entire transcript. The PK-1 subdomain in these RNAs plays an accessory role by supporting high-affinity SAM binding but does not play a direct role in the regulatory switch. In the SAM-IV and SAM-I/IV families, PK-2 is proposed not only to support high-affinity SAM binding but also to act as an integral part of the alternative structural switch that instructs the expression machinery (7). Thus, PK-2 plays a direct role in both ligand binding and regulation.
Conservation of a central core containing the key activity with variable peripheral domains playing supporting roles is observed in other biologically important RNAs. For example, although all group I introns share a conserved catalytic core composed of three domains (P4–P6, P3–P9, and P1–P2), they vary considerably in peripheral subdomains defining 13 structural subgroups (15). The peripheral P5abc subdomain of the Tetrahymena thermophila group I intron, although not essential for catalytic function, forms a series of tertiary interactions with elements of the core that serve to stabilize the RNA’s fold. Similarly, RNase P and ribosomal RNAs exhibit diverse nonessential peripheral architecture that support of a common core containing the catalytic active site (16, 17). The limited size and scope of alternative peripheral elements of the SAM clan make this RNA ideal for understanding how peripheral architecture is used to augment core function.
We present the structure of the aptamer domain of a member of the SAM-I/IV family that contains the alternative PK-2 subdomain to reveal how this alternative peripheral element facilitates both effector recognition and the regulatory switch. This structure reveals that the PK-2 subdomain forms extensive interactions with P3, along with the predicted pseudoknot with L3. Ligand-dependent chemical probing analysis reveals that SAM binding significantly stabilizes these interactions, including PK-2, which is part of the regulatory switch. Further, we show that the function of the switch in vivo is dependent on the strength of PK-2. The PK-2 subdomain is positioned on the opposite side of the SAM-binding core from the PK-1 subdomain. Although these two complete domains are never found together in biological RNAs, “hybrid” aptamers containing both domains are capable of binding SAM, demonstrating that the full PK-1 and PK-2 are not mutually exclusive.
Results
RNA Crystallization and Structure Determination.
To obtain crystals of a SAM clan member containing the PK-2 subdomain, several of the smallest member sequences of the SAM-I/IV family were screened against commercially available sparse matrices. To promote lattice contacts, nonconserved terminal loops (L2, L4, and L5) of each variant were converted to GAGA tetraloops (sequences and secondary structures of all RNAs used in this study are presented in Table S1 and Fig. S1). Of these RNAs, the env87 variant from a Pacific Ocean metagenome (accession number ABEF01012528.1) found in the 5′-leader of an mRNA encoding homoserine acetyltransferase (COG2021) crystallized in a number of conditions. Further variation in the lengths of P2, P4, and P5, along with mutagenesis or deletion of nonconserved nucleotides within the sequence, yielded crystals suitable for structural analysis. In particular, deletion of a single residue in J5/PK-2, U92, yielded crystals that diffracted X-rays to 3.2 Å resolution [this RNA is referred to as env87(∆U92); Fig. S1C]. The ∆U92 mutation was crucial for obtaining diffraction-quality crystals; addition of this nucleotide back to the RNA and extensive rescreening yielded crystals that diffracted X-rays to no greater than 6 Å resolution. The quality of the resulting electron density maps from diffraction data were sufficient to unambiguously observe features of the SAM-binding core and novel PK-2 subdomain (Fig. S2 A–C).
