Significance
Cytochrome c is essential to two important biochemical pathways, the electron transport chain and the intrinsic pathway of apoptosis. The heme crevice loop, which provides the Met80 ligand to the heme cofactor, is the most highly conserved segment of the cytochrome c sequence. The dynamics of this loop are likely important for both functions. Cytochrome c-mediated peroxidation of cardiolipin in the inner mitochondrial membrane is an early signal in apoptosis. We show that mutation of trimethyllysine 72 to alanine in yeast iso-1-cytochrome c allows formation of a conformer of the protein with Met80 displaced from the heme and enhances peroxidase activity. Thus, this residue is likely an important modulator of the peroxidase function of cytochrome c.
Abstract
At the onset of apoptosis, the peroxidation of cardiolipin at the inner mitochondrial membrane by cytochrome c requires an open coordination site on the heme. We report a 1.45-Å resolution structure of yeast iso-1-cytochrome c with the Met80 heme ligand swung out of the heme crevice and replaced by a water molecule. This conformational change requires modest adjustments to the main chain of the heme crevice loop and is facilitated by a trimethyllysine 72-to-alanine mutation. This mutation also enhances the peroxidase activity of iso-1-cytochrome c. The structure shows a buried water channel capable of facilitating peroxide access to the active site and of moving protons produced during peroxidase activity to the protein surface. Alternate positions of the side chain of Arg38 appear to mediate opening and closing of the buried water channel. In addition, two buried water molecules can adopt alternate positions that change the network of hydrogen bonds in the buried water channel. Taken together, these observations suggest that low and high proton conductivity states may mediate peroxidase function. Comparison of yeast and mammalian cytochrome c sequences, in the context of the steric factors that permit opening of the heme crevice, suggests that higher organisms have evolved to inhibit peroxidase activity, providing a more stringent barrier to the onset of apoptosis.
Mitochondrial cytochrome c (Cytc) plays a pivotal role in energy storage in living organisms, providing a critical link between complex III and complex IV of the electron transport chain (1). More recently, the role of Cytc as an initiator of the intrinsic pathway of apoptosis has been elucidated (2). Release of Cytc from mitochondria into the cytoplasm is a conserved step in apoptosis from yeast up through mammals (3). However, subsequent assembly with Apaf-1 to form the apoptosome is unique to metazoan animals (4, 5). Mitochondrial cytochromes c contain a c-type heme with axial His18 and Met80 ligands (6). However, when Cytc binds to the inner mitochondrial membrane lipid cardiolipin (CL), Met80 ligation is lost (7, 8). In this state, Cytc catalyzes CL peroxidation, which provides an early signal for initiation of apoptosis (7).
Despite advances in our understanding of the role of Cytc in apoptosis, our knowledge of the structural factors that facilitate the peroxidase activity of Cytc remains rudimentary. In the structure of a domain-swapped dimer of horse Cytc, Met80 is replaced by water as a heme ligand (9), causing a fourfold increase in peroxidase activity relative to monomeric Cytc (10). However, evidence for dimerization of Cytc on CL vesicles is lacking. Fluorescence methods provide evidence for an equilibrium between compact and extended conformers on the surface of CL vesicles (11–13), with the extended conformer linked to higher peroxidase activity (13). However, other studies suggest that compact conformers of Cytc are also competent for peroxidase activity (14).
The heme crevice loop of Cytc (residues 70–85) is the most highly conserved segment of the primary structure of Cytc (15, 16). This surface loop contains the Met80 heme ligand and is likely important for both electron transfer function in electron transport and peroxidase activity in apoptosis. Our knowledge of the sequence constraints operating in the heme crevice loop that modulate the dynamics necessary for peroxidase activity remains sparse. We have shown recently that the dynamics of the heme crevice loop are enhanced when lysine 72 of yeast iso-1-cytochrome c (iso-1-Cytc) is mutated to alanine (17, 18). When synthesized in its native host (Saccharomyces cerevisiae), but not in a heterologous Escherichia coli expression system, lysine 72 of iso-1-Cytc is trimethylated (tmK72) (19). Structural studies on yeast-expressed iso-1-Cytc (hereafter tmK72Cytc[Sc]) show that tmK72 lies across the surface of the heme crevice loop (20). Here we show by high-resolution X-ray crystallography that mutation of lysine 72 to alanine produces a variant of iso-1-Cytc (hereafter K72ACytc[Sc]) that permits ejection of Met80 from the heme-binding pocket and its replacement by water. An extensive buried water channel results, a feature required for substrate access to the heme active site (14, 21) and for proton transport away from the active site during catalysis. As anticipated from the crystal structure, we show that mutation of residue 72 to alanine (K72ACytc[Sc] variant) enhances peroxidase activity of iso-1-Cytc near physiological pH.
Results and Discussion
Crystallization of K72ACytc[Sc].