To validate that the crystallized RNA retains all the necessary features for high-affinity ligand recognition, SAM binding to a series of RNAs was measured using isothermal titration calorimetry (ITC). Ligand binding was tested under physiological monovalent cation concentrations (135 and 15 mM NaCl) and 10 mM magnesium chloride, a divalent cation concentration that promotes RNA folding in vitro to a similar extent as observed in vivo for the purine riboswitch aptamer domain (18). Wild-type and a minimized aptamer containing truncations in P2 and P4 [env87(minimal)] display ∼100 nM affinity for SAM (Table 1, Table S2, and Fig. S3) comparable to that observed for some SAM-I variant riboswitches (9, 19). The single-point deletion on the 3′-side of PK-2 (∆U92) results in a ∼fourfold reduction in SAM binding affinity, presumably because of the destabilizing effect on this peripheral element. Further degradation of PK-2 by introducing the point deletions ∆U92,G93 and ∆U92-U94 (Table 1) further reduces affinity by 100–1000-fold, clearly revealing the essential role of the PK-2 subdomain. Analysis of base pairing in PK-2 of the SAM-I/IV family indicates that although full pairing and a single unpaired nucleotide on the 3′-side of L3 are prevalent in natural sequences (63.9% and 22.4% of total sequences, respectively), two or more unpaired nucleotides on the 3′-side of L3 (equivalent to ∆U92,G93 and ∆U92-U94) is less tolerated. Thus, the ∆U92 mutation is representative of a significant fraction of the SAM-I/IV family. Finally, this deletion does not substantially affect in vivo activity.
Table 1.
Affinities of SAM for wild-type and mutant env87 SAM-I/IV aptamers
| RNA* | KD, µM | N |
| env87 (wild-type) | 0.091 ± 0.021 | 1.1 ± 0.1 |
| env87 (minimal) | 0.14 ± 0.02 | 0.85 ± 0.03 |
| env87 (∆U92) | 0.41 ± 0.05 | 1.1 ± 0.1 |
| env87 (∆U92 G93) | 7.3 ± 0.4 | 1.0 ± 0.1 |
| env87 (∆U92, G93, U94) | 100 ± 10 | 0.72 ± 0.04 |
| hybrid 1 | 0.27 ± 0.03 | 0.81 ± 0.02 |
| hybrid 2 | 0.08 ± 0.01 | 0.84 ± 0.03 |
| env87 (∆P4) | 5.3 ± 0.5 | 1.10 ± 0.02 |
Binding buffer was 10 mM Na-Hepes at pH 8.0, 135 mM KCl, 15 mM NaCl, and 10 mM MgCl2.
Structure of the SAM-I/IV Aptamer in Complex with SAM.
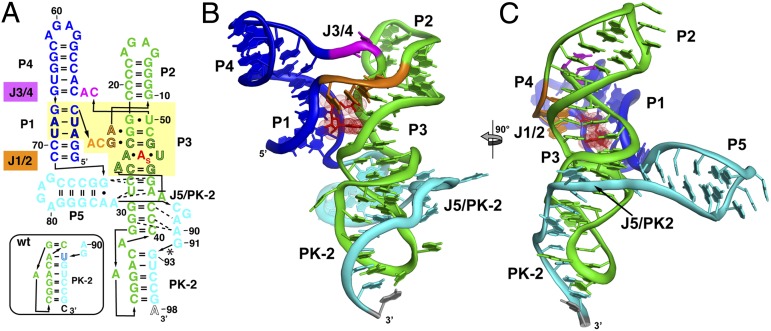
The architecture of the SAM-I/IV [env87(∆U92)] aptamer domain is similar to that of the SAM-I aptamers that have been previously determined (10, 11). The core of the aptamer is a four-way junction flanked by helices P1–P4 (Fig. 2 A–C) that organize into two sets of coaxial stacks, P1/P4 and P2/P3. These two coaxial stacks are tied together by the J1/2 and J3/4 joining regions. The SAM-binding core of the env87 aptamer superimposes nearly perfectly with that of the Thermoanearobacter tengcongensis (Tte) metF SAM-I aptamer (11) (rmsd, 0.50 Å; Fig. 3). In both structures, SAM is bound by a nearly universally conserved set of nucleotides in the SAM clan. The only exception is the A3-U69 base pair (env87 numbering; Fig. 2A), which is conserved in SAM-I and SAM-I/IV but is a G-C pair in SAM-IV. Mutation of this A-U pair to G-C in the TteSAM-I and Bacillus subtilis yitJ SAM-I aptamer domains results in a moderate loss in affinity (10, 20).
Fig. 2.