The K72A variant of yeast iso-1-Cytc, K72ACytc[Sc], was expressed from E. coli (19, 22). This variant carries an additional C102S mutation to eliminate disulfide dimerization (23). Crystals of oxidized (Fe3+–heme) K72ACytc[Sc] grown from 90% saturated ammonium sulfate at pH 8.8 diffracted to 1.45 Å. Refinement yielded a structural model with Rwork/Rfree = 0.145/0.156 (Table S1). Two molecules of K72ACytc[Sc] (chains A and B) are contained in the asymmetric unit of the crystal lattice. The rmsd between chains A and B is 0.25 Å. Heme proteins are susceptible to reduction to the Fe(II) state by synchrotron radiation (24). Therefore, we cannot eliminate the possibility that some reduction of the K72ACytc[Sc] heme occurs during data collection. However, treatment of crystals with sodium dithionite leads to crystal cracking, which is not observed following data collection. Also, because the data were collected at 100 K, large structural rearrangements due to heme reduction should be minimal.
Structural Consequences of the Trimethyllysine 72-to-Alanine Substitution.
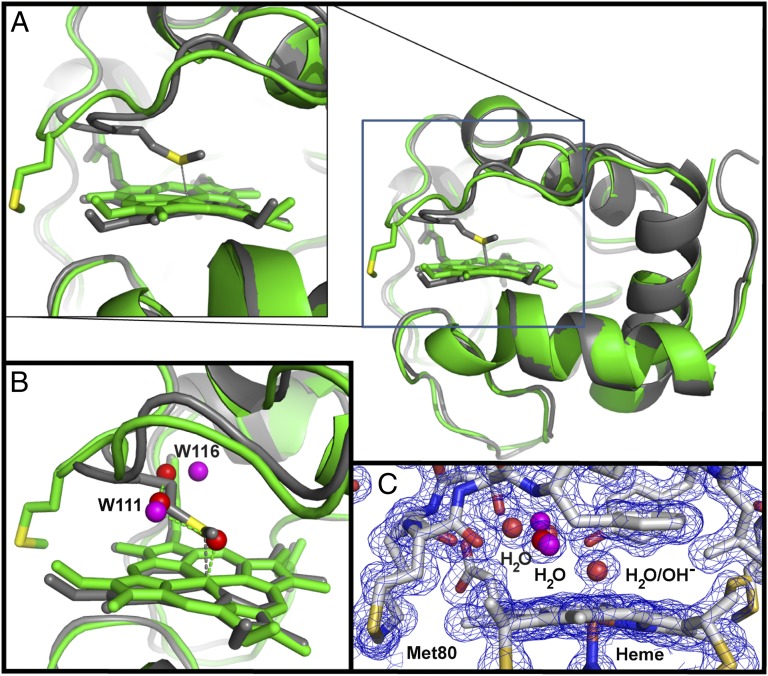
The structure of K72ACytc[Sc] superimposed on that of oxidized tmK72Cytc[Sc] crystallized near pH 6.5 (20) shows that the two proteins are quite similar (rmsd at Cα positions: 0.64 Å) (Fig. 1). The key difference is that Met80 has swung out of the heme crevice in K72ACytc[Sc] (Fig. 1A). Most previous structures of monomeric mitochondrial cytochromes c have Met80 bound to the heme iron (6). Exceptions include NMR structures of an alkaline conformer of K79ACytc[Sc] obtained at pH 10 where Met80 is displaced by Lys73 (25) and of horse Cytc and an M80A variant of iso-1-Cytc with exogenous ligands bound in place of Met80 (26–28). In K72ACytc[Sc], water (W)113 (probably hydroxide at pH 8.8) is bound to the heme iron in place of Met80. The Fe–O distance of 2.00 Å is similar to that observed in the oxidized (Fe3+–heme) horse Cytc domain-swapped dimer and trimer structures (∼2.10 Å) (9) and the 2.01-Å Fe–O distance observed for two Geobacter sulfurreducens chemotaxis protein sensor domains (29). The presence of H2O/OH− as the axial ligand in place of Met80 does not affect the Fe–N bond distance (2.04 Å) of the transaxial His18 ligand, and is similar to that of tmK72Cytc[Sc] (Fig. S1A) and to the Fe–N bond distance of 2.0–2.1 Å observed for c-type cytochromes with a water bound trans to the histidine of an Fe3+–heme (9, 29). The orientation of the plane of the imidazole ring of His18 remains typical of c-type cytochromes (30) and is indistinguishable from that in tmK72Cytc[Sc] (Fig. S1A). Three more buried water molecules are located on the Met80-proximal side of the heme. Two of these waters sit near positions occupied by the Cβ and Cγ atoms of Met80 in the tmK72Cytc[Sc] structure (Fig. 1B and Movie S1) and have well-defined electron density (Fig. 1C).
Fig. 1.
Comparison of the overall structures of K72ACytc[Sc] and tmK72Cytc[Sc]. (A) Alignment of K72ACytc[Sc] (green; chain A of PDB ID code 4MU8) with tmK72Cytc[Sc] (gray; PDB ID code 2YCC; carries a C102T mutation). The heme and Met80 are shown as stick models. A close-up view of the heme and Met80 is shown (Left). (B) Close-up view of the heme crevice showing waters (red spheres) in K72ACytc[Sc] in the space occupied by Met80 in tmK72Cytc[Sc]. Low occupancy positions observed for two of the waters are shown in purple. (C) Heme crevice close-up view showing the 2|Fo| − |Fc| electron density map contoured at 1.2σ (blue wire) with the model used to fit the data.
Alternate Side-Chain Conformers and Buried Water Channels in K72ACytc[Sc].
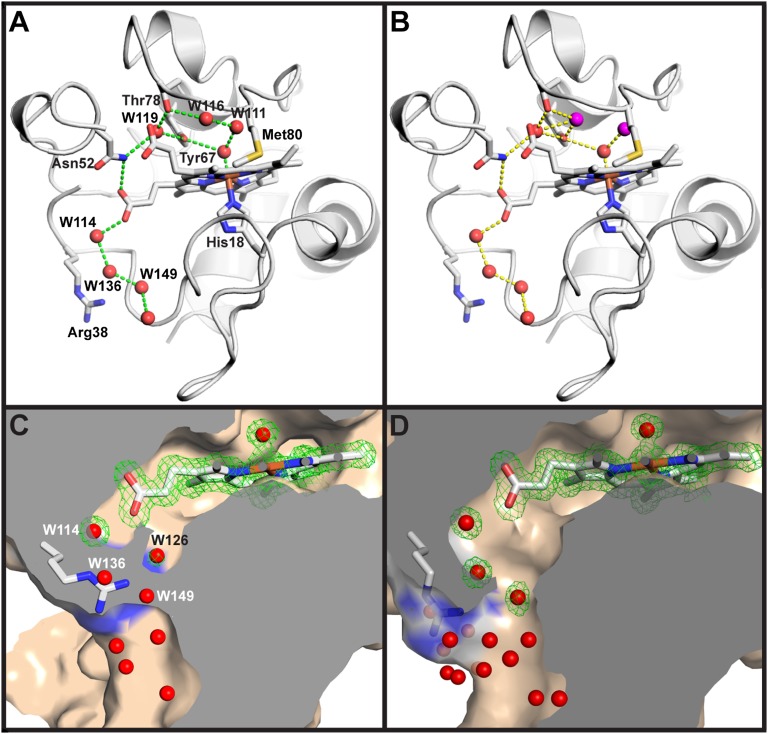
In oxidized tmK72Cytc[Sc], all residues are modeled as single conformers (20). However, in K72ACytc[Sc], several residues occupy two conformations (Table S2). Three of these residues, Asn52, Met64, and Leu85, are fully buried and pack against the heme (Fig. S2). Thus, invasion of water into the heme crevice when Met80 ligation is lost creates disorder around the heme. Both conformations of Asn52 can hydrogen-bond to W119, which is also hydrogen-bonded to Tyr67 (Fig. S1A). Two waters, W111 and W116, near the heme also adopt alternate high (∼80%) and low (∼20%) occupancy positions displaced from each other by 0.67 Å and 1.45 Å, respectively (Fig. 1B and Table S2). In their high occupancy positions, these two waters form a hydrogen-bonded chain emanating from the axial H2O/OH− heme ligand (Fig. 2A). In the low occupancy state, W116 breaks this chain and forms alternative hydrogen bonds (Fig. 2B and Movie S2). Thus, W111 and W116 could act as a transient proton shuttle in general acid/base catalysis during peroxidative turnover catalyzed by the heme of the K72ACytc[Sc] conformation reported here.
Fig. 2.
Water structure around the heme of K72ACytc[Sc]. (A) Hydrogen-bond network of buried water molecules (red spheres) showing high occupancy positions of W111 and W116 in the context of chain A. (B) Hydrogen-bond network of buried water molecules showing low occupancy positions (purple spheres) of W111 and W116. In A and B, residues 41–50 are removed to enhance the view of the buried water network. The heme and selected side chains are shown as stick models (labeled in A). (C) Section through the surface representation of chain B of K72ACytc[Sc] with Arg38 positioned to close the water channel to the protein surface. (D) Section through the surface representation of chain A of K72ACytc[Sc] with Arg38 positioned to open the water channel to the protein surface. In C and D, the heme and buried water molecules are shown with the 2|Fo| − |Fc| electron density (green wire). External waters and the partially occupied waters, W136 and W149, in C are shown without electron density.
The position of water molecules on the Met80-proximal side of the heme of tmK72Cytc[Sc] is sensitive to the redox state of the heme (20). In particular, W166 in the native structure of iso-1-Cytc (20) is 1.7 Å closer to the iron of the oxidized compared with the reduced heme. In the structure of K72ACytc[Sc], the corresponding water, W119, occupies the same position as W166 in reduced tmK72Cytc[Sc] (Fig. S1B). This observation could be taken as evidence for reduction of the heme by synchrotron radiation. However, the water bound to the iron atom of the heme is likely hydroxide at pH 8.8. Therefore, the oxidized form of K72ACytc[Sc] has a heme with no net charge, like the reduced form of tmK72Cytc[Sc] (31). We conclude that it is more likely that the position of W119 in K72ACytc[Sc] reflects a neutral heme rather than reduction of the heme during data collection.