Crystal structure of the env8 SAM-I/IV riboswitch aptamer. (A) Sequence of the crystallized RNA [env8[(∆U92); site of deletion denoted by asterisk] drawn to reflect the tertiary architecture of the RNA. The yellow box represents sequence (black outlined letters) and/or structural elements that are nearly universally conserved in the SAM clan, with the site of binding of the adenosyl moiety of SAM represented as “A”s. Coloring of the RNA is used to highlight the P1/P4 coaxial stack (blue), the P2/P3 coaxial stack (green), the joining regions between the stacks (orange and magenta), and the PK-2 subdomain (cyan). The box represents the most probable secondary structure of PK-2 for the wild-type sequence that was used for modeling. (B) Cartoon representation of the global architecture of the RNA, using the same coloring scheme as in A. (C) 90° clockwise rotation perspective of the structure.
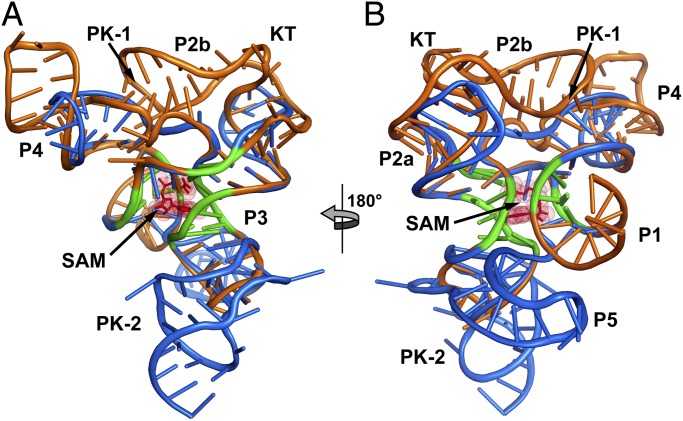
Fig. 3.
Superimposition of the TteSAM-I and env87SAM-I/IV aptamer domains. (A) The TteSAM-I riboswitch structure (Protein Data Bank accession code 2GIS) is shown in orange, and the env87SAM-I/IV structure in blue. The conserved core between the two RNAs is emphasized in green. This core represents the bases used to align the structures. (B) 180° clockwise rotation of the aligned structures.
Despite the identical SAM-binding cores (within the coordinate error of the known structures), the crystal structure of env87(∆U92) reveals a completely novel peripheral architecture surrounding the core in the SAM-I/IV aptamer. All known members of the SAM-I/IV family have no secondary structural features associated with the PK-1 subdomain (8), which is reflected in the tertiary architecture of the env87(∆U92) aptamer. Similar to SAM-I, P2 coaxially stacks on P3 but contains no internal loop motif present in the SAM-I and SAM-IV families that facilitates formation of PK-1 (Fig. 1). Thus, its terminal loop projects away from the core and makes no contacts to either J3/4 or J4/1. A second significant distinction in the SAM-I/IV family is the lack of J4/1. In SAM-I, and likely SAM-IV, there are two or three unpaired purine nucleotides that disrupt direct stacking between P1 and P4 and help mediate interactions in the PK-1 subdomain. In the SAM-I/IV aptamer, there are no non-Watson–Crick paired nucleotides between P1 and P4, allowing the two helices to directly stack on one another. Finally, the joining region J3/4 is severely truncated with respect to that found in the other two families, consisting of only two unpaired nucleotides. The two nucleotides (C51 and A52) reside in approximately the same spatial location as the first two adenosines of J3/4 in TteSAM-I that are used to form triple interactions with the minor groove of P2 but do not make any contacts with other regions of the RNA. Thus, the SAM-I/IV family is entirely devoid of the tertiary architectural features essential for organization of the SAM binding pocket in the SAM-I and SAM-I/IV families.