K72ACytc[Sc] harbors four buried waters on the His18-proximal side of the heme of chain A compared with two for tmK72Cytc[Sc] (6) (Fig. 2A and Fig. S1A). These buried waters, together with those located at positions vacated by the ejected Met80, occupy a channel that leads from the hydroxide-bound heme to the protein surface (Fig. 2 A and B and Movie S2), permitting them to exchange with bulk water. This water channel is similar to one identified by molecular dynamics (MD) simulations on iso-1-Cytc (21). Near the entrance of this channel in chain B of the asymmetric unit, Arg38 assumes multiple states. In an ordered, but partially occupied, state (60% occupancy) similar to that observed in tmK72Cytc[Sc] (Fig. S1A) (20), the side chain of Arg38 fully blocks the channel opening at the protein surface (Fig. 2C, Fig. S3, and Table S2). In chain B, but not in chain A, a partially occupied water, W126 (48% occupancy), is observed near the position of W168 in tmK72Cytc[Sc] within hydrogen-bonding distance of Arg38 (Fig. 2C and Fig. S1A). In an alternative, fully disordered state, Arg38 in chain B is replaced by W136 and W149, which also have counterparts in chain A (Fig. 2 C and D). In chain A, Arg38, stabilized by its interaction with a sulfate ion, adopts a single conformation in which the channel is open to bulk solvent (Fig. 2D and Fig. S3). In an X-ray structure of iso-1-Cytc with an R38A mutation, a similar water channel with a much larger opening at the protein surface is observed (32). Thus, it appears that the side chain of Arg38 can act as a conformational switch that gates access of buried waters in K72ACytc[Sc] to bulk water at the protein surface (Movie S3). By contrast, MD simulations suggested that access to buried water channels is mediated by main-chain, not side-chain, motions (21). This buried water channel (Fig. 2, Fig. S1A, and Movie S2) could afford substrate access to the heme active site and shuttle protons to the surface during Cytc-mediated peroxidation activity. Due to fluctuations in the conformations of Arg38, Asn52, and the positions of buried water molecules, the channel would be expected to flicker between low and high conductivity states. Previous work suggests that Arg38 plays a role in regulating heme redox potential (33), and thus this conserved residue may play a role in both functions of mitochondrial Cytc.
Arg38 is also unusual in that it is part of a short segment of tmK72Cytc[Sc] that becomes more rigid, rather than less rigid, in the oxidized state versus the reduced state of the protein (20). The thermal factors for residues near Arg38 are lower for chain A versus chain B (Fig. S4), and the pattern of variation of thermal factors near Arg38 is similar to oxidized tmK72Cytc[Sc] for chain A and similar to reduced tmK72Cytc[Sc] for chain B (20). This difference is likely attributable to the presence of sulfate ion interactions with Arg38 in chain A, which are absent in chain B.
Structural Constraints Mediating Ejection of Met80 from the Heme Crevice of Iso-1-Cytc.
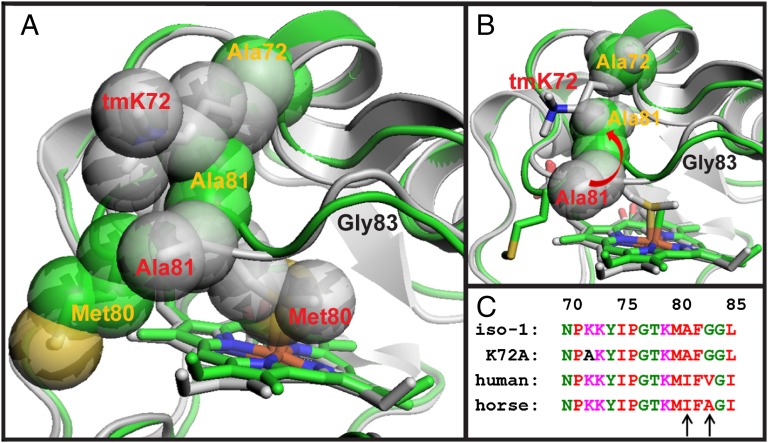
Relatively small backbone adjustments are sufficient to permit Met80 to swing out of the heme crevice of Cytc (Figs. 1 and 3 and Movie S4). Large displacements occur between the K72ACytc[Sc] and tmK72Cytc[Sc] structures at Met80 (Cα rmsd 3.59 Å) and Ala81 (Cα rmsd 2.18 Å), and to a lesser extent at Gly83 (Cα rmsd 1.26 Å) in the heme crevice loop (Fig. 3 A and B). Otherwise, Cα rmsd values for alignment of the two structures are mostly <1 Å (Fig. S5). The deviation of the main chain observed here for the heme crevice loop is considerably smaller than when Met80 is expelled from the heme crevice by exogenous ligands such as cyanide (28) and imidazole (27).
Fig. 3.
Steric stabilization of the native conformer of Cytc by the residue at position 72. (A) Overlay of the structures of K72ACytc[Sc] (green, yellow labels) and tmK72Cytc[Sc] (gray, red labels) with residues 72, 80, and 81 shown as space-filling models. The portion of the backbone corresponding to Gly83 is indicated with a black label. (B) Overlay of K72ACytc[Sc] and tmK72Cytc[Sc] showing the movement of Ala81 toward Ala72 when Met80 swings out of the heme crevice. (C) Alignment of yeast and mammalian Cytc sequences for the highly conserved heme crevice loop (residues 70–85). Residues 81 and 83 are marked with black arrows.