In SAM-I/IV, the PK-1 subdomain is replaced by the P5 stem-loop and a 3′-strand that forms a pseudoknot with L3 (PK-2), which together forms the PK-2 subdomain. These elements are placed on the opposite face of the core as the PK-1 subdomain in the SAM-I family (Fig. 3). P5, which is conserved in both the SAM-IV and SAM-I/IV families, is oriented perpendicular to the P2-P3 coaxial stack, with its 5′-side interacting with the minor groove of P3 (Fig. 2 B and C). Packing of P5 against P3 is mediated by three universally conserved nucleotides in the SAM-IV and SAM-I/IV families: G72, A85, and A86 (Fig. S2D). G72 and A85 form a purine–purine pair involving their Watson–Crick faces, whereas A86 remains unpaired and stacked directly below A85. This is the first known example, to the authors’ knowledge, of a highly conserved module that promotes a perpendicular helix–groove packing interaction but is similar to the HLout pseudoknot motif observed in the preQ1-II riboswitch (21, 22). The joining region J5/PK-2 makes further interactions with the minor groove of P3, primarily through a type I A-minor triple (23, 24) interaction between A90 and the G31-C41 base pair and a type II A-minor triple interaction between A89 and the G30-C42 base pair (Fig. S2E). These types of interactions are common in H-type pseudoknots, in which the strand equivalent to J5/PK-2 always crosses the minor groove of one of the helices (25).
The extreme 3′-terminal sequence forms five consecutive Watson–Crick base pairs with nucleotides in L3 to form PK-2 (Fig. 2). In this arrangement, the first and last nucleotides of L3 (A33 and A39) are unpaired and expelled from the helix. A39 is predicted from covariance models of both the SAM-IV and SAM-I/IV families to form a base pair with U92 that is deleted, which would enable P3 and PK-2 to coaxially stack (7, 8). Although this deletion likely slightly locally disrupts the PK-2 subdomain by disallowing perfect coaxial stacking of P3 and PK-2, it should be reiterated that the structure reflects a significant portion of the population of the SAM-I/IV riboswitch that has a similar unpaired nucleotide on the 3′-side of L3 (Table S3).
SAM Binding Stabilizes the PK-2 Subdomain Required for Regulatory Activity.
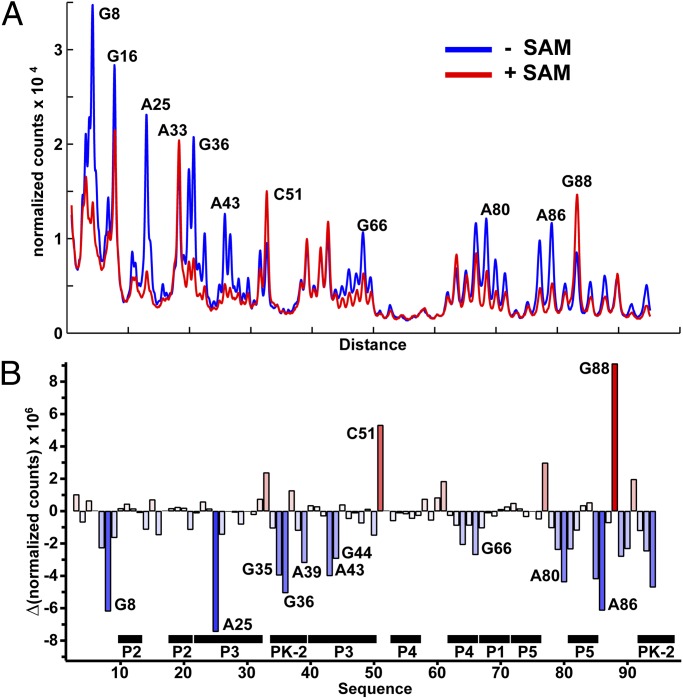
To assess the influence of SAM binding on the structure of the env87 SAM-I/IV aptamer domain, selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemical probing (26) was performed in the absence and presence of 1 mM SAM. SHAPE probing employs N-methylisotoic anhydride that generally reacts with 2′-hydroxyl groups of ribose sugars in conformationally dynamic regions of the backbone or where the hydroxyl group is locked into a conformation favoring in-line attack with the neighboring phosphodiester linkage (27). For each nucleotide, normalized reactivity in the presence of 1 mM SAM was subtracted from the normalized reactivity in the absence of SAM to yield a reactivity difference plot (Fig. 4 A and B and Fig. S4A). Importantly, chemical probing is consistent with the architecture of the env87(∆U92) crystal structure. Nucleotides directly interacting with SAM (G8 and A25) show the strongest degrees of protection, consistent with this experiment measuring SAM-dependent structural changes or stabilization. The majority of observed protections are distributed in and around the PK-2 subdomain. A set of strong protections are observed within the pseudoknot itself (G35, G36, A39, and C94), suggesting that this element, proposed to be part of the regulatory switch (7), is being stabilized by the aptamer domain on SAM binding. Another very strong SAM-dependent protection is observed at A86, reflecting the association of P5 with the minor groove of P3. A series of protections at the site of P3-J5/PK-2 (e.g., A44, A89, and A90) further indicates that SAM binding promotes the extensive interactions between P5 and J5/PK-2 along P3 between the adenine binding pocket and L3. Conversely, C51 and G88 show enhanced reactivity on binding, correlating well with the crystal structure, where these residues show high B-factors and are solvent-exposed in the structure.