Mutation of tmK72→Ala facilitates expulsion of Met80. In tmK72Cytc[Sc], tmK72 sterically blocks movement of Ala81, inhibiting release of Met80 from the heme (Fig. 3A and Movie S3). In horse Cytc, where Lys72 is not trimethylated, a similar situation exists. Lys72 lies across the heme crevice loop, forming hydrogen bonds to the carbonyls of Met80 and Phe82 (34). In K72ACytc[Sc], Ala81 is free to move toward Ala72 as Met80 swings out of the heme crevice (Fig. 3B and Movie S3). Our structural data demonstrate that the more than twofold increase we observe in the dynamics of the His79-mediated alkaline conformational transition of iso-1-Cytc in the presence of the tmK72→Ala mutation (18) results from relaxation of steric constraints in the heme crevice loop.
The most highly conserved portion of the Cytc sequence is the heme crevice loop. It encompasses both tmK72 and the mobile heme ligand, Met80 (15). Within this loop, yeast and mammals differ at positions 81 and 83 (Fig. 3C), two residues that move significantly when Met80 is displaced from the heme crevice. At both positions, more sterically demanding side chains are present in mammalian Cytc than in its yeast counterpart. Phylogenetic analysis shows that ancestral mitochondrial Cytc has alanine at position 81, as in yeast (35). Moving up the phylogenetic tree from yeast to mammals, position 81 first mutates to Val and then Ile (15, 16). These observations suggest that the heme crevice loop in mammals has evolved to more stringently minimize access to Cytc conformers capable of peroxidase activity than in yeast.
Peroxidase Activity of tmK72Cytc[Sc] Versus K72ACytc[Sc].
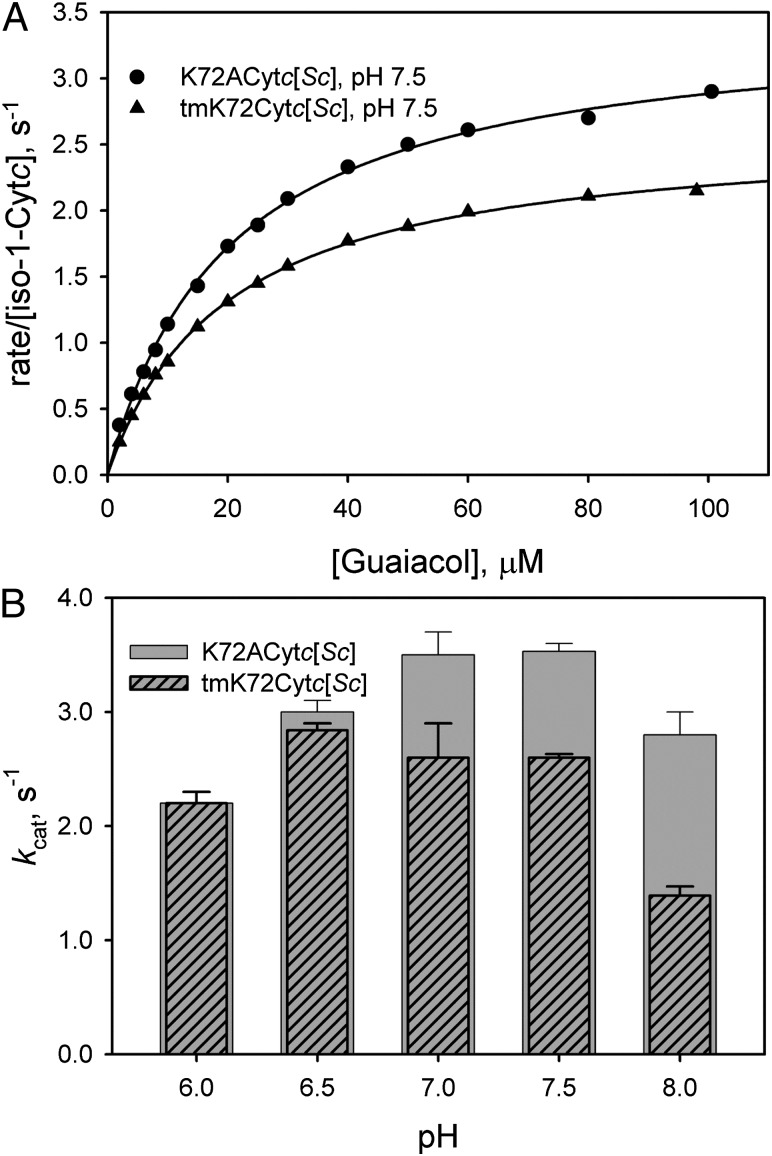
To test the effect of the tmK72→Ala mutation on the peroxidase activity of iso-1-Cytc, we compared the rate of oxidation of guaiacol to tetraguaiacol (10, 36, 37) for tmK72Cytc[Sc] versus K72ACytc[Sc] (Fig. S6). At pH 7.5 (Fig. 4A), kcat for peroxidase activity is 35% greater for K72ACytc[Sc] than for tmK72Cytc[Sc]. The pH dependence of kcat shows that peroxidase activity is similar at pH 6 and 6.5 for both proteins (Fig. 4B and Table S3). At these lower pH values, peroxidase activity is expected to be suppressed by the need to deprotonate H2O2 (36, 38, 39). However, from pH 7 to 8, kcat for K72ACytc[Sc] increases and remains high, whereas it declines for tmK72Cytc[Sc]. The pH dependence of the peroxidase activity of the yeast-expressed iso-1-Cytc, tmK72Cytc[Sc], mirrors that of horse Cytc (36), relatively constant below about pH 7 and then decreasing at higher pH. The similarity in the pH dependence of yeast and horse cytochromes c likely reflects the similar ordering of substructure stabilities for the two proteins (22, 40, 41).
Fig. 4.
Effect of the tmK72→Ala mutation on the peroxidase activity of iso-1-Cytc. (A) Michaelis–Menten plots of rate of consumption of guaiacol as a function of guaiacol concentration at 25 °C and pH 7.5 in 50 mM potassium phosphate buffer. Cytc concentration was 1 μM, and H2O2 concentration was 50 mM. (B) Plot of kcat (average of three independent experiments with SD) versus pH for K72ACytc[Sc] and tmK72Cytc[Sc].
At pH 8, the tmK72→Ala mutation leads to an enhancement of kcat by twofold (Fig. 4B). Furthermore, we observe that crystals of K72ACytc[Sc] crack and dissolve below pH 7, also suggesting that the Cytc conformer observed in the K72ACytc[Sc] structure is less stable at lower pH. Accordingly, the tmK72→Ala mutation favors a Cytc conformer with higher peroxidase activity above pH 6.5.
In mitochondria, the fatty acid chains of CL, which are subject to peroxidation, are believed to bind near Asn52 (site C) and lysines 72, 73, and 86 (site A) (42, 43). The channel of waters that fill the void left when Met80 is expelled could readily be displaced by a fatty acid chain entering from either site A or site C. The side chains of Asn52, Met64, and Leu85 also occupy two rotamers, indicating conformational plasticity on the Met80-proximal side of the heme that could facilitate binding of CL near the heme iron.
The peroxidase activity enhancement we observe compares well in magnitude to the increase in the dynamics of the His79-mediated alkaline conformational transition resulting from the tmK72→Ala mutation (18). Thus, residue 72 in the heme crevice loop significantly impacts the heme crevice dynamics necessary for peroxidase activity. Notably, kcat for the peroxidase activity of horse Cytc at pH 7 (10) determined under identical conditions is 20-fold and 27-fold less than for tmK72Cytc[Sc] and K72ACytc[Sc], respectively (Table S3). The additional steric congestion at positions 81 and 83 (Fig. 3C), in addition to the hydrogen-bond contacts that Lys72 makes with the Met80 and Phe82 carbonyls in mammalian Cytc (34), appears to diminish the heme crevice dynamics needed for peroxidase activity for mammalian versus yeast Cytc. We suggest that Cytc in higher organisms has evolved to limit peroxidase activity, leading to stress-induced release of Cytc from mitochondria, thereby imposing a more stringent barrier to the onset of apoptosis.
Methods
Protein Expression and Purification.
Expression of tmKCytc[Sc] was from S. cerevisiae GM-3C-2 cells (44) carrying the pRS425/CYC1 shuttle vector (45). K72ACytc[Sc] was expressed from E. coli BL21(DE3) cells carrying the pRbs_BTR1 vector (22), as described previously (18). Both proteins carry a C102S mutation to prevent disulfide dimerization during physical studies (23). K72ACytc[Sc] and tmKCytc[Sc] were purified as previously described (18, 46, 47). The procedures are outlined in brief in SI Methods.
Crystallization, Structure Determination, and Refinement.
A 1:1 mixture of oxidized K72ACytc[Sc] iso-1-Cytc at ∼6 mg/mL in 75% aqueous ammonium sulfate and a reservoir solution of 90% ammonium sulfate, 0.1 M Tris⋅HCl (pH 8.8), and 4% tert-butanol was crystallized at 20 °C by vapor diffusion from sitting drops. Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource. The structure was solved by molecular replacement and refined to 1.45-Å resolution. The coordinates and structure factors for K72ACytc[Sc] iso-1-Cytc at pH 8.8 have been deposited in the Protein Data Bank (PDB) (www.pdb.org) under ID code 4MU8. A more complete description of procedures may be found in SI Methods.
Guaiacol Assay of Peroxidase Activity.
Guaiacol assays were performed at 25 °C with an Applied Photophysics SX20 stopped-flow spectrophotometer. Peroxidase activity was monitored as the formation of tetraguaiacol at 470 nm. The rate of change of absorbance at 470 nm was converted to micromoles guaiacol consumed per second using an absorption coefficient of 26.6 mM−1 cm−1 for tetraguaiacol (36, 37). Michaelis–Menten plots were then used to extract kcat and Km for guaiacol from the data. Additional details on experimental procedures are provided in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (CHE-0910616 and CHE-1306903). The Macromolecular X-ray Diffraction Core Facility at the University of Montana was supported by a Centers of Biomedical Research Excellence grant from the National Institute of General Medical Sciences (P20GM10356). The Bruker microflex MALDI-TOF mass spectrometer was purchased with a grant from the National Science Foundation (CHE-1039814).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4MU8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323828111/-/DCSupplemental.