Fig. 4.
“SHAPE” probing of the env87SAM-I/IV RNA. (A) Normalized chromatogram of chemical probing of the env87(minimal) construct showing the RNA probed with N-methylisotoic anhydride in the presence and absence of SAM at 50 °C. Nucleotides showing significant changes in their reactivity are highlighted (raw gel in Fig. S4A). (B) Quantification of the reactivity differences observed in the gel shown in A. Bars above zero correspond to reactivity enhancements, whereas those below zero correspond to SAM-dependent protections of the RNA.
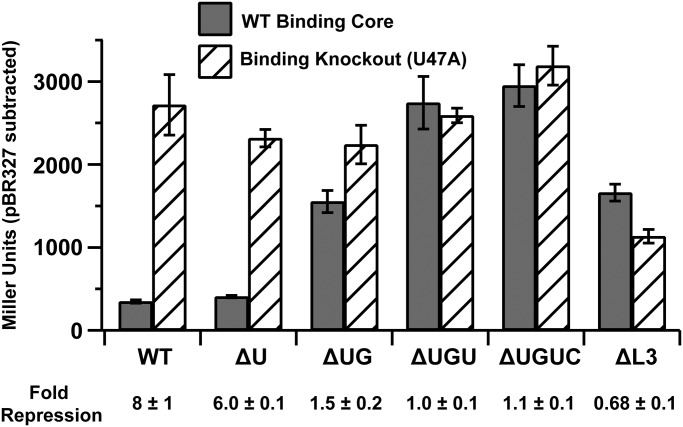
To correlate PK-2 formation with regulatory activity, a series of env87 SAM-I/IV riboswitches with deletions at the 3′-end that disrupt PK-2 were tested for their ability to repress expression of a lacZ reporter gene in Escherichia coli BW25113 [parental cell strain of the Keio knockout collection (28)]. β-galactosidase activity of log-phase cells was quantified using the Miller assay (29). For each mutant, a corresponding SAM-binding knockout was made in P3 (U47A). The wild-type riboswitch showed eightfold repression relative to the binding knockout (Fig. 5). Derepression of methionine and SAM biosynthesis in a metJ knockout strain (JW3909; derivative of BW25113) (30, 31) resulted in higher levels of repression, further indicating that the magnitude of repression correlates with intracellular SAM (Fig. S4B). Systematic weakening of PK-2 by deletion of nucleotides at the 3′-end of the aptamer that pair with L3 reduces the repression of lacZ. Loss of a single base pair at the interface between P3 and PK-2 (∆U92) has a small effect on repression (Fig. 5 and Fig. S4B), reflecting its ability to bind SAM with nearly wild-type affinity by ITC and its prevalence in phylogeny. Ablating two base pairs (∆U92,G93) resulted in weak regulatory activity, and further deletions resulted in complete loss of activity. Furthermore, conversion of L3 into a stable UUCG tetraloop that would be predicted to completely disrupt the L3-3′ tail interaction shows no regulatory activity. These trends are reflected in the metJ knockout cells, indicating that elevated SAM does not rescue the loss of PK-2.
Fig. 5.
In vivo lacZ reporter assay. Gray boxes represent the wild-type binding core of SAM-I/IV, and hashed boxes represent the U47A binding knockout. PK-2 destabilizing mutants are denoted as ∆U (∆U92), ∆UG (∆U92,G93), ∆UGU (∆U92-U94), and ∆UGUC (∆U92-C95). ∆L3 is a mutant that changes the sequence of L3 to a UUCG tetraloop. Fold repression is shown below the graph. All errors are the SD of three individual biological replicates.