References
- 1.Winge DR. Sealing the mitochondrial respirasome. Mol Cell Biol. 2012;32(14):2647–2652. doi: 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: Functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9(7):532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 3.Laun P, Buettner S, Rinnerthaler M, Burhans WC, Breitenbach M. Yeast aging and apoptosis. In: Breitenbach M, Jazwinski SM, Laun P, editors. Aging Research in Yeast, Subcellular Biochemistry. Vol 57. Dordrecht, The Netherlands: Springer; 2012. pp. 207–232. [DOI] [PubMed] [Google Scholar]
- 4.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome c binding to the apoptosis protease activation factor-1. J Biol Chem. 2001;276(16):13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- 5.Olteanu A, et al. Stability and apoptotic activity of recombinant human cytochrome c. Biochem Biophys Res Commun. 2003;312(3):733–740. doi: 10.1016/j.bbrc.2003.10.182. [DOI] [PubMed] [Google Scholar]
- 6.Brayer GD, Murphy MEP. Structural studies of eukaryotic cytochromes c. In: Scott RA, Mauk AG, editors. Cytochrome c: A Multidisciplinary Approach. Sausalito, CA: University Science Books; 1996. pp. 103–166. [Google Scholar]
- 7.Kagan VE, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1(4):223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 8.Kapetanaki SM, et al. Interaction of carbon monoxide with the apoptosis-inducing cytochrome c-cardiolipin complex. Biochemistry. 2009;48(7):1613–1619. doi: 10.1021/bi801817v. [DOI] [PubMed] [Google Scholar]
- 9.Hirota S, et al. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad Sci USA. 2010;107(29):12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Matsuo T, Nagao S, Hirota S. Peroxidase activity enhancement of horse cytochrome c by dimerization. Org Biomol Chem. 2011;9(13):4766–4769. doi: 10.1039/c1ob05552f. [DOI] [PubMed] [Google Scholar]
- 11.Muenzner J, Toffey JR, Hong Y, Pletneva EV. Becoming a peroxidase: Cardiolipin-induced unfolding of cytochrome c. J Phys Chem B. 2013;117(42):12878–12886. doi: 10.1021/jp402104r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong Y, Muenzner J, Grimm SK, Pletneva EV. Origin of the conformational heterogeneity of cardiolipin-bound cytochrome c. J Am Chem Soc. 2012;134(45):18713–18723. doi: 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanske J, et al. Conformational properties of cardiolipin-bound cytochrome c. Proc Natl Acad Sci USA. 2012;109(1):125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belikova NA, et al. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45(15):4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banci L, Bertini I, Rosato A, Varani G. Mitochondrial cytochromes c: A comparative analysis. J Biol Inorg Chem. 1999;4(6):824–837. doi: 10.1007/s007750050356. [DOI] [PubMed] [Google Scholar]
- 16.Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, Structural and Physicochemical Aspects. New York: Springer; 1990. [Google Scholar]
- 17.Cherney MM, Junior CC, Bergquist BB, Bowler BE. Dynamics of the His79-heme alkaline transition of yeast iso-1-cytochrome c probed by conformationally gated electron transfer with Co(II)bis(terpyridine) J Am Chem Soc. 2013;135(34):12772–12782. doi: 10.1021/ja405725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherney MM, Junior CC, Bowler BE. Mutation of trimethyllysine 72 to alanine enhances His79-heme-mediated dynamics of iso-1-cytochrome c. Biochemistry. 2013;52(5):837–846. doi: 10.1021/bi301599g. [DOI] [PubMed] [Google Scholar]
- 19.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of Lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37(17):6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 20.Berghuis AM, Brayer GD. Oxidation state-dependent conformational changes in cytochrome c. J Mol Biol. 1992;223(4):959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 21.Bortolotti CA, et al. The reversible opening of water channels in cytochrome c modulates the heme iron reduction potential. J Am Chem Soc. 2012;134(33):13670–13678. doi: 10.1021/ja3030356. [DOI] [PubMed] [Google Scholar]
- 22.Duncan MG, Williams MD, Bowler BE. Compressing the free energy range of substructure stabilities in iso-1-cytochrome c. Protein Sci. 2009;18(6):1155–1164. doi: 10.1002/pro.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutler RL, Pielak GJ, Mauk AG, Smith M. Replacement of cysteine-107 of Saccharomyces cerevisiae iso-1-cytochrome c with threonine: Improved stability of the mutant protein. Protein Eng. 1987;1(2):95–99. doi: 10.1093/protein/1.2.95. [DOI] [PubMed] [Google Scholar]
- 24.Can M, et al. Structural characterization of Nitrosomonas europaea cytochrome c-552 variants with marked differences in electronic structure. ChemBioChem. 2013;14(14):1828–1838. doi: 10.1002/cbic.201300118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assfalg M, et al. Structural model for an alkaline form of ferricytochrome c. J Am Chem Soc. 2003;125(10):2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 26.Banci L, et al. Three-dimensional solution structure of the cyanide adduct of a Met80Ala variant of Saccharomyces cerevisiae iso-1-cytochrome c. Identification of ligand-residue interactions in the distal heme cavity. Biochemistry. 1995;34(36):11385–11398. doi: 10.1021/bi00036a011. [DOI] [PubMed] [Google Scholar]
- 27.Banci L, et al. Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c. J Biol Inorg Chem. 2001;6(5-6):628–637. doi: 10.1007/s007750100240. [DOI] [PubMed] [Google Scholar]
- 28.Yao Y, et al. Solution structure of cyanoferricytochrome c: Ligand-controlled conformational flexibility and electronic structure of the heme moiety. J Biol Inorg Chem. 2002;7(4-5):539–547. doi: 10.1007/s00775-001-0334-y. [DOI] [PubMed] [Google Scholar]
- 29.Pokkuluri PR, et al. Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: Implications for signal transduction. J Mol Biol. 2008;377(5):1498–1517. doi: 10.1016/j.jmb.2008.01.087. [DOI] [PubMed] [Google Scholar]
- 30.Bren KL, Kellogg JA, Kaur R, Wen X. Folding, conformational changes, and dynamics of cytochromes c probed by NMR spectroscopy. Inorg Chem. 2004;43(25):7934–7944. doi: 10.1021/ic048925t. [DOI] [PubMed] [Google Scholar]
- 31.Louie GV, Brayer GD. High-resolution refinement of yeast iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J Mol Biol. 1990;214(2):527–555. doi: 10.1016/0022-2836(90)90197-T. [DOI] [PubMed] [Google Scholar]
- 32.Lo TP, Komar-Panicucci S, Sherman F, McLendon G, Brayer GD. Structural and functional effects of multiple mutations at distal sites in cytochrome c. Biochemistry. 1995;34(15):5259–5268. doi: 10.1021/bi00015a041. [DOI] [PubMed] [Google Scholar]
- 33.Cutler RL, et al. Role of arginine-38 in regulation of the cytochrome c oxidation-reduction equilibrium. Biochemistry. 1989;28(8):3188–3197. doi: 10.1021/bi00434a012. [DOI] [PubMed] [Google Scholar]
- 34.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214(2):585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 35.Fitch WM. The molecular evolution of cytochrome c in eukaryotes. J Mol Evol. 1976;8(1):13–40. doi: 10.1007/BF01738880. [DOI] [PubMed] [Google Scholar]
- 36.Diederix REM, Ubbink M, Canters GW. The peroxidase activity of cytochrome c-550 from Paracoccus versutus. Eur J Biochem. 2001;268(15):4207–4216. doi: 10.1046/j.1432-1327.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 37.DePillis GD, Sishta BP, Mauk AG, Ortiz de Montellano PR. Small substrates and cytochrome c are oxidized at different sites of cytochrome c peroxidase. J Biol Chem. 1991;266(29):19334–19341. [PubMed] [Google Scholar]
- 38.Diederix REM, Ubbink M, Canters GW. Peroxidase activity as a tool for studying the folding of c-type cytochromes. Biochemistry. 2002;41(43):13067–13077. doi: 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
- 39.Diederix REM, Ubbink M, Canters GW. Effect of the protein matrix of cytochrome c in suppressing the inherent peroxidase activity of its heme prosthetic group. ChemBioChem. 2002;3(1):110–112. doi: 10.1002/1439-7633(20020104)3:1<110::AID-CBIC110>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Krishna MM, Lin Y, Rumbley JN, Englander SW. Cooperative omega loops in cytochrome c: Role in folding and function. J Mol Biol. 2003;331(1):29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 41.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalanxhi E, Wallace CJA. Cytochrome c impaled: Investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem J. 2007;407(2):179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinibaldi F, et al. Extended cardiolipin anchorage to cytochrome c: A model for protein-mitochondrial membrane binding. J Biol Inorg Chem. 2010;15(5):689–700. doi: 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 44.Faye G, Leung DW, Tatchell K, Hall BD, Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci USA. 1981;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrmann LM, Flatt P, Bowler BE. Site-directed replacement of the invariant lysine 73 of Saccharomyces cerevisiae iso-1-cytochrome c with all ribosomally encoded amino acids. Inorg Chim Acta. 1996;242(1-2):97–103. [Google Scholar]
- 46.Redzic JS, Bowler BE. Role of hydrogen bond networks and dynamics in positive and negative cooperative stabilization of a protein. Biochemistry. 2005;44(8):2900–2908. doi: 10.1021/bi048218b. [DOI] [PubMed] [Google Scholar]
- 47.Wandschneider E, Hammack BN, Bowler BE. Evaluation of cooperative interactions between substructures of iso-1-cytochrome c using double mutant cycles. Biochemistry. 2003;42(36):10659–10666. doi: 10.1021/bi034958t. [DOI] [PubMed] [Google Scholar]