The Peripheral Subdomains Are Not Mutually Exclusive.
Superimposition of the SAM-I and SAM-I/IV aptamers suggests that the PK-1 (P2/PK-1/P4) and PK-2 (P5/PK-2) subdomains are mutually compatible (Fig. 3 A and B). However, in no natural sequence of any member of the SAM clan is there an RNA containing the full two subdomains. In the SAM-IV family, the PK-1 domain is significantly reduced by deletion of P4. No member of this family has this helix, suggesting the full PK-1 subdomain is incompatible with the PK-2 domain. Conversely, loss of P4 is tolerated by both SAM-I and SAM-I/IV. Within the SAM-I/IV family, only 31% of the members [144/470 sequences (8)] contain P4, with the rest having a longer J3/4 linker directly connecting the 3′-side of P3 with the 3′-strand of P1. Within the SAM-I family, loss of P4 is rarer [6% of total sequences (13)]. Deletion of this element in the B. subtilis metI riboswitch results in a reduction of both ligand-binding affinity and regulatory activity, reflecting its importance for organization of the PK-1 subdomain (13).
To examine the role of P4 in the residual PK-1 subdomain of the SAM-I/IV family, we examined the affinity of SAM for two variants of env87 that contain a four- and five-nucleotide J3/4 linker. An RNA in which P4 was replaced with the phylogenetically observed four-nucleotide linker 5′-GUAG yielded no detectable binding of SAM. However, replacement of P4 with a five-nucleotide linker (5′-AAAUA) could bind, albeit with a ∼460-fold reduction in affinity [Table 1; env87(∆P4)]. Given that P4 is frequently lost in the SAM-I/IV family, unknown sequence elements of these RNAs may serve to reduce the loss in SAM affinity.
Conversely, the full PK-1 and PK-2 subdomains might not be compatible, as no known natural sequence contains both. To test the ability of both to function in concert, we created a hybrid SAM aptamer with the P2, J3/4, and P4 sequences from the TteSAM-I RNA and the remainder from env87SAM-I/IV (Fig. S1D). This RNA has an affinity for SAM nearly equivalent with the wild-type SAM-I/IV RNA, revealing that the two subdomains are compatible. One hypothesis why the existence of these two domains is never found in concert is that it would overstabilize the aptamer, preventing switching, and result in constitutively repressed gene expression.
Evolutionary Relationship of the Three Families in the SAM Clan.
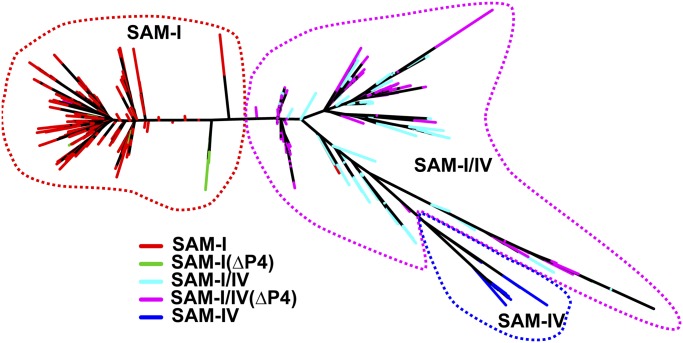
This structural and biochemical analysis of the SAM-I/IV reveals a clear structural relationship with SAM-I and further reinforces their assignment, along with SAM-IV, to the SAM clan in Rfam (32). Since the discovery of the SAM-I/IV family, there has not been a phylogenetic analysis of the clan to discern potential evolutionary relationship between the three families. To address this gap, we produced a phylogenetic tree of the three families, using a manually curated set of RNAs (removal of gap-rich and hypervariable regions) derived from the seed alignments of Rfam 11.0 (Fig. 6; full alignment in Dataset S1). This tree shows relationships among members of the SAM riboswitch clan. Although the root of the tree cannot be confidently located without an outgroup, it reveals several important features of the evolution of the SAM clan. First, it is apparent that SAM-IV (RF00634) evolved from SAM-I/IV (RF01725). A branch of the tree containing all members of the SAM-IV family is clearly embedded in the part of the tree comprising the SAM-I/IV family. Second, loss of the P4 helix evolved convergently and multiple times in both the SAM-I/IV and SAM-I families. Further, the relationship between SAM-I/IV and SAM-IV suggests that the loss of P4 may have been a preadaptation for the evolution of SAM-IV.
Fig. 6.
SAM clan phylogenetic tree. The three families are denoted by red (SAM-I), magenta (SAM-I/IV), and blue (SAM-IV) dashed lines. Colors are only given to terminal branches on the tree and distinguish sequences from each family, as well as differentiate with sequences that have lost the P4 helix in the SAM-I and SAM-I/IV families (green and cyan, respectively).
Discussion
In this study, we explored the structural and phylogenetic relationship of members of the SAM clan of riboswitches through the investigation of the SAM-I/IV family. Although extensive structural, biochemical, and genetic data have been accrued about the SAM-I family (33), the other two member families of the clan are almost completely unknown. The crystal structure of the env87 SAM-I/IV RNA has revealed the organization of the PK-2 subdomain and its relationship to the SAM-binding pocket. These data validate the hypothesis that the ligand-binding core of all members of the SAM clan are essentially identical but use differing combinations of peripheral subdomains to enhance ligand-binding affinity and/or communicate with the downstream regulatory switch (7). Notably, these two subdomains are found at opposite sides of the SAM-binding core and, from a ligand-binding perspective, are mutually compatible.
The structural and chemical probing data of the env87 aptamer strongly support a model of SAM-dependent regulation by the SAM-IV and SAM-I/IV members of the SAM clan. In the absence of ligand, PK-2 forms through Watson–Crick base pairing between nucleotides in L3 and the 3′-end of the aptamer domain, but this interaction is weak. Downstream sequences can readily form alternative hairpin structure with the 3′-end of the aptamer domain that disrupt PK-2, corresponding to the “ON” state of this riboswitch. On SAM binding, PK-2 is significantly stabilized via the formation of a supporting network of base-mediated interactions between the universally conserved G/AA motif at the base of P5 and J5/PK-2 with an extended region of P3 that extends from the SAM binding site to the PK-2. These interactions make PK-2 resistant to disruption by alternative structure formation and promotes the “OFF” state of the riboswitch.
Regulatory pseudoknots are a common theme among riboswitches. The most prevalent form is a simple H-type pseudoknot that encompasses both the aptamer domain and the expression platform. For example, the 3′-single-stranded tail SAM-II riboswitch contains the ribosome-binding site that becomes occluded on ligand binding (33). The SAM-V, preQ1-I, preQ1-II, and fluoride riboswitches also use this architecture (3, 33). The prevalence for the architectural theme is that the H-type pseudoknot may be the most parsimonious solution to creating a small-molecule responsive riboregulatory element. More rarely, the 3′-terminal pseudoknot does not fully incorporate the expression platform, such as the SAM-IV, SAM-I/IV, and the ydaO family (34). In these cases, the terminal pseudoknot participates in a classic secondary structural switch with downstream sequences, as observed in classes of riboswitches that do not contain a terminal pseudoknot. In these cases, ligand binding serves to stabilize the terminal helix (either the “P1” helix or 3′-terminal helix of the pseudoknot).
The structural data presented represent a substantial advance in our understanding of the diverse peripheral architecture of members of the SAM clan of riboswitches. The phylogenetic tree of the seed members of the three SAM clan families yields several intriguing models for how this diversity might have evolved. Because the root of the tree cannot be determined without a known outgroup sequence, several scenarios are consistent with the data. If a common ancestor is assumed, several models are possible. One model is that the ancestor is a SAM-I like RNA, from which SAM-I/IV and, subsequently, SAM-IV emerged, similar to a model proposed by Breaker (35). Our biochemical data support the possibility of a transitional RNA that contains both the PK-1 and PK-2 domains before loss of PK-1 to yield the modern SAM-I/IV variants. A second model would root the tree in the SAM-I/IV family, with SAM-I and SAM-IV independently emerging. It is clear from this tree that the PK-1 domain has evolved independently twice and that this domain in the SAM-I and SAM-IV families is not evolutionarily related, despite some secondary structural similarities. It must be emphasized that because the available data do not allow independent rooting of the tree, we cannot discriminate between the models that assume a single origin and a model in which members of this clan emerged independently. Even though the families are statistically significantly related to one another in terms of the stochastic context-free grammar models that underlie Rfam, these similarities might be a result of independent evolution to meet the same functional constraints, rather than descent from a single common ancestor. Nonetheless, the combined structural and phylogenetic analysis yields new insights into the diversification of RNA.
Materials and Methods
Detailed experimental methods used in this work are given in SI Materials and Methods, as well as crystallographic data and model refinement statistics (Table S4) and sequence alignments (Table S3 and Dataset S1).
Crystallographic Analysis.
RNA used in this study was prepared by in vitro transcription by T7 RNA polymerase, using standard methods (36). Diffraction data were collected on beamline 8.2.2 at the Advanced Light Source. An initial electron density map was calculated using molecular replacement with the conserved binding pocket (30% sequence composition) from the SAM-I riboswitch (2GIS) with PHASER (37). Iterative rounds of model building and refinement were performed in COOT (38) and PHENIX (39). Crystallographic data and model have been deposited in the Protein Data Bank under accession code 4OQU.
Isothermal Titration Calorimetry.
Equilibrium binding constants and binding stoichiometry for env87 SAM-I/IV riboswitch and variants were determined by measuring the heat released on SAM binding.
SHAPE Structure Probing.
The env87 SAM-I/IV riboswitch was probed using N-methylisotoic anhydride (26). The RNA was then reverse-transcribed, and the resulting DNA was resolved using a sequencing gel.
In vivo Reporter Assay.
The env87 SAM-I/IV riboswitch and variants were cloned upstream of the lacZ reporter gene (sequences of riboswitches used in these experiments given in Table S1). The Miller assay was used to measure lacZ expression (29), using a SAM binding knockout to mutant that does not disrupt riboswitch structure to represent expression at low SAM concentrations without having to alter the intracellular level of this essential metabolite.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant R01 GM083953 (to R.T.B.) and a Howard Hughes Medical Institute Early Career Scientist award (to R.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4OQU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312918111/-/DCSupplemental.
References
- 1.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2) doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garst AD, Edwards AL, Batey RT. Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3(6) doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45(3):345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385(3):938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci USA. 2007;104(41):16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100(6):3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg Z, et al. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA. 2008;14(5):822–828. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg Z, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11(3):R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10(9):701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 10.Lu C, et al. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J Mol Biol. 2010;404(5):803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441(7097):1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 12.Stoddard CD, et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18(7):787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heppell B, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7(6):384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 14.Grundy FJ, Henkin TM. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30(4):737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 15.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31(1):41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Mondragon A. Structural studies of RNase P. Ann Rev Biophysics. 2013;42:537–557. doi: 10.1146/annurev-biophys-083012-130406. [DOI] [PubMed] [Google Scholar]
- 17.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci. 2012;37(5):189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52(48):8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol. 2008;190(3):823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montange RK, et al. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol. 2010;396(3):761–772. doi: 10.1016/j.jmb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat Chem Biol. 2013;9(6):353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang M, Eichhorn CD, Feigon J. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1400126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8(4):339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 24.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98(9):4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3(6):e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127(12):4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 27.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc. 2012;134(15):6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Sys Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JH. A Short Course in Bacterial Genetics — A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Marincs F, Manfield IW, Stead JA, McDowall KJ, Stockley PG. Transcript analysis reveals an extended regulon and the importance of protein-protein co-operativity for the Escherichia coli methionine repressor. Biochem J. 2006;396(2):227–234. doi: 10.1042/BJ20060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5(7):1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 32.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(Database issue):D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batey RT. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip Rev RNA. 2011;2(2):299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol. 2010;192(15):3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2) doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards AL, Garst AD, Batey RT. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol Biol. 2009;535:135–163. doi: 10.1007/978-1-59745-557-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.